ABSTRACT
We recently identified an interaction between Atg12–Atg3, a complex between 2 core autophagy regulators, and the ESCRT-associated protein Pdcd6ip (programmed cell death 6 interacting protein, commonly known as Alix), which coordinately regulates basal autophagy, late endosome-to-lysosome trafficking, and exosome release. Because these processes all serve fundamental roles in cancer progression and therapy, Atg12-Atg3 may be an attractive anticancer target.
KEYWORDS: Atg12, Atg3, basal autophagy, exosome, late endosome, Pdcd6ip/Alix
Autophagy is a highly conserved pathway that sequesters cytoplasmic contents in a double-membrane structure called the autophagosome and delivers them to the lysosome for degradation. Autophagy is induced as a cytoprotective response to cell stresses such as starvation and hypoxia. Cells also exhibit low levels of basal autophagy independent of nutrient and stress status that controls homeostatic turnover of proteins and organelles. Autophagy is functionally connected to endosomal trafficking, another lysosomal degradation pathway in the cell. Autophagosomes can fuse with late endosomes rather than directly to the lysosome,1 and recent work has demonstrated that endosomal sorting complexes required for transport (ESCRT) function is required for efficient autophagosome maturation.2 However, until now, clear molecular interactions between these 2 pathways had not been established. In our recent study, we identified a novel interaction between components of the core autophagy and ESCRT machinery that controls basal autophagosome maturation and late endosome function.3
Autophagosome formation is controlled by 2 ubiquitin-like conjugation systems that attach the ubiquitin-like molecule Atg12 to Atg5 to form Atg12–Atg5 complexes and microtubule-associated protein 1 light chain 3 (MAP1LC3, best known as LC3) to the lipid phosphatidylethanolamine (PE) to produce LC3–PE. Although Atg5 is the principle conjugation target for Atg12, our laboratory previously identified Atg3, an E2-like enzyme that mediates LC3–PE conjugation, as an additional substrate for Atg12 conjugation.4 Interestingly, although Atg12 and Atg3 each control essential steps in autophagosome formation, cells lacking the Atg12–Atg3 complex have no observable defects in starvation-induced autophagy. However, in nutrient-rich conditions, cells lacking Atg12–Atg3 exhibit increased numbers of autophagosomes.4 Based on these results, we hypothesized that Atg12–Atg3 controls basal autophagic flux.
In addressing this question, we found that cells lacking Atg12–Atg3 have impaired basal autolysosome formation and display a striking accumulation of perinuclear late endosomes, both of which arise from impaired endolysosomal trafficking.3 Intriguingly, the role of Atg12–Atg3 in late endosome function is independent of the canonical role of either protein in autophagosome formation. Furthermore, we identified an interaction between Atg12–Atg3 and the ESCRT-associated protein Pdcd6ip (programmed cell death 6 interacting protein, commonly referred to as Alix). The binding of Atg12–Atg3 to Alix relieves the intramolecular inhibition of Alix by its proline-rich domain and promotes multiple Alix-dependent processes, including late endosome distribution, exosome biogenesis, and viral budding (Fig. 1). Lastly, similar to Atg12–Atg3, Alix is required for efficient basal, but not starvation-induced, autophagy, revealing novel functional differences between homeostatic and stress-induced autophagy in mammalian cells. These results identify a direct connection between the core autophagy and endosomal sorting machineries that coordinately regulate basal autophagy and late endosome function to maintain cellular homeostasis.
Figure 1.
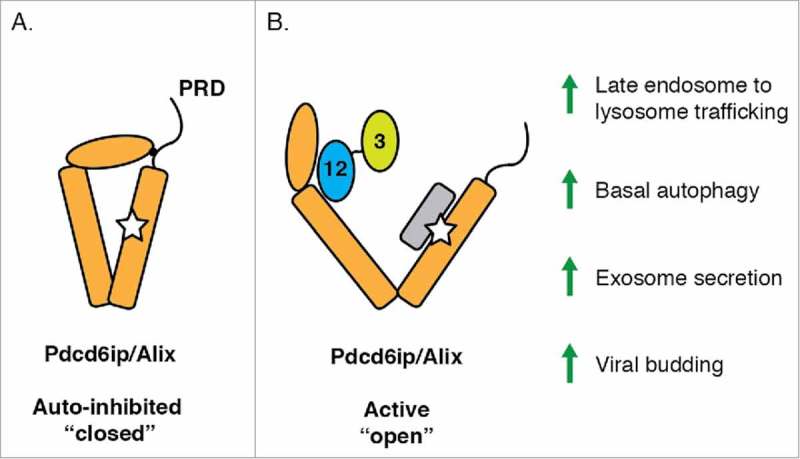
Putative mechanism of Alix activation by Atg12-Atg3. (A) Intramolecular interaction of the proline-rich domain (PRD) maintains Pdcd6ip (programmed cell death 6 interacting protein, commonly known as Alix) in an inhibitory, closed conformation. (B) Atg12–Atg3 binding to Alix displaces the PRD, leading to an open Alix conformation. Alix activation promotes multiple related pathways, including late endosome-to-lysosome trafficking, basal autophagy, exosome secretion, and viral budding.
