Abstract
Background
Tumor-infiltrating lymphocytes (TIL) in colorectal tumor tissue are significantly correlated with a favorable prognosis, such as CD8+ lymphocytes, which are also called tumor-reactive lymphocytes. However, not all tumor-infiltrating T cells confer benefit to patients. Therefore, it is of substantial benefit to identify a biomarker to demarcate these tumor-reactive lymphocytes.
Methods
We investigated whether ITGAE could be used to discriminate reactive CD8+ lymphocytes in colorectal cancer (CRC). TCGA colorectal cancer data sets (n1 = 492, n2 = 386) and FUSCC set (n3 = 276) were used in this study. Further phenotyping of ITGAE+ cells and the mechanistic basis were investigated.
Findings
In the training and testing sets from TCGA, ITGAE expression, which is strongly correlated with cytotoxic T cell markers (CD8/CD3/PD1), independently predicted longer disease-free survival (DFS) and overall survival (OS). In line with this, the association between ITGAE+ lymphocytes and survival has been confirmed in the FUSCC cohort for validation (P = .026). ITGAE + cells in the series always co-stained with CD8 were preferentially located in the tumor. Interestingly, ITGAE+ lymphocytes tended to associate with the epithelial–mesenchymal transition (EMT) with decreased Snail and increased E-cadherin expression accompanied. Finally, gene set enrichment analysis showed that immune activation was significantly enriched in the high ITGAE+ TIL group, accompanied by enriched EMT-related pathways.
Interpretation
Because of the specified expression of tumor-reactive CD8+ T-cells, ITGAE may be a promising biomarker for the rapid identification of immune infiltration in CRC.
Keywords: ITGAE, Tumor-infiltrating lymphocytes, Colorectal cancer, Prognosis
Highlights
-
•
ITGAE expression independently predicted longer disease-free survival (DFS) and overall survival (OS) in colorectal cancers.
-
•
ITGAE could be used to discriminate CD8+ TIL populations
We demonstrate here in colorectal cancers for the first time that ITGAE+ CD8+ lymphocytes infiltration plays a vital role in the antitumor immune response and ITGAE has been identified as a biomarker of tumor-reactive CD8+ TIL. In mechanism, ITGAE+ lymphocytes may even associate with interferon-response chemokines and EMT signaling, then serves as an independent predictor in colorectal cancers.
Research in context.
Evidence before this study
It have been previously demonstrated that an improved prognosis for patients was associated with accumulation of CD8+ TIL in several malignances. By contrast, presence of CD8+ CTL was not consistently found to confer survival benefit to patients. Therefore, it is of substantial benefit to identify a biomarker for demarcating tumor-reactive cells.
Added value of this study
The association between ITGAE+ lymphocytes and survival has been confirmed in the TCGA and FUSCC cohorts. ITGAE + cells in the series always co-stained with CD8 were preferentially located in the tumor. Interestingly, ITGAE+ lymphocytes tended to associate with the epithelial–mesenchymal transition (EMT) with decreased Snail and increased E-cadherin expression accompanied.
Implications of all available evidence
ITGAE+ CD8+ lymphocyte infiltration plays a vital role in the antitumor immune response, and ITGAE has been identified as a biomarker of tumor-reactive CD8+ TIL.
Alt-text: Unlabelled Box
1. Introduction
Colorectal cancer is the fourth most common cancer worldwide, and its overall 5-year survival rate is approximately 50% [1]. In addition to pathological staging, histological subtype, and the extent of the surgical procedure, the presence of tumor-infiltrating lymphocytes (TIL) in surgically resected tumor tissue is also significantly correlated with favorable prognosis [2]. Indeed, the accumulation of CD8+ TIL has been demonstrated to associate with an improved prognosis for patients in several malignances [3, 4]. In contrast, the presence of CD8+ TIL has not been consistently found to confer a survival benefit. Herein, a distinction has frequently focused on CD8+ TIL infiltrating the epithelial nests versus the surrounding stroma, while this approach has not been determined to identify the localization-related prognosis for CD8+ TIL [3, 5, 6]. In other words, the approach depending on the demarcation of epithelial and stromal regions is inaccurate and non-trivial in many malignances. Therefore, it is of substantial benefit to identify a biomarker for demarcating tumor-reactive cells.
Recent reports have suggested that ITGAE (integrin, alpha E), also named CD103, plays a vital role in anti-cancer immunity. For example, ITGAE can recognize aberrant cells via cytotoxic T cells in colon, lung and pancreatic cancer [[7], [8], [9]]. In addition, ITGAE+ T cells were identified as an effector memory phenotype [10]. Ultimately, ITGAE expression was profoundly upregulated in lymphocytes when TIL migrated to the tumor and co-engaged with TCR and TGF-β receptors at the site of the lesion [11]. Together, these observations led us to hypothesize that ITGAE expression might identify tumor-reactive cells that confer a favorable prognosis in colorectal cancers. Therefore, the aim of our study was to determine whether ITGAE+ TIL were associated with prolonged prognosis and whether the expression of ITGAE defined tumor-reactive CD8+ cells in colorectal cancers. Furthermore, we explored the mechanistic basis of our findings and whether the ITGAE+ TIL behaviors affected the epithelial–mesenchymal transition (EMT) to influence the patient's outcome in CRC.
2. Methods
2.1. Study Population
This study included three independent cohorts of CRC patients. The discovery cohort enrolling 492 CRC and testing sets including 386 consecutive patients, were obtained from the Cancer Genome Atlas (TCGA) database available from Cancer Genomics Browser of the University of California Santa Cruz (https://genome-cancer.ucsc.edu/). The pattern of ITGAE expression relative to that of other immunomarkers [12] was directly visualized with a heatmap using GraphPad Prism. The validation set enrolling 276 CRC patients was obtained from Fudan University Shanghai Cancer Center (FUSCC) between 2007 and 2009. Only patients with fully characterized tumors, intact overall survival (OS) or disease-free survival (DFS) were included. All patient data were collected, such as: age at the diagnosis, race, tumor localization, diagnostic year, tumor diameter, histological grade, number of lymph nodes retrieved, post-operative multimodal treatment (adjuvant chemotherapy or radiation), details of surgical procedures, complications rate, postoperative histopathology, and follow-up information (date of last visit, tumor relapse, tumor-related or unrelated death, overall survival: OS and disease-free survival: DFS). All patients from the FUSCC dataset provided written informed consent. The research protocol was reviewed and approved by the institutional review board of the FUSCC.
2.2. Tissue Microarray (TMA) Construction and Immunohistochemistry (IHC) Staining
In all, 276 unselected, non-consecutive, primary colorectal cancers were enrolled form January 2007 to November 2009 in FUSCC to construct the tissue microarray(TMA). Construction of this TMA has been previously described in detail [13]. Briefly for IHC, after deparaffinization of tissue blocks, antigen retrieval was performed in antigen retrieval solution at 100 °C for 15 min. Then primary antibody incubation were performed with one appropriate antibody [anti-ITGAE antibody (1:200), Snail antibody (1:400) from Abcam; anti-CD3e(1:200), CD8a(1:200), FOXP3(1:400), E-cadherin(1:400), CD56(1:800) from Cell Signaling Technology] overnight at 4 °C, separately. Secondary antibody, anti-rabbit/mouse horse radish peroxidase (HRP) (Gene Tech, China), was applied and hematoxylin was used for counterstaining for 1 h. The number of positive cells per field was estimated using Image-Pro Plus 6.0 (Media Cybernetics Inc., Bethesda, MD, USA). The immunostaining was evaluated by two oncologists blinded to the clinical information. The cutoff values for survival analysis were determined by X-Tile 3.6.1 software 11 (Yale University, New Haven, CT, USA).
