Abstract
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal cancers, with a high mortality rate and poor prognosis. However, little is known concerning the molecular mechanism of PDAC at the proteomics level. Here we report a proteomics analysis of PDAC tumor and adjacent tissues by shotgun proteomics followed by label-free quantification, and in total, 3031 and 3306 proteins were identified in three pairs of PDAC tumor and adjacent tissues, respectively; 40 of them were differentially expressed for at least three-fold in PDAC tumor tissues. Ontological and interaction network analysis highlighted the dysregulation of a set of four proteins in the carboxypeptidase family: carboxypeptidase A1 (CPA1), A2 (CPA2), B1 (CPB1), and chymotrypsin C (CTRC). Western blotting confirmed the downregulation of the carboxypeptidase network in PDAC. Immunohistochemistry of tissue microarray from 90 PDAC patients demonstrated that CPB1 was downregulated 7.07-fold (P < .0001, n = 81) in tumor comparing with the peritumor tissue. Further 208 pancreatic tissues from PDAC tumor, peritumor, and pancreatis confirmed the downregulation of CPB1 in the PDAC patients. In summary, our results displayed that the expression of carboxypeptidase is significantly downregulated in PDAC tumor tissues and may be novel biomarker in the patient with PDAC.
Introduction
Pancreatic cancer, with its extremely high mortality and poor prognosis, is one of the most lethal cancers worldwide. Pancreatic ductal adenocarcinoma (PDAC) accounts for approximately 85% to 90% of all pancreatic cancers. Although resection remains the best treatment option for patients, most cases are diagnosed at late stages and therefore miss the opportunity for surgery. Chemotherapy, radiation, or combinations of these approaches have had little effect on the course of this malignant neoplasm. Indeed, the 5-year survival rate of patients with advanced-stage disease is less than 2% after diagnosis [1]. Therefore, there is an urgent need for novel treatment targets based on the underlying molecular mechanisms of pancreatic cancer progression.
Proteomic technologies link protein expression patterns with the developmental process of pancreatic cancer. Thus, this approach may help to discover novel specific biomarkers for the diagnosis, treatment, or prognosis of this disease [2] even to illuminate underlying molecular mechanisms. Currently, various approaches combined with mass spectrometry (MS) are available to identify differentially expressed proteins in pancreatic cells, tissues, and biologic fluids [3]. Recently, the use of label-free protein profiling to examine differentially expressed proteins in tissue specimens has gained considerable attention from investigators because it does not require special reagents or extra experimental steps, so it largely eliminates variances and biases between replicate MS measurements [4], [5]. Application of label-free proteomics may help us to explore the mechanism of tumorigenesis and discover novel biomarkers of diverse diseases [6].
To study the novel specific biomarkers may provide more precise diagnosis and treatment with PDAC. We analyzed the differential protein profiles among three pairs of PDAC tissues and adjacent tissues using the MS approach, and a total of 3031 and 3306 proteins were identified in three pairs of PDAC tumor and adjacent tissues, respectively. The further combination of label-free analysis highlighted 40 proteins that were differentially expressed in at least 2 pairs of tissues, and the expression difference exceeded three-fold. Furthermore, we carried out an analysis of the functional interactions linking these proteins, showing that they were mainly involved in cellular metabolism, location, and development processes. Among the identified proteins, we focused on a set of pancreatic tissue-specific proteins: carboxypeptidase A1 (CPA1), carboxypeptidase A2 (CPA2), carboxypeptidase B1 (CPB1), and chymotrypsin C (CTRC). These proteins are carboxypeptidase family members secreted by the pancreas that exist as inactive proenzymes. Once activated, they are involved in the process of pancreatitis. For instance, previous studies have shown that CTRC is a potential key regulator of digestive zymogen activation, and it can activate proCPA1 and proCPA2 [7]. However, the roles of these proteins in PDAC have not been elucidated. Here, we found that these proteins displayed low abundance in tumor tissues compared with control samples. Furthermore, immunohistochemistry (IHC) experiments showed that the expression level of CPB1 displayed the most significant difference between the experimental groups and control groups. Next, we investigated the expression level of CPB1 using a tissue microarray and found that this protein was expressed in almost all peritumoral normal tissues but not in all tumor samples.
Methods and Materials
Patient Samples
Ten pairs of PDAC tissues and peritumoral tissues were obtained from PDAC patients undergoing surgical resection at Xijing Hospital of Digestive Diseases or Tangdu Hospital. All of the PDAC patients met the 2011 revised American Joint Committee on Cancer diagnostic criteria. Ethics approval was granted from the Ethics Committee of the Fourth Military Medical University. All of the patients gave their informed consent to participate in this study. The parameters of clinical pathology are displayed in Table S1, Supplementary Material. The tissues were immediately frozen and stored in liquid nitrogen until they were used in experiments.
Protein Extraction
Fifty-milligram tumor tissues and peritumoral tissues were cut into pieces and homogenized, using a glass pestle, in the RIPA lysis buffer containing some protease inhibitor at a ratio of 1:10 (w/v). Each lysate was centrifuged at 4°C for 15 minutes at 12,000 ×g and then was dialyzed for 5 hours with 3500 MWCO membrane. The protein concentration was measured with BCA Kit. The proteins were deposited at −80°C.
