Abstract
Clinical prediction of advanced hepatic fibrosis (HF) and cirrhosis has long been challenging due to the gold standard, liver biopsy, being an invasive approach with certain limitations. Less invasive blood test tandem with a cutting-edge machine learning algorithm shows promising diagnostic potential.
In this study, we constructed and compared machine learning methods with the FIB-4 score in a discovery dataset (n = 490) of hepatitis B virus (HBV) patients. Models were validated in an independent HBV dataset (n = 86). We further employed these models on two independent hepatitis C virus (HCV) datasets (n = 254 and 230) to examine their applicability.
In the discovery data, gradient boosting (GB) stably outperformed other methods as well as FIB-4 scores (p < .001) in the prediction of advanced HF and cirrhosis. In the HBV validation dataset, for classification between early and advanced HF, the area under receiver operating characteristic curves (AUROC) of GB model was 0.918, while FIB-4 was 0.841; for classification between non-cirrhosis and cirrhosis, GB showed AUROC of 0.871, while FIB-4 was 0.830. Additionally, GB-based prediction demonstrated good classification capacity on two HCV datasets while higher cutoffs for both GB and FIB-4 scores were required to achieve comparable specificity and sensitivity.
Using the same parameters as FIB-4, the GB-based prediction system demonstrated steady improvements relative to FIB-4 in HBV and HCV cohorts with different cutoff values required in different etiological groups. A user-friendly web tool, LiveBoost, makes our prediction models freely accessible for further clinical studies and applications.
Keywords: Hepatic fibrosis, FIB-4, Hepatitis B, Hepatitis C, Machine learning, Gradient boosting
Research in context.
Evidence before this study
We searched the PubMed database according to the terms [(“prediction” OR “risk prediction” OR “prediction model” OR “predictive” OR “predictive modeling”) AND (“FIB-4” OR “Fibrosis-4” OR “FIB4”) AND (“machine learning” OR “ensemble learning” OR “gradient boosting”) AND (“liver fibrosis” OR “hepatic fibrosis”)] among English-language articles before March 4th, 2018. We identified two studies using genotype-based decision tree models to predict advanced liver fibrosis in patients with chronic hepatitis C viral infection (HCV) and non-alcoholic fatty liver disease in patients with chronic hepatitis B viral infection (HBV). Neither of these studies attempted to use machine learning methods to augment the diagnostic performance based on commonly used clinical indicators, nor discussed the performance between different viral etiologies. We hypothesized that applying cutting-edge machine learning algorithms to existing blood-test scoring system (FIB-4) can augment the detection of advanced hepatic fibrosis and cirrhosis in chronic liver disease patients.
Added value of this study
Our study constructed and compared machine learning methods based on the same clinical parameters of the FIB-4 scoring system in an HBV cohort for detecting advanced hepatic fibrosis and cirrhosis. We validated our models in three independent cohorts including both HBV and HCV. Our machine learning-based prediction system, a less-invasive approach, demonstrated steady diagnostic improvements, which could overcome certain limitations of the gold standard (i.e., liver biopsy), facilitate medical decision making, and enhance long-term clinical surveillance of chronic liver disease.
Implications of all the available evidence
To fill in gaps between machine learning algorithms and real-world clinical studies, we built a user-friendly web tool (LiveBoost) that makes our prediction models easily accessible for further studies and applications.
Alt-text: Unlabelled Box
1. Introduction
Every year, chronic liver disease (CLD) and its complications lead to approximately 2 million deaths globally [1]. Hepatitis B and C virus infections, chronic alcohol consumption and immune system abnormalities are leading causes of liver injury, with the wound-healing response from liver injury leading to hepatic fibrosis (HF), cirrhosis, and ultimately organ failure or liver cancer [2]. Evidence suggests that HF is reversible in many cases of CLD, but the clinically-significant regression of cirrhosis is still controversial [3], thus highlighting the necessity of a clinical tool for early detection of HF, differentiation of cirrhosis, and longitudinal surveillance of therapeutic responses. The gold standard for clinical measurement of HF is the liver biopsy, which is associated with both significant complications [4] and limitations [5] (e.g., pain, bleeding, infection, perforation of nearby organs, sampling errors, inter-observer and intra-observer variability). A less invasive and more reproducible approach of assessing HF severity and progression would be of great value in the clinical setting.
The development of scoring systems based on simple clinical parameters and blood tests (e.g., FIB-4) is one important step towards non-invasive CLD monitoring and diagnosis [6]. FIB-4 was introduced as a non-invasive method to predict HF stages among Caucasian patients with hepatitis C virus (HCV) and human immunodeficiency virus co-infection [7]. Since then, this method has been independently validated in multiple HCV infected,and hepatitis B virus (HBV) infected patient cohorts [[8], [9], [10]]. FIB-4 provides an attractive alternative for biopsy due to its affordable price, objective measurements, and avoidance of complications. The formula of FIB-4 is defined as:
AST: aspartate transaminase; ALT: alanine transaminase; PLT: platelet count.
This relatively simple formula was originally derived from a multiple logistic regression (LR) model with odds ratios considered [7]. However, this statistical approach ignores more complex non-linear interactions between variables that might play significant roles in determining HF severity, and which could be captured using more sophisticated modeling approaches. In recent years, machine learning along with the explosive growth of biomedical big-data has generated much interest in developing clinical informatics tools for disease diagnosis, staging, and prognosis [[11], [12], [13]]. Machine learning, especially ensemble learning, has been successfully applied for recognizing hidden patterns in complex data, allowing for better predictions of clinical outcomes than traditional statistical models, especially when applied to large-scale datasets [14]. Unlike conventional regression-based approaches, ensemble learning algorithms such as random forest (RF) and gradient boosting (GB), are capable of capturing higher-order, non-linear interactions between predictors [15]. For HBV and HCV patients, pathology and genetic data have been successfully used for the implementation of predictive models [[16], [17], [18], [19]]. Here, for the first time, we propose to reconstruct an existing blood test-based clinical scoring system, FIB-4, with cutting-edge machine learning approaches for improved detection and classification of advanced HF and cirrhosis, validating models in multiple independent datasets from patients with CLD of different viral etiologies.
2. Materials and methods
2.1. Data and ethics
An HBV discovery dataset included a total of 490 HBV infected subjects recruited from Shuguang Hospital in affiliation with Shanghai University of Traditional Chinese Medicine (Shanghai, China) from April 2013 to June 2015. Patients were included after providing informed consent and meeting inclusion and exclusion criteria as described in the appendix (Text S1). An independent HBV dataset (validation-1) included a total of 86 HBV infected subjects recruited from Xiamen Hospital of Traditional Chinese Medicine (Xiamen, China). Recruitment and eligibility criteria were the same as those established for the discovery dataset. These studies were approved by the institutional review board of the Shanghai University of Traditional Chinese Medicine and Xiamen Hospital of Traditional Chinese Medicine. All participants signed informed consent forms for the study.
