Abstract
Xanthomonas massiliensis strain SN6T is a Gram-negative bacterium which is aerobic, motile and nonsporulating. This new species isolated from human faeces exhibited the characteristic traits of members of this genus, such as yellow pigmentation and viscosity. Here we present the main phenotypic characteristics and the taxonogenomics description of this strain. The genome is 3 690 720 bp long with DNA G + C content of about 70.52%.
Keywords: Culturomics, genome, human gut microbiota, taxonogenomics, Xanthomonas massiliensis
Introduction
The first member of the genus Xanthomonas was described by Dowson [1], and the genus [2] contains plant-associated bacteria that establish neutral, commensal or pathogenic relationships with plants. Taxonomically, the members of this genus were revised several times because the taxonomy had been previously based on host specificity. Each bacterium isolated from a new host was considered as a new species. Dye and Lelliott [2] reduced the number of species from about 120 to the following five: Xanthomonas campestris, Xanthomonas albilineans, Xanthomonas axonopodis, Xanthomonas fragariae and Xanthomonas ampelina. The others were grouped together as nomenspecies in the X. campestris group (pathovar). In 1995, Vauterin et al. [3] partially clarified the classification and described 20 species among the three former species, X. axonopodis, X. fragariae and X. albilineans, and 62 pathovars of X. campestris, on the basis of DNA ± DNA hybridization data and biochemical and physiological tests. However, members of the genus Xanthomonas can be differentiated from members of the phylogenetically closest genus Pseudoxanthomonas by the absence of reduction of nitrates to nitrites and the presence of C13:0 iso 3-OH fatty acid [4].
Members of the Xanthomonas genus were known exclusively as plant-associated organisms and did not durably colonize other niches [5]. However, during the study of the bacterial diversity of the human microbiota by culturomics [6], a strain of Xanthomonas was isolated from the stool sample of an obese French patient. It is the first Xanthomonas isolate identified in humans to date. Here we report the characterization of strain SN6 as a novel species of the genus Xanthomonas, Xanthomonas massiliensis strain SN6 (= CSUR P2129 = DSM 100900), with a description of the complete genomic sequence and its annotation.
Materials and methods
Organism information and strain isolation
Strain SN6 was discovered in the context of a study on the microaerophilic bacteria of the human digestive microbiota by culturomics in September 2015. The strain was isolated from a 41-year-old obese Frenchwoman hospitalized in September 2012 at the La Timone Hospital in Marseille, France. This study and the assent procedure were validated by the ethics committee of the IFR48 Federative Research Institute Marseille under number 09-022, and we obtained the signed consent of the patient.
After collecting the stool, a portion of a sample was stored at −80°C until use. In June 2015, the stool sample was cultivated as part of an exploration of the human microbiome centred on microaerophilic bacteria the. Part of the frozen aliquot of the specimen (approximately 1 g) was taken out and diluted in 900 μL of phosphate-buffered saline (Life Technologies, Carlsbad, CA, USA) following ten serial dilutions to obtain 1/10. Inoculum (50 μL) was seeded on Columbia agar supplemented with 5% sheep's blood (bioMérieux, Marcy l’Etoile, France) and incubated under microaerophilic conditions (7% O2, 5% H2, 10% CO2, 85% N2) using the generator CampyGen (Thermo Scientific, Villebon-sur-Yvette, France) at 37°C for 48 hours.
Strain identification
After 48 hours of incubation in microaerophilic conditions, pure colonies were isolated on Columbia agar and identified by proteomic analysis using matrix-assisted desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) following the same protocol as previously described by Seng et al. [7] with a Microflex spectrometer (Bruker Daltonics, Leipzig, Germany). All obtained spectra of strain SN6 were imported into the MALDI BioTyper software (version 2.0; Bruker) and analysed by standard pattern matching (with default parameter settings) and compared to those of the BioTyper database and our own collection. Thus, a score of >2 allowed identification at the species level, and a score of <1.7 did not allow any identification.
If the identification of the spectrum from colonies selected and purified several times by subculturing on Columbia agar failed, then the 16S rRNA gene was amplified and complete genome sequencing was carried out as previously described [8].
The nucleotide sequence obtained was corrected using Chromas Pro 1.34 software (Technelysium, Tewantin, Australia) and compared to the nucleotide database using the BLAST similarities web services in the online PubMed National Center for Biotechnology Information (NCBI) database (http://blast.ncbi.nlm.nih.gov.gate1.inist.fr/Blast.cgi). As suggested previously, similarity level thresholds of 98.65% and 95% allowed the definition of a new species or a new genus, respectively [9], [10].
Growth conditions and morphologic characterization
Strain SN6 was isolated at first under microaerophilic conditions (CampyGen) at 37°C for 48 hours on Columbia medium supplemented with 5% sheep's blood (COS) agar (bioMérieux) and we also tested its growth under aerobic and anaerobic conditions generated by AnaeroGen (bioMérieux). The minimum and maximum growth temperature ranges (28°C, 37°C, 45°C, 55°C) were determined as well as maximum salinity levels (0–5, 50–75, 100 g/L NaCl). The ability of the strain to grow on media with different pH was also tested. The colonies appeared on day 3 after culture on Columbia agar, and their diameter was measured. Cell morphology, Gram staining and motility were observed in fresh colonies using a DM1000 photonic microscope (Leica Microsystems, Nanterre, France) with a 40 × objective lens. Sporulation was tested by thermal shock, which consists of exposing the bacterium to a temperature of 60°C for 20 minutes and then watching its growth after 48 hours. Negative staining was carried out with detection Formvar-coated grids placed on a drop of 40 μL of bacterial suspension (after an overnight fixation in glutaraldehyde 2.5%) and incubated at 37°C for 30 minutes, followed by a 10-second incubation in 1% ammonium molybdate. The grids were dried on blotting paper and finally observed using a Tecnai G20 transmission electron microscope (FEI Company, Limeil-Brévannes, France).