In-Solution Trypsin Digestion
For quantitative proteomics analysis, 50-μg proteins were digested of each sample with sequencing-grade modified trypsin (1:50 w/w; Sigma) at 37°C for 24 hours. The solution was desalted using the C18 columns (Sigma), then were dried and deposited at −80°C until ready for use.
LC-MS/MS Analysis
The peptides of each sample were fractionated using the high-pressure liquid chromatography (Thermo EASY-nLC System) and were analyzed by Orbitrap Elite mass spectrometer (Thermo LTQ Orbitrap Velos Elite, Thermo Scientific). The solution of washing column contains buffer A (0.1% FA in deionized water) and buffer B (0.1% FA in acetonitrile). The peptides were eluted from the column using the washing buffer A and B at flow rates 200 nL/min for about 120 minutes. Then, the peptides were analyzed using mass spectrometer with the collision-induced dissociation mode. Each sample was analyzed three times using the same procedure.
Protein Identification
The MS/MS data were analyzed using Proteome Discoverer 1.4 software. Diverse proteins were identified using the human protein database (UniProtKB, 72,275 sequences). Mass tolerances were set to 10 ppm in MS modes, fragment mass tolerances were set to 0.5 Da in MS/MS modes, and the missed cleavages were set to 2. Carbamidomethyl (C) was set as a static modification, and oxidation (M) was set as a variable modification. The threshold of false discovery rate was 0.01.
Label-Free Quantification
We used the Progenesis LC-MS software (version 4.1; Nonlinear Dynamics, UK) to analyze three paired samples. The following parameters were used when analyzing: the missed cleavages were set to 2, and the human proteome from Swiss-Prot was used as a background proteome. Carbamidomethyl (C) was set as a static modification, and oxidation (M) was set as a variable modification. The peritumoral sample was set as the reference, and data processing was aligned. The analysis of variance (P < .05) and q values (P < .05) were used to detect statistically significant proteins.
Western Blot Analysis
The protein concentrations of the lysates were determined using the bicinchoninic acid assay (Thermo, US, 23227). Fifty micrograms of each lysate was separated by reducing SDS-PAGE and was transferred onto a PVDF membrane (Roche, Swiss, 03010040001). The membrane was then blocked in TBS buffer containing 5% skim milk for 1 hour. The target bands were blotted with various primary antibodies overnight, including anti-CPA1 (abcam, UK; ab173283), anti-CPA2 (Proteintech, US; 15,626-1-AP), anti-CPB1 (Proteintech, US; 12,600-1-AP) and CTRC (abcam, UK; ab35694), and horseradish peroxidase–conjugated secondary antibodies were used to develop the membrane.
Immunohistochemistry and Tissue Microarray Analysis
Paraffin sections of tumor tissues and peritumoral tissues were used to examine the different expression levels of CPA1, CPA2, CPB1, and CTRC. After deparaffinization in xylene and rehydration through graded ethanol solutions, the tissue slices were incubated in 3% hydrogen peroxide at room temperature for 15 minutes to inactivate endogenous peroxidases. Next, antigen retrieval was performed by heating in 10 mM sodium citrate buffer (pH 6.0) for 15 minutes. The sections were blocked with 10% normal goat serum for 1 hour and were incubated with the appropriate primary antibodies overnight at 4°C. The next day, the tissue slices were incubated with horseradish peroxidase–labeled secondary antibody for 30 minutes at room temperature, and protein detection was performed using 3,3′-diaminobenzidine in substrate chromogen solution (Genetex, China). The slices were counterstained with hematoxylin prior to dehydration and mounting. The CPB1 expression pattern was examined using PDAC tissue microarrays, one of which (Biomax, US, HPan-Ade180Sur-01) contained 90 pairs of PDAC tissues and adjacent nontumor tissues, and the other contained 42 PDAC tissues, 3 pancreatic adenosquamous carcinoma tissues, 1 pancreatic islet cell carcinoma tissue, 6 pancreatic metastatic carcinoma tissues, 10 pancreatic islet cell tumor tissues, 11 pancreatic inflammation tissues, 21 adjacent normal pancreatic tissues, and 10 normal pancreatic tissues. Staining of the tissue microarrays was performed according to the protocols mentioned above.
Statistical Analysis
The slides were scanned using a high-resolution scanner (ScanScope XT; Aperio) at 20× magnification. Three surgical pathologists blinded to the clinical variables and outcomes evaluated the slides independently. Both the intensity (0 = absent, 1 = weak, 2 = moderate, and 3 = strong expression) and percentage of positive cells (0 = 0%, 1 = 1%-10%, 2 = 11%-49%, 3 = 50%-74%, 4 = 75%-100%) were assessed, and the scores were multiplied. Scores of 0 to 3 were considered negative, and scores of 4 to 12 were considered positive. SPSS version 19.0 software was used to conduct the statistical analyses. The statistical significance of differences between two groups was assessed by χ2 test, and a P value < .05 was accepted as statistically significant. All of the statistical tests were two-sided.
The digital whole slide images were analyzed using Aperio ImageScope software. An region of interest for individual tumor cores was selected and analyzed using the positive pixel count algorithm. The data were analyzed using paired and unpaired Student t tests by SPSS version 19.0 software. P < .05 was considered significant. The receiver operating characteristic analysis (ROC) curve was generated to evaluate the sensitivity and the specificity of CPB1 between peritumor tissues and tumor tissues.