Two additional retrospective anonymous datasets from existing studies were used to further evaluate the prediction models in HCV infected patients. Validation-2 (HCV), comprised of a total of 254 HCV infected subjects, was recruited from China Medical University Hospital, Taiwan. Detailed information about this cohort was provided in the original study publication [20]. Another independent dataset, validation-3 (HCV), comprised of a total of 230 samples from 115 HCV infected patients, was recruited from Komaki City Hospital (Komaki, Japan). In this cohort, biopsy results, clinical parameters, and blood samples were available from before and after antiviral treatment. Detailed information about this study is provided in the original study publication [21].
2.2. Liver biopsy
An ultrasound-guided liver biopsy was performed on all patients in both the discovery and HBV validation datasets. All liver biopsies were performed within one week after study recruitment. Liver specimens were placed in 10% neutral buffered formalin and embedded in paraffin for histologic processing. Tissue sections were stained with Masson's trichrome staining and hematoxylin and eosin (H&E). The histologic staging was based on Scheuer's classification using a 5-point scale for HF severity ranging from S0 (non-fibrosis) to S4 (cirrhosis) [22]. The staging was performed by three independent pathologists from Shanghai Medical College of Fudan University who were blinded to patient clinical information. In cases of discordant staging, specimens were re-examined until consensus was reached.
2.3. Serum sample collection and test
Overnight fasting (12h) blood samples were collected from all discovery and validation-1 subjects within one week after recruitment. Blood specimens were placed on ice, processed by centrifugation, and stored in a − 80 °C freezer until analysis. Hematological and standard biochemical tests were performed according to the manufacturers' protocols using an LH750 Hematology Analyzer and Synchron DXC800 Clinical System (Beckman Coulter, USA).
2.4. Machine learning and statistics
The original formula for FIB-4 suggested potential interactions between predictors. Thus we began our machine learning modeling with a decision tree (DT) considering its intrinsic capacity for interaction detection. We applied DT, along with two ensemble learning models, RF and GB, to reconstruct the four individual components (Age, AST, ALT, and PLT) of the FIB-4 score.
DT is a flowchart-like prediction model that depicts a complete decision-making process where each internal node represents a decision point on a single attribute, and each leaf represents a single assigned class label [23]. The structure of the DT model is similar to the clinical decision-making process, providing a sound rationale for its application to clinical problems [24]. Unfortunately, it often suffers from over-fitting and is considered a high-variance model [25]. The RF model, on the other hand, is an ensemble method which aggregates a large number of DTs using bootstrap resampling and often yields lower variances and better model generalization than single DT [26]. The GB model goes one step further, instead of averaging prediction results from all DTs in RF, it grows a new DT based on old trees by decreasing prediction errors that the old trees made [27].
To optimize the model hyper-parameters, 10-fold cross-validation was performed with different hyper-parameter settings in the discovery set. We optimized the complexity parameter for DT and the mtry parameter for RF. For GB, we tuned parameters including, interaction.depth, n.trees, shrinkage, and n.minobsinnode in a grid search manner. Receiver operating characteristic (ROC) curves were used as evaluation metrics. The R package caret was applied for the hyper-parameter optimization [28]. The details of tuned hyper-parameters can be found in https://github.com/elise-is/LiveBoost.
To determine the final model and test its robustness, we randomly split the discovery set into training (70%) and testing sets (30%) 100 times. Each time, we trained the three different machine learning models on the training set using fixed hyper-parameters based on previous model tuning results and lastly, compared these results with the FIB-4 score on the testing set. Area under ROC curves (AUROC) and area under precision-recall curves (AUPR) were calculated to compare the four methods (Step 1 in Fig. 1). The R packages rpart, randomForest, and gbm were applied for the DT, RF, and GB model training, respectively [27, 29, 30].
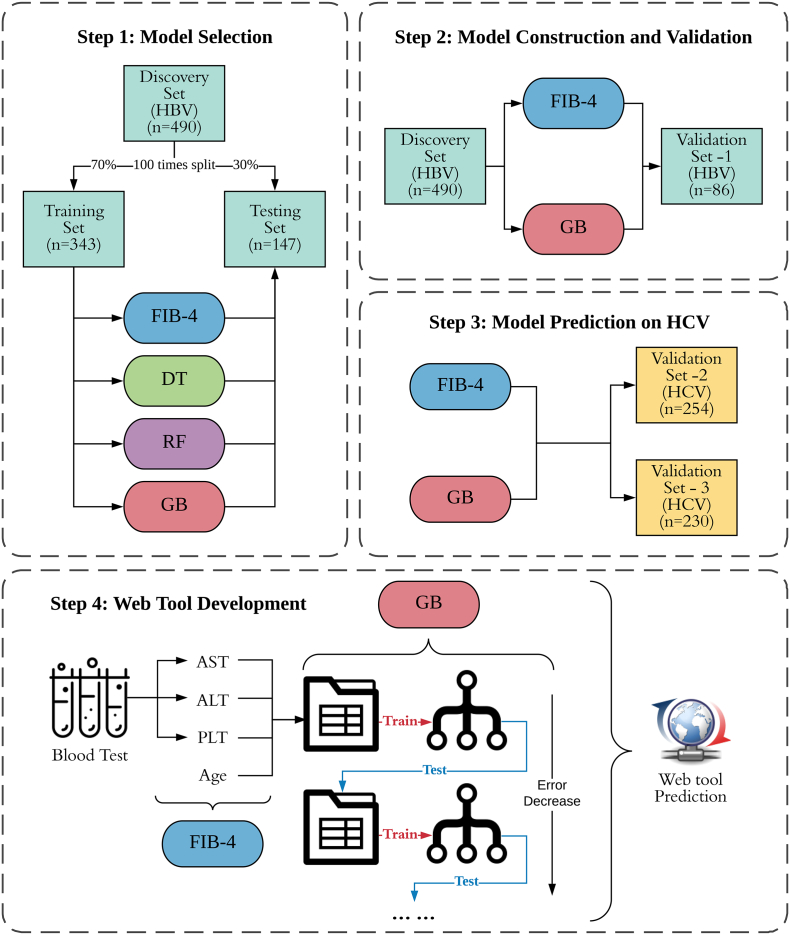
Fig. 1.