Regarding EMT markers, the staining intensity was scored as 0 (negative), 1 (weak), 2 (medium) or 3 (strong). The extent of staining was scored as 0 (<5%), 1(5–25%), 2 (26–50%), 3 (51–75%) and 4 (>75%) according to the percentage of the positive staining area in relation to the whole carcinoma area. We multiplied the percentage score by the staining intensity score to generate the immunoreactivity score (IRS). E-cadherin was scored as negative (final score ≤ 2) and positive (final score ≥ 3), and Snail was scored as negative (final score ≤ 2) and positive (final score ≥ 3), respectively.
2.3. Immunofluorescent Analysis of ITGAE+ cells' Phenotype
Antigen retrieval and incubation with the primary antibody of the full tumor were performed as described for TMA above. Then, Alexa Fluor 488 and 594 or goat anti-rabbit or mouse (1:200, Invitrogen) was used as the secondary antibody for an hour at room temperature. Ultimately, nuclei were stained with DAPI for 5 min when necessary. Fluorescence images were photographed with a fluorescence microscope. Within each tumor core, single positive (ITGAE or CD8) cells as well as double-positive (ITGAE and CD8) cells were counted manually.
2.4. Gene Set Enrichment Analysis (GSEA) and Differential Expression Analysis
GSEA was carried out to determine the pathways that were significantly enriched between low and high ITGAE+ lymphocyte infiltration samples [14]. One thousand random sample permutations were performed. If we identified a gene set with a positive enrichment score, the set was defined as “enriched” when the majority of its members were highly expressed accompanied by a higher risk score.
We used edge R to analyze the differential gene expression. Significantly altered genes between low and high ITGAE+ lymphocyte infiltration samples were defined as expressed genes with FDR-adjusted p-value<.05 and a change of at least two-fold.
2.5. Statistical Analysis
Statistical evaluation was performed using IBM SPSS statistics Version 22 (SPSS Inc.; IBM Corporation Software Group, Somers, NY, USA). All quantification data are presented as mean ± s.d. Statistically significant differences were determined by one-way ANOVA test and Student's t-test. All statistical tests were two-sided, and P-values <.05 were considered to be statistically significant. OS and DFS were estimated with the Kaplan-Meier method, and the log-rank (Mantel-Cox) test was used to compare independent subgroups. Cox proportional hazard models were used to investigate the effect on survival of multivariable relationships among covariates, including the age at diagnosis, gender, stage at diagnosis, histological type, histological grade and treatment. Stage, status of perineural and vascular invasion or any known clinical characteristics that could affect the prognosis were the stratified variables. Hazard ratios (HRs) and 95% confidence intervals (CIs) for multivariate analyses were computed using the Cox proportional hazard regression models.
3. Results
3.1. Predictive Value of ITGAE and Association with an Immune Signature in TCGA
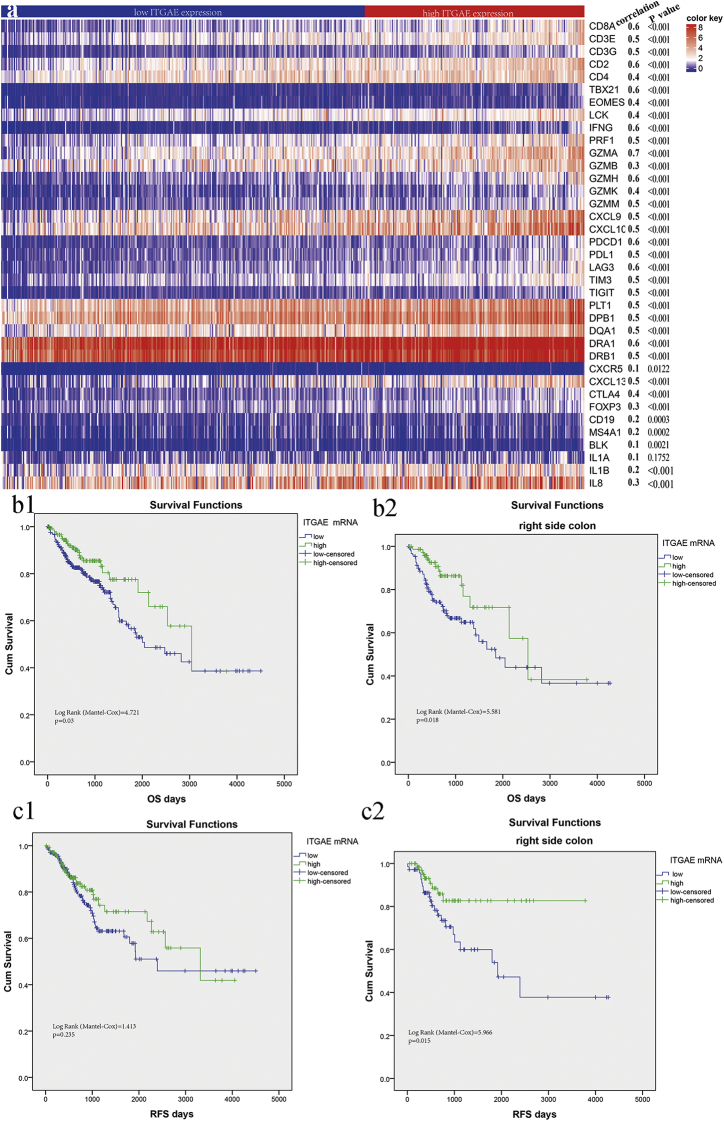
To explore the utility of ITGAE as a biomarker of a tumor-reactive lymphocyte response in colorectal cancer, we first assessed the distribution pattern of ITGAE mRNA in the Cancer Genome Atlas (TCGA) colorectal cancer data set. ITGAE expression was found to be closely correlated with the expression levels of reactive tumor lymphocyte markers (CD3e-g, CD2), exhaustion molecules (PD1, TIGIT), antigen-presenting molecules (HLA-DRA1, HLA—B1, HLA-DQA1) and B cell biomarkers (CD19). Therefore, increased ITGAE expression stratified the immunologically “hot” tumors in the colorectal cancer cohort (Fig. 1a).
Fig. 1.
ITGAE-associated immune responses and prognostic role in TCGA colorectal cancers.
(a) Heatmap showing expression of immunologic genes according to ITGAE expression. For pairwise analysis, the correlation with ITGAE expression was calculated by spearman. b: Kaplan–Meier curves demonstrating overall survival of patients in the discovery set (TCGA) divided by ITGAE mRNA expression for the total cohort (b1) and for the right-side colon cancer sub-group (b2). c: Kaplan–Meier curves demonstrating relapse-free survival of patients in the testing set (TCGA) divided by ITGAE mRNA expression for the total cohort (c1) and for the right-side colon cancer sub-group (c2).