The scores of tumor tissues and control tissues were normalized. The expression levels of CPB1 were remarkably higher in peritumor tissues than in tumor tissues. The ROC curves were established to assess the potential value of CPB1 for distinguishing peritumor tissues and tumor tissues. The analysis discovered that the area under the ROC curve (AUC) was 0.9851, which was greater than 0.9 (P < .0001) [95% confidence interval 0.9704-0.9991].
Results
Label-Free Quantitation of Differentially Expressed Proteins in Tumor and Peritumoral Tissues
We performed protein extraction from the tumor and peritumoral tissues of three patients with pancreatic cancer. Next, the lysates were digested using the trypsin, and the peptides were fractionated and analyzed using HPLC and mass spectrometer, respectively. The peptide MS/MS spectra were searched against the human protein database using Proteome Discoverer 1.4 software to identify the proteins in the tumor and peritumoral tissues, and in total, 3031 and 3306 proteins were identified using Proteome Discoverer 1.4 in three pairs of PDAC tumor and adjacent tissues (Table S1, Supplementary Material). The label-free quantitation analysis of proteins from tumor and peritumoral tissue was performed with Progenesis LC-MS software; 220, 222, and 303 differentially expressed proteins (≥3-fold) were identified in three PDAC patients (Table S1, Supplementary Material), while 40 proteins were significantly differentially expressed (expression differed more than 3-fold in at least 2 of 3 pairs of tissue samples). Statistical analysis showed that 17 proteins were upregulated and 23 proteins were downregulated in the tumor tissue (Table 1). Among the upregulated proteins, the expression differences among F13A1, THBS2, POSTN/OSF2, and HPX in tumor tissues and control tissues were greater than 10-fold. Conversely, the carboxypeptidase family members (CPA1, CPA2, CPB1, and CTRC) were among the main significantly downregulated proteins, and CPA1 showed the greatest fold change of 0.022.
Table 1.
The different expression proteins in PDAC tissues
| Gene Name | Uniprot Accession | Protein Name | Peptides Used for Quantitation | Mascot Score | Anova (P)* | t/n |
|---|---|---|---|---|---|---|
| CPA1 | P15085 | Carboxypeptidase A1 | 6 | 1281.75 | 2.1E-07 | 0.02 |
| HSPA9 | P38646 | Stress-70 protein, mitochondrial | 3 | 407.04 | 9.3E-06 | 0.03 |
| PNLIPRP2 | P54317 | Pancreatic lipase-related protein 2 | 4 | 495.47 | 7.5E-06 | 0.04 |
| NUCB2/NEFA | D3DQX5 | Nucleobindin-2 | 3 | 235.82 | 1.2E-05 | 0.04 |
| ACAT1 | P24752 | Acetyl-CoA acetyltransferase, mitochondrial | 4 | 277.73 | 4.7E-05 | 0.06 |
| PRDX4 | Q13162 | Peroxiredoxin-4 | 3 | 623.1 | 1.6E-06 | 0.06 |
| CTRC | Q99895 | Chymotrypsin-C | 5 | 388.35 | .00024 | 0.08 |
| RPS11 | P62280 | 40S ribosomal protein S11 | 3 | 92.24 | .00018 | 0.09 |
| CPB1 | P15086 | Carboxypeptidase B | 6 | 1127.54 | 4.3E-05 | 0.1 |
| RPL35 | P42766 | 60S ribosomal protein L35 | 2 | 132.09 | .00017 | 0.1 |
| CPA2 | P48052 | Carboxypeptidase A2 | 7 | 811.78 | 1.2E-05 | 0.11 |
| RPL38 | P63173 | 60S ribosomal protein L38 | 2 | 295.06 | 7.9E-05 | 0.11 |
| PARK7 | Q99497 | Protein DJ-1 | 2 | 158.81 | .00021 | 0.11 |
| AKR7A3 | O95154 | Aflatoxin B1 aldehyde reductase member 3 | 3 | 334.45 | 8.2E-05 | 0.14 |
| P4HB | P07237 | Protein disulfide-isomerase | 16 | 2156.05 | 5.4E-06 | 0.14 |
| LRRC59 | Q96AG4 | Leucine-rich repeat-containing protein 59 | 5 | 621.89 | 2.7E-05 | 0.14 |
| PDIA4 | P13667 | Protein disulfide-isomerase A4 | 19 | 1654.58 | 1.41E-08 | 0.15 |
| RPL12 | P30050 | 60S ribosomal protein L12 | 2 | 197.63 | .00012 | 0.16 |
| PPIB | P23284 | Peptidyl-prolyl cis-trans isomerase B | 3 | 540.2 | 2.4E-06 | 0.18 |
| CPT2 | P23786 | Carnitine O-palmitoyltransferase 2, mitochondrial | 3 | 75.18 | .00293 | 0.18 |
| RPS18 | P62269 | 40S ribosomal protein S18 | 4 | 500.82 | 3.4E-05 | 0.19 |
| ENPP1 | P22413 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 1 | 2 | 304.67 | .00137 | 0.2 |
| COPB1 | P53618 | Coatomer subunit beta | 6 | 219.23 | 6.5E-06 | 0.22 |
| CAT | P04040 | Catalase | 4 | 303.86 | 1.1E-05 | 3.18 |
| VCP | P55072 | Transitional endoplasmic reticulum ATPase | 9 | 692.07 | 2E-05 | 3.21 |
| ANXA2 | P07355 | Annexin A2 | 8 | 1382.27 | 2.3E-06 | 3.22 |
| PLEC | Q15149 | Plectin | 29 | 2488.63 | 1.7E-06 | 3.78 |
| RPL8 | P62917 | 60S ribosomal protein L8 | 4 | 244.97 | .00026 | 3.9 |
| APOA1 | P02647 | Apolipoprotein A-I | 10 | 777.02 | 7.4E-05 | 4.27 |
| LYST | Q99698 | Lysosomal-trafficking regulator | 3 | 91.02 | .0002 | 4.58 |
| TPM4 | P67936 | Tropomyosin alpha-4 chain | 2 | 750.3 | 1.2E-05 | 4.7 |
| C3 | P01024 | Complement C3 | 29 | 2407.93 | 3.2E-06 | 5.07 |