Flowchart of the study design. In step 1 of model selection, we performed training-testing splitting 100 times on the discovery set and trained DT, RF, GB models on the training sets, then compared these results with FIB-4 on testing sets. In step 2, we constructed final GB models and compared results with FIB-4 on the whole discovery set and then validated on the HBV validation set. In step 3, GB models and FIB-4 were used to predict the risks for two extra HCV cohorts. In step 4, we developed a user-friendly web-tool for clinical practices.
After selecting GB as our preferred reconstruction approach, we trained the final GB models on the full discovery set using previously optimized hyper-parameters. Then, we compared our GB prediction scores with FIB-4 scores on both discovery set and validation set-1 (HBV) using ROC and PR curves. The best GB cutoff points were selected using Youden's index, maximizing the sum of sensitivity and specificity [31]. For the FIB-4 score, we applied two previously reported clinical cutoff points (1.45 and 3.25) [[6], [7], [8]]. We calculated specificity and sensitivity at these cutoff points and their 95% confidence intervals (CIs) using 500 times bootstrap resampling (Step 2 in Fig. 1). ROC and PR calculation were conducted with the R packages pROC and PRROC, respectively [32, 33].
To further assess the classification robustness of the FIB-4 reconstruction models for staging HF related to HCV, we applied our trained GB models on two independent HCV validation cohorts (Step 3 in Fig. 1). We employed t-tests to compare FIB-4 and GB scores in early vs. advanced HF and fibrosis vs. cirrhosis within both HCV cohorts. ROC curves, sensitivity, specificity and 95% CIs for predicting advanced HF were calculated for the FIB-4 and GB scores.
We additionally included two extra blood test-based clinical indicators (i.e., albumin (ALB) and gamma-glutamyl transpeptidase (GGT)) to check whether classification performances could be further improved. We rebuilt new GB models on six predictors (Age, AST, ALT, PLT, ALB, and GGT) and LR models based on FIB-4, ALB and GGT in the discovery set and compared with the original models in two HBV cohorts using ROC curves.
Datasets and R-code related to this study can be found at https://github.com/elise-is/LiveBoost.
2.5. Web-tool development
To develop a tool for HF staging that is amenable to use in clinical practices, we designed a web-based application, LiveBoost, providing a graphical user interface (GUI) to access our final trained GB models (Step 4 in Fig. 1). This application is hosted on our server which is publicly accessible via https://metabolomics.cc.hawaii.edu/software/LiveBoost/. The web-tool development was conducted using the R package shiny.
3. Results
3.1. Machine learning model selection
For our first aim (i.e., finding a machine learning approach that robustly improves the original FIB-4 score), we compared the original FIB-4 scoring system with different models using a 100-times jackknife resampling approach by randomly splitting the discovery set into 70% training set and 30% testing set. We built each model on the training set with optimized hyper-parameters and compared results to FIB-4 on the testing set. For differentiating between early (S0–2) and advanced (S3–4) fibrosis, we found that compared to FIB-4 score, DT was associated with comparable AUPR (0.67 vs. 0.68, p = .15) but significantly lower AUROC (0.79 vs. 0.82, p < .001). The RF approach was associated with significantly higher AUPR (0.73 vs. 0.68, p < .001) and comparable AUROC (0.82 vs. 0.82, p = .59). The GB approach was associated with significantly higher AUPR (0.77 vs. 0.68, p < .001) and AUROC (0.85 vs. 0.82, p < .001) (left side panels of Fig. 2). Similarly, for the identifying of cirrhosis cases (S4) (right side panels of Fig. 2), we found that compared to FIB-4 scoring, the DT based approach was associated with significantly lower AUPR (0.60 vs. 0.66, p < .001) and AUROC (0.81 vs. 0.87, p < .001). The RF-based approach was associated with significantly higher AUPR (0.70 vs. 0.66, p = .0047) and AUROC (0.89 vs. 0.87, p = .0023). The GB approach was associated with even greater significant differences in AUPR (0.72 vs. 0.66, p < .001) and AUROC (0.90 vs. 0.87, p < .001). Descriptive statistics of the AUROCs and AUPRs for these approaches are summarized in Table S1. Altogether, the GB approach provided the greatest improvements in classification capacity over FIB-4 scoring system among the three machine learning methods. Additionally, higher variances associated with the DT approach indicated less robustness than the other approaches. In addition to showing significantly better classification performance relative to FIB-4, two ensemble learning approaches were associated with smaller variances than DT.
Fig. 2.