In the discovery set (TCGA), we used the X-tile program to determine the cut-off value of the ITGAE mRNA levels in tumor tissue: 1.847. Then, patients with high and low levels of ITGAE expression were identified for further analysis (≤1.847 for the low ITGAE group and > 1.847 for the high ITGAE group). Kaplan–Meier survival analysis was carried out to compare overall survival (OS) according to ITGAE expression, and significantly improved OS was found in the high ITGAE group (Fig. 1 b1, log-rank test = 4.721, P = .03). Furthermore, a much stronger association of ITGAE expression with regard to patient survival and prognosis was observed in the right-side colon cancer group (Fig. 1 b2, log-rank test = 5.581, P = .018).
In the testing set (TCGA), we observed a prolonged relapse-free survival of 2718 days in the high ITGAE expression group versus 2653 days in the low ITGAE expression group, although no significance was seen (Fig. 1 c1, log-rank test = 1.413, P = .235). Notably, in the right-side colon cancer group, high ITGAE expression was significantly associated with significantly improved relapse-free survival (Fig. 1 c2, log-rank test = 5.966, P = .015). The baseline characteristics of the patients in TCGA were found in supplementary Table 1.
3.2. Association of ITGAE+ and CD8+ T Cells with Survival in FUSCC
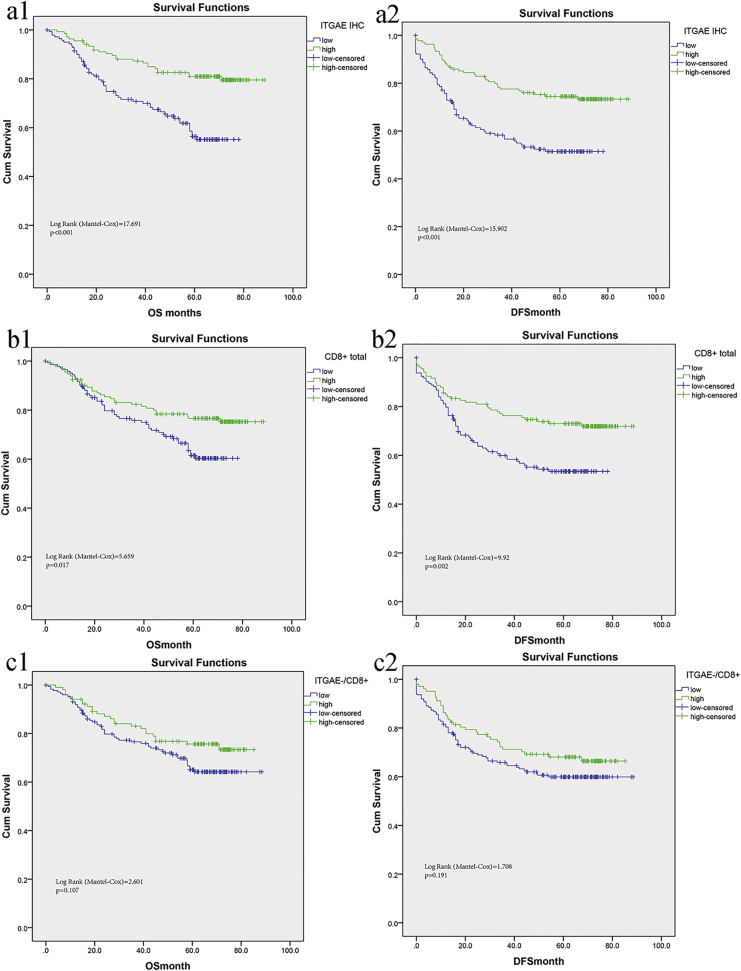
To consolidate our findings in the discovery and testing sets from TCGA data, we identified the infiltration of ITGAE+ cells by TMA in a FUSCC cohort with colorectal cancer patients (Supplementary Fig. 1 displaying representative image of high and low infiltration). Table 1 shows the baseline characteristics of the patients with high and low ITGAE+ cell quantification. The number of ITGAE+ lymphocytes was evaluable, and the cutoff number was identified as 31 per tissue core. A high infiltration group of total ITGAE+ lymphocytes was defined as >31 ITGAE+ lymphocytes per tissue core, while others exhibiting lower numbers were defined as the low infiltration group. Overall survival (OS) analysis based on the infiltration of ITGAE+ cells revealed significantly improved survival in the high infiltration group (Fig. 2 a1; p < .001). Consistent results were obtained when disease-free survival was analyzed (Fig. 2 a2). Univariate and multivariate analyses for OS and DFS were carried out (Table 2 and Table 3). Finally, multivariate analysis of ITGAE+ cell infiltration and clinicopathological factors revealed that ITGAE was an independent prognostic factor for OS and DFS.
Supplementary Fig. 1.
ITGAE+ TIL in colorectal cancers from FUSCC for validation. Representative images of tissue cores as high (a), or low infiltration (b).
Table 1.
Baseline characteristics in patients with high and low ITGAE+ TIL in FUSCC.
| Variables, N (%) | ITGAE+ TIL |
P value | |
|---|---|---|---|
| Low infiltration (n = 141) | High infiltration (n = 135) | ||
| Gender | |||
| Male | 60(42.6) | 52(38.5) | 0.495 |
| Female | 81(57.4) | 83(61.5) | |
| Age, years | 57.6 ± 11 | 56.7 ± 11 | 0.531 |
| TNM stage | 0.001 | ||
| I | 3(2.1) | 18(13.3) | |
| II | 37(26.2) | 44(32.6) | |
| III | 74(52.5) | 58(43.0) | |
| IV | 27(19.1) | 15(11.1) | |
| N stage | 0.107 | ||
| N0 | 52(36.9) | 68(50.4) | |
| N1 | 45(31.9) | 37(27.4) | |
| N2 | 43(30.5) | 30(22.2) | |
| M stage | 0.063 | ||
| M0 | 114(80.9) | 120(88.9) | |
| M1 | 27(19.1) | 15(11.1) | |
| Grade | 0.805 | ||
| Well/moderate | 113(80.1) | 107(79.3) | |
| poor | 28(19.9) | 28(20.7) | |
| T stage | <0.001 | ||
| T2 | 9(6.4) | 34(25.2) | |
| T3 | 23(16.3) | 31(23.0) | |
| T4 | 109(77.3) | 70(51.9) | |
| Histological type | 0.478 | ||
| Adenocarcinoma | 132(93.6) | 129(95.6) | |
| Mucinous | 9(6.4) | 6(4.4) | |
| Lymph node examined | 0.543 | ||
| Median | 15 ± 5 | 15 ± 6 | |
| Perineural invasion | 0.747 | ||
| Negative | 119(84.4) | 112(83.0) | |
| Positive | 22(15.6) | 23(17.0) | |
| Vascular invasion | 0.296 | ||
| Negative | 92(65.2) | 96(71.1) | |
| Positive | 49(34.8) | 39(28.9) | |
| MS status/MMR status | 0.466 | ||
| MSS/MMR-proficient | 87(61.7) | 89(65.9) | |
| MSI/MMR-deficient | 54(38.3) | 46(34.1) | |
| CEA (ng/ml) | 49.2 ± 14 | 24.2 ± 10 | 0.033 |
TIL indicates tumor infiltrating lymphocytes; MMR indicates mismatch repair; MS, microsatellite; MSS, microsatellite stability; MSI, microsatellite instability.
Fig. 2.
ITGAE+ infiltration is closely associated with survival in validation set from FUSCC.
