ABSTRACT
Maintaining cellular redox is critical for growth, metabolism and survival in response to changing environments. How nutrients regulate this process is a long-standing fundamental question in cell biology. Our recent study revealed a conserved mechanism by which eukaryotes, particularly cancer cells, couple nutrient signaling to dynamically regulate redox homeostasis.
Abbreviations: ATP: adenosine triphosphate; Ala: alanine; C6H12O6: glucose; OH−: hydroxyl radical; Glu: glutamate; mRNA: messenger RNA; mTOR: mechanistic/mammalian target of rapamycin; OXYPHOS: oxidative phosphorylation; Ser: serine; ROS: reactive oxygen species; O2−: superoxide; SOD1: superoxide dismutase 1; Thr: threonine
Keywords: mTOR, SOD1, ROS, nutrient, ischemia
Living organisms have evolved mechanisms to respond to changing environmental nutrients in order to maximize their growth and survival. For example, in the presence of glucose, budding yeast uses glycolysis to generate energy and basic building blocks such as nucleosides for supporting rapid growth and proliferation.1 Eukaryotic cells rapidly adjust growth and metabolic capacities according to nutrient conditions. The expression of an estimated 40% genes encoded by the yeast genome is significantly altered within minutes after the culture medium is changed from starvation or a non-fermentable carbon source to glucose.1 These genes include those involved in protein synthesis such as components of the ribosomes, mitochondrial respiration and biosynthesis, and biosynthetic pathways.1 Other cellular programs such as messenger RNA (mRNA) translation and autophagy, which regulates protein synthesis and degradation, are also affected.1 At the center of nutrient sensing and signaling is mechanistic target of rapamycin (mTOR, also known as mammalian TOR). mTOR is a highly conserved protein kinase whose activity rapidly responds to the quantity and quality of nutrients, and regulates diverse cellular processes ranging from transcription, translation to autophagy.2–4
Eukaryotic cells have two distinct energetic pathways, glycolysis in the cytosol and oxidative phosphorylation (OXYPHOS) in the mitochondria, to generate adenosine triphosphate (ATP) depending on the available nutrients. Yeast and cancer cells predominantly use glycolysis when glucose is available, known as Crabtree effect and Warburg effect, respectively.5,6 When non-fermentable carbon source (e.g., glycerol) is the only available nutrient source, yeast cells switch to OXYPHOS.1 For mammalian cells, when carbohydrates are limited, they produce energy through β-oxidation and mitochondrial OXYPHOS by utilizing free fatty acids derived from food, intracellular lipid storage or adipose tissue. Mitochondrial respiration not only produces ATP, but also generates superoxide free radicals, a major source of cellular reactive oxygen species (ROS). ROS plays important regulatory roles in normal physiology, controlling growth, proliferation, survival, differentiation and regeneration.7 Nutrients/growth factors-stimulated production of ROS serves as proliferative signals to regulate cell growth and metabolism. For example, hydrogen peroxide oxidizes and inactivates protein tyrosine phosphatases, restraining their negative role in growth signal transduction.7
Oncogenic transformation or certain pathological conditions such as ischemia leads to robust production of ROS. Excessive ROS oxidizes cellular protein, lipid and nucleic acids, causing oxidative damage and cell death, especially under the ischemic tumor microenvironment. Therefore, cells must up-regulate anti-oxidative capacity to gain more resistance to oxidative damage and enhance survival ability. To gain a comprehensive view of the global nutrient signaling network, we carried out a genomic rapamycin sensitivity screen in yeast to systematically identify new components of mTOR pathway.8 Superoxide dismutase 1 (SOD1) was a top gene identified in this screen. SOD1 is the major cytosolic form of superoxide dismutase and a key conserved antioxidant enzyme that catalyzes the superoxide to hydrogen peroxide reaction.9 Hydrogen peroxide is further converted to oxygen and water by catalase. Because most cellular ROS is originated in the form of superoxide, SOD1 provides the first line of defense against excessive ROS and ROS-mediated cellular damages. Indeed, we subsequently demonstrated that mTOR negatively regulates SOD1 activity in both yeast and human cells in response to nutrient signals: SOD1 is activated when mTOR is inhibited by starvation or rapamycin.10
We next showed that mTORC1 inhibits SOD1 activity by phosphorylating SOD1 at serine 39 (Ser-39) in yeast and threonine 40 (Thr-40) in humans in a nutrient-dependent manner. Interestingly, the phosphorylation site is located at the gateway for the superoxide substrates to enter a positively charged catalytic tunnel that attracts the negatively charged substrates. When phosphorylated, gateway becomes negatively charged, which hinders the entry of superoxide anions into the catalytic tunnel, thereby restraining SOD1 activity. The reversible phosphorylation of SOD1 is crucial for controlling cellular ROS level in response to changing nutrient conditions. When Ser-39/Thr-40 is mutated to glutamate (Glu), a mutation that locks SOD1 at the low activity state, cells are unable to reduce the excessive ROS and suffer extensive oxidative damages and cell death during starvation and hypoxia. On the other hand, when Ser-39/Thr-40 is mutated to alanine (Ala), a residue that locks SOD1 at the high activity state, although cells are more protected from oxidative damages, their rapid cell growth is hindered when nutrient is abundant. Collectively, our findings revealed a conserved mechanism by which eukaryotic cells dynamically control cellular ROS level to ensure adequate proliferation under nutrient-rich condition, while minimizing oxidative damages under starvation and ischemic conditions (Figure 1).