Boxplots of AUPR and AUROC on testing sets for four different methods. P-values were calculated using Student's t-tests.
3.2. Model construction and validation
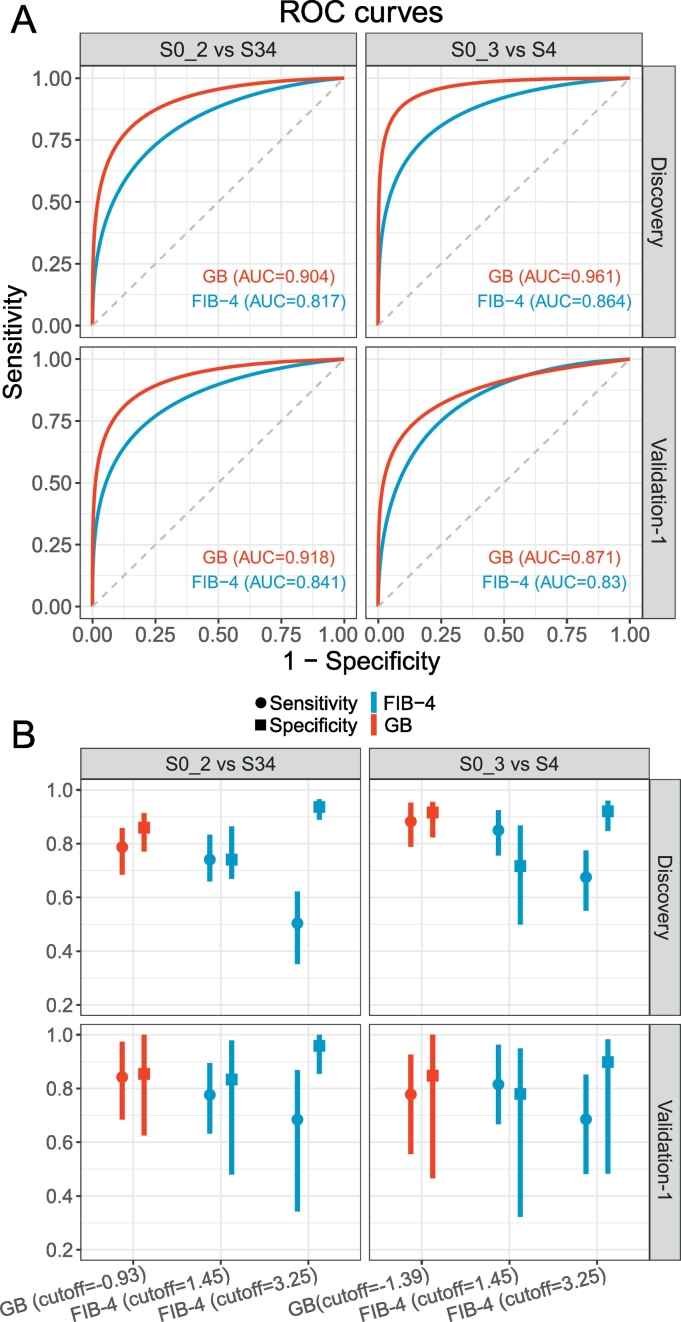
After we selected the GB model as our reconstruction approach for FIB-4, we finalized our prediction models for detecting advanced HF (discriminating S0–2 from S3–4) and cirrhosis (discriminating S0–3 from S4) by training GB models on the full discovery set with optimized hyper-parameters, to produce GB-based risk scores. To validate this GB-based scoring system, we applied it to our first validation set which was derived from an independent HBV cohort. Table 1 summarizes the four clinical indicators and other demographic information for the discovery and validation-1 datasets. Applying our final trained GB models to generate risk scores (in log-odds scale) for all the samples in both the discovery and validation-1 sets, we again found higher AUROC (Fig. 3A) and AUPR (Fig. S1) values for the GB-based scoring relative to FIB-4 scoring for both datasets. For classification between S0-2 and S3-4, GB showed an AUROC of 0.904 and 0.918, AUPR of 0.836 and 0.925 in the discovery set and validation set-1, respectively while FIB-4 showed an AUROC of 0.817 and 0.841, AUPR of 0.688 and 0.844, respectively. For classification between S0–3 and S4, GB showed an AUROC of 0.961 and 0.871, AUPR of 0.891 and 0.833 in the discovery set and validation set-1, respectively while FIB-4 showed an AUROC of 0.864 and 0.830, AUPR of 0.671 and 0.738, respectively. We then compared the specificity and sensitivity at the best cutoff values for GB scores and two recommended cutoffs for FIB-4 (Fig. 3B), finding that the GB prediction model produced higher specificity (0.86 and 0.85 in the discovery set and validation set-1, respectively) and sensitivity (0.79 and 0.84 in the discovery set and validation set-1, respectively) than FIB-4 (specificity = 0.74 and 0.83, sensitivity = 0.74 and 0.78 in the discovery set and validation set-1, respectively) with cutoff = 1.45 for discriminating stages S0-2 from S3-4. While FIB-4 scoring with cutoff = 3.25 resulted in higher specificity (0.94 and 0.96 in the discovery set and validation set-1, respectively), it suffered from lower sensitivity (0.50 and 0.68 in the discovery set and 0.68 in the validation-1 set) (left panels of Fig. 3B). Similarly, for detecting cirrhosis (S4), the GB prediction model showed higher and more stable (with smaller CIs) specificity (0.92) and sensitivity (0.88) in the discovery set than FIB-4 using cutoff = 1.45 (specificity = 0.72 and sensitivity = 0.85) (upper right panel of Fig. 3B). Again, FIB-4 with cutoff = 3.25 showed high specificity (0.92) with much lower sensitivity (0.68) (upper right panel of Fig. 3B). In the validation-1 set, GB still demonstrated more balanced specificity (0.85) and sensitivity (0.78) while FIB-4 with cutoff = 1.45 showed lower specificity (0.78) with larger CI and cutoff = 3.25 and showed lower sensitivity (0.69) (lower right panel of Fig. 3B).
Table 1.
Clinical and demographical characteristics of the HBV cohorts.
| Data | HF stage | Total Num | Num of M | Num of F | BMI (kg/m^2) | Age (years) | AST (U/L) | ALT (U/L) | PLT (10^9/L) |
|---|---|---|---|---|---|---|---|---|---|
| Discovery Set (HBV) | 0 | 46 | 39 | 7 | 22.1 (20.3–23.5) | 32 (27–40) | 49 (35–66) | 106 (58–171) | 190 (161–215) |
| 1 | 169 | 125 | 44 | 21.2 (19.5–24.1) | 30 (25–38) | 58 (39–99) | 114 (65–190) | 179 (155–214) | |
| 2 | 134 | 93 | 41 | 21.6 (20.1–24.0) | 31 (27–39) | 74 (43–138) | 155 (80–267) | 176 (150–210) | |
| 3 | 56 | 47 | 9 | 22.5 (20.9–25.0) | 39 (29–47) | 62 (44–112) | 90 (56–250) | 148 (108–182) | |
| 4 | 85 | 53 | 32 | 22.5 (20.9–24.5) | 50 (40–58) | 45 (31–77) | 45 (28–100) | 86 (43–121) | |
| Validation Set (HBV) | 0 | 15 | 7 | 8 | 23.2 (21.2–24.0) | 35 (28–40) | 40 (23–67) | 65 (33−100) | 173 (152–193) |
| 1 | 21 | 14 | 6 | 22.5 (21. 3–24.8) | 31 (26–45) | 67 (36–128) | 98 (77–183) | 193 (174–221) | |
| 2 | 12 | 7 | 5 | 22.4 (21.5–23.3) | 39 (34–43) | 50 (40–97) | 76 (52–357) | 161 (145–178) | |
| 3 | 11 | 8 | 3 | 21.5 (20.5–22.8) | 40 (31–49) | 35 (33–53) | 40 (31–95) | 108 (93–118) | |
| 4 | 27 | 18 | 9 | 22.4 (20.1–23.9) | 45 (37–56) | 43 (32–70) | 35 (28–82) | 74 (40–98) |
Continuous variables are displayed as median value (25% - 75% quantile values), Num (number), F (female) M (male).
Fig. 3.
Classification performances of GB and FIB-4 on the discovery set and the HBV validation set. (A) ROC curves of GB and FIB-4 in advanced HF detection (left-panel) and cirrhosis detection (right-panel). (B) Specificity, sensitivity and their 95% CIs of GB and FIB-4 scores in advanced HF detection (left-panel) and cirrhosis detection (right-panel). We selected the best GB cutoff based on the Youden index for the discovery set and two commonly applied FIB-4 cutoffs (1.45 and 3.25).