(a1) Overall survival (OS) and (a2) disease-free survival (DFS) of patients in FUSCC according to high or low infiltration of ITGAE+ cells. (b1) OS and (b2) DFS of patients in FUSCC according to high or low infiltration of CD8+ cells. (c1) OS and (c2) DFS of patients in FUSCC according to high or low infiltration of CD8+/ ITGAE- cells.
Table 2.
Univariate and multivariate analysis for overall survival in FUSCC.
| Variables | Univariate Analysis |
Multivariate Analysis |
||||
|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | P value | Hazard ratio | 95% CI | P value | |
| ITGAE+TIL | 0.379 | 0.237–0.607 | <0.001 | 0.57 | 0.346–0.938 | 0.027 |
| Male gender | 1.255 | 0.797–1.976 | 0.328 | |||
| Age > 70 | 1.581 | 0.915 -2.734 | 0.101 | |||
| TNM stage IV | 5.118 | 3.574–7.330 | <0.001 | 4.097 | 1.739 -9.649 | 0.001 |
| T stage, T4 | 10.531 | 2.580–42.981 | 0.001 | 1.632 | 0.983–2.709 | 0.058 |
| N stage, N2 | 4.139 | 2.332–7.346 | <0.001 | 0.861 | 0.612–1.212 | 0.391 |
| Grade, poor | 1.36 | 0.880–2.103 | 0.166 | |||
| M1 stage | 9.06 | 5.735–14.314 | <0.001 | 1.254 | 0.414–3.804 | 0.689 |
| Lymph node examined | 0.72 | 0.465–1.114 | 0.14 | |||
| Mucinous Adenocarcinoma | 0.865 | 0.316 -2.364 | 0.77 | |||
| Perineural invasion | 1.639 | 0.980–2.740 | 0.06 | |||
| Vascular invasion | 2.44 | 1.578–3.776 | <0.001 | 1.293 | 0.772–2.164 | 0.329 |
| MSI | 0.923 | 0.589–1.444 | 0.725 | |||
| CEA >10 ng/ml | 2.705 | 1.713–4.273 | <0.001 | 1.004 | 0.591–1.708 | 0.987 |
TIL indicates tumor infiltrating lymphocytes; MMR indicates mismatch repair; MS, microsatellite; MSS, microsatellite stability; MSI, microsatellite instability; LN, lymph nodes.
Table 3.
Univariate and Multivariate analyses of prognostic factors for disease-free survival in FUSCC.
| Univariate Analysis |
Multivariate Analysis |
|||||
|---|---|---|---|---|---|---|
| Variables | Hazard ratio | 95% CI | P value | Hazard ratio | 95% CI | P value |
| ITGAE+TIL | 0.443 | 0.293–0.670 | <0.001 | 0.5491 | 0.355–0.849 | 0.007 |
| Male gender | 1.322 | 0.883–2.009 | 0.172 | |||
| Age > 70 | 1.214 | 0.711–2.072 | 0.478 | |||
| TNM stage IV | 5.363 | 3.837–7.496 | <0.001 | 3.921 | 1.885–8.157 | <0.001 |
| T stage, T4 | 2.401 | 1.630–3.537 | 0.001 | 1.456 | 0.966–2.197 | 0.073 |
| N stage, N2 | 1.886 | 1.498–2.374 | <0.001 | 0.831 | 0.603–1.147 | 0.261 |
| Grade, poor | 1.168 | 0.788–1.730 | 0.439 | |||
| M1 stage | 10.069 | 6.567–15.439 | <0.001 | 1.426 | 0.538–3.777 | 0.476 |
| LN examined >12 | 0.662 | 0.447 0.981 | 0.04 | 0.684 | 0.453–1.032 | 0.07 |
| Mucinous Adenocarcinoma | 0.872 | 0.355–2.144 | 0.765 | |||
| Perineural invasion | 1.892 | 1.203 -2.977 | 0.006 | 1.326 | 0.818–2.149 | 0.252 |
| Vascular invasion | 2.223 | 1.500–3.296 | <0.001 | 0.918 | 0.566–1.489 | 0.729 |
| MSI | 0.946 | 0.631–1.416 | 0.786 | |||
| CEA >10 ng/ml | 3.318 | 2.211 -4.981 | <0.001 | 1.3 | 0.812–2.080 | 0.275 |
TIL indicates tumor infiltrating lymphocytes; MMR, mismatch repair; MS, microsatellite; MSS, microsatellite stability; MSI, microsatellite instability; LN, lymph nodes.
Meanwhile, we compared the overall survival and disease-free survival rates in patients with different levels of infiltrating CD8+ cells. An increased infiltration of CD8+ cells (high score) was significantly associated with an improved prognosis (p = .017, Fig. 2 b1). A similar association was seen in the disease-free survival rates (p = .02, Fig. 2 b2). Previous studies have shown that only tumor-reactive CD8+ lymphocytes are associated with survival prognosis in cancers, while the locations (epithelium or stroma) of these anti-tumor CD8+ lymphocytes were not specified accordingly. Herein, biomarkers were used to demarcate anti-tumor CD8+ lymphocytes, which obviated the need to precisely account for their localizations within the tumor.
Finally, we attempted to determine whether ITGAE+ cells primarily accounted for the observed prognostic effect conferred by CD8+ lymphocytes in CRC. Positive staining for both ITGAE+ and CD8+ TIL was present in the majority of species, which was associated with an improved prognosis. However, tumors with CD8+ TIL but lacking ITGAE expression (CD8+/ITGAE-) were not associated with improved survival (p > .05, Fig. 2 c1-c2), suggesting that it was the ITGAE+ TIL that primarily accounted for improved prognosis in CRC.