Biochemical characterization
The biochemical properties of strain SN6 were characterized using API ZYM, API 20NE and API 50CH strips, according to the manufacturer's instructions (bioMérieux) for testing of carbon source utilization and enzyme activity. The presence of catalase and oxidase activities was tested by using a BBL DrySlide (Becton, Le Pont de Claix, France) according to the manufacturer's instructions. The analysis of cellular fatty acid methyl ester composition was performed by gas chromatography/mass spectrometry (GC/MS). Two samples of approximately 100 mg of bacterial biomass per tube collected from five fresh culture plates were used for the extraction of cellular fatty acid methyl esters with the protocol described by Sasser [11]. GC/MS analyses were carried out as described by Dione et al. [12].
Antibiotic susceptibility
The sensitivity to classical antibiotics was tested to determine the antibiogram profile of strain SN6 using the disc diffusion method following the European Committee on Antimicrobial Susceptibility Testing 2016 recommendations (http://www.eucast.org). A suspension of 0.5 McFarland of the species was grown on Mueller-Hinton agar in a petri dish (bioMérieux), and the discs used were provided by i2a (Montpellier, France). The reading of inhibition diameters according to manual measurement by using a ruler was done after 48 hours of incubation at 37°C under aerobic conditions with the Sirscan system (i2a).
Genome sequencing and assembly
A EZ1 DNA tissue kit was used to extract the DNA of strain SN6 on the EZ1 biorobot (Qiagen, Courtaboeuf, France) after pretreatment by lysozyme incubation at 37°C as previously described [13].
Genomic DNA (gDNA) was quantified by a Qubit assay with the high sensitivity kit (Life Technologies) and sequenced on the MiSeq Technology (Illumina, San Diego, CA, USA) with the mate-pair strategy as previously described [14]. The Nextera Mate sample collection kit (Illumina) was used to mix DNA previously barcoded with 11 other projects. An assembly of six different software packages (Velvet [15], Spades [16] and Soap Denovo [17]), on trimmed (MiSeq and Trimmomatic [18] or untrimmed data (only MiSeq software) was created from a pipeline and allowed to perform genome assembly. GapCloser was used to reduce the gaps of each of the six assemblies performed [17]. The contamination with Phage Phix was identified by Blastn against Phage Phix174 DNA sequence and then eliminated. Finally, all scaffolds smaller than 800 bp or with a depth value lower than 25% of the mean depth were removed (identified as possible contaminants). On the basis of different criteria (number of scaffolds, N50, number of N), the best assembly was selected. For strain SN6, Spades gave the best assembly, with a depth coverage of 267.
Genome annotation and comparison
Prodigal allowed to predict open reading frames (ORFs) using default parameters [19] and those that were spanning a sequencing gap region (contained N) were excluded. BLASTP with an E value of 1e-03, coverage of 0.7 and 30% identity was used to search the predicted bacterial protein sequences against the Clusters of Orthologous Groups (COGs) database.
If no hit was found, it searched against the NR database using BLASTP (E value of 1e-03, coverage of 0.7 and 30% identity) and an E value of 1e-05 was used if the sequence's length was shorter than 80 aa. The tRNAScanSE [20] and RNAmmer [21] tools were used to find transfer RNA genes and ribosomal RNA genes, respectively. The number of transmembrane helices and the lipoprotein signal peptides were predicted using Phobius [22]. ORFans were identified if all the BLASTP performed did not yield positive results (E value smaller than 1e-05 for ORFs with sequence length inferior to 80 aa or E value smaller than 1e-03 for ORFs with sequence size larger than 80 aa). These different parameter thresholds had already been used in previous works to define ORFans.
The genomes of each species from the 16S RNA tree used in the comparison were automatically retrieved using Xegen software (PhyloPattern), and the NCBI FTP was used to recover the complete genome sequence, proteome sequence and Orfeome [23]. When the complete genome of one specific strain was not available, we used the complete genome of the same species. All proteomes were analysed with proteinOrtho [24]. For each couple of genomes, a similarity score defined by the mean value of nucleotide similarity between all couples of orthologous genes was computed by average genomic identity of orthologous gene sequences (AGIOS) software. The AGIOS values were calculated from the genome of Xanthomonas and Stenotrophomonas genera. The genome of Xanthomonas massiliensis strain SN6 (FCOY00000000) was compared to that of Xanthomonas vesicatoria ATCC_35937_LMG_911T (AEQV00000000), Xanthomonas gardneri DSM_19127 (AEQX00000000), Xanthomonas axonopodis LMG_538T (JPYE00000000), Xanthomonas sacchari LMG_471T(CP010409), Xanthomonas campestris ATCC_33913 (AE008922), Stenotrophomonas acidaminiphila AMX19 (CP012900) and Stenotrophomonas maltophilia IAM_12423(CP008838). Genome-to-Genome Distance Calculator (GGDC) analysis was also performed using the GGDC web server as previously reported by Meier-Kolthoff et al. [25].