To verify whether the classification performances could be further improved by introducing extra clinical parameters, ALB and GGT, which were reported in previous studies [21, 34], we additionally rebuilt GB models based on six predictors (Age, AST, ALT, PLT, ALB, and GGT) and LR models based on FIB-4, ALB and GGT. Comparing to our original GB models, new GB models slightly improved the AUROC (0.929 and 0.974 for S0–2 vs. S34 and S0–3 vs. S4, respectively, Fig. S2 upper panel) in the discovery set, and showed similar results in the HBV validation set-1 (0.91 and 0.874 for S0–2 vs. S34 and S0–3 vs. S4, respectively, Fig. S2 lower panel). When we compared the new FIB-4 models (FIB-4, ALB, and GGT) to the original FIB-4 score, we found although new FIB-4 models displayed similar AUROC in the discovery set (0.818 and 0.842 for S0–2 vs. S34 and S0–3 vs. S4, respectively, Fig. S3 upper panel), showed much lower AUROC in the HBV validation set-1 (0.738 and 0.757 for S0–2 vs. S34 and S0–3 vs. S4, respectively, Fig. S3 lower panel). Thus, we did not include these two parameters in the following analyses.
3.3. Model prediction on HCV cohorts
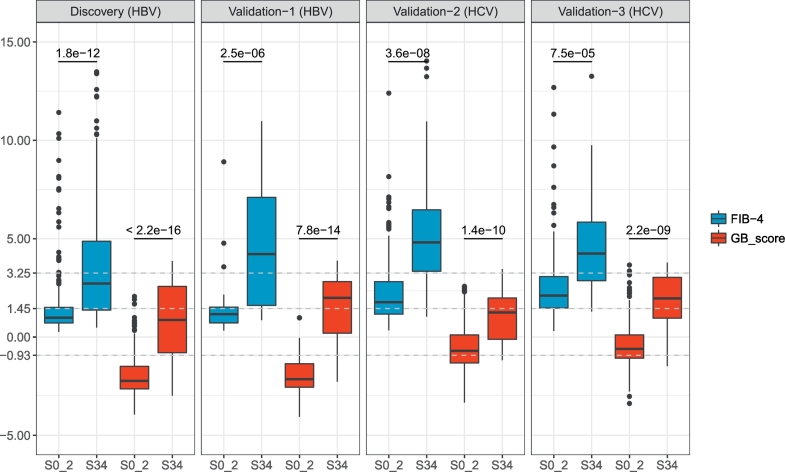
To investigate potential differences in GB risk score and FIB-4 score between HBV-related and HCV-related CLD cohorts, we applied our prediction models on two independent HCV validation data sets. We found significant differences in both FIB-4 and GB scores between groups staged S0–2 and S3–4, with more significant differences in GB scores than FIB-4 scores in both HBV (p < 2.2e-16 vs. p = 1.8e-12 in the discovery set and p = 7.8e-14 vs. p = 2.5e-6 in the validation set-1) and HCV cohorts (p = 1.4e-10 vs. p = 3.6e-8 in the validation set-2 and p = 2.2e-9 vs. p = 7.5e-5 in the validation set-3) (Fig. 4). Similar results were observed when discriminating cirrhosis (S4) from HF (S0–3) (Fig. S4). The GB and FIB-4 scores in HCV-related cohorts performed with AUROC = 0.797 and 0.816, respectively in validation set-2 and AUROC = 0.849 and 0.795, respectively in validation set-3. Thus, classification performance with the GB model was improved relative to FIB-4 in validation set-3 set (HCV), but not in validation set-2 dataset (HCV) (Fig. S5A).
Fig. 4.
FIB-4 and GB scores for four independent cohorts between S0–2 and S3–4. P-values were calculated using Student's t-tests.
When HCV-infected cohorts were compared to HBV-infected cohorts, higher mean FIB-4 scores and GB scores were noted in both the S0–2 and S0–3 groups (Fig. 4 and Fig. S4), a finding which suggested that cutoff values built on one cohort might not be ideal for other cohorts with different etiological causes for CLD. Measurement of the FIB-4 score in the HCV cohorts for staging advanced HF revealed that a cutoff value of 3.25 resulted in a better specificity and sensitivity trade-off point relative to a cutoff value of 1.45 which resulted in more false positive findings due to low specificity (Fig. S5B). In contrast, a FIB-4 cutoff value of 1.45 exhibited more balance between sensitivity and specificity values in the HBV cohorts while a cutoff value of 3.25 yielded false negative findings due to low sensitivity (Fig. 3B). Correspondingly, we assessed the sensitivity and specificity of a GB cutoff value of −0.93 (which produced optimal results on the HBV discovery set) for HCV in differentiating early and advanced HF. Applying this cutoff value to the validation-2 and 3 HCV datasets produced a classification performance associated with higher sensitivity, but at the expense of much lower specificity (Fig. S5B). A new GB cutoff value of −0.14 was calculated based on the Youden's index on the ROC curve of the validation-2 (HCV) dataset. This higher cutoff value yielded improvements on the point and interval estimations of specificity in both HCV cohorts while maintaining a reasonable balance with specificity and sensitivity for discriminating S0–2 vs. S3–4. We did not assess cirrhosis detection performances on these two HCV datasets due to the small number of cirrhosis samples. Table S2 includes all point and interval estimations of AUROC, specificity, and sensitivity of both the FIB-4 and GB scores.
3.4. Web-tool development
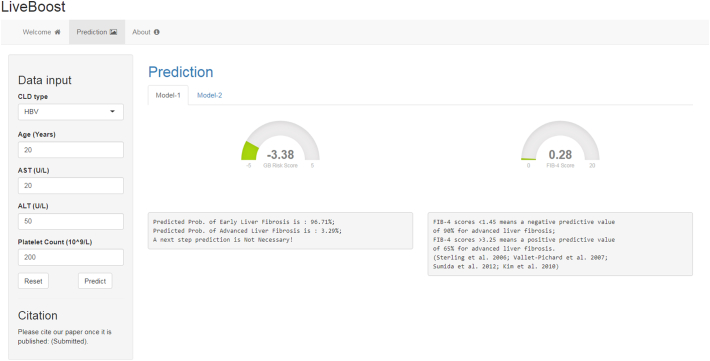
To encourage further study of the clinical application of HF staging using cutting-edge machine learning approaches, we packaged our trained GB models into a free, accessible web-tool (LiveBoost: https://metabolomics.cc.hawaii.edu/software/LiveBoost/). Fig. 5 shows the GUI of the web-tool. To use this web-tool, one can simply input values for the four clinical indicators along with the disease etiology followed by a click of the “Predict” button. Two gauge plots will be generated at the right panel of the interface showing the GB risk score and the FIB-4 score for this subject. Corresponding descriptions with calculated disease probabilities will appear below the plots. The next step for prediction of liver cirrhosis is provided in the “Model-2” tab. One clicks the “Reset” button to bring values back to their default settings.
Fig. 5.
A screenshot of the web-tool (LiveBoost).