3.3. ITGAE Demarcates CD8+ TIL
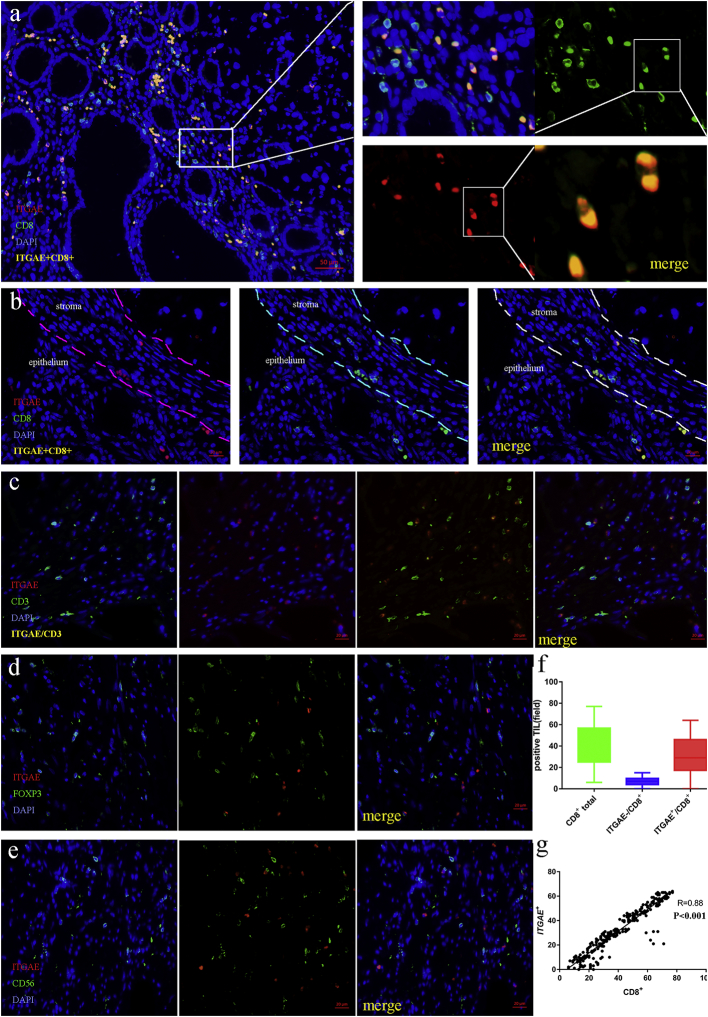
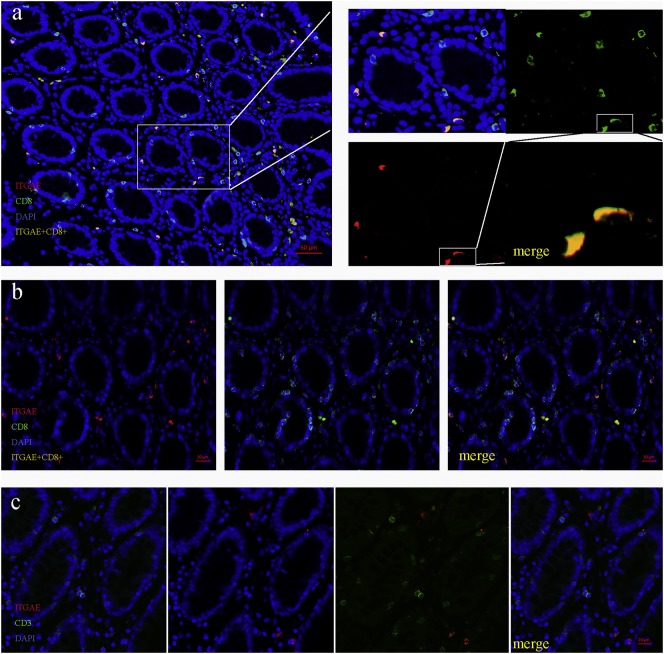
To identify the phenotype of ITGAE+ TIL in colorectal cancer, CD3, CD8, FoxP3, CD56 and ITGAE were stained. Multi-colour fluorescence staining of the cancer tissue showed that ITGAE+ cells always co-stained with CD8, and CD3 (a lymphocyte marker), suggesting a CD8+ ITGAE+ lymphocyte phenotype (Fig. 3a). Furthermore, ITGAE+ TIL were resided in both the tumor stroma and the tumor epithelium (Fig. 3b). Above all, no ITGAE+ TILs that presented in the tissue were negative for CD8 and CD3, indicating lymphocyte phenotype (Fig. 3b-3c). In addition, ITGAE+ lymphocytes in the cancer tissue were negative for FOXP3 and CD56 (Fig. 3d-3e). At last, the correlation between ITGAE+ and CD8+ cells was strongly close, demonstrated in Fig.3f-3g.
Fig. 3.
ITGAE demarcates CD8+ TIL in colorectal cancer tissue.
(a) Representative image of tissue from a colorectal cancer stained with DAPI (DNA, blue), anti-CD8 (green), anti-ITGAE (red) and double CD8/ITGAE+ (orange). (b) Representative images of CD8+ and ITGAE+ cells in the epithelial or stromal areas. (c-d) Representative image of tissue from a colorectal cancer stained with DAPI (DNA, blue), anti-CD3, anti-Foxp3 (green), anti-ITGAE (red). (e) Representative image of tissue from a colorectal cancer stained with DAPI (DNA, blue), anti-CD56 (green), anti-ITGAE (red); (f) Quantification of total CD8+, CD8+/ITGAE+ and CD8+ /ITGAE- cells in the tumor tissue (n = 276). The Quantification was determined by one-way ANOVA. (g) The correlation of CD8+ and ITGAE+ cells. The statistical test was used for linear logistic regression analyses.
To validate these findings, we analyzed the interaction between ITGAE+ and CD8+ cells in the adjacent normal epithelium. The number of CD8+ cells in normal epithelium was significantly higher than the total number of ITGAE+ lymphocytes, and we also could observe the ITGAE co-stained with CD8 in the lymphocytes, in which ITGAE+ lymphocytes appeared to distribute exclusively. What is more, ITGAE cells in the healthy normal epithelium were negative for CD3, suggesting a non-T-cell origin. Taken together, these findings demonstrated that the ITGAE+ cells in colorectal cancer tissue were predominantly CD8+ T cells (Fig. 4a-4c).
Fig. 4.
ITGAE demarcates CD8+ TIL in normal colorectal epithelial tissue.
(a) Representative image of normal colorectal epithelial tissue stained with DAPI (DNA, blue), anti-CD8 (green), anti-ITGAE (red) and double CD8/ITGAE+ (orange). (b) Representative images of CD8+ and ITGAE+ cells in the epithelial areas. (c) Representative image of tissue from a colorectal cancer stained with DAPI (DNA, blue), anti-CD3 (green), anti-ITGAE (red).
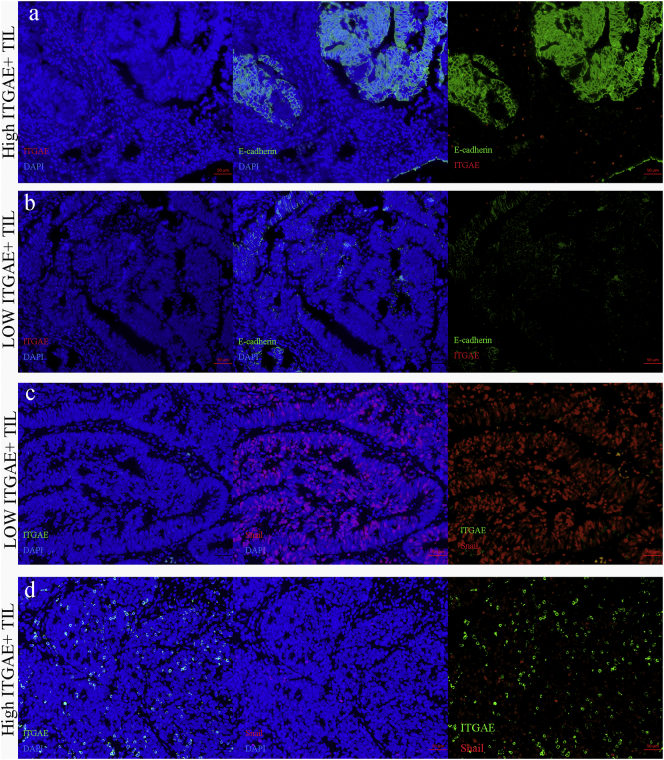
3.4. Correlation of ITGAE+ TIL with EMT Markers
The epithelial–mesenchymal transition marker E-cadherin and the representative transcriptional factor Snail were also detected. The relationships between ITGAE+ cells and EMT markers were analyzed in TMA. ITGAE+ lymphocytes tended to decrease Snail expression and increase E-cadherin expression (Table 4). Fluorescent microscopy demonstrated that ITGAE+ TILs in colorectal cancer were accompanied by high E-cadherin expression, a hallmark of the epithelial–mesenchymal transition. To confirm signs of the inhibited EMT signaling induced by ITGAE+ TIL from CRC, the simultaneous expression of ITGAE and nuclear Snail was examined. It was found that tumor islets surrounded by ITGAE+ cells were all characterized by weak nuclear Snail staining (Fig. 5).