Results
Strain identification and phylogenetic analyses
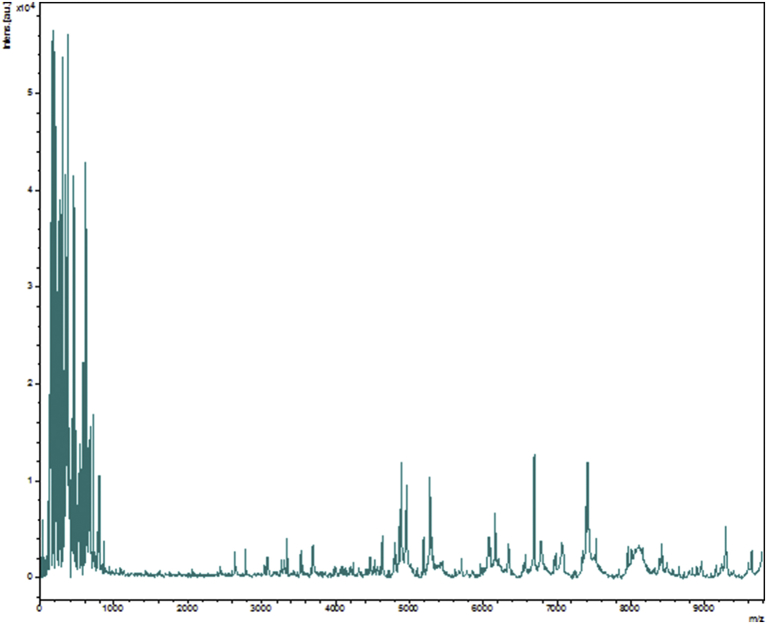
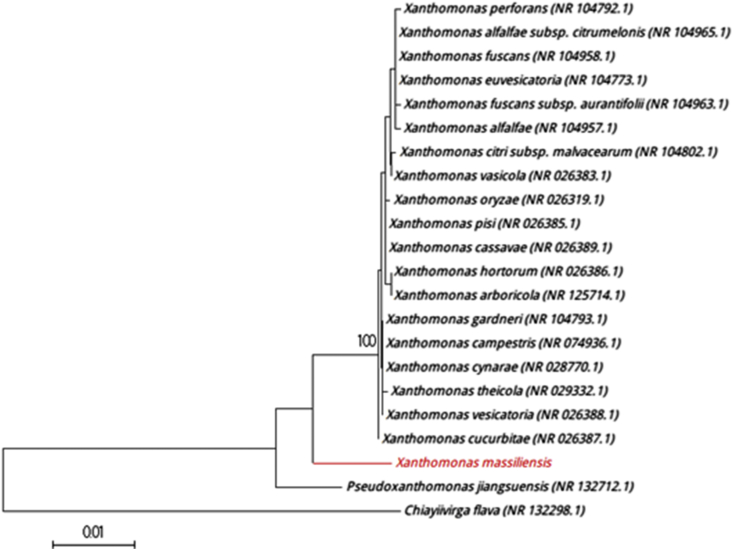
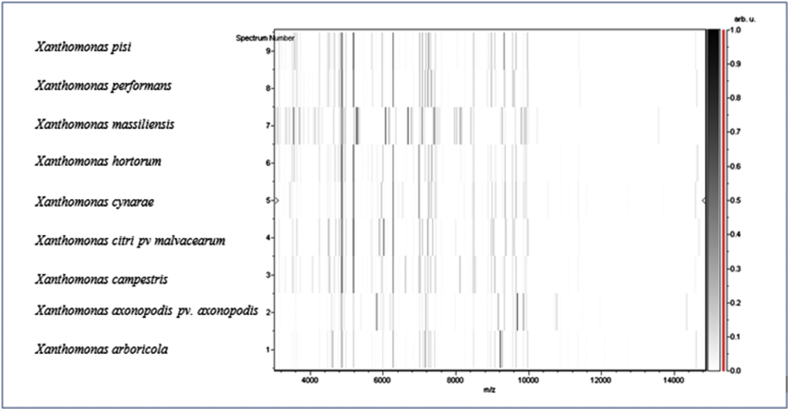
The first colonies of strain SN6 were isolated after direct inoculation of the stool sample on Columbia agar plates under microaerobic condition at 37°C for 48 hours. The bacterial spectrum obtained by MALDI-TOF MS did not match against the Bruker or our own database. Thus, it was incremented in our database (Fig. 1). The 16S rRNA sequenced showed that strain SN6 was phylogenetically clustered in the genus of Xanthomonas and presented a sequence identity of 98.08% with Xanthomonas campestris strain ATCC33913 (NR_074936), the phylogenetically closest species with standing in nomenclature [3] (Fig. 2), which putatively classifies strain SN6 as a member of a new species within the genus Xanthomonas in the phylum Proteobacteria. Thus, we propose the creation of the new species Xanthomonas massiliensis (Table 1). The 16S rRNA gene of Xanthomonas massiliensis strain SN6 is 1508 bp long and was deposited with the accession number AA00102 in the 16S IHU bank and LN881611 in GenBank. A comparison between the spectrum of the strain's protein level and that of the closely related species on the 16S rRNA tree and present in our database was performed in a gel view (Fig. 3).
Fig. 1.
Reference mass spectrum from Xanthomonas massiliensis strain SN6T.
Fig. 2.
Phylogenetic tree showing position of Xanthomonas massiliensis strain SN6T relative to other close species. Sequences were aligned using CLUSTALW and phylogenetic inferences were obtained with Kimura two-parameter models using maximum-likelihood method with 1000 bootstrap replicates, within MEGA software. Scale bar indicates 1% nucleotide sequence divergence.
Table 1.
Classification and general features of Xanthomonas massiliensis strain SN6T
| Property | Term |
|---|---|
| Current classification | Domain: Bacteria |
| Phylum: Proteobacteria | |
| Class: Gammaproteobacteria | |
| Order: Xanthomonadales | |
| Family: Xanthomonadaceae | |
| Genus: Xanthomonas | |
| Species: massiliensis | |
| Type strain: sn8T | |
| Gram stain | Negative |
| Cell shape | Rod |
| Motility | Motile |
| Sporulation | Nonsporulating |
| Temperature range | Mesophilic |
| Optimum temperature | 37°C |
| Oxygen requirement | Aerobic/microaerobic |
| Salinity | 0–5 g/L |
| pH | 7–8.5 |
| Optimum pH | 7 |
| Energy source | Chemoorganotrophic |
| Pathogenicity | Unknown |
| Isolation | Human faeces |
| Habitat | Host associated |
| Biosafety level | 2 |
Fig. 3.
Gel view comparing Xanthomonas massiliensis strain SN6T to other close species. Gel view displays raw spectra of strain SN6T of loaded spectrum files arranged in a pseudo–gel like look. X-axis records m/z value. Left y-axis displays running spectrum number originating from subsequent spectra loading. Peak intensity is expressed by greyscale scheme code. Colour bar and right y-axis indicate relation between colour of peak and its intensity, in arbitrary units. Displayed species are indicated at left.