| NIN | C9J066 | Ninein | 2 | 152.64 | .00022 | 5.12 |
| LAMC1 | P11047 | Laminin subunit gamma-1 | 5 | 333.95 | 1.4E-05 | 6.6 |
| PRELP | Q6FG38 | Prolargin | 6 | 826.17 | 4.3E-05 | 9.17 |
| LUM | P51884 | Lumican | 8 | 762.91 | .0006 | 9.43 |
| HPX | P02790 | Hemopexin | 2 | 282.23 | .0005 | 11.2 |
| POSTN/ OSF2 | C0IMJ1 | Periostin | 3 | 369.81 | 3.1E-05 | 12.4 |
| THBS2 | P35442 | Thrombospondin-2 | 3 | 122.02 | .00012 | 13.7 |
| F13A1 | P00488 | Coagulation factor XIII A chain | 2 | 223.84 | .00236 | 19.4 |
Ontological Classification and Functional Interaction Networks of the Differentially Expressed Proteins
All of the 40 differentially expressed proteins were classified using the PANTHER (http://www.pantherdb.org/) database according to “Molecular function” and “Biological process.” The classification of proteins according to their molecular function showed a repartition mainly involved in catalytic activity (44.2%), binding activity (20.9%), and structural molecule activity (19.0%) (Figure 1A). The biological process repartition analysis showed that the identified proteins were mainly involved in metabolic (42.4%) and cellular processes (11.9%) (Figure 1B).
Figure 1.
(A-B) Identified proteins from the tumor and peritumor tissues of three PDAC patients analyzed with the PANTHER bioinformatics tool (http://www.pantherdb.org) using the Gene Ontology categories “Molecular Function” and “Biological Process.” For each category, the number of differentially expressed proteins and their percentage of the total number of proteins in the pie chart were marked for each Gene Ontology term. (C-D) Functional interaction network linking the significantly differentially expressed proteins between the tumor and peritumor tissues from PDAC patients (String database). K-means classification revealed three groups mainly represented by pancreatic-specific proteins: ribosomal proteins, mitochondrial proteins, and isomerases (D).
The proteins involved in the network are marked by the following abbreviations: C3: complement component3; HPX: hemopexin; APOA1: apolipoprotein A-1; VCP: valosin-containing protein; P4HB: prolyl-4-hydroxylase; PRDX4: peroxiredoxin 4; PPIB: peptidylprolyl isomerase B; PDIA4: protein disulfide isomerase family A member 4; CPA1: carboxypeptidase A1; CPA2: carboxypeptidase A2; CPB1: carboxypeptidase B1; CTRC: chymotrypsin C; RPS11: 40S ribosomal protein S11; RPL12: ribosomal protein L12; RPS18: 40S ribosomal protein S18; RPL35: ribosomal protein L35; RPL38: ribosomal protein L38; RPL8: ribosomal protein L8; PLEC: plectin 1; PARK7: Protein deglycase DJ-1; POSTN: periostin; THBS2: thrombospondin2; LUM: lumican; PRELP: proline/arginine-rich end leucine-rich repeat protein; NIN: ninein; ENO2: enolase2; and HSPA9: heat shock 70-kDa protein 9.
Then, we performed an analysis to determine the functional interaction network linking the 40 differentially expressed proteins between tumor and peritumoral tissues using the String database (Figure 1C). Further K-means analysis was performed to examine the associations of these proteins, and the result indicated that 27 of the 40 proteins showed good correlation, and the proteins were classified into 3 groups (Figure 1D). The first group (proteins in the red circle) was mainly composed of specific proteins in the pancreas that function as regulators of the secretion of pancreatic juices and the activation of various enzymes: CPA1, CPA2) CPB1, and CTRC. The second group (blue cluster) contained ribosomal proteins and mitochondrial proteins that may be involved in metabolism: 40S ribosomal protein S11 (RPS11), 40S ribosomal protein S18 (RPS18), ribosomal protein L12 (RPL12), ribosomal protein L35 (RPL35), ribosomal protein L38 (RPL38), ribosomal protein L8 (RPL8), protein deglycase DJ-1 (PARK7), and plectin 1 (PLEC). The third group (proteins in the green circle) included some isomerases with potential roles in protein formation, isomerism, and folding: valosin-containing protein (VCP), peptidylprolyl isomerase B (PPIB), protein disulfide isomerase family A member 4 (PDIA4), apolipoprotein A-1 (APOA1), prolyl-4-hydroxylase (P4HB), and peroxiredoxin 4 (PRDX4). Other linked proteins included complement component 3 (C3), hemopexin (HPX), periostin (POSTN), thrombospondin2 (THBS2), proline/arginine-rich end leucine-rich repeat protein (PRELP), lumican (LUM), ninein (NIN), enolase2 (ENO2), and heat shock 70-kDa protein 9 (HSPA9). The functional interaction network indicated that the differences between PDAC and the adjacent normal tissues were mainly determined by proteins involved in pancreas-specific enzyme function, metabolism and protein formation, and isomerism and folding.