Table 4.
Correlation between ITGAE+ TIL and expression of EMT markers in colorectal cancer (Spearman rank test).
| variables | ITGAE+ TIL |
|||
|---|---|---|---|---|
| Low infiltration | High infiltration | P value | ||
| E-cadherin | low | 72 | 45 | 0.003 |
| high | 69 | 90 | ||
| Snail | low | 77 | 99 | 0.001 |
| high | 64 | 36 | ||
Fig. 5.
The association between ITGAE+ TIL and EMT markers in CRC.
Representative image of colorectal cancer tissue in (a) high ITGAE+ T cells (red) with high level of E-cadherin (green), compared with the low ITGAE+ T cells (red) with low expression of E-cadherin (green) (b). (c-d) Representative image of ITGAE+ T cells (green) negatively associated with level of Snail (red).
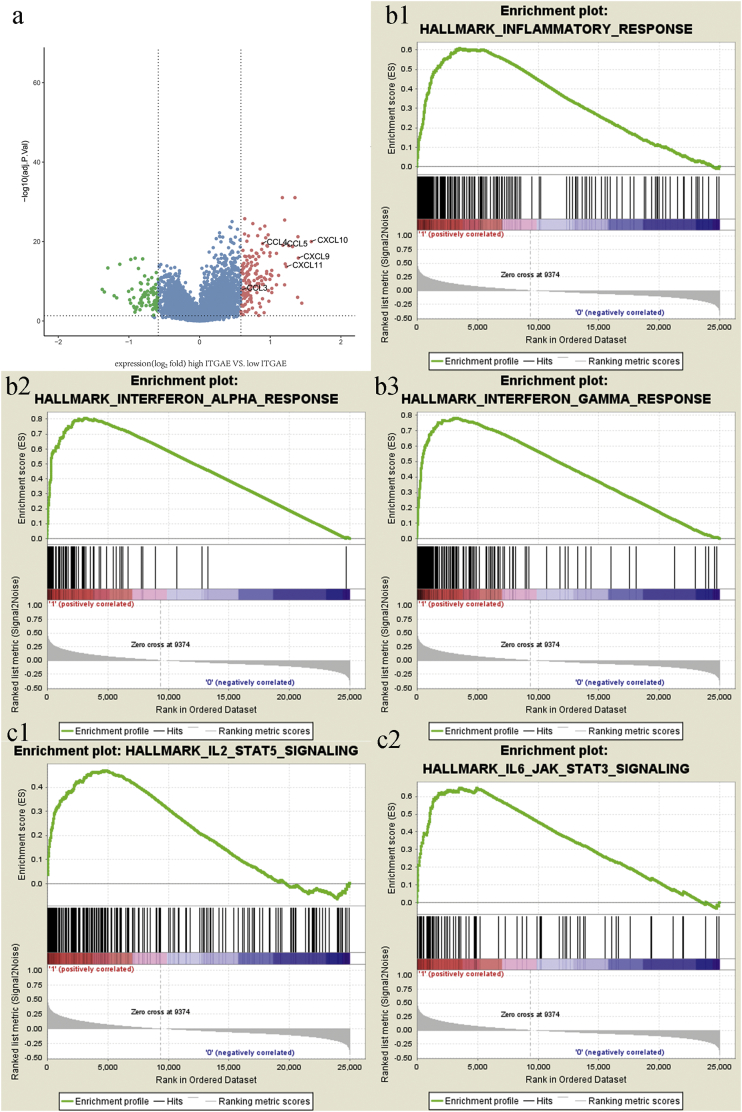
3.5. Identification of ITGAE+ Lymphocyte-Associated Biological Pathways and Processes
Gene set enrichment analysis (GSEA) [15] was performed to determine the related biologic processes and signaling pathways. First, the differential gene expression profiles of colorectal cancer patients with high and low ITGAE+ lymphocytes were compared. We found that numerous cytokines and chemokines relative to immune filtration were significantly elevated in the ITGAE+ lymphocyte group (Fig. 6a). Interferon-response chemokines (CXCL9–10, and CXCL11), which accounted for the migration of activated T cells and enhanced the anti-tumor activity, increased significantly in the high ITGAE+ lymphocyte infiltration group. These results validated that high ITGAE+ lymphocytes correlated with immune activation in the niche of CRC.
Fig. 6.
ITGAE+ lymphocytes associated biologic pathways and processes.
(a) Volcano plot comparing the FDR versus fold-change for genes from high- ITGAE+ TIL group relative to low- ITGAE+ TIL group. (b1-b3, c1-c2) lymphocyte activation and EMT-related pathways were enriched in the ITGAE+ lymphocytes group.
Meanwhile, several immune-related pathways, such as inflammatory response, interferon alpha and gamma activation, were enriched in the high ITGAE+ lymphocyte infiltration group (Fig. 6 b1-b3). These findings indicated that the presence of high ITGAE+ lymphocytes was associated with the positive regulation of lymphocyte activation in CRC, which also explained that patients with high ITGAE+ lymphocyte infiltration had longer survival than those with low ITGAE+ lymphocyte infiltration. Interestingly, additional EMT-related pathways were associated with the ITGAE+ lymphocytes group (Fig. 6 c1-c2), including IL2-STAT5 and IL6-JAK-STAT3 signaling. JAK/STAT signaling is essential for the epithelial-mesenchymal transition in cancer cells. These results suggested that the recruitment of ITGAE+ lymphocytes was associated with the modulation of EMT in CRC, which was consistent with altered E-cadherin expression.
4. Discussion
Utilizing these three independent sets of colorectal cancers, we demonstrated that infiltrating ITGAE+ lymphocytes were closely associated with survival in colorectal cancer patients. Moreover, ITGAE can be regarded as a potential biomarker to discriminate the active phenotype of CD8+ lymphocytes in CRC. Using correlation analysis, ITGAE+ lymphocyte infiltration was not only accompanied by active CD8+ lymphocyte recruitment but also inherently associated with EMT. These results explained that patients with high ITGAE+ lymphocyte infiltration had longer survival times than those with low ITGAE+ lymphocyte infiltration.