Phenotypic characteristics
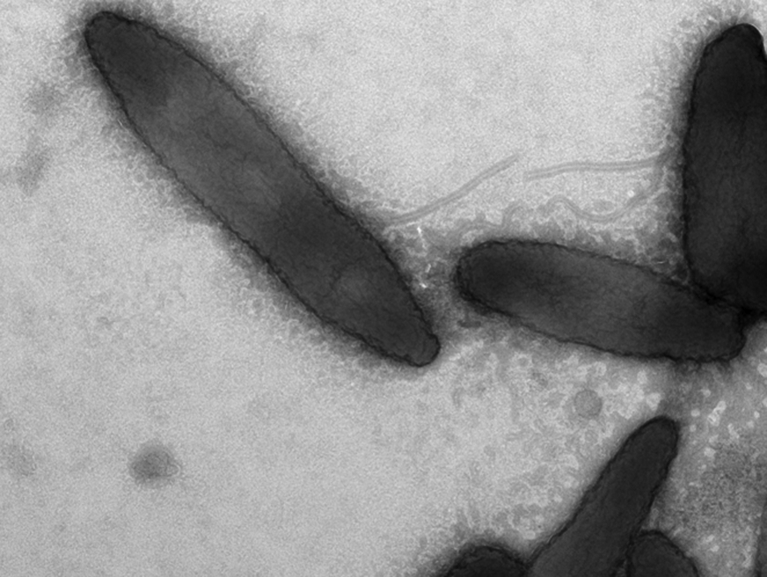
Xanthomonas massiliensis strain SN6 grows between 28°C and 42°C; optimal growth was observed under aerobic conditions on COS at 37°C and pH7 after 48 hours of incubation. A smaller growth rate was observed under microaerobic atmosphere after 48 hours of incubation, and no growth was observed under anaerobic conditions. Also, a smaller growth rate was observed at pH 7 and 8.5, and no growth was observed above 5% salinity. Colonies of the strain were yellowish, circular, viscous and smooth, nonhaemolytic and approximately 1 to 2 mm in diameter on Columbia agar under aerobic conditions after 48 hours. The colonies became khaki green after 4 days of incubation. The yellow pigments, which are mono- or dibromo arylpolyenes called xanthomonadins [26], are characteristic of this genus. Bacterial cells were Gram negative, rod shaped, motile and non–spore forming. Observed under electron microscopy, they occur singly or in chains and measure 0.6 μm in diameter and 1.8 to 2.0 μm in length (Fig. 4).
Fig. 4.
Electron microscopy of Xanthomonas massiliensis strain SN6T.
Biochemical analysis
The catalase activity test was positive, but the oxidase test was negative. Using API ZYM strip for the research of enzymatic activities of strain SN6, positive reactions were detected for alkaline phosphatase, esterase lipase (C8), leucine arylamidase, valine arylamidase, trypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, β-galactosidase, β-glucosidase, N-acetyl-β-glucosaminidase and α-mannosidase. Esterase (C4), lipase (C14), cystine arylamidase, α-chymotrypsin, α-galactosidase, β-glucuronidase, α-glucosidase and α-fucosidase activities did not show any sign of activity. The study of carbohydrate and its derivatives metabolism using API 50CH showed no fermentation of substrates, except for esculin. API 20NE strip revealed that there was neither nitrate reduction nor indole production, and urease was also negative. The reduction of nitrate to nitrite also makes it possible to differentiate the genus Xanthomonas from the genus Pseudoxanthomonas. On the same strip, positive reactions were observed for gelatin hydrolysis, malate and N-acetylglucosamine, and it also allowed to confirm the assimilation of esculin and β-galactosidase. A panel of 15 antibiotics was tested, and strain SN6 was sensitive to vancomycin, ceftriaxone, ciprofloxacin, clindamycin, doxycycline, erythromycin, gentamicin, penicillin, rifampicin, colistin, fosfomycin and trimethoprim/sulfamethoxazole but resistant to oxacillin, teicoplanin and metronidazole. Table 2 compares the phenotypic characteristics of strain SN6 with those of closely related species.
Table 2.
Differential characteristics of Xanthomonas massiliensis strain SN6, Xanthomonas campestris. pv. campestris ATCC33913, Xanthomonas sacchari LMG471, Xanthomonas vesicatoria ATCC35937_LMG911, Xanthomonas gardneri DSM 19127, Xanthomonas axonopodis LMG538 and Pseudoxanthomonas suwonensis 4M1
| Property | X. massiliensis | X. campestris | X. sacchari | X. vesicatoria | X. gardneri | X. axonopodis | P. suwonensis |
|---|---|---|---|---|---|---|---|
| Cell diameter (μm) | 0.5–0.6 | 0.4–0.6 | 0.4–0.6 | 0.4–0.6 | 0.4–0.6 | 0.4–0.6 | 0.3–0.5 |
| Motility | + | + | + | + | + | + | + |
| Indole | − | − | − | − | − | − | − |
| Catalase | + | + | + | + | + | + | + |
| Oxidase | − | − | − | − | − | − | + |
| Nitrate reductase | − | − | − | − | − | − | + |
| Urease | − | − | − | − | − | − | − |
| N-Acetyl-glucosamine | + | − | − | + | − | ++/− | + |
| Acid from: | |||||||
| l-Arabinose | − | − | + | − | − | − | + |
| d-Mannose | − | + | + | + | + | + | − |
| d-Mannitol | − | − | +/− | − | − | − | |
| d-Trehalose | + | + | + | + | + | + | NA |
| d-Glucose | − | + | + | + | + | + | + |
| d-Fructose | − | + | + | + | + | + | NA |
| d-Maltose | − | + | + | + | + | + | + |
| d-Lactose | − | − | + | − | − | − | NA |
| d-Raffinose | − | +/− | − | − | − | −/+ | NA |
| Habitat | Human gut | Tomato/pepper | Tomato/pepper | Tomato/pepper | Tomato/pepper | Pasturage | Cotton waste compost |
+, positive result; −, negative result; NA, data not available.
According to the cellular fatty acid methyl ester analysis, the most abundant fatty acid by far was branched 13-methyl-tetradecanoic acid (58%). Many other branched structures were also described for this strain. Several specific 3-OH structures were detected. Minor amounts of unsaturated and saturated fatty acids were also identified. Regarding the differentiation between the species of the genera Pseudoxanthomonas and Xanthomonas, Xanthomonas massiliensis strain SN6 contains up to 4.5 ± 0.2% of 3-hydroxy-11-methyl-dodecanoic acid (C13:0 iso 3-OH) compared to the other species, which have none or only traces (Table 3).
Table 3.