Western Blotting and Immunohistochemical Staining
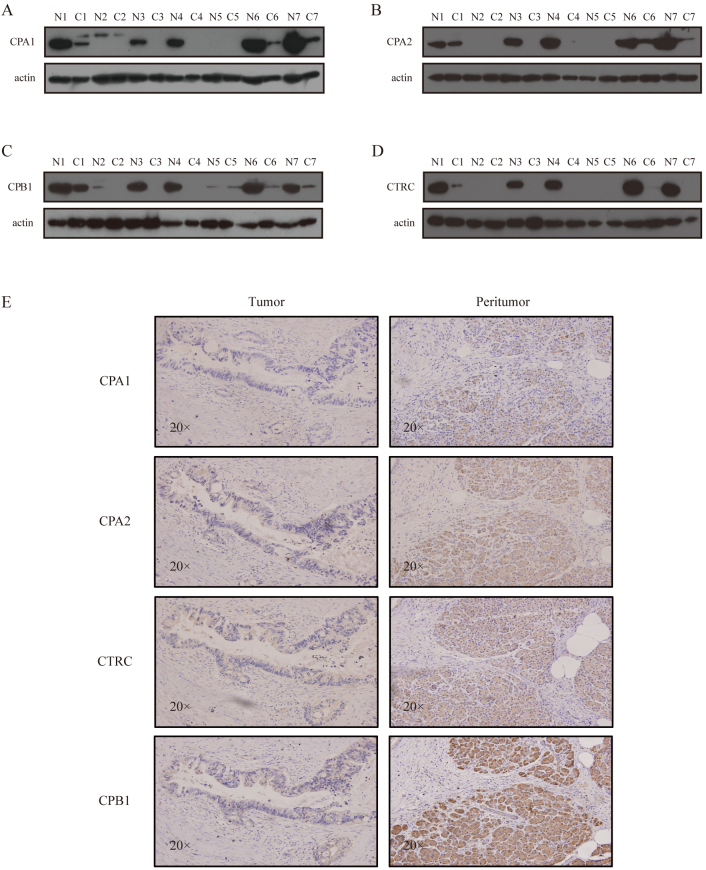
To select candidate proteins for Western blot analysis, we focused on proteins with biological functions that may be involved in the process of PDAC, such as “pancreatic-specific proteins” and “regulators of pancreatic secretion.” The above ontological and interaction network analysis highlighted a set of proteins including CPA1, CPA2, CPB1, and CTRC, which were specifically expressed in pancreatic tissues. These proteins were pancreatic-specific enzymes and were all less abundant in tumor tissues. We tested CPA1, CPA2, CPB1, and CTRC expression in tumor and peritumoral tissues. All four of these proteins were validated by Western blot analysis (Figure 2, A-D).
Figure 2.
(A-D) Representative Western blot of CPA1, CPA2, CPB1, and CTRC between tumor and peritumor tissues. (E) Representative IHC staining for CPA1, CPA2, CPB1, and CTRC in tumor and peritumor tissues.
IHC staining was performed to investigate the expression level of CPA1, CPA2, CPB1, and CTRC in tumor and peritumoral tissues. The results showed that the cellular localization of these four proteins was in the cytoplasm. In addition, all of these proteins were more abundant in peritumoral tissue, as shown by Western blot analysis, and CPB1 was the most significantly downregulated protein in tumor tissues (Figure 2E). However, no previous reports have investigated the correlation between the expression of CPB1 and PDAC; therefore, we selected this protein for further tissue microarray study.