Autophagy is proposed to play context-dependent roles in tumor initiation and progression.5 Autophagy suppresses tumor initiation by degrading protein aggregates and damaged organelles that cause oxidative stress and DNA damage. Conversely, tumor cells commonly exhibit high basal levels of autophagy, which supports the survival and metabolic adaptation of established tumors in response to microenvironmental or therapeutic stresses. As a result, the lysosomal inhibitor hydroxychloroquine (HCQ) is being tested as a chemosensitizing agent in clinical oncology trials.5 Nevertheless, we have an incomplete understanding of whether HCQ and similar agents exert their anticancer effects by inhibiting autophagy versus other lysosomal degradation pathways. In particular, alterations in endosomal trafficking have also been implicated in cancer progression.6 Following their endocytosis, signaling molecules can either be recycled back to the plasma membrane or targeted to late endosomes for subsequent degradation. Sorting to late endosomes is controlled by the ESCRT machinery, which mediates ubiquitination of cargo targeted for downregulation, membrane remodeling, and budding of intralumenal vesicles. Many cancer-promoting mutations bias endosomal trafficking toward recycling of integrins and receptor tyrosine kinases rather than degradation.6 Our data identify the Atg12–Atg3–Alix complex as a direct link between the autophagy and endosomal sorting machineries.3 Thus, dissecting the functional consequences of Atg12–Atg3 conjugation on oncogenic signaling and cancer progression represents an exciting avenue of research.
In association with accessory proteins such as Alix, the ESCRT machinery also participates in topologically similar membrane budding events involving membrane deformation away from the cytoplasm. In a poorly understood process, late endosomes can fuse with the plasma membrane rather than with the lysosome, leading to secretion of intralumenal vesicles as exosomes.7 Exosome signaling influences multiple aspects of tumor biology, including priming of distant metastatic sites and resistance to chemotherapy and radiation therapy.8 During exosome biogenesis, Alix forms a complex with the heparan sulfate-containing coreceptor syndecan 1 and the scaffolding protein Sdcbp (syndecan binding protein, also known as syntenin), mediating the loading of signaling cargo into exosomes and promoting exosome release.9 We demonstrate that Atg12–Atg3 relieves the intramolecular inhibition of Alix and promotes exosome release.3 Interestingly, previous work in yeast has implicated core autophagy components in the unconventional secretion of the long-chain fatty acid transporter ACB1 through an exosome-like intermediate.10 Our data now identify Atg12–Atg3 as a novel regulator of exosome release in mammalian cells. Future studies are required to ascertain the physiological significance of Atg12–Atg3 in exosome-dependent processes during cancer metastasis.
As increasing evidence points to important roles for basal autophagy, endosomal trafficking, and exosome secretion in cancer progression and response to cancer therapy, our work reveals unique and timely new interconnections between these fundamental cell biological pathways. Further mechanistic and in vivo studies are required to determine the precise contributions of Atg12–Atg3 and Alix to each of these pathways during cancer progression in order to inform the rational design of future therapeutic strategies.
Funding Statement
HHS | NIH | National Cancer Institute (NCI) CA1884040 CA126792; NSF | EHR | Division of Graduate Education (DGE) 1144247
Acknowledgments
This work was supported by the NIH (CA126792, CA188404) and a Howard Hughes Medical Institute Physician-Scientist Early Career Award to JD and a NSF Graduate Research Fellowship (DGE-1144247) to LM.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Berg TO, Fengsrud M, Stromhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes - Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem 1998; 273:21883-92; PMID:9705327; https://doi.org/ 10.1074/jbc.273.34.21883 [DOI] [PubMed] [Google Scholar]
- 2.Lee J-A, Beigneux A, Ahmad ST, Young SG, Gao F-B. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol 2007; 17:1561-7; PMID:17683935; https://doi.org/ 10.1016/j.cub.2007.07.029 [DOI] [PubMed] [Google Scholar]
- 3.Murrow L, Malhotra R, Debnath J. ATG12–ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat Cell Biol 2015; 17:300-10; PMID:25686249; https://doi.org/ 10.1038/ncb3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radoshevich L, Murrow L, Chen N, Fernandez E, Roy S, Fung C, Debnath J. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell 2010; 142:590-600; PMID:20723759; https://doi.org/ 10.1016/j.cell.2010.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer 2007; 7:961-7; PMID:17972889; https://doi.org/ 10.1038/nrc2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mosesson Y, Mills GB, Yarden Y. Derailed endocytosis: an emerging feature of cancer. Nat Rev Cancer 2008; 8:835-50; PMID:18948996; https://doi.org/ 10.1038/nrc2521 [DOI] [PubMed] [Google Scholar]
- 7.Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002; 2:569-79; PMID:12154376 [DOI] [PubMed] [Google Scholar]
- 8.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Meta Rev 2013; 32:623-42; PMID:23709120; https://doi.org/ 10.1007/s10555-013-9441-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, et al.. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol 2012; 14:677-85; PMID:22660413; https://doi.org/ 10.1038/ncb2502 [DOI] [PubMed] [Google Scholar]
- 10.Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V. Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol 2010; 188:527-36; PMID:20156967; https://doi.org/ 10.1083/jcb.200911154 [DOI] [PMC free article] [PubMed] [Google Scholar]