ITGAE+ lymphocytes are an important component of the colorectal cancer microenvironment and are positively associated with survival, as demonstrated in this study. To explain this, the increase in ITGAE showed simultaneous T cell activation. With regard to the phenotype and ontogeny of ITGAE+ lymphocytes in human malignancies, diversity remains. In lung cancer patients, the role of ITGAE integrin was demonstrated in lung tumor-infiltrating lymphocyte (TIL) clones to enhance specific TCR-mediated tumor cell cytotoxicity [16]. ITGAE+ lymphocytes were characterized by tissue-resident memory T cell transcriptomic and phenotypic signatures. More importantly, PD-1 and Tim-3 checkpoint receptors were frequently co-expressed simultaneously with ITGAE, thus increasing immune-induced cell death and modulating specific cytolytic activity toward autologous tumor cells upon blocking the PD-1/PD-L1 interaction. In pancreatic cancer, ITGAE+ lymphocytes were defined as activated cytolytic lymphocytes contributing to tumor lysis, as well as the maturation of the cytotoxic immune synapsis [8, 9, 17]. In addition, cytotoxic T lymphocytes from CD103 elevated tumors displayed features of enhanced cytotoxicity, then predicted a better survival prognosis in lung cancer [30]. In this study, we also demonstrated dominant ITGAE and CD8 co-staining in colorectal cancer and normal epithelium, while the precise differentiation status has not been explored. ITGAE may play a role in programing CD8+ T cell residency [31]. As the colorectal epithelium functions as a barrier against pathogens, we extrapolated that tissue-resident memory T cells may also be characterized by ITGAE. However, in the niche of tumor-specific immune responses, the majority of ITGAE+ lymphocytes are likely tumor-reactive and potentially tumorlytic-infiltrating lymphocytes.
More importantly in this study, a close correlation between elevated ITGAE+ lymphocyte infiltration and improved survival of CRC was demonstrated with no need to discriminate their location in epithelial or stromal regions. Our findings implied that active CD8+ TIL in CRC was demarcated by the expression of ITGAE and suggested that these cells were induced and recruited as a result of the tumor immune response. Consistent results were observed in ovarian cancer, and ITGAE+ lymphocytes were also linked with prolonged patient survival, serving as a predicting marker to enrich the most beneficial subsets of TILs for the immuno-response [18]. In cervical cancer, dense ITGAE+ lymphocyte infiltration was strongly associated with a better prognosis than low ITGAE + lymphocyte infiltration, and ITGAE seemed to be a more precise predictor of outcome than CD8 [19]. Meanwhile, increased numbers of ITGAE+/CD8+ TIL were resided frequently in microsatellite instability (MSI) colorectal cancers, which may contribute improved prognosis [32]. These results suggested that ITGAE alone may be considered an excellent biomarker for tumor-reactive T cells in CRC. Conversely, not all correlations between total CD8+, CD3+, CD45+ or FOXP3+ TIL and patient survival were observed, unless they were located in specific tumor regions, even though inconsistency remained. Thus, ITGAE identified a subpopulation of CD8+ T cells with a reactive phenotype that more accurately predicted patient outcome. In addition, through GSEA, we found that the filtration of dense ITGAE+ lymphocytes was associated with the positive regulation of the inflammatory response, interferon alpha and gamma activation in CRC patients. Furthermore, cancers with high ITGAE+ lymphocytes exhibited significantly increased levels of interferon-response chemokines (CXCL9–10, and CXCL11), which contributed to the modulation of the migration of activated T cells, NK cells and enhanced the anti-tumoral response [[20], [21], [22]]. These results suggested that high ITGAE+ lymphocytes correlated with immune activation in the niche of CRC. Therefore, ITGAE+ lymphocytes might serve as an independent prognostic factor in stratifying and selecting colorectal patients with prolonged survivals.
The tumor-associated immune niche was significantly correlated with EMT. As in gastric cancer, an inflammatory microenvironment that recruits macrophages can contribute to EMT by secreting transforming inducers such as TNF-α, TGF-β, TGF-α, and IL-6 [23, 24]. However, the modulation of EMT by the tumor immune microenvironment was poorly explored. The majority of reports so far have focused on the M2-like macrophage phenotype, which was first proven to induce tumor metastasis and EMT [25, 26]. In this study, ITGAE+ lymphocytes were found to be associated with increased E-cadherin expression and accompanied by decreased Snail expression. E-cadherin belongs to the cell adhesion molecule, whose expression on the membrane has been regarded as the hallmark of EMT [27]. It demonstrated that ITGAE + lymphocytes had the potential to fight against malignant cells with EMT traits. However, it is still unclear whether immune cells could eliminate aberrant cells with EMT traits or support tumor cells to become more EMT-like.
Our study also has limitations. The primary limitation concerns the TMA approach, which may only partly evaluate tissue and tumor heterogeneity. However, our analyses have shown that duplicate TMA cores are sufficient to overcome sampling bias [28, 29]. In addition, two independent TCGA sets were utilized to find an almost perfect agreement concerning ITGAE + lymphocyte infiltration. Still, variations in the amount of tumor stroma and epithelium among different tumors might have confounded the results. Our study included only patients with resected cancers, which might also induce the biased exclusion of unresectable patients.
Taken together, we demonstrated for the first time that ITGAE+ CD8+ lymphocyte infiltration plays a vital role in the antitumor immune response, and ITGAE has been identified as a biomarker of tumor-reactive CD8+ TIL. In this mechanism, ITGAE+ lymphocytes may even associate with interferon-response chemokines and EMT signaling and then serve as an independent predictor in colorectal cancers.
The following are the supplementary data related to this article.
Supplementary material
Representative images of tissue cores as high (a), or low infiltration (b).
Acknowledgments
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. NSFC1631).
Funding
No
Availability of Data and Materials
Not applicable.
Authors' Contributions
Conceived and designed the experiments: H.X. and C.S.J. Analyzed the data: H.X., L.Y.Q., C.S.J., L.Q.G. Contributed reagents/materials/analysis tools: H.X., P.J.J and M.Y.L. Wrote the paper: H.X. and L.Y.Q.
Competing Interests
The authors declare that they have no competing interests.
Funding
No
Sources of Funding for Research
No
Contributor Information
Jun-Jie Peng, Email: junjiepeng67@gmail.com.
San-Jun Cai, Email: caisanjun_sh@163.com.