4. Discussion
An affordable, reproducible, objective and non-invasive method for predicting the severity of HF is needed to support longitudinal surveillance and clinical decision making. In this study, we aimed to reconstruct the current FIB-4 scoring system to improve the sensitivity and specificity of classifications between early and advanced HF and for the detection of cirrhosis. To our knowledge, this is the first time that machine learning algorithms have been employed to improve the staging of CLD by building on an existing clinical scoring system. Furthermore, this algorithm has been implemented into a user-friendly web-tool to support further independent explorations of its clinical utility.
We compared the original FIB-4 score with three machine learning methods: DT, RF, and GB. The results showed that GB outperformed FIB-4 and other methods regarding AUPR and AUROC. Applying GB to an independent HBV dataset, we observed consistently superior performance to FIB-4 scoring. On two independent HCV validation sets, the trained GB model also showed good classification performance with more significant group-differences compared to FIB-4 scoring (Fig. 4, Fig. S4). Although the GB model produced similar AUROC values to the FIB-4 scores in validation set-2 (HCV) (Fig. S5), this might due to group imbalance and a lack of S0 group in this dataset. The GB model was associated with narrower CIs of specificity and sensitivity, supporting its potential for robust classification. Also, GB showed better classification performance than FIB-4 in validation set-3 (HCV) (Fig. S5). In this validation set, each patient underwent serial liver biopsies before and after antiviral therapy along with the corresponding blood tests. To avoid potential confounding factors caused by the therapy, we performed additional validation on the pre-treatment data and achieved consistent results with our previous analyses (Fig. S6).
In addition to FIB-4 parameters, other clinical indicators (e.g., ALB, GGT) were discovered as significant diagnostic predictors of liver disease [21, 34]. To assess whether ALB and GGT values could augment the performance of GB models and FIB-4 for the staging of CLD, we rebuilt GB and FIB-4 models with these two indicators added to the existing panel. The inclusion of ALB and GGT values did not produce significant improvements over our current models. Notably, the new FIB-4 LR models displayed even worse classification performances than the original FIB-4 score. These results might suggest a potential problem of overfitting in LR models when we include redundant predictors, while GB models did not particularly suffer from overfitting issues. What's more, high-order non-linear interactions between predictors may be better captured by innovative machine learning approaches [15] which might explain why the GB model outperformed FIB-4 for classifying CLD for the HBV cohorts. Thus, it will be worth trying this approach with other clinical predictors such as, the AST/platelet ratio index (APRI) [35], the AST/ALT ratio [36], the AST/ALT ratio/platelet ratio index (AARPRI) [37], the age-platelet index [38] or the FibroScan score [39]).
In addition to conventional clinical indicators, serum metabolomics could also serve as a potential source of biomarkers for assessing CLD. The liver is the principal organ for lipid metabolism in the manufacture of fatty acids from excess acetyl-CoA along with transportation and storage of lipid metabolites [40]. Bile acids are originally synthesized by liver cells and fibrosis-related changes in their enterohepatic circulation may be reflected as serum biomarkers [41]. Additionally, the liver performs a significant role in the degradation of amino acids [42]. CLD with progressive HF should therefore lead to alterations of various metabolites and indeed, previous studies demonstrated that changes in levels of amino acids [43], free fatty acids [44], and bile acids [45, 46] were highly correlated with progression of liver disease. Thus, metabolic alterations might serve as complementary information to existing clinical indicators for HF staging, making it worth exploring whether including metabolic markers into our CLD prediction models will improve classification performances, especially for the early fibrosis stratifications.
A potential caveat worth discussing is the performance of different cutoff values for FIB-4 and GB scores for different etiological CLD cohorts (i.e., HBV and HCV cohorts). Compared to HBV cohorts, there was a trend for higher FIB-4 and GB scores in early stage fibrosis of HCV patients (Fig. 4) which prompted us to apply a higher cutoff value to achieve more reasonable classification performances. Age, one of the parameters used to calculate the FIB-4 score, is also on average higher in HCV patients than HBV patients as shown in our study (Fig. S7A). HCV infections which are commonly acquired later in life than HBV infections, likely produce age at exposure differences and age-related prevalence differences between HBV and HCV induced liver injuries [47, 48]. Thus, etiologic and epidemiologic differences between HBV and HCV patients may both be contributing to differences in optimal FIB-4 and GB scores cutoffs between these groups. AST and ALT were found at lower levels in S0 and S4 groups versus intermediate groups (Fig. S7B and C), which is consistent with former studies [49]. PLT levels decreased with HF progression in both HBV and HCV cohorts (Fig. S7D). Thus, the natural progression of liver injuries induced by different hepatitis etiologies may be reflected in different cutoffs of FIB-4 and GB scores. The first cutoff of FIB-4 (1.45) is a better trade-off point with specificity and sensitivity in HBV cohorts (Fig. 3) while the second cutoff (3.25) showed more consistent classification performances in HCV cohorts (Fig. S5). For GB scores, we found that the best cutoff (−0.93) for the HBV discovery set showed biased classifications in HCV cohorts (Fig. S5B). After changing the cutoff from −0.93 to −0.14, we observed better specificity without drastically decreasing the sensitivity (Fig. S5B). Thus, different etiologies of CLD may need to be directly factored into models, or etiology-specific cutoff values should be considered. However, in this study, HCV datasets suffered from small sample sizes and a limited number of cirrhosis subjects. In the future, we propose re-training new machine learning models using GB with larger HCV cohorts to achieve better staging performances.
Certain limitations of this study and the results need to be discussed. First, the training sample size remains limited and individual cohorts were drawn from only Asian ethnic cohorts. We are planning to collect more samples from multiple sites in the future which will be necessary to further establish the robustness of these predictive models. Second, we trained the GB models on HBV cohorts and recognized the need to recruit additional cohorts to examine models trained on groups affected by specific etiologies such as HCV patients. Based on the differences in model performance that we have preliminarily observed between HBV and HCV-related CLD patients, we believe it is possible to achieve further refinements of the models through the incorporation of etiology-related parameters. Third, limitations in clinical data infrastructure and mechanisms to support data sharing for biomedical research currently poses a severe bottleneck in validating cutting-edge machine learning techniques [50, 51]. We have implemented our GB prediction model as an online tool to support its dissemination for independent testing in other cohorts. We hope that independent researchers can share their results and data to help expedite this and other potential clinical applications of machine learning.
In conclusion, we employed a cutting-edge machine learning algorithm (GB) to reconstruct a well-studied clinical scoring system (FIB-4) for better detection of advanced HF and cirrhosis in HBV cohorts with CLD. We validated the prediction capacity of our models in multiple independent groups of HBV and HCV patients. Due to the etiological differences between HBV and HCV, we proposed that different cutoff values for GB and FIB-4 scoring should be applied. Particular machine learning models could be trained on larger HCV cohort in future studies. The idea of using machine learning to reconstruct existing clinical scoring systems could be applied to other indicators in other disease cohorts.
