Abstract
Pancreatic cancer is the fourth leading cause for cancer-related death, and early diagnosis is one key to improve the survival rate of this disease. Molecular biomarkers are an important method for diagnostic use in pancreatic cancer. We used data from three mRNA microarray datasets and a microRNA dataset (GSE16515, GSE15471, GSE28735, and GSE41372) to identify potential key genes. Differentially expressed genes (DEGs) and microRNAs (DEMs) were identified. Functional, pathway enrichment, and protein-protein interaction analyses were performed on common DEGs across all datasets. The target genes of the DEMs were identified. DEMs targets that were also DEGs were further scrutinized using overall survival analysis. A total of 236 DEGs and 21 DEMs were identified. There were a total of four DEGs (ECT2, NR5A2, NRP2, and TGFBI), which were also predicted target genes of DEMs. Overall survival analysis showed that high expression levels of three of these genes (ECT2, NRP2, and TGFBI) were associated with poor overall survival for pancreatic cancer patients. The basic expression of DEGs in pancreas stood lower level in various organ tissues. The expression of ECT2 and NRP2 was higher in different pancreatic cancer cell lines than normal pancreas cell line. Knockout of ECT2 by Crispr Cas9 gene editing system decreased proliferation and migration ability in pancreatic cancer cell line MiaPaCa2. In conclusion, we think that data mining method can do well in biomarker screening, and ECT2 and NRP2 can play as potential biomarker or therapy target by Crispr Cas9 in pancreatic cancer.
Introduction
Pancreatic cancer is one of the few cancer types where the 5-year survival rate shows no improvement [1]. In 2016 in the United States, pancreatic cancer was diagnosed in approximately 53,070 patients and caused an estimated 41,780 deaths [2]. In China, the incidence of pancreatic cancer was in an upward trend from 2000 to 2011, and pancreatic cancer is one of the top 10 most common cancers in both men and women [3]. In Europe, pancreatic cancer is currently one of the most lethal types of cancer and has a 5-year survival of approximately 7% [2]. It is expected to rise to second place behind lung cancer by 2030 [4]. Surgery is currently the only potentially curative treatment for pancreatic cancer [5]. However, pancreatic surgery in early state is associated with significant morbidity and mortality [6]. The poor prognosis of pancreatic cancer is mainly attributed to rapid disease progression; late diagnosis at advanced, unresectable stages; and inadequate response to current adjuvant or palliative regimens. Indeed, there is no effective diagnostic method at early stage, and most pancreatic cancer patients are first diagnosed at advanced stages [7], [8].
High-throughput technologies combined with vast amounts of publicly available data and sophisticated online analysis tools enable the scientific community to mine knowledge about genes related to cancer development in an unprecedented manner. Different types of molecular biomarkers can be used for diagnosis, prognosis, and prediction and monitoring of response to treatment. For example, the presence or absence or changes in the level of specific biomarkers in a cell are routinely used as an indication of cancer development. Molecular biomarkers are an important diagnosis tool in pancreatic cancer [9]. Thus, multiple studies have confirmed that carbohydrate antigen 19-9 (CA 19-9) and carcinoembryonic antigen (CEA) can be used to predict the outcomes of multiple diseases, including pancreatic cancer [10], [11], [12]. However, these biomarkers are not sufficiently specific or sensitive for use in pancreatic cancer [13], [14]. The identification of novel pancreatic cancer-specific molecular biomarkers is crucial for early effective diagnosis [15].
Microarrays, including DNA, microRNA, protein, and antibody microarrays, are a multiplex lab-on-a-chip system. More specifically, a microarray is a 2D array on a solid substrate for assaying large amounts of biological material using miniaturized, multiplexed, and parallel processing and detection methods. Microarrays are widely used in biomedical research. For instance, they have been used to infer general genetic alterations during tumorigenesis [16]. Microarray data are also the main resources of biomarker candidates [17]. In this study, we integrated data from three mRNA microarray datasets and a microRNA dataset with pathway, functional, and overall survival analysis to identify putative novel pancreatic cancer biomarkers among differentially expressed genes (DEGs) and microRNAs (DEMs) between normal and pancreatic cancer tissue. Crispr Cas9 system has led a revolution in gene editing. Our laboratory has done a lot of work on this field with wealth of experience. In this study, we verified the data mining results by Crispr Cas9 gene editing system and other experiments.
Materials and Methods
Microarray Datasets and Data Processing
We selected and downloaded three human pancreatic cancer gene expression (GSE16515 [18], GSE15471 [19], and GSE28735 [20], [21]) and one human pancreatic cancer miRNA expression (GSE41372 [22]) datasets from the Expression Omnibus database (GEO) [23]. The three gene expression datasets include 117 cancer tissue samples and 97 normal tissue samples from 117 pancreatic cancer patients. The miRNA dataset comprises pancreatic ductal adenocarcinoma tumor samples from nine patients and normal pancreas samples from nine healthy individuals. All these datasets were generated on Affymetrix chips and uploaded by independent groups. The GSE16515 and GSE15471 datasets are based on the GPL570 platform; the GSE28735 and GSE41372 datasets are based on the GPL16142 platform. These platforms are the mainstream choice for biomarker research [24].
We used GEO2R [25] to identify DEGs and DEMs between pancreatic cancer and normal tissue samples. GEO2R is an online tool which can compare different groups in a GEO series. The pancreatic cancer samples served as the “cancer group,” and the normal samples as the “control group.” We selected genes whose adjusted (Benjamini & Hochberg or false discovery rate [26]) P value was smaller than .01 and with an absolute log fold-change greater than 1.
We used the online tool “Calculate and draw custom Venn diagrams” (http://bioinformatics.psb.ugent.be/webtools/Venn/) to identify genes that were up- or downregulated in all three GEO datasets.
Gene Function and Pathway Enrichment Analysis
We used the Database for Annotation, Visualization, and Integrated Discovery (DAVID [27]) to perform a functional enrichment analysis of the DEGs. DAVID is an online program to help understand biological meaning behind plenty of genes with comprehensive function, including the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. We set the cutoff criterion P < .05 of false discovery rate.
Protein-Protein Interaction (PPI) Network Construction and Module Analysis
We used the Search Tool for the Retrieval of Interacting Genes (STRING) database to analyze functional interactions between proteins [28]. We retrieved interactions with confidence scores greater or equal to 0.7 and screened the resulting PPI network for modules with Cytoscape-MCODE [29] and the following parameters: degree cutoff=2, node score cutoff=0.2, k-core=0.2, and maximum depth=100.
Prediction of miRNA Targets
We used GEO2R to identify DEMs between pancreatic cancer and normal tissue samples. The pancreatic cancer samples served as the “cancer group” and the normal samples as the “control group.” We selected miRNAs whose adjusted (Benjamini & Hochberg or false discovery rate [26]) P value was smaller than .01 and with an absolute log fold-change greater than 1. We used the “miRecords” tool to predict miRNA target genes [30]. miRecords integrates the results of more than 10 miRNA target prediction programs. No target was predicted for more than six miRNA target prediction programs, so we only considered DEM target genes predicted by exactly six prediction programs. Finally, we used the online tool “Calculate and draw custom Venn diagrams” (http://bioinformatics.psb.ugent.be/webtools/Venn/) to identify common target DEGs of for the DEMs.
Survival Analysis of DEGs
We used the OncoLnc tool (www.oncolnc.org) [31] to perform overall survival analysis in pancreatic cancer. OncoLnc is an online tool for interactively exploring survival correlations. It contains survival data for 8647 patients from 21 cancer studies in The Cancer Genome Atlas (TCGA), along with RNA-seq data for mRNA and miRNA expression from TCGA, and lncRNA expression from MiTranscriptome beta. The tool gives the user the ability to separate patients based on the gene expression level of a specific gene and create Kaplan-Meier plots. We recorded hazard ratios with their 95% confidence intervals and log-rank P values. Top 10, 20, and 40 and bottom 90, 80, and 60 percentiles of expression values, respectively, were considered as high and low groups. Additionally, OncoLnc was used to do the overall survival analysis for other cancer types, including breast, lung, gastric, and liver cancer.