CPB1 Expression Level in PDAC Tissue Microarray
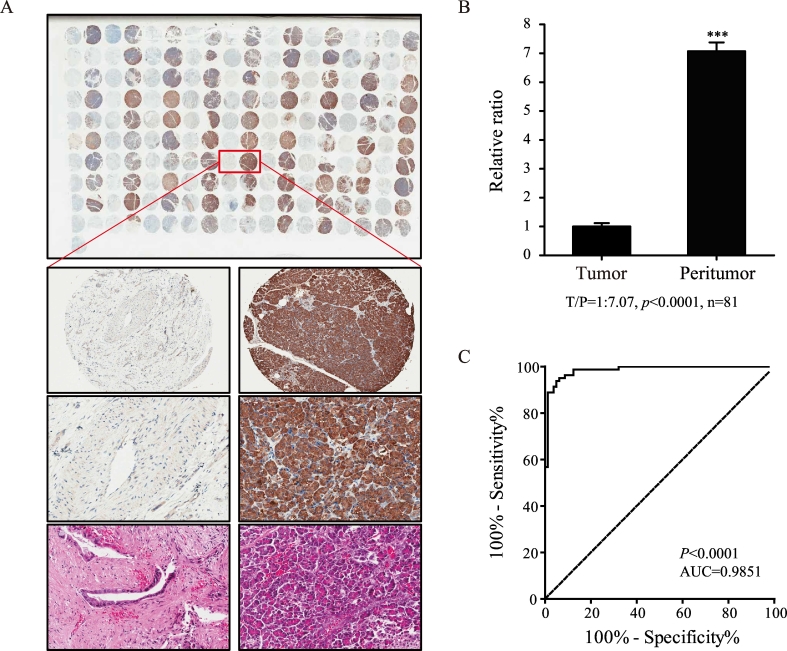
First, we analyzed CPB1 expression in tissue microarrays by IHC staining. The evaluable percent of CPB1 IHC staining was 93.3% (n = 84/90) for all PDAC tissues and 96.7% (n = 87/90) for the control parts. The remaining cases were not evaluable mainly due to the lack of adequate tissues. The cellular localization of CPB1 was in the cytoplasm, and representative histological images are depicted in Figure 3A. Further quantitative analysis revealed that the mean cytoplasmic CPB1 expression in peritumor tissue was 7.07-fold higher than the PDAC tissue, as shown in the histogram of Figure 3B (P < .0001). ROC of the 81 samples revealed that expression level of CPB1 can be used to distinguish PDAC tissues from controls, and the AUC was 0.9851(Figure 3C). Staining score analysis revealed that the positive expression rate of CPB1 in tumor tissue was significantly lower than peritumoral tissue (7.41% vs 98.77%, P < .0001) (Table 2).
Figure 3.
(A) Representative images of tumor and peritumor tissues with CPB1 staining. (B) The expression of CPB1 in tumor tissue was lower than that in peritumor tissue (P < .0001). (C) An ROC curve was generated according to the CPB1 staining. The dashed reference line represents the ROC curve for a test with no discriminatory ability. The AUC is shown on the graph with the 95% confidence interval shown between the parentheses (0.9704-0.9997), P < .0001. No possible cutoff value was derived from the analysis.
Table 2.
Expression of CPB1 in Tissue Microarray
| - | + | Total | Positive Rate | |
|---|---|---|---|---|
| Tumor | 75 | 6 | 81 | 7.41% |
| Peritumor | 1 | 80 | 81 | 98.77% |
χ2 test (two-sided), P < .0001.
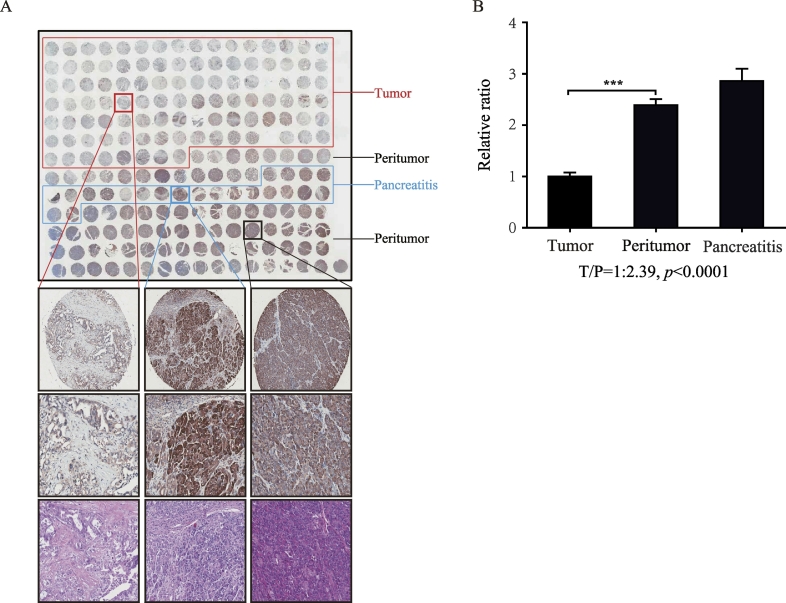
Next, we increased the sample number to 208 dots for repeated analysis, and a total of 22 pancreatitis samples were included in this tissue microarray plate. A similar result was observed in tumor and peritumor tissues. Meanwhile, pancreatitis tissue showed an even higher level of CPB1 expression than peritumoral tissue (Figure 4A). The mean cytoplasmic CPB1 expression in tumor tissue was 2.39-fold lower than peritumoral tissue (Figure 4B). The relative lower score ratio compared with the upper microarray in Figure 3 was mainly attributed to the differences in the tissue origins. That is, the Figure 3 microarray was the paired tissue slices, while the Figure 4 was not. Staining score analysis showed that expression of CPB1 was higher in pancreatitis tissue (P < .0001) and lower in tumor tissue (P < .0001) compared with peritumoral tissue (Table 3).
Figure 4.
(A) Representative images of tumor, peritumor, and pancreatitis tissues with CPB1 staining. (B) The expression of CPB1 in tumor tissue was lower than that in peritumor tissue (P < .0001).
Table 3.