Cellular fatty acid composition (%)
| Fatty acid | Name | Mean relative %a |
|---|---|---|
| 15:0 iso | 13-Methyl-tetradecanoic acid | 57.6 ± 0.4 |
| 11:0 iso | 9-Methyl-decanoic acid | 10.2 ± 0.5 |
| 17:1 iso | 15-Methylhexadecenoic acid | 4.9 ± 0.2 |
| 13:0 3-OH iso | 3-hydroxy-11-methyl-Dodecanoic acid | 4.5 ± 0.2 |
| 16:1n7 | 9-Hexadecenoic acid | 3.7 ± 0.2 |
| 17:0 iso | 15-methyl-Hexadecanoic acid | 3.5 ± 0.1 |
| 12:0 3-OH | 3-Hydroxydodecanoic acid | 3.3 ± 0.1 |
| 16:0 9,10-methylene | 2-hexyl-Cyclopropaneoctanoic acid | 2.3 ± 0.2 |
| 15:1 iso | 13-Methyltetradecenoic acid | 2.2 ± 0.3 |
| 16:0 | Hexadecanoic acid | 1.7 ± 0.1 |
| 11:0 3-OH iso | 3-hydroxy-9-Methyl-decanoic acid | 1.4 ± 0.2 |
| 15:0 anteiso | 12-methyl-Tetradecanoic acid | 1.3 ± 0.1 |
| 16:1n9 | 7-Hexadecenoic acid | TR |
| 18:1 iso | 16-Methylheptadecenoic acid | TR |
| 13:0 iso | 11-methyl-Dodecanoic acid | TR |
| 18:1n9 | 9-Octadecenoic acid | TR |
| 10:0 | Decanoic acid | TR |
| 17:0 anteiso | 14-methyl-Hexadecanoic acid | TR |
| 14:0 | Tetradecanoic acid | TR |
Mean peak area percentage; TR, trace amounts < 1 %.
Genome properties
The genome of Xanthomonas massiliensis strain SN6 is 3 690 720 bp long with 70.52% GC content (Table 4, Fig. 5). It is composed of four scaffolds (composed of seven contigs). Of the 3196 predicted genes, 3137 were protein-coding genes and 59 were RNAs (two were 5S rRNA, two were 16S rRNA, two were 23S rRNA, 53 were tRNA genes). A total of 2533 genes (80.75%) were assigned as putative function (by COGs or by NR BLAST). A total of 104 genes were identified as ORFans (3.32%). The remaining genes were annotated as hypothetical proteins (350 genes, 11.16%). The distribution of genes into the different COGs functional categories is provided in Table 5.
Table 4.
Nucleotide content and gene count levels of genome
| Attribute | Genome (total) |
|
|---|---|---|
| Value | % of totala | |
| Size (bp) | 3 690 720 | 100 |
| G + C content (%) | 2 602 093 | 70.52 |
| Coding region (bp) | 3 265 075 | 88.46 |
| Total genes | 3196 | 100 |
| RNA genes | 59 | 1.84 |
| Protein-coding genes | 3137 | 100 |
| Genes with function prediction | 2533 | 80.74 |
| Genes assigned to COGs | 2202 | 70.19 |
| Genes with peptide signals | 779 | 24.83 |
| Genes with transmembrane helices | 634 | 20.21 |
| Genes associated to virulence | 687 | 21.89 |
| ORFan genes | 104 | 3.31 |
| Genes associated with PKS or NRPS | 19 | 0.60 |
| Genes associated to toxin/antitoxin | 99 | 3.15 |
COGs, Clusters of Orthologous Groups database; NRPS, nonribosomal peptide synthase; PKS, polyketide synthase.
Total is based on either size of genome in base pairs or total number of protein-coding genes in annotated genome.
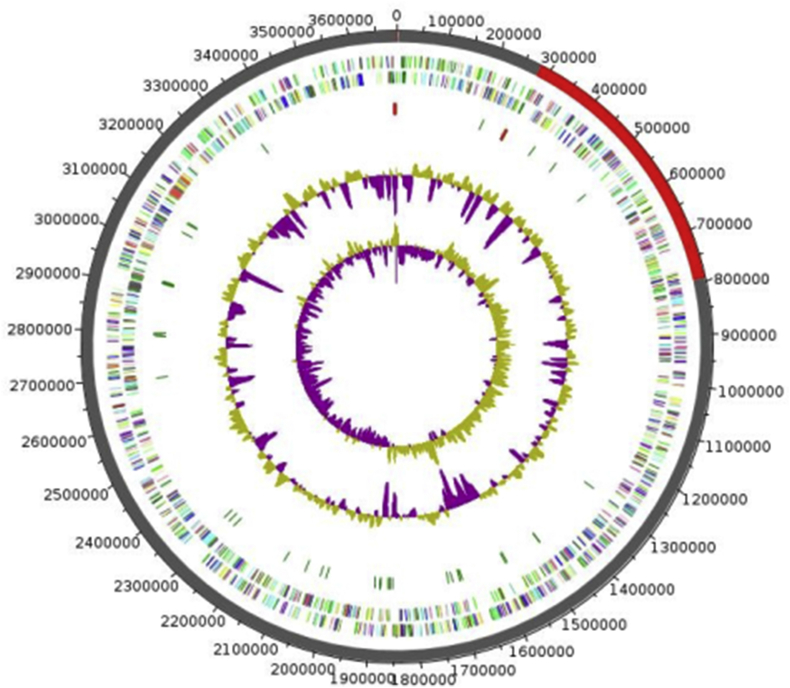
Fig. 5.
Graphical circular map of genome of Xanthomonas massiliensis strain SN6T. From outside to center: Genes on forward strand coloured by COGs categories (only genes assigned to COGs), genes on reverse strand coloured by COGs categories (only gene assigned to COGs), RNA genes (tRNAs green, rRNAs red), GC content and GC skew. COGs, Clusters of Orthologous Groups database.
Table 5.