References
- 1.Siegel R., Desantis C., Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Marisa L., Svrcek M., Collura A., Becht E., Cervera P., Wanherdrick K. The balance between cytotoxic T-cell lymphocytes and immune checkpoint expression in the prognosis of Colon tumors. J Natl Cancer Inst. 2018;110 doi: 10.1093/jnci/djx136. [DOI] [PubMed] [Google Scholar]
- 3.Nakano O., Sato M., Naito Y., Suzuki K., Orikasa S., Aizawa M. Proliferative activity of intratumoral CD8(+) T-lymphocytes as a prognostic factor in human renal cell carcinoma: clinicopathologic demonstration of antitumor immunity. Cancer Res. 2001;61:5132–5136. [PubMed] [Google Scholar]
- 4.Kondratiev S., Sabo E., Yakirevich E., Lavie O., Resnick M.B. Intratumoral CD8+ T lymphocytes as a prognostic factor of survival in endometrial carcinoma. Clin Cancer Res. 2004;10:4450–4456. doi: 10.1158/1078-0432.CCR-0732-3. [DOI] [PubMed] [Google Scholar]
- 5.Donnem T., Hald S.M., Paulsen E.E., Richardsen E., Al-Saad S., Kilvaer T.K. Stromal CD8+ T-cell density-a promising supplement to TNM staging in non-small cell lung Cancer. Clin Cancer Res. 2015;21:2635–2643. doi: 10.1158/1078-0432.CCR-14-1905. [DOI] [PubMed] [Google Scholar]
- 6.Lohneis P., Sinn M., Bischoff S., Juhling A., Pelzer U., Wislocka L. Cytotoxic tumour-infiltrating T lymphocytes influence outcome in resected pancreatic ductal adenocarcinoma. Eur J Cancer. 2017;83:290–301. doi: 10.1016/j.ejca.2017.06.016. [DOI] [PubMed] [Google Scholar]
- 7.Ling K.L., Dulphy N., Bahl P., Salio M., Maskell K., Piris J. Modulation of CD103 expression on human colon carcinoma-specific CTL. J Immunol. 2007;178:2908–2915. doi: 10.4049/jimmunol.178.5.2908. [DOI] [PubMed] [Google Scholar]
- 8.Le Floc'h A., Jalil A., Vergnon I., Le Maux Chansac B., Lazar V., Bismuth G. Alpha E beta 7 integrin interaction with E-cadherin promotes antitumor CTL activity by triggering lytic granule polarization and exocytosis. J Exp Med. 2007;204:559–570. doi: 10.1084/jem.20061524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.French J.J., Cresswell J., Wong W.K., Seymour K., Charnley R.M., Kirby J.A. T cell adhesion and cytolysis of pancreatic cancer cells: a role for E-cadherin in immunotherapy? Br J Cancer. 2002;87:1034–1041. doi: 10.1038/sj.bjc.6600597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webb J.R., Wick D.A., Nielsen J.S., Tran E., Milne K., McMurtrie E. Profound elevation of CD8+ T cells expressing the intraepithelial lymphocyte marker CD103 (alphaE/beta7 integrin) in high-grade serous ovarian cancer. Gynecol Oncol. 2010;118:228–236. doi: 10.1016/j.ygyno.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 11.Franciszkiewicz K., Le Floc'h A., Jalil A., Vigant F., Robert T., Vergnon I. Intratumoral induction of CD103 triggers tumor-specific CTL function and CCR5-dependent T-cell retention. Cancer Res. 2009;69:6249–6255. doi: 10.1158/0008-5472.CAN-08-3571. [DOI] [PubMed] [Google Scholar]
- 12.van Gool I.C., Eggink F.A., Freeman-Mills L., Stelloo E., Marchi E., de Bruyn M. POLE proofreading mutations elicit an antitumor immune response in endometrial Cancer. Clin Cancer Res. 2015;21:3347–3355. doi: 10.1158/1078-0432.CCR-15-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noordhuis M.G., Eijsink J.J., Ten Hoor K.A., Roossink F., Hollema H., Arts H.J. Expression of epidermal growth factor receptor (EGFR) and activated EGFR predict poor response to (chemo)radiation and survival in cervical cancer. Clin Cancer Res. 2009;15:7389–7397. doi: 10.1158/1078-0432.CCR-09-1149. [DOI] [PubMed] [Google Scholar]
- 14.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 15.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Djenidi F., Adam J., Goubar A., Durgeau A., Meurice G., de Montpreville V. CD8+CD103+ tumor-infiltrating lymphocytes are tumor-specific tissue-resident memory T cells and a prognostic factor for survival in lung cancer patients. J Immunol. 2015;194:3475–3486. doi: 10.4049/jimmunol.1402711. [DOI] [PubMed] [Google Scholar]
- 17.Franciszkiewicz K., Le Floc'h A., Boutet M., Vergnon I., Schmitt A., Mami-Chouaib F. CD103 or LFA-1 engagement at the immune synapse between cytotoxic T cells and tumor cells promotes maturation and regulates T-cell effector functions. Cancer Res. 2013;73:617–628. doi: 10.1158/0008-5472.CAN-12-2569. [DOI] [PubMed] [Google Scholar]
- 18.Webb J.R., Milne K., Watson P., Deleeuw R.J., Nelson B.H. Tumor-infiltrating lymphocytes expressing the tissue resident memory marker CD103 are associated with increased survival in high-grade serous ovarian cancer. Clin Cancer Res. 2014;20:434–444. doi: 10.1158/1078-0432.CCR-13-1877. [DOI] [PubMed] [Google Scholar]
- 19.Komdeur F.L., Prins T.M., van de Wall S., Plat A., Wisman G.B.A., Hollema H. CD103+ tumor-infiltrating lymphocytes are tumor-reactive intraepithelial CD8+ T cells associated with prognostic benefit and therapy response in cervical cancer. Oncoimmunology. 2017;6 doi: 10.1080/2162402X.2017.1338230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bindea G., Mlecnik B., Tosolini M., Kirilovsky A., Waldner M., Obenauf A.C. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Neville L.F., Mathiak G., Bagasra O. The immunobiology of interferon-gamma inducible protein 10 kD (IP-10): a novel, pleiotropic member of the C-X-C chemokine superfamily. Cytokine Growth Factor Rev. 1997;8:207–219. doi: 10.1016/s1359-6101(97)00015-4. [DOI] [PubMed] [Google Scholar]
- 22.Farber J.M. Mig and IP-10: CXC chemokines that target lymphocytes. J Leukoc Biol. 1997;61:246–257. [PubMed] [Google Scholar]
- 23.Ma H.Y., Liu X.Z., Liang C.M. Inflammatory microenvironment contributes to epithelial-mesenchymal transition in gastric cancer. World J Gastroenterol. 2016;22:6619–6628. doi: 10.3748/wjg.v22.i29.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuxe J., Karlsson M.C. TGF-beta-induced epithelial-mesenchymal transition: a link between cancer and inflammation. Semin Cancer Biol. 2012;22:455–461. doi: 10.1016/j.semcancer.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Dehai C., Bo P., Qiang T., Lihua S., Fang L., Shi J. Enhanced invasion of lung adenocarcinoma cells after co-culture with THP-1-derived macrophages via the induction of EMT by IL-6. Immunol Lett. 2014;160:1–10. doi: 10.1016/j.imlet.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Lakshmi Narendra B., Eshvendar Reddy K., Shantikumar S., Ramakrishna S. Immune system: a double-edged sword in cancer. Inflamm Res. 2013;62:823–834. doi: 10.1007/s00011-013-0645-9. [DOI] [PubMed] [Google Scholar]
- 27.Rosso M., Majem B., Devis L., Lapyckyj L., Besso M.J., Llaurado M. E-cadherin: a determinant molecule associated with ovarian cancer progression, dissemination and aggressiveness. PLoS One. 2017;12 doi: 10.1371/journal.pone.0184439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoos A., Urist M.J., Stojadinovic A., Mastorides S., Dudas M.E., Leung D.H. Validation of tissue microarrays for immunohistochemical profiling of cancer specimens using the example of human fibroblastic tumors. Am J Pathol. 2001;158:1245–1251. doi: 10.1016/S0002-9440(10)64075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henriksen K.L., Rasmussen B.B., Lykkesfeldt A.E., Moller S., Ejlertsen B., Mouridsen H.T. Semi-quantitative scoring of potentially predictive markers for endocrine treatment of breast cancer: a comparison between whole sections and tissue microarrays. J Clin Pathol. 2007;60:397–404. doi: 10.1136/jcp.2005.034447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganesan A.P., Clarke J., Wood O., Garrido-Martin E.M., Chee S.J., Mellows T. Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat Immunol. 2017;18(8):940–950. doi: 10.1038/ni.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, et al. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature. 14;(7684):253–257. [DOI] [PMC free article] [PubMed]
- 32.Quinn E, Hawkins N, Yip YL, Suter C, Ward R. CD103+ intraepithelial lymphocytes--a unique population in microsatellite unstable sporadic colorectal cancer. Eur J Cancer. 39(4):469–75. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Not applicable.