Figure 1.
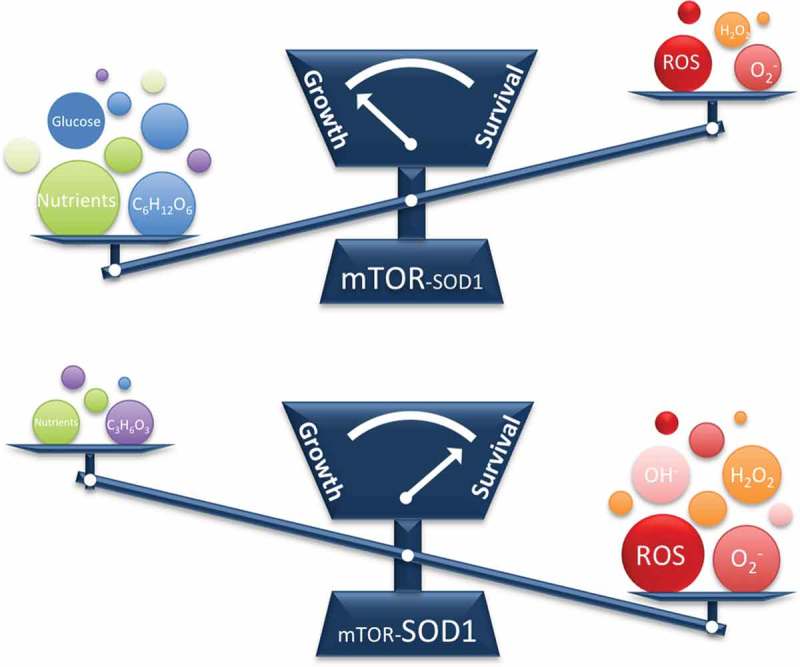
A working model for this study. The conserved mechanistic target of rapamycin (mTOR)- superoxide dismutatse 1 (SOD1) axis balances growth and survival by modulating cellular reactive oxygen species (ROS) contents in response to nutrient signals. C6H12O6, glucose; O2−, superoxide; OH−, hydroxyl radical.
Our finding has several important implications. First, the mTOR-SOD1 axis serves as a survival mechanism to protect tissues and organs from oxidative damages under ischemia, a pathological condition seen with ischemic heart disease and stroke due to blockage of blood supply. Similarly, this signaling axis could also be important for brain protection during hypoglycemia that is known to elicit mitochondrial ROS production in the central nervous system neurons. On the other hand, cancer cells in solid tumors are often deprived of nutrients and oxygen due to poor angiogenesis and ischemic tumor microenvironment. These cancer cells, though are largely dormant, have enhanced survival ability and are resistant to many cancer therapies. Our results suggest that targeting SOD1 is a promising approach to improve therapeutic outcomes. Further exploring the mTOR-SOD1 signaling axis may lead to new treatments for ischemia and hypoglycemia-related diseases.
Funding Statement
This work was supported by the National Institutes of Health [R01CA166575];National Institutes of Health [R01CA123391];
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Zaman S, Lippman SI, Zhao X, Broach JR.. How saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- 2.Li H, Tsang CK, Watkins M, Bertram PG, Zheng XFS. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature. 2006;442:1058–1061. doi: 10.1038/nature05020. [DOI] [PubMed] [Google Scholar]
- 3.Tsang CK, Qi H, Liu L, Zheng XF. Targeting mammalian target of rapamycin (mTOR) for health and diseases. Drug Discov Today. 2007;12:112–124. doi: 10.1016/j.drudis.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Wei Y, Tsang CK, Zheng XF. Mechanisms of regulation of RNA polymerase III-dependent transcription by TORC1. EMBO J. 2009;28:2220–2230. doi: 10.1038/emboj.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crabtree HG. Observations on the carbohydrate metabolism of tumours. Biochem J. 1929;23:536–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 7.Schieber M, Chandel Navdeep S. ROS function in redox signaling and oxidative stress. Curr Biol. 2014;24:R453–62. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan T-F, Carvalho J, Riles L, Zheng XFS. A chemical genomics approach toward understanding the global functions of the target of rapamycin protein (TOR). Proc Natl Acad Sci. 2000;97:13227–13232. doi: 10.1073/pnas.240444197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Che M, Wang R, Li X, Wang H-Y, Zheng XFS. Expanding roles of superoxide dismutases in cell regulation and cancer. Drug Discov Today. 2016;21:143–149. doi: 10.1016/j.drudis.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsang CK, Chen M, Cheng X, Qi Y, Chen Y, Das I, Li X, Vallat B, Fu LW, Qian CN, et al. SOD1 phosphorylation by mTORC1 couples nutrient sensing and redox regulation. Mol Cell. 2018;70:502–515.e508. doi: 10.1016/j.molcel.2018.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]