Number of genes associated with 25 general COGs functional categories
| Code | Value | % of total | Description |
|---|---|---|---|
| J | 199 | 6.3436403 | Translation |
| A | 1 | 0.031877592 | RNA processing and modification |
| K | 142 | 4.5266175 | Transcription |
| L | 93 | 2.9646158 | Replication, recombination and repair |
| B | 1 | 0.031877592 | Chromatin structure and dynamics |
| D | 32 | 1.020083 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0 | Nuclear structure |
| V | 83 | 2.64584 | Defense mechanisms |
| T | 104 | 3.3152692 | Signal transduction mechanisms |
| M | 158 | 5.0366592 | Cell wall/membrane biogenesis |
| N | 37 | 1.1794709 | Cell motility |
| Z | 1 | 0.031877592 | Cytoskeleton |
| W | 34 | 1.0838381 | Extracellular structures |
| U | 72 | 2.2951865 | Intracellular trafficking and secretion |
| O | 119 | 3.7934332 | Posttranslational modification, protein turnover, chaperones |
| X | 37 | 1.1794709 | Mobilome: prophages, transposons |
| C | 164 | 5.227925 | Energy production and conversion |
| G | 130 | 4.144087 | Carbohydrate transport and metabolism |
| E | 193 | 6.152375 | Amino acid transport and metabolism |
| F | 65 | 2.0720434 | Nucleotide transport and metabolism |
| H | 103 | 3.283392 | Coenzyme transport and metabolism |
| I | 146 | 4.654128 | Lipid transport and metabolism |
| P | 157 | 5.0047817 | Inorganic ion transport and metabolism |
| Q | 80 | 2.5502074 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 227 | 7.236213 | General function prediction only |
| S | 150 | 4.7816386 | Function unknown |
| — | 935 | 29.805548 | Not in COGs |
COGs, Clusters of Orthologous Groups database.
Genome comparison
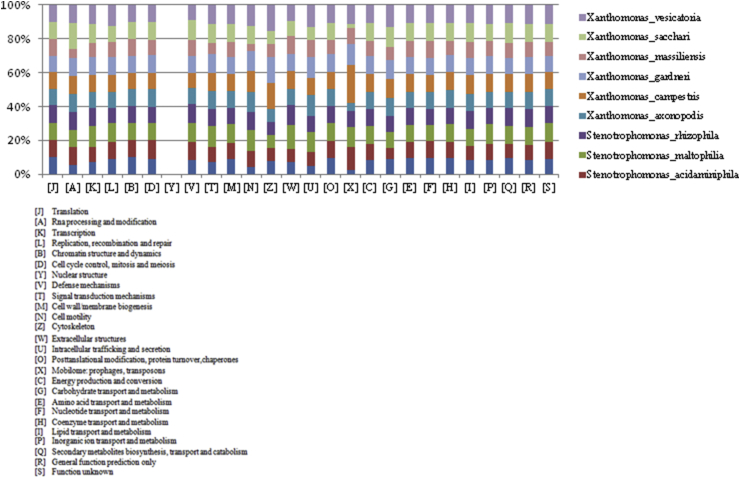
The draft genome sequence of Xanthomonas massiliensis is smaller than that of Stenotrophomonas acidaminiphila, Xanthomonas vesicatoria, Stenotrophomonas maltophilia, Stenotrophomonas rhizophila, Xanthomonas gardneri, Xanthomonas sacchari, Xanthomonas campestris and Xanthomonas axonopodis (3.69, 4.14, 5.53, 4.93, 4.65, 5.31, 4.93, 5.08 and 5.03 MB, respectively), but larger than the genome of Pseudoxanthomonas suwonensis (3.53 MB). The G + C content of Xanthomonas massiliensis is larger than that of Stenotrophomonas acidaminiphila, Xanthomonas vesicatoria, Stenotrophomonas maltophilia, Stenotrophomonas rhizophila, Pseudoxanthomonas suwonensis, Xanthomonas gardneri, Xanthomonas sacchari, Xanthomonas campestris and Xanthomonas axonopodis (70.523, 68.48, 64.07, 66.23, 67.30, 70.515, 63.53, 69.04, 65.07 and 64.89%, respectively). The gene content of Xanthomonas massiliensis is smaller than that of Stenotrophomonas acidaminiphila, Xanthomonas vesicatoria, Stenotrophomonas maltophilia, Stenotrophomonas rhizophila, Xanthomonas gardneri, Xanthomonas sacchari, Xanthomonas campestris and Xanthomonas axonopodis (3137, 3617, 4927, 4565, 3938, 4228, 4168, 4181 and 3904, respectively), but larger than the genome of Pseudoxanthomonas suwonensis (3132). This comparison of genomes between X. massiliensis and the other genetically closest species is shown in Table 6. In all genomes compared, the distribution of genes into COGs categories is identical (Fig. 6).
Table 6.
Genome comparison of closely related species to Xanthomonas massiliensis strain SN6T.
| Organism | INSDC | Size (Mbp) | G + C % | Protein-coding genes |
|---|---|---|---|---|
| Xanthomonas_ massiliensis | FCOY00000000.1 | 3.69 | 70.52 | 3137 |
| Xanthomonas_vesicatoria_ATCC_35937_LMG_911_T | AEQV00000000.1 | 5.53 | 64.06 | 4927 |
| Xanthomonas_gardneri_type_strain__DSM_19127 | AEQX00000000.1 | 5.30 | 63.53 | 4228 |
| Xanthomonas_axonopodis_LMG_538-T | JPYE00000000.1 | 5.02 | 64.88 | 3904 |
| Xanthomonas_sacchari_LMG_471_T | CP010409.1 | 4.92 | 69.04 | 4168 |
| Xanthomonas_campestris_pv. _campestris_str. _ATCC_33913_ATCC_33913 | AE008922.1 | 5.07 | 65.06 | 4181 |
| Stenotrophomonas_acidaminiphila_AMX19 | CP012900.1 | 4.13 | 68.48 | 3617 |
| Stenotrophomonas_maltophilia_IAM_12423 | CP008838.1 | 4.93 | 66.22 | 4565 |
INSDC, International Nucleotide Sequence Database Collaboration.
Fig. 6.
Distribution of functional classes of predicted genes according to clusters of orthologous groups of proteins.
Among Xanthomonas species with standing in nomenclature, AGIOS values ranged from 64.76% between Xanthomonas campestris pv. campestris and Stenotrophomonas acidaminiphila to 79.65% between Xanthomonas sacchari and Pseudoxanthomonas suwonensis. When comparing Xanthomonas massiliensis sp. nov. to other species, AGIOS values were in the same range, from 66.21% with Xanthomonas vesicatoria to 80.88% with Xanthomonas sacchari (Table 7). Among the species with standing in nomenclature, we found that by using the digital DNA-DNA hybridization (dDDH) with the GGDC software, values ranged from 20.8% between Xanthomonas vesicatoria and Pseudoxanthomonas suwonensis to 32.1% between Xanthomonas gardneri and Xanthomonas axonopodis. When comparing Xanthomonas massiliensis to other species, the dDDH value ranged from 21.3% with Stenotrophomonas maltophilia to 23.5% with Xanthomonas sacchari (Table 8).