Acknowledgments
Acknowledgements
We would like to acknowledge all participants of this project for providing samples and experts of local hospitals for collecting data. Also, we wish to thank Dr. Cheng-Yuan Peng for sharing data to support this study.
Funding sources
This work was financially supported by the National Institutes of Health/National Cancer Institute Grant 1U01CA188387-01A1, Key Program of National Natural Science Foundation of China (8153000502), General Program of National Natural Science Foundation of China (81573873, 81774196), China Postdoctoral Science Foundation funded project, China (2016T90381, 2015M581652), E-institutes of Shanghai Municipal Education Commission, China (E03008). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. W.J. had full access to all of the data in the study and had final responsibility for the decision to submit for publication.
Declaration of interests
The authors declare that they have no competing interests.
Authors' contributions
W.J. and P. L. lead the study. R.W. and J.W. performed the data analysis, implemented the methodology, and generated the web-tool; X.W., G.X., Y.W., H.Z., and C.Y.P. collected the data and discussed the results; R.W., J.W, and S.W. prepared the original draft; W.J., P.L., R.W., J.W, S.W., G.X., C.R., and C.Y.P. reviewed and edited the final manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.07.041.
Contributor Information
Ping Liu, Email: liuliver@vip.sina.com.
Wei Jia, Email: wjia@cc.hawaii.edu.
Appendix A. Supplementary data
Supplementary material
References
- 1.Rowe I.A. Lessons from epidemiology: the burden of liver disease. Dig Dis. 2017;35:304–309. doi: 10.1159/000456580. [DOI] [PubMed] [Google Scholar]
- 2.Bataller R., Brenner D.A. Liver fibrosis. J Clin Invest. 2005;115:209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellis E.L., Mann D.A. Clinical evidence for the regression of liver fibrosis. J Hepatol. 2012;56:1171–1180. doi: 10.1016/j.jhep.2011.09.024. [DOI] [PubMed] [Google Scholar]
- 4.Thampanitchawong P., Piratvisuth T. Liver biopsy: complications and risk factors. World J Gastroenterol. 1999;5:301–304. doi: 10.3748/wjg.v5.i4.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Regev A., Berho M., Jeffers L.J., Milikowski C., Molina E.G., Pyrsopoulos N.T. Sampling error and intraobserver variation in liver biopsy in patients with chronic HCV infection. Am J Gastroenterol. 2002;97:2614–2618. doi: 10.1111/j.1572-0241.2002.06038.x. [DOI] [PubMed] [Google Scholar]
- 6.Tapper E.B., Lok A.S.-F. Use of liver imaging and biopsy in clinical practice. N Engl J Med. 2017;377:756–768. doi: 10.1056/NEJMra1610570. [DOI] [PubMed] [Google Scholar]
- 7.Sterling R.K., Lissen E., Clumeck N., Sola R., Correa M.C., Montaner J. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology. 2006;43:1317–1325. doi: 10.1002/hep.21178. [DOI] [PubMed] [Google Scholar]
- 8.Vallet-Pichard A., Mallet V., Nalpas B., Verkarre V., Nalpas A., Dhalluin-Venier V. FIB-4: an inexpensive and accurate marker of fibrosis in HCV infection. Comparison with liver biopsy and FibroTest. Hepatology. 2007;46:32–36. doi: 10.1002/hep.21669. [DOI] [PubMed] [Google Scholar]
- 9.Sumida Y., Yoneda M., Hyogo H., Itoh Y., Ono M., Fujii H. Validation of the FIB4 index in a Japanese nonalcoholic fatty liver disease population. BMC Gastroenterol. 2012;12 doi: 10.1186/1471-230X-12-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim B.K., Kim D.Y., Park J.Y., Ahn S.H., Chon C.Y., Kim J.K. Validation of FIB-4 and comparison with other simple noninvasive indices for predicting liver fibrosis and cirrhosis in hepatitis B virus-infected patients. Liver Int. 2010;30:546–553. doi: 10.1111/j.1478-3231.2009.02192.x. [DOI] [PubMed] [Google Scholar]
- 11.Xu R., Wei W., Krawczyk M., Wang W., Luo H., Flagg K. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017 doi: 10.1038/nmat4997. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J.-X., Song W., Chen Z.-H., Wei J.-H., Liao Y.-J., Lei J. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295–1306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 13.Zak D.E., Penn-Nicholson A., Scriba T.J., Thompson E., Suliman S., Amon L.M. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet. 2016;387:2312–2322. doi: 10.1016/S0140-6736(15)01316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Z.-H. 2012. Ensemble Methods: Foundations and Algorithms. [Google Scholar]
- 15.Zeevi D., Korem T., Zmora N., Israeli D., Rothschild D., Weinberger A. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163:1079–1095. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Eslam M., Hashem A.M., Romero-Gomez M., Berg T., Dore G.J., Mangia A. FibroGENE: a gene-based model for staging liver fibrosis. J Hepatol. 2016;64:390–398. doi: 10.1016/j.jhep.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 17.Shousha H.I., Awad A.H., Omran D.A., Elnegouly M.M., Mabrouk M. Data mining and machine learning algorithms using IL28B genotype and biochemical markers best predicted advanced liver fibrosis in chronic hepatitis C. Jpn J Infect Dis. 2018;71:51–57. doi: 10.7883/yoken.JJID.2017.089. [DOI] [PubMed] [Google Scholar]
- 18.Shang G., Richardson A., Gahan M.E., Easteal S., Ohms S., Lidbury B.A. Predicting the presence of hepatitis B virus surface antigen in Chinese patients by pathology data mining. J Med Virol. 2013;85:1334–1339. doi: 10.1002/jmv.23609. [DOI] [PubMed] [Google Scholar]
- 19.Tsipouras M.G., Giannakeas N., Tzallas A.T., Tsianou Z.E., Manousou P., Hall A. A methodology for automated CPA extraction using liver biopsy image analysis and machine learning techniques. Comput Methods Programs Biomed. 2017;140:61–68. doi: 10.1016/j.cmpb.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 20.Chen S.-H., Lai H.-C., Chiang I.-P., Su W.-P., Lin C.-H., Kao J.-T. Changes in liver stiffness measurement using acoustic radiation force impulse elastography after antiviral therapy in patients with chronic hepatitis C. PLoS One. 2018;13 doi: 10.1371/journal.pone.0190455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tachi Y., Hirai T., Toyoda H., Tada T., Hayashi K., Honda T. Predictive ability of laboratory indices for liver fibrosis in patients with chronic hepatitis C after the eradication of hepatitis C virus. PLoS One. 2015;10 doi: 10.1371/journal.pone.0133515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheuer P.J., Standish R.A., Dhillon A.P. Scoring of chronic hepatitis. Clin Liver Dis. 2002;6:335–347. doi: 10.1016/s1089-3261(02)00009-0. [DOI] [PubMed] [Google Scholar]