The Basic Expression of DEGs in Different Organs and Cancer Tissues
We used the online tool “ The Human Protein Atlas ” to perform the basic expression level of DEGs in different human organs. The Human Protein Atlas is a Swedish-based program started in 2003. We worked with version 17 in this study, which was launched in August 2017 [32]. In the Tissue Atlas part, it shows the distribution of the proteins across all major tissues and organs in human. There were three datasets in the website: HPA dataset (RNA-seq tissue data are reported as mean transcripts per million, corresponding to mean values of the different individual samples from each tissue), GTEx dataset (RNA-seq data are reported as median reads per kilobase per million mapped reads, generated by the Genotype-Tissue Expression project), and FANTOM5 dataset (tissue data obtained through cap analysis of gene expression are reported as tags per million, generated by the FANTOM5 project). We used them together in this study. In additional, we also conducted the different expression between normal pancreas and pancreatic cancer tissues. UALCAN is an easy-to-use online tool for in-depth analyses of TCGA gene expression data. It uses TCGA level 3 RNA-seq and clinical data from 31 cancer types. In this study, we used it to identify the up- or downregulated of DEGs in pancreatic cancer.
Cell Culture
In this study, we used human pancreatic cancer lines ASPC1, BXPC3, PANC1, HS766T, and MiaPaCa 2; primary cell lines PaCaDD119, PaCaDD137, and PaCaDD159; and normal pancreas cell line HDPE. ASPC1, BXPC3, PANC1, HS766T, MiaPaCa 2, and HDPE were purchased from ATCC, and all the cells were grown in monolayer culture in a humidified atmosphere containing 5% CO2 at 37°C. The culture medium for ASPC1, BXPC3, and PANC1 consists of RPMI Medium 1640 (Gibco, 21875-034) with 10% FBS (Gibco, A31608-01). HS766T cell was cultured in DMEM (Gibco, 30966-021) with 10% FBS. MiaPaCa 2 cell was cultured in DMEM with 10% FBS and 2% horse serum (Gibco, 16050-130). HDPE cell was cultured in Keratinocyte-SFM (Gibco, 10724-011). PaCaDD119, PaCaDD137, and PaCaDD159 were cultured in mixed medium with 53.5% DMEM, 13.2% FBS, and 33.3% Keratinocyte-SFM. All cells were harvested by 0.25% Trypsin-EDTA (Gibco, 25200-072).
Western Blot
Cells were lysed with RIPA buffer. Gel electrophoresis was performed under reducing conditions with acrylamide gel, and proteins were transferred to a nitrocellulose membrane. As primary antibodies, ECT2 (Merck Millipore, 07-1364), NRP2 (Cell Signaling Technology, 3366), and TGFBI (Thermo Fisher Scientific, PA5-19242) were used for detection. The GAPDH (Cell Signaling Technology, 5175S) served as loading control. HRP-linked anti-rabbit IgG (Cell Signaling Technology, 7074) was used as second antibody. Quantification of signal was performed by Amersham Imager 600 with SignalFire Elite ECL Reagent (Cell Signaling Technology, 12757S). All Western blot results were done at least two times.
Quantitative RT-PCR
The isolation of RNA was performed with NucleoSpin RNA Plus kit (Macherey-Nagel, #740984.250), and the cDNA was synthesized with High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, 00364942). The quantitative RT-PCR was performed with PowerUP SYBR Green Master Mix (Applied Biosystems, A25741) and analyzed by 2−△△Ct method. The GAPDH was used as reference value. Primer sequences for amplification were as follows: ECT2-forward (5′- AGGAGTCGGCGTTTGAAGAG-3′), ECT2-reverse (5′- GGTCCAATAACTCTACAATCAGCC-3′), NRP2-forward (5′- CGCGGAGGAGACAGTATCAC-3′), NRP2-reverse(5′- CCGAAGAGGAGGCCACAGAG-3′), TGFBI-forward (5′- AAGGTAACGGCCAGTACACG-3′), TGFBI-reverse(5′- GAGATGATCGCCTTCCCGTT-3′), GAPDH-forward (5′-CTTTGGTATCGTGGAAGGACTC-3′), GAPDH-reverse(5′-AGTAGAGGCAGGGATGATGT-3′). All primers were synthesized by Eurofins.
CRISPR/Cas9 Gene Editing
In this study, ECT2 was knocked out by CRISPR/Cas9 gene editing in the MiaPaCa2 cell line. The pSpCas9(BB)-2A-GFP (PX458, Plasmid #48138) plasmid was purchase from Addgene. HGLibB-14402-ECT2 sgRNA (forward: 5′- CACCTGTCTTTAATGACCTCTACA -3′ and reverse: 5′-AAACTGTAGAGGTCATTAAAGACA-3′) and Hu-64433nc-sgRNA (forward: 5′-CACCATCGTATCATCAGCTAGCGC-3′ and reverse: 5′-AAACGCGCTAGCTGATGATACGAT-3′) were synthesized by Eurofins. Plasmid construction was done as described in the protocol and confirmed by sequencing. MiaPaCa cells were transfected with ECT2 knockout plasmid by Lipofectamine 3000 Transfection Reagent (Invitrogen, L3000015) as protocol for 72 hours. Then, all the GFP-positive cells were sorted by fluorescence activated cell sorter. Sixty GFP-positive cells were plated in one 96-well plate for selecting single clones. After a few days of culture, Western blot and sequencing were used to make sure the knockout effect. Sequencing was done by Eurofins, and all selected clones were sequenced at least five times.
Proliferation Assay
Cell counting was used to analyze the proliferation ability between ECT2 knockout clones and negative control clone. Cells were plated as 3 × 103 cells per well in grown medium on 96-well black plates (Costar, #3603) for 4 days. After 24, 48, and 72 hours of proliferation, cells were stained with NucBlue Live Cell Stain ReadyProbes reagent ( Invitrogen, R37605) and imaged in 38 fields for each well by Evos FL Auto 2 imaging system (Invitrogen, AMAFD2000). Images were counted by HCS studio cell analysis software (Thermo, SX000041A). All experiments were done in duplicate and repeated at least three times.
Migration Assay
FluoroBlok Insert system (Corning, REF351152) was used to analyze the migration ability between ECT2 knockout clones and negative control clone. After incubation with medium without FCS for 24 hours cells were plated as 25,000 cells per well in FluoroBlokTM Insert, and put FluoroBlokTM Insert into Multiwell 24 Well (Corning, REF353504). The medium in the upper compartment was without FCS, in the lower compartment the growth medium contained 20% FCS. After 16 hours of migration in a cell culture incubator, the NucSpot Live 488 Nuclear Stain (Biotium, Cat40081) was used, and the cells attached on center of the membrane were imaged in random four fields for each well by Evos FL Auto 2 imaging system (Invitrogen, AMAFD2000).The positive point in all images was manually counted.
Statistical Analysis
Statistical analysis was performed with GraphPad Version 7 and Microsoft Excel Version 2017. For cell culture experiments, t test was used. For interpretation of differences, a P value < .05 was considered as statistically significant.
Results
Identification of DEGs in Gene Expression Microarrays
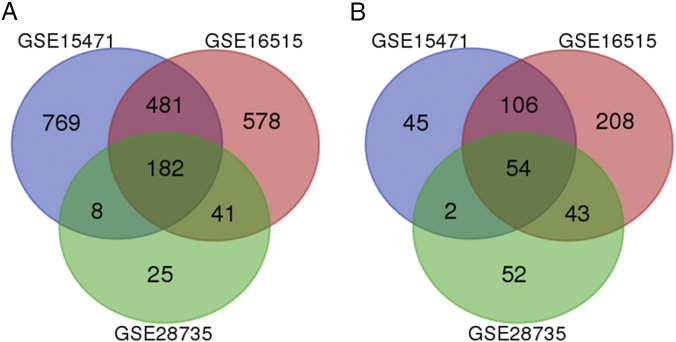
We identified 1647, 1693, and 407 DEGs in pancreatic cancer tissue compared to normal tissue in the GSE15471, GSE16515, and GSE28735 datasets, respectively. A total of 236 genes were considered DEG in all the 3 datasets (Figure 1 and Table 1, see Materials and Methods). In particular, 182 genes were upregulated and 54 genes were downregulated.
Figure 1.
Identification of DEGs in mRNA expression profiling datasets GSE15471, GSE16515, and GSE28735. (A) The upregulated genes in the profiling datasets. (B) The downregulated genes in the profiling datasets.
Table 1.
Total 236 Genes Which Showed Consistent Trend in Up- or Downregulation in Three Microarrays Were Considered DEG in GSE15471, GSE16515, and GSE28735 Datasets
| Upregulated DEGs | ||||||
|---|---|---|---|---|---|---|
|
GSE15471 |
GSE16515 |
GSE28735 |
||||
| Adj P | Log FC | Adj P | Log FC | Adj P | Log FC | |
| ABHD17C | 6.41e-05 | −1.10562734 | 1.60e-08 | −2.895825 | 3.31e-12 | −1.128362 |
| ACSL5 | 2.43e-03 | −1.18356539 | 1.96e-04 | −1.9244909 | 8.95e-08 | −1.4634847 |
| ACTA2 | 9.91e-06 | −1.56347311 | 4.02e-03 | −1.3521614 | 1.73e-05 | −1.1454927 |
| ADAM12 | 4.08e-08 | −1.81573918 | 2.20e-02 | −1.2155662 | 6.89e-04 | −1.0717007 |
| ADAM28 | 1.00e-07 | −1.86173177 | 3.35e-04 | −1.5977978 | 5.10e-06 | −1.141082 |
| ADAM9 | 2.19e-10 | −1.71106472 | 8.41e-07 | −1.8466136 | 1.74e-07 | −1.1530387 |
| ADAMTS12 | 5.56e-17 | −2.42053266 | 2.20e-04 | −1.8683158 | 3.77e-09 | −1.4163264 |
| ADAMTS6 | 1.14e-08 | −1.08645267 | 2.58e-05 | −1.1985481 | 2.89e-08 | −1.038462 |
| ADGRF1 | 4.41e-06 | −1.13978583 | 2.11e-03 | −1.1893599 | 1.24e-08 | −1.6035156 |
| AEBP1 | 2.36e-16 | −2.70716438 | 1.99e-03 | −1.7154245 | 1.80e-06 | −1.1198558 |
| AGR2 | 1.28e-04 | −2.04964413 | 3.29e-05 | −2.7721982 | 2.25e-10 | −2.1373789 |
| AHNAK2 | 2.75e-13 | −2.53976184 | 2.20e-04 | −1.6346964 | 5.51e-13 | −1.7455516 |
| ANKRD22 | 1.07e-05 | −1.30858993 | 2.94e-07 | −2.1128529 | 1.35e-08 | −1.5765009 |
| ANLN | 4.03e-06 | −1.3095382 | 1.35e-07 | −2.231518 | 7.00e-09 | −1.6620173 |
| ANO1 | 6.05e-15 | −2.97053176 | 1.59e-04 | −1.7574292 | 2.55e-09 | −1.2739089 |
| ANTXR1 | 1.83e-12 | −1.49081539 | 8.76e-04 | −1.218539 | 7.64e-09 | −1.5904338 |
| ANXA10 | 9.12e-05 | −2.00705104 | 2.49e-04 | −3.2509851 | 7.58e-09 | −2.3579367 |
| ANXA3 | 2.08e-07 | −1.49208035 | 1.47e-06 | −1.9301469 | 4.51e-06 | −1.0462073 |
| ANXA8L1///ANXA8 | 1.24e-05 | −1.29098707 | 1.50e-04 | −2.8578011 | 1.45e-07 | −1.4583824 |
| APOL1 | 2.98e-10 | −1.11856761 | 1.82e-06 | −1.9469703 | 7.27e-10 | −1.5050227 |
| AREG | 2.33e-06 | −1.95574758 | 2.20e-04 | −1.9826671 | 3.11e-03 | −1.0105213 |
| ARNTL2 | 1.55e-09 | −1.43839367 | 1.05e-06 | −1.8453465 | 1.15e-08 | −1.2634424 |
| ASAP2 | 5.40e-12 | −1.27142048 | 1.25e-06 | −1.4437181 | 1.38e-11 | −1.0555331 |
| ASPM | 1.34e-05 | −1.38313105 | 4.95e-06 | −2.1664253 | 2.91e-07 | −1.2180584 |
| BGN | 2.63e-12 | −1.61702033 | 4.83e-03 | −1.2980139 | 5.11e-06 | −1.2178456 |
| CAPG | 1.74e-16 | −2.23018436 | 8.56e-06 | −2.2290411 | 8.56e-10 | −1.409224 |
| CCL18 | 4.24e-05 | −1.50008642 | 3.09e-03 | −1.7869004 | 1.20e-03 | −1.1312298 |
| CCL20 | 2.65e-06 | −2.18836311 | 9.93e-06 | −3.3482898 | 3.16e-08 | −1.3648089 |
| CD109 | 7.32e-12 | −2.35811014 | 1.96e-04 | −1.9508099 | 2.18e-07 | −1.2498704 |
| CDH11 | 2.09e-11 | −1.62227423 | 8.02e-04 | −1.2080804 | 2.35e-07 | −1.4890444 |
| CDH3 | 4.98e-09 | −1.50129524 | 7.33e-07 | −2.6825856 | 1.81e-12 | −1.6362931 |
| CEACAM1 | 4.36e-05 | −1.41621839 | 2.67e-06 | −1.635777 | 3.37e-09 | −1.292694 |
| CEACAM5 | 7.46e-06 | −2.76526848 | 2.33e-07 | −6.2525663 | 1.07e-10 | −3.1812869 |
| CEACAM6 | 3.73e-08 | −3.19875853 | 2.11e-06 | −4.2600263 | 3.78e-10 | −2.6271224 |
| CEMIP | 2.81e-07 | −2.45896857 | 4.01e-06 | −2.7846301 | 9.49e-11 | −1.8033796 |
| CLDN18 | 2.91e-04 | −1.61076389 | 2.73e-05 | −4.0364872 | 2.41e-08 | −2.1265416 |
| COL10A1 | 4.79e-18 | −4.11443223 | 1.37e-05 | −3.3673523 | 2.59e-11 | −1.5692449 |
| COL11A1 | 1.24e-07 | −1.88113667 | 1.64e-04 | −2.2589512 | 1.42e-10 | −2.1946749 |
| COL12A1 | 7.24e-08 | −1.52434785 | 3.78e-04 | −1.6268338 | 3.71e-07 | −1.732026 |
| COL1A1 | 8.67e-08 | −1.33504418 | 1.18e-03 | −2.0914504 | 1.80e-05 | −1.5020929 |
| COL3A1 | 5.36e-10 | −1.47689589 | 2.61e-03 | −1.2776155 | 7.92e-05 | −1.3701904 |
| COL5A2 | 4.21e-16 | −3.44422932 | 7.75e-04 | −1.8667315 | 8.85e-06 | −1.3867727 |
| COL8A1 | 1.60e-14 | −2.08659948 | 1.46e-02 | −1.0532559 | 2.56e-07 | −1.4414827 |
| COMP | 1.60e-14 | −3.57453998 | 3.99e-03 | −2.155856 | 2.08e-06 | −1.4673376 |
| CORIN | 6.57e-09 | −1.80041916 | 9.81e-04 | −1.4909615 | 7.13e-07 | −1.0800867 |
| CP | 3.17e-05 | −1.38183429 | 2.07e-02 | −1.3531284 | 3.29e-05 | −1.767232 |
| CST1 | 4.28e-13 | −3.34647655 | 4.61e-04 | −3.0382825 | 2.25e-08 | −2.0248356 |
| CST2 | 4.08e-10 | −1.80213342 | 2.41e-03 | −1.2489128 | 9.70e-09 | −1.2321847 |
| CTHRC1 | 6.72e-17 | −4.43075384 | 2.67e-04 | −2.7308879 | 7.77e-09 | −1.0049256 |
| CTSE | 3.35e-08 | −2.7257767 | 2.67e-07 | −4.6208679 | 6.93e-12 | −2.6841011 |
| CTSK | 2.95e-13 | −2.61423967 | 7.83e-03 | −1.3986692 | 2.43e-04 | −1.0460687 |
| CXCL5 | 7.79e-08 | −2.4255966 | 1.03e-04 | −3.6610098 | 1.74e-07 | −1.5615738 |
| DCBLD2 | 1.10e-09 | −1.34056587 | 3.01e-03 | −1.111635 | 3.46e-07 | −1.0321687 |
| DDX60 | 2.29e-08 | −1.55703481 | 3.20e-05 | −1.5469602 | 6.06e-07 | −1.0535862 |
| DGKH | 5.53e-09 | −1.51658317 | 2.00e-04 | −1.3050633 | 7.09e-07 | −1.0501716 |
| DHRS9 | 1.83e-07 | −1.25123427 | 5.22e-04 | −2.4339574 | 2.72e-06 | −1.2735449 |
| DKK1 | 3.90e-10 | −2.30661726 | 1.03e-06 | −3.8761945 | 2.17e-07 | −1.2162724 |
| DLGAP5 | 1.41e-04 | −1.03619236 | 7.33e-07 | −2.0790401 | 6.75e-07 | −1.1127349 |
| DPCR1 | 2.17e-03 | −1.16453163 | 8.01e-05 | −3.7233879 | 5.55e-08 | −2.1386271 |
| DPYSL3 | 8.50e-12 | −2.01701306 | 4.72e-03 | −1.3999587 | 2.12e-05 | −1.1362667 |
| ECT2 | 2.12e-10 | −2.18367354 | 1.04e-04 | −1.2024523 | 4.55e-10 | −1.3293738 |
| EDIL3 | 3.14e-11 | −2.55947048 | 1.10e-03 | −1.5476678 | 8.64e-07 | −1.5138422 |
| EDNRA | 1.24e-14 | −1.83821405 | 1.84e-03 | −1.3556256 | 2.52e-07 | −1.3127658 |
| EFNA5 | 1.66e-10 | −1.35782403 | 6.99e-06 | −1.3028334 | 7.69e-10 | −1.1553482 |
| EFNB2 | 1.18e-07 | −1.32743105 | 3.28e-05 | −2.038012 | 2.49e-09 | −1.0293451 |
| ENO2 | 5.56e-10 | −1.6129782 | 9.21e-05 | −1.6946701 | 1.25e-07 | −1.1134007 |
| EPHA4 | 1.75e-08 | −1.01758354 | 2.29e-03 | −1.0761163 | 1.88e-07 | −1.1711916 |
| EPYC | 1.32e-04 | −1.73276174 | 8.42e-03 | −1.9365965 | 9.64e-04 | −1.0430022 |
| ERO1A | 1.11e-05 | −1.0560388 | 8.59e-07 | −1.9237551 | 1.17e-09 | −1.219264 |
| ETV1 | 9.65e-14 | −2.3034463 | 3.70e-04 | −1.270131 | 2.53e-06 | −1.0579796 |
| FAP | 1.77e-16 | −3.38881049 | 2.81e-03 | −1.9046404 | 1.33e-05 | −1.4972633 |
| FBXO32 | 9.65e-14 | −1.72359572 | 2.45e-03 | −1.3835639 | 2.13e-11 | −1.4711818 |
| FCGR3B///FCGR3A | 7.76e-08 | −2.41674826 | 6.89e-03 | −1.6425105 | 6.57e-05 | −1.0251151 |
| FERMT1 | 1.64e-06 | −1.35306111 | 4.34e-06 | −1.3772776 | 7.21e-10 | −1.5010753 |
| FGD6 | 1.59e-09 | −1.06124145 | 2.87e-07 | −1.6101424 | 7.37e-10 | −1.2339911 |
| FN1 | 2.54e-10 | −1.50889045 | 2.98e-03 | −1.4748314 | 8.61e-10 | −2.2111 |
| FNDC1 | 2.35e-13 | −2.72629222 | 8.25e-03 | −1.6330736 | 1.79e-06 | −1.4763207 |
| FOXQ1 | 1.99e-07 | −1.95639952 | 2.26e-07 | −2.5948572 | 5.81e-09 | −1.1159918 |
| FXYD3 | 9.25e-07 | −1.80033698 | 2.60e-06 | −2.0743889 | 9.49e-11 | −1.5452418 |
| GABRP | 4.28e-09 | −3.01817945 | 1.64e-04 | −3.6208516 | 2.95e-06 | −2.0308344 |
| GCNT3 | 5.28e-05 | −1.84782101 | 1.03e-06 | −3.3547133 | 3.12e-06 | −1.4315447 |
| GJB2 | 3.55e-13 | −3.69109958 | 1.68e-07 | −3.5861489 | 2.21e-12 | −1.0717644 |
| GPRC5A | 1.73e-13 | −2.71073028 | 2.33e-07 | −3.7777146 | 7.88e-10 | −1.1751238 |
| GPX2 | 6.17e-06 | −2.06753479 | 7.71e-03 | −1.0570848 | 1.44e-05 | −1.1878256 |
| GPX8 | 6.62e-13 | −1.75319459 | 4.07e-03 | −1.0746134 | 5.69e-06 | −1.0094489 |
| GREM1 | 8.23e-09 | −3.20668556 | 1.89e-03 | −2.0067714 | 2.56e-07 | −1.0603713 |
| HEPH | 1.09e-06 | −2.04989436 | 9.21e-04 | −2.1788215 | 2.74e-07 | −1.2034709 |
| HK2 | 7.54e-10 | −1.54961514 | 1.25e-08 | −2.7161319 | 7.91e-09 | −1.3526864 |
| IFI27 | 1.87e-09 | −2.24277682 | 2.33e-07 | −3.3295833 | 3.18e-09 | −1.3512633 |
| IFI44L | 9.20e-10 | −1.99737328 | 1.04e-03 | −2.0539791 | 1.51e-04 | −1.2289118 |
| IGF2BP3 | 1.63e-07 | −1.58746323 | 1.67e-05 | −3.0528705 | 1.76e-07 | −1.1658791 |
| IGFBP5 | 4.44e-10 | −1.00217799 | 1.31e-03 | −1.3627484 | 2.24e-05 | −1.3557069 |
| IL1R2 | 4.90e-07 | −1.65352388 | 3.00e-04 | −2.0617929 | 1.07e-06 | −1.0359142 |
| INPP4B | 2.09e-10 | −1.61525246 | 1.07e-03 | −1.4033133 | 5.46e-09 | −1.12227 |
| ITGA2 | 3.03e-12 | −1.38190767 | 1.13e-07 | −2.6491086 | 4.28e-11 | −2.2658244 |
| ITGA3 | 7.36e-09 | −1.32864472 | 1.25e-06 | −2.2800449 | 7.10e-10 | −1.5188551 |
| ITGB4 | 6.66e-09 | −1.23313595 | 2.66e-05 | −1.9259157 | 2.30e-11 | −1.303354 |
| KCNN4 | 1.15e-07 | −1.11084866 | 4.45e-07 | −2.2400205 | 1.59e-14 | −1.0487336 |
| KRT19 | 3.16e-11 | −3.71089111 | 1.61e-08 | −4.472929 | 1.21e-11 | −2.0580204 |
| KRT7 | 1.31e-13 | −3.10730417 | 6.96e-07 | −3.2872588 | 5.98e-07 | −1.6835824 |
| KYNU | 3.47e-08 | −1.17046153 | 1.57e-04 | −1.3876635 | 1.61e-06 | −1.2622391 |
| LAMA3 | 4.56e-09 | −2.25796154 | 2.83e-03 | −1.3755993 | 8.50e-12 | −1.3547947 |
| LAMB3 | 1.58e-08 | −1.7945134 | 2.96e-10 | −3.6738991 | 1.59e-14 | −2.3442298 |
| LAMC2 | 3.83e-09 | −1.59628476 | 4.14e-07 | −3.0935974 | 1.59e-14 | −2.9016536 |
| LCN2 | 8.46e-09 | −2.89069735 | 6.50e-06 | −3.7867526 | 6.00e-05 | −1.1424984 |
| LEF1 | 2.24e-10 | −2.75342766 | 1.16e-04 | −2.4864991 | 6.33e-09 | −1.3104127 |
| LOXL2 | 1.75e-09 | −1.61216046 | 5.53e-06 | −2.2926783 | 1.28e-08 | −1.2380353 |
| LRRN1 | 2.66e-12 | −1.68745871 | 4.99e-04 | −1.5438467 | 3.21e-06 | −1.1022207 |
| MALL | 4.20e-05 | −1.44176733 | 4.37e-05 | −2.5583935 | 5.55e-08 | −1.0471758 |
| MBOAT2 | 1.30e-10 | −1.12972612 | 1.27e-06 | −1.9018549 | 1.76e-11 | −1.5977576 |
| MELK | 4.19e-06 | −1.21151988 | 1.50e-06 | −2.0270539 | 1.38e-07 | −1.1532836 |
| MET | 2.66e-09 | −1.50207236 | 6.50e-04 | −1.1683551 | 1.20e-09 | −1.4803842 |
| MICAL2 | 1.38e-10 | −1.20391622 | 6.07e-04 | −1.1188393 | 7.99e-11 | −1.2684082 |
| MLPH | 2.74e-07 | −1.47717238 | 4.75e-07 | −2.748077 | 1.63e-14 | −1.4629327 |
| MMP1 | 2.56e-08 | −3.49148002 | 4.63e-04 | −2.9607088 | 1.36e-03 | −1.2251593 |
| MMP11 | 2.98e-09 | −1.96177983 | 1.92e-05 | −2.6722979 | 2.28e-09 | −1.5014524 |
| MMP12 | 2.52e-06 | −2.66435443 | 5.23e-04 | −3.1168124 | 2.42e-07 | −1.7443562 |
| MMP14 | 9.90e-11 | −1.19811673 | 6.29e-05 | −1.222454 | 9.99e-09 | −1.325874 |