Table 7.
Number of orthologous proteins shared between genomes (upper right) and AGIOS values obtained (lower left)
| P. suwonensis | X. gardneri | X. campestris | S. maltophilia | X. massiliensis | X. sacchari | X. vesicatoria | S. acidaminiphila | X. axonopodis | S. rhizophila | |
|---|---|---|---|---|---|---|---|---|---|---|
| P. suwonensis | 3132 | 1898 | 1925 | 1895 | 1678 | 1858 | 1914 | 1858 | 1902 | 1901 |
| X. gardneri | 66.11 | 4228 | 2945 | 2231 | 1859 | 2390 | 2962 | 2032 | 2880 | 2219 |
| X. campestris | 66.12 | 72.68 | 4181 | 2234 | 1848 | 2425 | 2970 | 2018 | 2909 | 2234 |
| S. maltophilia | 77.64 | 66.59 | 65.63 | 4565 | 1865 | 2137 | 2239 | 2174 | 2195 | 2466 |
| X. massiliensis | 79.82 | 67.19 | 66.64 | 78.60 | 3137 | 1788 | 1835 | 1778 | 1834 | 1854 |
| X. sacchari | 79.65 | 67.94 | 67.01 | 79.13 | 80.88 | 4168 | 2440 | 1973 | 2427 | 2127 |
| X. vesicatoria | 65.61 | 69.79 | 68.62 | 65.75 | 66.21 | 66.85 | 4927 | 2024 | 2934 | 2223 |
| S. acidaminiphila | 69.27 | 64.47 | 64.76 | 69.67 | 69.57 | 69.80 | 67.53 | 3617 | 1983 | 2125 |
| X. axonopodis | 66.01 | 70.07 | 63.74 | 66.33 | 66.92 | 67.70 | 71.02 | 67.35 | 3904 | 2197 |
| S. rhizophila | 66.12 | 65.42 | 61.96 | 68.84 | 66.81 | 67.04 | 66.54 | 67.77 | 66.98 | 3938 |
Numbers of proteins per genome are indicated in bold. AGIOS, average genomic identity of orthologous gene sequences.
Pseudoxanthomonas suwonensis 4M1, Xanthomonas gardneri DSM 19127, Xanthomonas campestris pv. campestris ATCC33913, Stenotrophomonas maltophilia IAM12423, Xanthomonas massiliensis SN6T, Xanthomonas sacchari LMG471T, Xanthomonas vesicatoria ATCC35937_LMG911T, Stenotrophomonas acidaminiphila AMX19, Xanthomonas axonopodis LMG538T, Stenotrophomonas rhizophila ep10.
Table 8.
Pairwise comparison of Xanthomonas massiliensis with other species using GGDC, formula 2 (DDH estimates based on identities/HSP length),a upper right
| XM | XV | XS | XG | XCC | XA | SR | SM | SA | PS | |
|---|---|---|---|---|---|---|---|---|---|---|
| XM | 100 | 22 (19.7–24.4%) | 23.5 (21.2–26%) | 21.9 (19.6–24.3%) | 21.9 (19.7–24.4%) | 22 (19.8–24.5%) | 21.9 (19.7–24.4%) | 21.3 (19.1–23.7%) | 22.7 (20.4–25.2%) | 22.2 (20–24.7%) |
| XV | 100 | 23.1 (20.8–25.6%) | 32 (29.6–34.6%) | 29.8 (27.4–32.3%) | 31.4 (29–33.9%) | 22 (19.8–24.5%) | 21.6 (19.4–24.1%) | 22.1 (19.8–24.5%) | 20.8 (18.6–23.3%) | |
| XS | 100 | 23.3 (21–25.8%) | 23.3 (21–25.8%) | 23.6 (21.3–26%) | 23.6 (21.3–26%) | 22.7 (20.5–25.2%) | 23.7 (21.4–26.2%) | 22.3 (20–24.7%) | ||
| XG | 100 | 31.1 (28.7–33.6%) | 32.1 (29.7–34.6%) | 22.2 (19.9–24.6%) | 21.6 (19.3–24%) | 22 (19.8–24.5%) | 21 (18.7–23.4%) | |||
| XCC | 100 | 29.6 (27.2–32.1%) | 22.2 (19.9–24.6%) | 21.6 (19.3–24%) | 22.1 (19.8–24.5%) | 21.1 (18.8–23.5%) | ||||
| XA | 100 | 22.5 (20.2–24.9%) | 22.1 (19.8–24.5%) | 22.3 (20.1–24.8%) | 21 (18.8–23.5%) | |||||
| SR | 100 | 24 (21.7–26.5%) | 23.9 (21.5–26.3%) | 21.6 (19.3–24%) | ||||||
| SM | 100 | 23.1 (20.9–25.6%) | 21.1 (18.8–23.5%) | |||||||
| SA | 100 | 22.2 (20–24.7%) | ||||||||
| PS | 100 |
Bold indicates comparison between strain and itself.
DDH, DNA-DNA hybridization; GGDC, Genome-to-Genome Distance Calculator; HSP, high-scoring segment pairs; PS, Pseudoxanthomonas suwonensis; SA, Stenotrophomonas acidaminiphila; SM, Stenotrophomonas maltophilia; SR, Stenotrophomonas rhizophila; XA, Xanthomonas axonopodis; XCC, Xanthomonas campestris pv. Campestris; XG, Xanthomonas gardneri; XM, Xanthomonas massiliensis; XS, Xanthomonas sacchari; XV, Xanthomonas vesicatoria.
Confidence intervals indicate inherent uncertainty in estimating DDH values from intergenomic distances based on models derived from empirical test data sets (which are always limited in size).