Expression of CPB1 in Tissue Microarray
| - | + | Total | Positive Rate | |
|---|---|---|---|---|
| Tumor | 44 | 10 | 54 | 18.52% |
| Peritumor | 5 | 36 | 41 | 87.80% |
| Pancreatitis | 0 | 11 | 11 | 100.00% |
χ2 test (two-sided), P < .0001.
Discussion
PDAC carries one of the highest mortality rates of all malignant cancers worldwide. Only 23.4% of patients survive more than 5 years after diagnosis even when they have the opportunity to receive surgical treatment [8]. The poor prognosis of this disease can mainly be attributed to the lack of knowledge of the underlying pathological disease mechanisms. Thus, although researchers have sought to identify biomarkers for PDAC diagnosis, prognosis evaluation, and treatment assessment, no effective markers have been introduced into the clinic. In the current study, we identified a series of differentially expressed proteins in PDAC tissues versus matched peritumoral tissues, some of which have previously showed a high expression level, including PRELP [3], POSTN/OSF2 [9], C3 [10], annexin A2 (ANXA2) [11], LUM [12], and the transitional endoplasmic reticulum ATPase VCP [13]. In addition, APOAI was previously found to present a lower expression level in tumor tissues [14]. The above results suggest that our findings are in accordance with previous studies and imply that, through analyzing data from paired samples of tumor and peritumoral tissues, tumor characteristic protein profiles can be readily identified from a small set of patients.
Although various proteins have been found to be differentially expressed in PDAC cells or tissues, most of them are not specific to pancreatic tissues. One interesting finding of this study was that a set of pancreatic-specific proteins (CTRC, CPA1, CPA2, and CPB1) was downregulated in pancreatic cancer. Western blot analysis in seven paired tumor and peritumoral tissues of PDAC patients revealed that the expression levels of these proteins in tumor tissues were lower than those in control tissues, a finding that was in accordance with the label-free results. Further IHC experiments showed that the expression level of CPB1 displayed the most significant difference between the experimental group and control group. IHC staining of the tissue microarray indicated that CPB1 presented an “all or none” expression pattern. Therefore, we could not perform correlated prognostic analysis. Previous studies have shown that these carboxypeptidase family members are digestive enzymes secreted by pancreatic acinar cells as inactive precursors. The precursor enzymes are activated by trypsin through proteolytic cleavage of the propeptide at the C-terminal end and play fundamental roles in the pathogenesis of acute or chronic pancreatitis (CP). In addition, these enzymes were found to be upregulated and activated in response to inflammation in acute pancreatitis [15], [16], [17]. Genetic studies have further indicated that variants of CTRC and CPA1, but not CPA2 and CPB1, are associated with CP [18]. In recent years, accumulating evidence has suggested that CP is a risk factor for pancreatic cancer, resulting in a high likelihood of PDAC progression [19], [20], [21], [22]. However, the underlying molecular mechanisms remain unclear. CTRC was reported to be involved in pancreatic cancer migration; however, these mechanisms are also unknown [23]. Total CPA and the active form of CPA in serum were also found to be effective for the surveillance of early-stage pancreatic carcinoma [24]. Thus, we speculate that the carboxypeptidase family plays an important role in the malignant transformation process from pancreatitis to pancreatic cancer. Based on the above statements, the following hypothesis was proposed: downregulated levels of carboxypeptidase lead to acinar cell death, inducing CP and inflammatory cell proliferation; additionally, reduced expression of functional carboxypeptidase releases the apoptosis-inducing effect and causes malignant proliferation, promoting PDAC development.
In conclusion, the label-free proteomic approach to identify differential protein profiles between PDAC and adjacent normal tissues showed great application potential. Specifically, we identified a group of pancreatic-specific proteins using this method, and our results shed light on the pathogenesis of PDAC. The expression of carboxypeptidase is significantly downregulated in PDAC tumor tissues and may be novel biomarkers in the patient with PDCA. However, further functional investigations are required to determine the roles of these proteins in the process of PDAC.
Acknowledgments
Acknowledgement
This work was supported by grants from National Natural Science Foundation of China to Haichuan Su (#31571414) and Jinghua yang (#31471322 and #30928031). We thank Li Gong, Shumei Wang, and Lu Yu for the evaluation of tissue microarray. It was the result of work partially supported with resources and the use of facilities at Cancer Research Center, Shandong University, China, and VA Boston Healthcare System, USA.
Author Contributions
Haichuan Su and Jing-Hua Yang designed the study and performed data analysis and interpretation. Yang Song, Desheng Wang, and Qing Wang performed experiments. Hong Li, Jing Yang, Junqiang Li, and Ruirui Jing collected data. Xiang Wang, Xuerong Jin, assisted in data analysis and interpretation. Yang Song and Hong Li wrote the manuscript. Haichuan Su and Jing-Hua Yang reviewed and revised the manuscript.
Conflicts of Interest
The authors have declared no conflict of interest.
Ethics Statement
All procedures were consistent with the National Institutes of Health guide and approved by the institutional board with patients' written consent. This study was evaluated and approved by the Ethics Committee of Tangdu Hospital and Shandong Provincial Hospital Affiliated to Shandong University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2018.03.005.
Contributor Information
Jing-Hua Yang, Email: jyang@bu.edu.
Haichuan Su, Email: suhc@fmmu.edu.cn.