- 23.Breiman L., Friedman J.H., Olshen R.A., Stone C.J. vol. 19. 1984. Classification and Regression Trees. [Google Scholar]
- 24.Fonarow G.C., Adams K.F., Abraham W.T., Yancy C.W., Boscardin W.J. Risk stratification for in-hospital mortality in acutely decompensated heart failure: classification and regression tree analysis. JAMA. 2005;293:572–580. doi: 10.1001/jama.293.5.572. [DOI] [PubMed] [Google Scholar]
- 25.Hastie T., Tibshirani R., Friedman J. The elements of statistical learning. Elements. 2009;1:337–387. [Google Scholar]
- 26.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 27.Greg R. gbm . R Packag Version 16–31. 2010. Generalized Boosted Regression Models.http://CRAN.R-project.org/package=gbm [Google Scholar]
- 28.Kuhn M. Building predictive models in R using the caret package. J Stat Softw. 2008;28:1–26. [Google Scholar]
- 29.Therneau T., Atkinson B., Ripley B., Ripley M.B. R Packag Version 41–10. 2015. rpart: recursive partitioning and regression trees. https://CR. [Google Scholar]
- 30.Liaw A., Wiener M. Classification and regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- 31.Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006;27:861–874. [Google Scholar]
- 32.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12 doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grau J., Grosse I., Keilwagen J. PRROC: computing and visualizing precision-recall and receiver operating characteristic curves in R. Bioinformatics. 2015;31:2595–2597. doi: 10.1093/bioinformatics/btv153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Attallah A.M., Abdallah S.O., Attallah A.A., Omran M.M., Farid K., Nasif W.A. Diagnostic value of fibronectin discriminant score for predicting liver fibrosis stages in chronic hepatitis C virus patients. Ann Hepatol. 2013;12:44–53. [PubMed] [Google Scholar]
- 35.Wai C.T., Greenson J.K., Fontana R.J., Kalbfleisch J.D., Marrero J.A., Conjeevaram H.S. A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C. Hepatology. 2003;38:518–526. doi: 10.1053/jhep.2003.50346. [DOI] [PubMed] [Google Scholar]
- 36.Williams A.L.B., Hoofnagle J.H. Ratio of serum aspartate to alanine aminotransferase in chronic hepatitis relationship to cirrhosis. Gastroenterology. 1988;95:734–739. doi: 10.1016/s0016-5085(88)80022-2. [DOI] [PubMed] [Google Scholar]
- 37.Tseng P.-L., Wang J.-H., Hung C.-H., Tung H.-D., Chen T.-M., Huang W.-S. Comparisons of noninvasive indices based on daily practice parameters for predicting liver cirrhosis in chronic hepatitis B and hepatitis C patients in hospital and community populations. Kaohsiung J Med Sci. 2013;29:385–395. doi: 10.1016/j.kjms.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poynard T., Bedossa P. Age and platelet count: a simple index for predicting the presence of histological lesions in patients with antibodies to hepatitisC virus. J Viral Hepat. 1997;4:199–208. doi: 10.1046/j.1365-2893.1997.00141.x. [DOI] [PubMed] [Google Scholar]
- 39.Foucher J., Chanteloup E., Vergniol J., Castéra L., Le Bail B., Adhoute X. Diagnosis of cirrhosis by transient elastography (FibroScan): a prospective study. Gut. 2006;55:403–408. doi: 10.1136/gut.2005.069153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Canbay A., Bechmann L., Gerken G. Lipid metabolism in the liver. Z Gastroenterol. 2007;45:35–41. doi: 10.1055/s-2006-927368. [DOI] [PubMed] [Google Scholar]
- 41.Roberts M.S., Magnusson B.M., Burczynski F.J., Weiss M. Enterohepatic circulation: physiological, pharmacokinetic and clinical implications. Clin Pharmacokinet. 2002;41:751–790. doi: 10.2165/00003088-200241100-00005. [DOI] [PubMed] [Google Scholar]
- 42.Campollo O., Sprengers D., McIntyre N. The BCAA/AAA ratio of plasma amino acids in three different groups of cirrhotics. Rev Invest Clin. 1992;44:513–518. [PubMed] [Google Scholar]
- 43.Zhang Q., Takahashi M., Noguchi Y., Sugimoto T., Kimura T., Okumura A. Plasma amino acid profiles applied for diagnosis of advanced liver fibrosis in patients with chronic hepatitis C infection. Hepatol Res. 2006;34:170–177. doi: 10.1016/j.hepres.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J., Zhao Y., Xu C., Hong Y., Lu H., Wu J. Association between serum free fatty acid levels and nonalcoholic fatty liver disease: a cross-sectional study. Sci Rep. 2014;4 doi: 10.1038/srep05832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen T., Xie G., Wang X., Fan J., Qiu Y., Zheng X. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.004945. M110.004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X., Wang X., Xie G., Zhou M., Yu H., Lin Y. Urinary metabolite variation is associated with pathological progression of the post-hepatitis B cirrhosis patients. J Proteome Res. 2012;11:3838–3847. doi: 10.1021/pr300337s. [DOI] [PubMed] [Google Scholar]
- 47.El-Serag H.B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142 doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poynard T., Mathurin P., Lai C.-L., Guyader D., Poupon R., Tainturier M.-H. A comparison of fibrosis progression in chronic liver diseases. J Hepatol. 2003;38:257–265. doi: 10.1016/s0168-8278(02)00413-0. [DOI] [PubMed] [Google Scholar]
- 49.Tai D.-I., Tsay P.-K., Jeng W.-J., Weng C.-C., Huang S.-F., Huang C.-H. Differences in liver fibrosis between patients with chronic hepatitis B and C. J Ultrasound Med. 2015;34:813–821. doi: 10.7863/ultra.34.5.813. [DOI] [PubMed] [Google Scholar]
- 50.Manamley N., Mallett S., Sydes M.R., Hollis S., Scrimgeour A., Burger H.U. Data sharing and the evolving role of statisticians. BMC Med Res Methodol. 2016;16:37–43. doi: 10.1186/s12874-016-0172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geifman N., Bollyky J., Bhattacharya S., Butte A.J. Opening clinical trial data: are the voluntary data-sharing portals enough? BMC Med. 2015;13 doi: 10.1186/s12916-015-0525-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material