Conclusion
Phenotypic characteristics as well as phylogenetic and genomic analyses of strain SN6 suggest that it represents a novel species within the Xanthomonas genus, for which the name Xanthomonas massiliensis sp. nov. is proposed. This bacterial strain was isolated from the faecal flora of an obese Frenchwoman, and the description was based on a single isolate.
Description of Xanthomonas massiliensis sp. nov.
Xanthomonas massiliensis (mas.si.li.en'sis, L. fem. adj. massiliensis, ‘of Massilia,’ the Latin name of Marseille where strain SN6T was first cultivated).
X. massiliensis is a rod-shaped (0.6 × 1.8–2.0 μm), aerobic and Gram-negative bacterium occurring singly or in chains. Growth was also observed under microaerophilic conditions. Cells are motile with a flagellum and nonsporulating. Fresh colonies were yellow, circular, smooth and viscous with a diameter of 1 to 2 mm on COS. Optimal growth of strain SN6 occurred at 37°C under aerobic atmosphere with a pH of 7 but did not grow at 5% of salinity or under anaerobic conditions. The strain was catalase positive. Tests for nitrate reduction, indole production and urease were negative. API 50CH showed that the only substrate used as a carbon source was esculin. Positive reactions were detected for alkaline phosphatase, esterase lipase (C8), leucine arylamidase, valine arylamidase, trypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, β-galactosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, gelatin hydrolysis, malate and N-acetylglucosamine. The strain was sensitive to ceftriaxone, ciprofloxacin, clindamycin, doxycycline, erythromycin, gentamicin, penicillin, rifampicin, colistin, vancomycin, fosfomycin and trimethoprim/sulfamethoxazole but resistant to metronidazole, oxacillin and teicoplanin. Predominant fatty acids were 13-methyl-tetradecanoic acid followed by 9-methyl-decanoic acid.
The DNA G + C content is about 70.52%. The 16S rRNA gene and genome sequences were deposited in GenBank under accession number LN881611 and FCOY00000000, respectively. The type strain is Xanthomonas massiliensis strain SN6T (= CSUR P2129 = DSM 100900) and was isolated from human faeces.
Acknowledgements
The authors thank the Xegen Company (http://www.xegen.fr/) for automating the genomic annotation process, and M. Lardiere (MEPHI) for English-language editorial review. This study was supported by the Fondation Méditerranée Infection and the French government under the ‘Investissements d'avenir’ with the reference Mediterranée Infection 10-IAHU-3.
Conflict of Interest
None declared.
References
- 1.Dowson D. On the systematic position and generic names of the Gram negative bacterial plant pathogens. Zbl Bakteriol Parasitenkd Infekt Hyg Abt. 2 100 1939;100:177–193. [Google Scholar]
- 2.Dye D.W., Lelliott R.A. Genus 11. Xanthomonas Dowson 1939. In: Buchanan R.E., Gibbons N.E., editors. Bergey’s manual of determinative bacteriology. 8th ed. Williams & Wilkins; Baltimore: 1974. pp. 243–249. [Google Scholar]
- 3.Vauterin L., Hoste B., Kersters K., Swings J. Reclassification of Xanthomonas. Int J Syst Bacteriol. 1995;45:472–489. [Google Scholar]
- 4.Yang P., Vauterin L., Vancanneyt M., Swings J., Kersters K. Application of fatty acid methyl esters for the taxonomic analysis of the genus Xanthomonas. Syst Appl Microbiol. 1993;16:47–71. [Google Scholar]
- 5.Darrasse A., Carrère S., Barbe V., Boureau T., Arrieta-Ortiz M.L., Bonneau S. Genome sequence of Xanthomonas fuscans subsp. fuscans strain 4834-R reveals that flagellar motility is not a general feature of xanthomonads. BMC Genomics. 2013;14:761. doi: 10.1186/1471-2164-14-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lagier J.C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P.E., Rolain J.M. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 8.Morel A.S., Dubourg G., Prudent E., Edouard S., Gouriet F., Casalta J.P. Complementarity between targeted real-time specific PCR and conventional broad-range 16S rDNA PCR in the syndrome-driven diagnosis of infectious diseases. Eur J Clin Microbiol Infect Dis. 2015;34:561–570. doi: 10.1007/s10096-014-2263-z. [DOI] [PubMed] [Google Scholar]
- 9.Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006:152–155. [Google Scholar]
- 10.Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 11.Sasser M. Microbial ID; Newark, NY: 2006. Bacterial identification by gas chromatographic analysis of fatty acids methyl esters (GC-FAME) [Google Scholar]
- 12.Dione N., Sankar S.A., Lagier J.C., Khelaifia S., Michele C., Armstrong N. Genome sequence and description of Anaerosalibacter massiliensis sp. nov. New Microbes New Infect. 2016;10:66–76. doi: 10.1016/j.nmni.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lagier J.C., Ramasamy D., Rivet R., Raoult D., Fournier P.E. Non contiguous-finished genome sequence and description of Cellulomonas massiliensis sp. nov. Stand Genomic Sci. 2012;7:258–270. doi: 10.4056/sigs.3316719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagier J.C., Bibi F., Ramasamy D., Azhar E.I., Robert C., Yasir M. Non contiguous-finished genome sequence and description of Clostridium jeddahense sp. nov. Stand Genomic Sci. 2014;9:1003–1019. doi: 10.4056/sigs.5571026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012 27;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyatt D., Chen G.L., Locascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagesen K., Hallin P., Rødland E.A., Stærfeldt H.H., Rognes T., Ussery D.W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Käll L., Krogh A., Sonnhammer E.L. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 23.Gouret P., Thompson J.D., Pontarotti P. PhyloPattern: regular expressions to identify complex patterns in phylogenetic trees. BMC Bioinform. 2009;10:298. doi: 10.1186/1471-2105-10-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lechner M., Findeiß S., Steiner L., Marz M., Stadler P.F., Prohaska S.J. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinform. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meier-Kolthoff J.P., Auch A.F., Klenk H.P., Göker M. Genome sequence–based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrewes A.G., Jenkins C.L., Starr M.P., Shepherd J., Hope H. Structure of xanthomonadin I, a novel dibrominated arylpolyene pigment produced by the bacterium Xanthomonas juglandis. Tetrahedron Lett. 1976;45:4023–4024. [Google Scholar]