Appendix A. Supplementary data
Supplementary tables
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.McKinney KQ. Discovery of putative pancreatic cancer biomarkers using subcellular proteomics. J Proteome. 2011;74:79–88. doi: 10.1016/j.jprot.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Iuga C. Proteomic identification of potential prognostic biomarkers in resectable pancreatic ductal adenocarcinoma. Proteomics. 2014;14:945–955. doi: 10.1002/pmic.201300402. [DOI] [PubMed] [Google Scholar]
- 4.Cho WC. Proteomics technologies and challenges. Genomics Proteomics Bioinformatics. 2007;5:77–85. doi: 10.1016/S1672-0229(07)60018-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffin NM. Label-free, normalized quantification of complex mass spectrometry data for proteomic analysis. Nat Biotechnol. 2010;28:83–89. doi: 10.1038/nbt.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atrih A, Mudaliar MA, Zakikhani P, Lamont DJ, Huang JT, Bray SE, Barton G, Fleming S, Nabi G. Quantitative proteomics in resected renal cancer tissue for biomarker discovery and profiling. Br J Cancer. 2014;110:1622–1633. doi: 10.1038/bjc.2014.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szmola R. Chymotrypsin C is a co-activator of human pancreatic procarboxypeptidases A1 and A2. J Biol Chem. 2011;286:1819–1827. doi: 10.1074/jbc.M110.187369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oettle H. Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer: a randomized controlled trial. JAMA. 2007;297:267–277. doi: 10.1001/jama.297.3.267. [DOI] [PubMed] [Google Scholar]
- 9.Erkan M. Periostin creates a tumor-supportive microenvironment in the pancreas by sustaining fibrogenic stellate cell activity. Gastroenterology. 2007;132:1447–1464. doi: 10.1053/j.gastro.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 10.Chen J. Expression and clinical significance of complement C3, complement C4b1 and apolipoprotein E in pancreatic cancer. Oncol Lett. 2013;6:43–48. doi: 10.3892/ol.2013.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang CY, Lin CF. Annexin A2: its molecular regulation and cellular expression in cancer development. Dis Markers. 2014;2014:308976. doi: 10.1155/2014/308976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SH. Analysis of proteoglycan gene messages in human nasal mucosa and nasal polyp using dot blot hybridization. Acta Otolaryngol. 2001;121:398–402. doi: 10.1080/000164801300102914. [DOI] [PubMed] [Google Scholar]
- 13.Lague MN. Proteomic profiling of a mouse model for ovarian granulosa cell tumor identifies VCP as a highly sensitive serum tumor marker in several human cancers. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padoan A. Usefulness of MALDI-TOF/MS identification of low-MW fragments in sera for the differential diagnosis of pancreatic cancer. Pancreas. 2013;42:622–632. doi: 10.1097/MPA.0b013e318273096c. [DOI] [PubMed] [Google Scholar]
- 15.Deng L. Prediction of the severity of acute pancreatitis on admission by carboxypeptidase-B activation peptide: a systematic review and meta-analysis. Clin Biochem. 2015;48:740–746. doi: 10.1016/j.clinbiochem.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 16.Kemik O. Serum procarboxypeptidase A and carboxypeptidase A levels in pancreatic disease. Hum Exp Toxicol. 2012;31:447–451. doi: 10.1177/0960327111405864. [DOI] [PubMed] [Google Scholar]
- 17.Muller CA, Appelros S, Uhl W, Buchler MW, Borgstrom A. Serum levels of procarboxypeptidase B and its activation peptide in patients with acute pancreatitis and non-pancreatic diseases. Gut. 2002;51:229–235. doi: 10.1136/gut.51.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakano E. Variants in pancreatic carboxypeptidase genes CPA2 and CPB1 are not associated with chronic pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2015;309:G688–694. doi: 10.1152/ajpgi.00241.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bracci PM. Pancreatitis and pancreatic cancer in two large pooled case-control studies. Cancer Causes Control. 2009;20:1723–1731. doi: 10.1007/s10552-009-9424-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duell EJ. Pancreatitis and pancreatic cancer risk: a pooled analysis in the International Pancreatic Cancer Case-Control Consortium (PanC4) Ann Oncol. 2012;23:2964–2970. doi: 10.1093/annonc/mds140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malka D. Risk of pancreatic adenocarcinoma in chronic pancreatitis. Gut. 2002;51:849–852. doi: 10.1136/gut.51.6.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munigala S, Kanwal F, Xian H, Scherrer JF, Agarwal B. Increased risk of pancreatic adenocarcinoma after acute pancreatitis. Clin Gastroenterol Hepatol. 2014;12:1143–1150. doi: 10.1016/j.cgh.2013.12.033. e1141. [DOI] [PubMed] [Google Scholar]
- 23.Wang H, Sha W, Liu Z, Chi CW. Effect of chymotrypsin C and related proteins on pancreatic cancer cell migration. Acta Biochim Biophys Sin. 2011;43:362–371. doi: 10.1093/abbs/gmr022. [DOI] [PubMed] [Google Scholar]
- 24.Matsugi S. Serum carboxypeptidase A activity as a biomarker for early-stage pancreatic carcinoma. Clin Chim Acta. 2007;378:147–153. doi: 10.1016/j.cca.2006.11.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables