ABSTRACT
Homologous recombination is initiated by nucleolytic degradation (resection) of DNA double-strand breaks (DSBs), which involves different nucleases including the Mre11-Rad50-Xrs2 (MRX) complex and the Exonuclease 1 (Exo1). The characterization of a novel mutation in Mre11 causing accelerated DSB resection has allowed to show that MRX facilitates DNA end processing by Exo1 through local unwinding of double-stranded DNA ends.
Keywords: double-strand break, Exo1, MRX, resection, S. cerevisiae
The maintenance of genome stability requires the action of nucleases that cleave the DNA phosphodiester bonds. Among them, Exonuclease 1 (Exo1), an evolutionarily conserved member of the Xeroderma Pigmentosum group G (XPG) nuclease family, is implicated in a multitude of DNA metabolic pathways, including double-strand break (DSB) repair by homologous recombination (HR), DNA mismatch repair, DNA replication and telomere maintenance. Exo1 possesses 5′-3′ double-stranded DNA (dsDNA) exonuclease activity in vitro, and is able to release mononucleotide products from dsDNA ends or internal nicks1.
During HR, research from several laboratories has established that Exo1 is involved in the nucleolytic processing of DSB DNA ends to generate single-stranded DNA (ssDNA), which invades and pairs with the intact homologous DNA sequence and serves as a primer for DNA synthesis in the downstream HR steps. In the current model for resection, the Sporulation in the Absence of spo Eleven (Sae2) protein stimulates the endonuclease activity of Mre11, one of the subunits of the evolutionarily conserved Mre11-Rad50-Xrs2 (MRX) complex, to cleave the 5ʹ-terminated strands at both DNA ends. The resulting nicks are entry sites for Exo1 and DNA replication helicase/nuclease 2 (Dna2) nucleases, which degrade long tracts of DNA in the 5ʹ-3ʹ direction, while Mre11 degrades back toward the DNA end in the 3ʹ-5ʹ direction1.
Given the important role of Exo1 in HR and other DNA transactions, its activity is positively and negatively regulated at multiple levels. In yeast, the nucleolar protein 3 (Npl3), related to the metazoan heterogeneous nuclear ribonucleoproteins (hnRNPs), promotes proper biogenesis of EXO1 mRNA2. Furthermore, the checkpoint protein kinase Rad53 inhibits Exo1 activity through phosphorylation events3, whereas human EXO1 is phosphorylated and possibly activated by cyclin-dependent kinases (CDKs)4. Finally, the access of Exo1 to the dsDNA ends is counteracted by the heterodimer autoantigen Ku70-Ku80 protein complex1, whereas the heterotrimeric Replication Protein A (RPA) complex, which rapidly binds ssDNA, limits the processive activity of Exo1 by physically displacing this nuclease from DNA5.
Exo1-catalyzed DNA degradation is known to be stimulated also by the MRX complex, which enhances both the processivity and the affinity of Exo1 to DNA ends through a poorly understood mechanism1,6,7. On the one hand, MRX could act as a platform to facilitate Exo1 association to DNA ends. On the other hand, MRX could enhance Exo1 activity by competing with the Ku70-Ku80 complex for DNA binding and/or by generating a recessed DNA substrate less suitable for Ku70-Ku80 binding1.
Interestingly, the MRX complex was shown to possess an ATP-dependent unwinding activity capable of releasing a short oligonucleotide from dsDNA8. Exo1 is able to degrade the 5ʹ-terminated strand within dsDNA and, therefore, it in principle does not require an helicase activity to unwind the dsDNA1. However, our recent characterization of the hypermorphic Saccharomyces cerevisiae Mre11-R10T mutant variant has allowed to demonstrate that the strand-separation function of MRX is important to stimulate Exo1 resection activity9. In particular, the mre11-R10T mutation accelerates DSB resection compared to wild type Mre11 by potentiating the processing activity of Exo1, whose association to DSBs is increased in mre11-R10T cells. This increased Exo1 activity is accompanied by decreased association of the Ku70-Ku80 complex to DNA ends, which is not due to either enhanced Mre11 nuclease activity or increased MRX association to DSBs. Rather, the lack of Exo1 or of its nuclease activity restores Ku70-Ku80 persistence at DSBs in mre11-R10T cells, indicating that MRX can restrict Ku70-Ku80 association to DNA ends by promoting the resection activity of Exo1.
The DNA unwinding activity of MRX has been hypothesized to stem from a conformational change of the complex10. Molecular dynamic simulations on wild type Mre11 dimer with dsDNA show that the DNA ends rapidly interact with the two capping domains of the Mre11 dimer and such arrangements causes a strain and partial unwinding of the dsDNA molecule (Figure 1, left)9. The same experiment performed on Mre11-R10T dimer shows that the capping domain of one Mre11-R10T subunit undergoes an abnormal rotation that leads this domain to wedge in between the two DNA strands and to persistently melt the dsDNA ends (Figure 1, right)9. This persistent unwinding can account for both the enhanced access to DNA ends and processing activity of Exo1 by Mre11-R10T. In fact, easily melted dsDNA ends facilitate in vitro the resection activity of Exo1 and its stimulation by MRX9. Furthermore, substitution with alanine of both arginine 368 and 412 residues, through which the Mre11-R10T capping domain contacts the dsDNA, reduces the ability of Mre11-R10T to promote Exo1-mediated DSB resection9, indicating that the increased Exo1 activity depends on the interaction between the Mre11 capping domain and the dsDNA.
Figure 1.
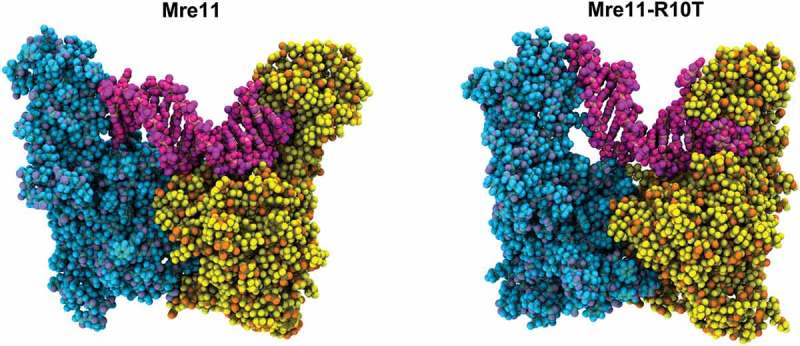
Mre11 dimer contacting DNA. Atomic representation of Mre11-DNA complex. Picture is a snapshot of molecular dynamic simulation and it shows the interaction of double-strand DNA (dsDNA) molecule (pink) with Mre11 dimer (light blue and yellow) during double-strand break (DSB) resection.
Altogether, these findings support a model in which MRX can directly control the resection activity of Exo1 by promoting a change in DNA end structure that helps the retainment of Exo1 on DNA. Although Exo1 is a processive nuclease on its own, it becomes distributive in the presence of RPA that physically removes this nuclease from DNA, resulting in multiple cycles of Exo1 rebinding at the same DNA end5. If, on the one hand, this RPA-mediated control results in a reduction of non-productive Exo1 binding to DNA, on the other hand, the support of MRX in the stimulation of Exo1 association and activity at DNA ends can be of benefit to increase the processivity of this nuclease in the presence of RPA and, therefore, to ensure an efficient long-range resection.
Funding Statement
This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC) under IG grant 19783 and Progetti di Ricerca di Interesse Nazionale (PRIN) 2015 to MPL.
Disclosure statement
No potential conflicts of interest were disclosed.
References
- 1.Symington LS. Mechanism and regulation of DNA end resection in eukaryotes. Crit Rev Biochem Mol Biol. 2016;51(3):195–212. doi: 10.3109/10409238.2016.1172552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colombo CV, Trovesi C, Menin L, Longhese MP, Clerici M. The RNA binding protein Npl3 promotes resection of DNA double-strand breaks by regulating the levels of Exo1. Nucleic Acids Res. 2017;45(11):6530–6545. doi: 10.1093/nar/gkx347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morin I, Ngo HP, Greenall A, Zubko MK, Morrice N, Lydall D. Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J. 2008;27(18):2400–2410. doi: 10.1038/emboj.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomimatsu N, Mukherjee B, Catherine Hardebeck M, Ilcheva M, Vanessa Camacho C, Louise Harris J, Porteus M, Llorente B, Khanna KK, Burma S. Phosphorylation of EXO1 by CDKs 1 and 2 regulates DNA end resection and repair pathway choice. Nat Commun. 2014;5:3561. doi: 10.1038/ncomms4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Myler LR, Gallardo IF, Zhou Y, Gong F, Yang SH, Wold MS, Miller KM, Paull TT, Finkelstein IJ. Single-molecule imaging reveals the mechanism of Exo1 regulation by single-stranded DNA binding proteins. Proc Natl Acad Sci USA. 2016;113(9):E1170–9. doi: 10.1073/pnas.1516674113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicolette ML, Lee K, Guo Z, Rani M, Chow JM, Lee SE, Paull TT. Mre11-Rad50-Xrs2 and Sae2 promote 5ʹ strand resection of DNA double-strand breaks. Nat Struct Mol Biol. 2010;17(12):1478–1485. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cannavo E, Cejka P, Kowalczykowski SC. Relationship of DNA degradation by Saccharomyces cerevisiae exonuclease 1 and its stimulation by RPA and Mre11-Rad50-Xrs2 to DNA end resection. Proc Natl Acad Sci USA. 2013;110(18):E1661–8. doi: 10.1073/pnas.1305166110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13(10):1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gobbini E, Cassani C, Vertemara J, Wang W, Mambretti F, Casari E, Sung P, Tisi R, Zampella G, Longhese MP. The MRX complex regulates Exo1 resection activity by altering DNA end structure. EMBO J. 2018;pii:e98588. doi: 10.15252/embj.201798588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Sung S, Kim Y, Li F, Gwon G, Jo A, Kim AK, Kim T, Song OK, Lee SE, et al. ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex. EMBO J. 2016;35(7):743–758. doi: 10.15252/embj.201592462. [DOI] [PMC free article] [PubMed] [Google Scholar]