| MMP7 | 6.50e-11 | −3.37196074 | 8.22e-04 | −2.4186142 | 1.57e-03 | −1.2129338 |
| MMP9 | 7.76e-04 | −1.25841013 | 5.39e-04 | −1.7459301 | 2.83e-07 | −1.1166764 |
| MTMR11 | 2.59e-08 | −1.73578547 | 4.71e-06 | −2.3963792 | 9.96e-11 | −1.1160298 |
| MXRA5 | 6.59e-14 | −2.3876267 | 3.45e-04 | −1.7945193 | 1.56e-06 | −1.1398482 |
| MYOF | 1.50e-15 | −2.31380533 | 1.80e-05 | −1.5493516 | 6.87e-09 | −1.3488062 |
| NMU | 1.21e-07 | −1.741711 | 7.71e-07 | −3.4424442 | 2.18e-08 | −1.0295189 |
| NOX4 | 4.79e-18 | −3.02767059 | 9.50e-05 | −2.3154453 | 2.09e-10 | −1.6264947 |
| NPR3 | 2.04e-08 | −1.42535705 | 6.59e-05 | −1.8797023 | 1.10e-06 | −1.2844418 |
| NQO1 | 1.99e-06 | −1.55021834 | 1.51e-07 | −2.8756253 | 1.46e-10 | −1.4664629 |
| NRP2 | 3.65e-09 | −1.00101663 | 7.92e-04 | −1.0036763 | 2.27e-07 | −1.0398169 |
| NT5E | 1.64e-05 | −1.57562593 | 2.94e-03 | −1.0329535 | 7.10e-06 | −1.208268 |
| OAS1 | 9.55e-07 | −1.21738702 | 4.37e-05 | −2.3434841 | 3.69e-08 | −1.130268 |
| OAS2 | 1.22e-07 | −1.39078244 | 7.96e-04 | −1.063974 | 3.18e-07 | −1.0571731 |
| OLR1 | 1.05e-13 | −2.99889051 | 7.02e-04 | −2.2324636 | 1.91e-04 | −1.2763264 |
| OSBPL3 | 5.07e-09 | −1.43808519 | 1.23e-06 | −1.8104033 | 3.59e-08 | −1.1831684 |
| PCDH7 | 9.65e-07 | −1.22868649 | 2.91e-04 | −1.4454759 | 8.62e-10 | −1.0136969 |
| PGM2L1 | 9.65e-14 | −1.61020343 | 1.50e-04 | −1.4075755 | 3.30e-09 | −1.0538702 |
| PKM | 1.15e-12 | −1.53410463 | 1.49e-05 | −1.1332567 | 2.28e-09 | −1.021846 |
| PLA2R1 | 2.50e-08 | −1.20222447 | 2.37e-04 | −1.2000276 | 2.51e-07 | −1.1070027 |
| PLAC8 | 7.70e-04 | −1.69996462 | 4.67e-06 | −3.5791825 | 6.80e-08 | −1.8359027 |
| PLAT | 1.12e-11 | −2.53990857 | 4.05e-03 | −1.757376 | 8.54e-07 | −1.3462227 |
| PLAU | 2.69e-11 | −2.05029247 | 4.78e-05 | −2.2217329 | 2.68e-08 | −1.3743673 |
| PLPP4 | 1.80e-11 | −2.2263166 | 2.18e-05 | −2.4051361 | 6.88e-12 | −1.1414518 |
| POSTN | 1.94e-07 | −1.33350561 | 1.09e-04 | −2.7902679 | 2.53e-10 | −2.6298373 |
| PXDN | 6.18e-07 | −1.36897353 | 3.37e-03 | −1.2099297 | 1.04e-05 | −1.1653407 |
| RAI14 | 2.21e-09 | −1.43740669 | 2.98e-05 | −1.2529126 | 1.57e-07 | −1.0131847 |
| RHBDL2 | 1.29e-07 | −1.17563806 | 7.96e-05 | −1.1628286 | 9.96e-11 | −1.1258644 |
| RUNX2 | 9.52e-11 | −2.92838651 | 8.05e-04 | −1.9477636 | 1.96e-10 | −1.2121193 |
| S100A16 | 1.59e-08 | −1.1650921 | 9.73e-08 | −2.110029 | 3.77e-10 | −1.1656542 |
| S100P | 9.23e-11 | −3.66658089 | 4.81e-09 | −6.2580134 | 6.89e-14 | −1.2260793 |
| SCEL | 4.29e-06 | −1.26569808 | 2.44e-03 | −1.1355372 | 6.18e-11 | −1.6301907 |
| SCNN1A | 6.02e-04 | −1.08886497 | 1.59e-03 | −1.5464286 | 6.28e-06 | −1.0022056 |
| SDR16C5 | 7.10e-08 | −2.31520076 | 2.32e-08 | −4.1847444 | 3.80e-12 | −1.5155709 |
| SEMA3C | 7.54e-09 | −1.22426469 | 2.11e-03 | −1.6503623 | 4.02e-04 | −1.0999293 |
| SERPINB3 | 3.59e-05 | −1.6054987 | 4.16e-03 | −1.8953011 | 6.28e-04 | −1.2747167 |
| SERPINB5 | 1.31e-07 | −2.32054066 | 3.79e-08 | −4.392844 | 1.21e-11 | −2.1830693 |
| SLC22A3 | 7.77e-06 | −1.09781014 | 1.34e-04 | −1.6463225 | 4.44e-06 | −1.14787 |
| SLC2A1 | 7.88e-08 | −1.41478683 | 2.96e-10 | −3.0533566 | 3.62e-12 | −1.8381029 |
| SLC44A4 | 1.94e-04 | −1.27453752 | 3.13e-05 | −2.3995452 | 9.50e-08 | −1.2232882 |
| SLC6A14 | 1.67e-09 | −3.00993894 | 9.65e-08 | −4.6265813 | 3.82e-12 | −3.075768 |
| SLC6A6 | 3.25e-14 | −2.68187019 | 7.62e-05 | −1.825323 | 3.86e-08 | −1.2712856 |
| SLPI | 2.79e-14 | −2.67459132 | 1.24e-08 | −3.3129465 | 8.66e-09 | −1.7899024 |
| SRPX2 | 8.92e-16 | −2.47287599 | 9.36e-04 | −1.6544553 | 3.38e-05 | −1.1052856 |
| ST6GALNAC1 | 1.22e-03 | −1.36196224 | 1.41e-04 | −2.8412139 | 1.24e-07 | −1.028552 |
| STYK1 | 6.76e-06 | −1.25356262 | 4.62e-06 | −1.6294317 | 6.56e-13 | −1.2155107 |
| SULF1 | 1.31e-19 | −3.68940574 | 1.45e-04 | −2.4181396 | 1.52e-08 | −1.9594916 |
| SULF2 | 5.19e-10 | −2.10614738 | 6.89e-04 | −1.6246188 | 3.13e-05 | −1.0889349 |
| SULT1C2 | 2.42e-03 | −1.3303751 | 3.50e-03 | −1.7726936 | 4.11e-04 | −1.0128167 |
| SYTL2 | 2.17e-09 | −1.66532807 | 3.23e-04 | −1.7670993 | 1.40e-08 | −1.0928962 |
| TCN1 | 1.36e-07 | −2.90090404 | 1.95e-04 | −3.3290883 | 1.94e-04 | −1.5896369 |
| TFF1 | 1.06e-04 | −2.39909674 | 1.28e-05 | −4.6834284 | 4.87e-07 | −1.9432884 |
| TGFBI | 7.08e-10 | −1.89345065 | 4.10e-04 | −1.6460046 | 1.92e-06 | −1.1341551 |
| TGM2 | 5.08e-09 | −1.12833345 | 2.32e-03 | −1.7831911 | 2.70e-06 | −1.205276 |
| THBS2 | 1.26e-17 | −3.98457919 | 4.24e-04 | −2.181901 | 2.56e-07 | −1.6264449 |
| TMC5 | 2.08e-04 | −1.27698246 | 1.10e-05 | −2.6388004 | 3.41e-07 | −1.7178244 |
| TMEM45B | 1.56e-03 | −1.4088581 | 9.04e-05 | −2.1279175 | 5.75e-07 | −1.3963273 |
| TMPRSS4 | 2.04e-07 | −1.9637276 | 1.14e-09 | −4.5041304 | 7.48e-13 | −2.2573618 |
| TNFAIP6 | 7.96e-11 | −2.48765791 | 1.06e-03 | −1.5405643 | 1.75e-04 | −1.30502 |
| TOP2A | 7.40e-06 | −1.48965862 | 7.55e-07 | −2.3640636 | 1.27e-07 | −1.3128398 |
| TRIM29 | 2.88e-10 | −2.00085653 | 8.29e-09 | −3.5036033 | 5.15e-11 | −1.4077371 |
| TSPAN1 | 1.16e-07 | −1.78293915 | 2.62e-08 | −3.6882971 | 1.63e-14 | −2.7266844 |
| TSPAN8 | 1.36e-02 | −1.3032792 | 2.05e-07 | −2.4782873 | 1.15e-04 | −1.4074942 |
| VCAN | 5.20e-17 | −3.73740193 | 9.41e-04 | −1.9461635 | 1.73e-06 | −1.7040513 |
| VSIG1 | 1.80e-05 | −1.7832075 | 1.95e-03 | −2.200699 | 8.04e-08 | −1.9952567 |
| Downregulated DEGs | ||||||
| ABAT | 7.74e-08 | 1.04113554 | 1.53e-04 | 1.7414842 | 5.42e-07 | 1.0759596 |
| ACADL | 1.67e-04 | 1.309813 | 3.68e-05 | 2.0830065 | 8.80e-07 | 1.6686389 |
| ALB | 2.22e-07 | 1.46077833 | 7.79e-05 | 1.4892635 | 4.12e-06 | 2.6310982 |
| ANPEP | 1.53e-05 | 2.42621752 | 1.10e-03 | 2.4850007 | 4.96e-06 | 1.8945569 |
| AOX1 | 5.78e-04 | 1.06899195 | 1.86e-05 | 2.3824797 | 1.56e-09 | 2.0361547 |
| AQP12B///AQP12A | 3.38e-07 | 1.77319312 | 4.00e-03 | 1.6068264 | 8.50e-06 | 1.1688947 |
| AQP8 | 7.06e-05 | 2.88068653 | 2.11e-03 | 3.5681394 | 2.55e-05 | 1.5447562 |
| BACE1 | 1.96e-04 | 1.33697382 | 1.34e-04 | 1.8149867 | 3.46e-06 | 1.2745662 |
| BNIP3 | 1.49e-05 | 1.31255588 | 6.37e-04 | 1.8414334 | 8.51e-05 | 1.093632 |
| BTG2 | 5.22e-07 | 1.01870823 | 5.01e-04 | 1.482411 | 1.47e-08 | 1.1710536 |
| C5 | 9.74e-08 | 1.78863462 | 1.65e-03 | 1.7982847 | 3.80e-06 | 1.1974638 |
| CHRM3 | 7.26e-07 | 1.21565011 | 1.97e-03 | 1.0395154 | 1.88e-04 | 1.0729804 |
| CTNND2 | 1.03e-04 | 1.1145235 | 8.06e-05 | 1.8025505 | 1.67e-06 | 1.3781816 |
| CTRL | 7.59e-04 | 3.00886301 | 5.20e-03 | 4.1102965 | 3.02e-05 | 2.2352664 |
| DPP10 | 1.09e-07 | 2.10610623 | 1.68e-03 | 1.8993883 | 4.07e-05 | 1.204828 |
| EGF | 4.57e-05 | 2.25361836 | 7.83e-04 | 2.8202033 | 4.66e-05 | 2.0791816 |
| EPB41L4B | 3.34e-08 | 1.27662689 | 4.04e-04 | 1.5076869 | 1.36e-05 | 1.0014458 |
| EPHX2 | 1.41e-08 | 1.42766027 | 2.27e-04 | 1.5900637 | 2.33e-09 | 1.309602 |
| ERO1B | 4.08e-05 | 1.62417116 | 8.13e-05 | 2.5162192 | 7.77e-09 | 1.9621349 |
| ERP27 | 2.00e-03 | 2.31577382 | 4.01e-03 | 3.4721533 | 2.88e-05 | 2.5885804 |
| F11 | 7.44e-07 | 1.32674611 | 8.59e-04 | 1.1769399 | 1.72e-06 | 2.006714 |
| F8 | 1.23e-04 | 1.11066971 | 1.02e-04 | 1.8290128 | 1.47e-05 | 1.1382302 |
| FAM129A | 1.30e-05 | 1.06645494 | 7.63e-05 | 1.6379657 | 1.14e-05 | 1.0048831 |
| FAM150B | 1.18e-03 | 1.23501259 | 2.08e-05 | 3.2500833 | 7.11e-07 | 1.1093278 |
| FGL1 | 1.35e-03 | 1.89436601 | 1.95e-03 | 2.8963528 | 2.10e-04 | 1.6669633 |
| GATM | 5.75e-05 | 1.25039334 | 1.30e-03 | 1.5289067 | 6.25e-04 | 1.6627798 |
| GNMT | 1.55e-06 | 2.74551014 | 5.24e-04 | 2.9488702 | 1.89e-04 | 1.3901202 |
| GP2 | 3.21e-02 | 1.92081986 | 1.82e-03 | 2.4593161 | 1.12e-04 | 2.793808 |
| GPHA2 | 6.69e-07 | 1.86728762 | 7.73e-04 | 1.4332783 | 7.22e-05 | 1.10197 |
| GSTA1 | 3.35e-04 | 1.87171685 | 3.97e-04 | 2.6806129 | 6.43e-05 | 1.1862356 |
| GUCA1C | 2.88e-07 | 1.07977556 | 2.27e-03 | 1.1651245 | 2.21e-05 | 1.321844 |
| HOMER2 | 8.96e-04 | 1.4104841 | 5.11e-04 | 2.007722 | 3.37e-05 | 1.248792 |
| KIAA1324 | 3.10e-05 | 1.58434185 | 2.54e-04 | 2.1736865 | 6.23e-07 | 2.0721882 |
| KLK1 | 5.08e-04 | 2.25690227 | 2.94e-03 | 3.1178643 | 6.33e-05 | 1.9187229 |
| LIFR | 3.01e-05 | 1.12632478 | 1.46e-05 | 1.319066 | 7.71e-09 | 1.3452642 |
| MCOLN3 | 3.64e-08 | 1.13701745 | 7.15e-05 | 1.1513915 | 1.49e-07 | 1.1641287 |
| MT1G | 1.95e-06 | 1.3722596 | 2.11e-04 | 1.8544499 | 7.22e-05 | 1.2238896 |
| NR5A2 | 1.24e-03 | 1.28135842 | 6.52e-04 | 2.2282414 | 4.57e-06 | 2.0610724 |
| NRG4 | 1.01e-06 | 1.60115287 | 2.26e-03 | 1.0302797 | 1.11e-04 | 1.6338384 |
| NUCB2 | 5.22e-07 | 1.0613891 | 5.30e-04 | 1.6431302 | 1.22e-05 | 1.0996724 |
| PAK3 | 2.28e-03 | 1.28384452 | 9.94e-05 | 1.9989158 | 1.89e-06 | 1.4388473 |
| PDIA2 | 4.07e-05 | 2.61613826 | 2.08e-03 | 3.7840517 | 9.44e-06 | 2.1084087 |
| PDK4 | 1.89e-05 | 1.37573077 | 1.56e-06 | 2.4030511 | 6.18e-07 | 1.61166 |
| PM20D1 | 2.63e-07 | 2.41556837 | 3.83e-03 | 1.8976107 | 7.82e-05 | 1.380062 |
| PNLIPRP1 | 3.32e-02 | 2.04233634 | 2.38e-03 | 4.0693577 | 3.86e-05 | 3.0266571 |
| RBPJL | 2.13e-06 | 2.35782027 | 1.36e-03 | 2.9782084 | 1.74e-05 | 1.7077773 |
| RGN | 1.21e-04 | 1.09262047 | 1.32e-04 | 2.1504185 | 3.81e-07 | 1.2929591 |
| SERPINI2 | 1.63e-03 | 2.97325268 | 1.78e-03 | 4.2512033 | 2.38e-05 | 2.4767376 |
| SLC16A10 | 1.50e-06 | 1.92460149 | 2.10e-05 | 1.9189034 | 4.74e-04 | 1.0443833 |
| SLC1A2 | 1.46e-06 | 1.01926645 | 8.71e-04 | 1.6966492 | 9.13e-05 | 1.0052447 |
| SLC39A5 | 1.85e-08 | 1.59792695 | 1.02e-03 | 1.5697093 | 2.57e-05 | 1.2896791 |
| SLC43A1 | 3.32e-06 | 1.66833588 | 6.63e-04 | 1.9614561 | 1.22e-05 | 1.1801271 |
| TMED6 | 9.89e-05 | 2.76897653 | 6.43e-04 | 3.5223984 | 2.09e-06 | 1.8966689 |
| TRHDE | 2.83e-08 | 2.25612973 | 2.00e-04 | 2.4896933 | 1.18e-05 | 1.680468 |
If the |Log FC| > 1, and Adj P < .05, it means that the gene has more than 10-fold change between pancreatic cancer and normal tissue, and the difference is statistically significant. The adjusted P values are listed in the Adj P value column of the results table. The Benjamini & Hochberg false discovery rate method is selected by default in GEO as it is the most commonly used adjustment for microarray data and provides a good balance between discovery of statistically significant genes and limitation of false positives.
Functional and Pathway Enrichment Analysis
DAVID was used to analyze the functions and pathways (see Materials and Methods) of identified DEGs. DEGs were mainly involved in biological processes associated integrin-mediated signaling pathway, proteolysis, and collagen catabolic process. Moreover, seven KEGG pathways were overrepresented among upregulated genes, including ECM-receptor interaction, focal adhesion, and PI3K-Akt signaling pathway (Table 2).
Table 2.
Functional and Pathway Enrichment Analysis of Upregulated and Downregulated Genes in Pancreatic Cancer with the DEGs in the Three mRNA Expression Profiling Datasets
| Term | Description | Count | P Value |
|---|---|---|---|
| Upregulated | |||
| GO:0030198 | integrin-mediated signaling pathway | 21 | 1.05E-14 |
| GO:0030574 | collagen catabolic process | 14 | 6.69E-14 |
| KEGG:hsa04512 | ECM-receptor interaction | 13 | 1.88E-10 |
| GO:0004222 | metalloendopeptidase activity | 12 | 1.65E-08 |
| GO:0030199 | collagen fibril organization | 8 | 1.17E-07 |
| KEGG:hsa04510 | Focal adhesion | 14 | 4.69E-07 |
| KEGG:hsa05146 | Amoebiasis | 10 | 2.82E-06 |
| GO:0005201 | extracellular matrix structural constituent | 8 | 4.49E-06 |
| GO:0004252 | serine-type endopeptidase activity | 13 | 9.59E-06 |
| KEGG:hsa04151 | PI3K-Akt signaling pathway | 15 | 2.96E-05 |
| GO:0005581 | collagen trimer | 8 | 3.05E-05 |
| GO:0031581 | hemidesmosome assembly | 4 | 2.13E-04 |
| KEGG:hsa04974 | protein digestion and absorption | 7 | 4.74E-04 |
| GO:0007229 | integrin-mediated signaling pathway | 7 | 5.24E-04 |
| GO:0005788 | endoplasmic reticulum lumen | 9 | 5.65E-04 |
| GO:0006508 | proteolysis | 15 | 6.11E-04 |
| GO:0007160 | cell-matrix adhesion | 6 | 0.00226 |
| KEGG:hsa05222 | small cell lung cancer | 6 | 0.002763 |
| KEGG:hsa05200 | Pathways in cancer | 12 | 0.004792 |
| GO:0043588 | skin development | 4 | 0.004925 |
| GO:0048333 | mesodermal cell differentiation | 3 | 0.005338 |
| Downregulated | |||
| GO:0003756 | protein disulfide isomerase activity | 3 | 0.001837 |
PPI Network Construction and Modules Selection
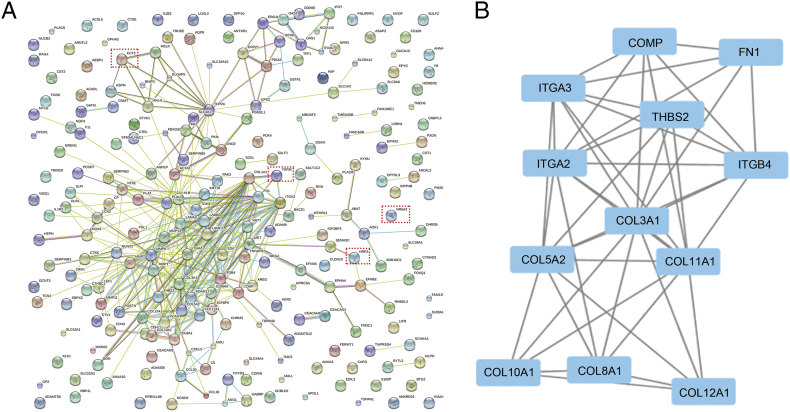
The PPI network (see Materials and Methods) included 182 upregulated (176 nodes and 282 edges) and 54 downregulated (52 nodes and 10 edges) DEGs (Figure 2A). A total of 12 genes, including THBS2and ITGA3, have more than 10 interaction partners (Figure 2B) and are potential hub genes. A significant module was observed in the PPI network of DEGs (Figure 2B). Overall survival analysis of the hub genes (see Materials and Methods) showed that the expression level of most of them (10/12) is strongly associated with the overall survival in pancreatic adenocarcinoma. In particular, low expression levels (bottom 10%) are associated with poor survival, while high expression levels (top 90%) are associated with improved survival (Table 3, 10% vs 90% part). However, the effect decreases when patients are split into the bottom 20% and upper 80% and into the bottom 40% and upper 60% according to their expression values for the corresponding genes, with 5/12 and 4/12 hub genes showing a significant correlation with the overall survival, respectively (Table 3, 20% vs 80% and 40% vs 60% part). (See Fig. 3.)
Figure 2.
PPI network and a significant module. (A) PPI network of total DEGs in the three mRNA expression profiling datasets. (B) A significant module selected from PPI network. All the genes are upregulated genes. The line represents interaction relationship between nodes.
Table 3.
Significant Prognostic Value of Hub Genes and Screening Out DEGs in Pancreatic Cancer Patients
| 10% vs 90% | |||||
|---|---|---|---|---|---|
| Gene name | COMP | FN1 | ITGB4 | COL8A1 | COL10A1 |
| Log-rank P value | .0193 | .0301 | .000194 | .00444 | .00158 |
| Gene name | COL5A2 | ITGA2 | ITGA3 | THBS2 | COL3A1 |
| Log-rank P value | .00416 | .0309 | .0252 | .0207 | .00386 |
| Gene name | ECT2 | NRP2 | TGFBI | ||
| Log-rank P value | .00124 | .0177 | .0199 | ||
| 20% vs 80% | |||||
| Gene name | ITGB4 | COL11A1 | ITGA2 | ITGA3 | THBS2 |
| Log-rank P value | .000842 | .0241 | .0152 | .000484 | .00866 |
| Gene name | ECT2 | NRP2 | TGFBI | ||
| Log-rank P value | .00484 | .00801 | .0448 | ||
| 40% vs 60% | |||||
| Gene name | ITGB4 | COL11A1 | ITGA2 | ITGA3 | |
| Log-rank P value | .00836 | .0113 | 2.91e-06 | 6.21e-05 | |
| Gene name | ECT2 | NRP2 | TGFBI | ||
| Log-rank P value | .000851 | .059 | .0343 | ||
Figure 3.
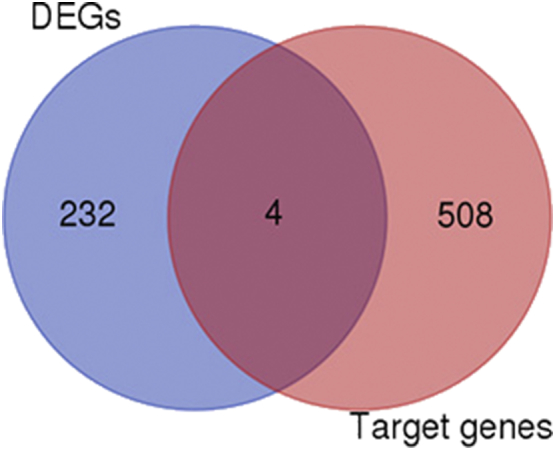
Identification of DEGs and paired miRNAs in the three mRNA expression and one miRNA expression profiling datasets. We found four specially DEGs and their miRNA, including ECT2-miR302c, NR5A2-miR27a, NRP2-miR27a, and miR331-3p, TGFBI-miR21.
MiRNA-DEG Pairs
We identified 21 DEMs in pancreatic cancer tissue compared to normal tissue in the GSE41372 dataset. In particular, 19 miRNAs were upregulated and 2 were downregulated (Table 4). Using miRecords (see Materials and Methods), we predicted 718 gene targets for these DEMs. Specifically, four predicted DEM targets were DEGs: NRP2 was among the predicted gene targets of miR27a, and miR331-3p, ECT2, NR5A2, and TGFBI were among the predicted gene targets of miR302c, miR27a, and miR21, respectively.
Table 4.
DEMs in Pancreatic Cancer Screened Out from miRNA Expression Microarray GSE41372 and Their Target Genes Predicted by miRecords
| miRNA | Adj.P | logFC | Target Genes |
|---|---|---|---|
| hsa-miR-630 | 0.002038 | −4.21846 | NA |
| hsa-miR-302c | 0.002434 | −1.37368 | E2F7, PARP8, NEUROD6, SLC22A23, CFL2, DMTF1, TNFAIP1, ZNFX1, TSHZ3, DCUN1D1 |
| hsa-miR-337-3p | 0.003922 | 1.307959 | NA |
| hsa-miR-190b | 0.005768 | 1.369034 | NA |
| hsa-miR-140-3p | 0.003638 | 1.504247 | NA |
| hsa-miR-331-3p | 0.003497 | 1.830589 | B4GALT2, ZNF513, RIC8B, BAIAP2, CPSF2, CAMK2A, SNX17, PHC2, DAG1, NRP2 |
| hsa-miR-484 | 0.002038 | 1.835517 | NA |
| hsa-miR-490-3p | 0.001403 | 1.888625 | NA |
| hsa-miR-342-3p | 0.002434 | 2.136132 | PPP3R1, ZAK, BTN2A1, MRFAP1, ZIC4, RSBN1, FAM53C, PDGFRA, FUT8, UBE2D2 |
| hsa-miR-221 | 0.000713 | 2.181648 | CDKN1B, ATXN1, TMCC1, NSMCE4A, GOLSYN, EIF5A2, DCUN1D1, DMRT3, C12orf30, MIER3 |
| hsa-miR-197 | 0.001103 | 2.192276 | NA |
| hsa-miR-1975 | 0.003424 | 2.291544 | NA |
| hsa-miR-125a-5p | 0.002574 | 2.483418 | MCL1, SLITRK6, OSBPL9, ELOVL6, NIN, ALPK3, CGN, GTPBP2, CCNJ, SYVN1 |
| hsa-miR-142-5p | 0.003922 | 2.625738 | BTBD7, RAB6B, FLJ10357, LRP1B, TRIM36, ALS2, EGLN3, CHD9, WDR26, SCOC |
| hsa-miR-135b | 3.37E-05 | 2.819987 | TSEN54, GGNBP2, C6orf106, NUCKS1, TMEM168, PELI2, LZTS1, NUDT4, HOXA10, ELOVL2 |
| hsa-miR-10a | 0.001103 | 2.841119 | TBX5, NARG1, HOXA3, USP46, BACH2, RAP2A, SLC38A2, ANKFY1, NCOA6, BAZ2B |
| hsa-miR-223 | 0.002038 | 2.930734 | NFIA, FBXO8, STK39, SACS, RBM16, PDS5B, PHF20L1, CRIM1, FBXW7, SLC8A1 |
| hsa-miR-21 | 0.001978 | 2.992536 | RP2, JAG1, SOX5, LEMD3, KIAA1012, BAHD1, ADNP, ASPN, CHD7, PELI1 |
| hsa-miR-145 | 0.001133 | 3.244704 | MDFIC, CACHD1, EPB41L5, SEMA6A, NUFIP2, SRGAP1, RIN2, CDC37L1, KIF21A, INOC1 |
| hsa-miR-199a-5p | 0.007061 | 3.264147 | ATXN7, C11orf9, CELSR1, RAB10, CCNJ, CCNL1, SLC24A3, ZNF512B, ALS2, CCDC43 |
| hsa-miR-27a | 0.003424 | 3.636233 | TMCC1, NXT2, HOXA10, DLL4, CCNJ, ANK1, SEMA6A, GPAM, OTX2, TEAD1 |
Overall Survival Analysis
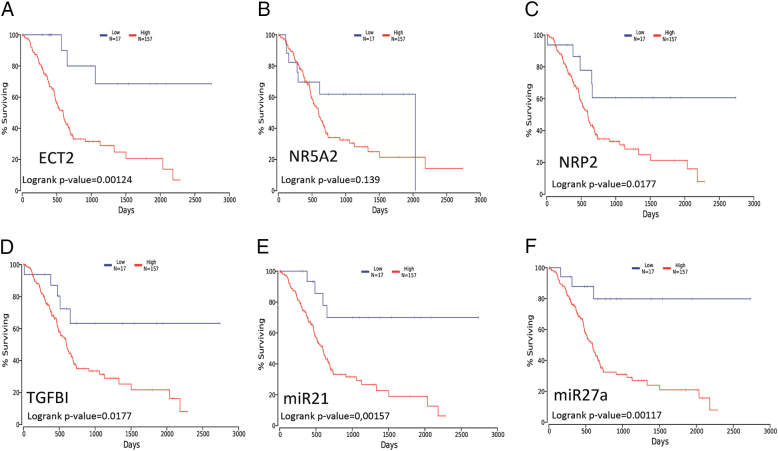
We used OncoLnc to conduct overall survival analysis of ECT2, NR5A2, NRP2, and TGFBI in pancreatic cancer. We found that high mRNA expression of ECT2, NRP2, or TGFBI was associated with worse overall survival (log-rank P value = .001, .02, and .02, respectively; Figure 4, A-D and Table 3). We also analyzed the predicted DEM regulators of the four DEGs and found that high expression of miR27a and miR21 was associated with worse overall survival (log-rank P values = .001 and .002, respectively; Figure 4, E and F). There was no significant association between miR302C expression and overall survival (data not shown). Then, we changed the percentage from 10% vs 90% to 20% vs 80%; the results also showed that high expression of ECT2, NRP2, or TGFBI was associated with worse overall survival (Table 3). Finally, we also investigated associations between the DEGs and other kinds of cancer, including breast, lung, colorectal, and prostate cancer (Table 5).
Figure 4.
Prognostic value of four DEGs and their miRNA in pancreatic cancer patients. Prognostic value of (A) ETC2 (log-rank P value = .00124), (B) NR5A2 (log-rank P value = .139), (C) NRP2 (log-rank P value = .0177), (D) TGFBI (log-rank P value = .0199), (E) miR21 (log-rank P value = .00157), and (F) miR27a (log-rank P value = .00117).
Table 5.
Prognostic Value of Four DEGs in Other Kinds of Cancer Patients
| Gene/Cancer Name | ECT2 | NR5A2 | NRP2 | TGFBI |
|---|---|---|---|---|
| Breast cancer | 0.956 | 0.254 | 0.57 | 0.869 |
| Gastric cancer | 0.983 | 0.794 | 0.091 | 0.00395 |
| Lung cancer | 0.0433 | 0.965 | 0.975 | 0.862 |
| Ovarian cancer | 0.193 | 0.622 | 0.992 | 0.611 |
| Liver cancer | 0.00295 | 0.614 | 0.00763 | 0.457 |
Expression of DEGs in Different Organs
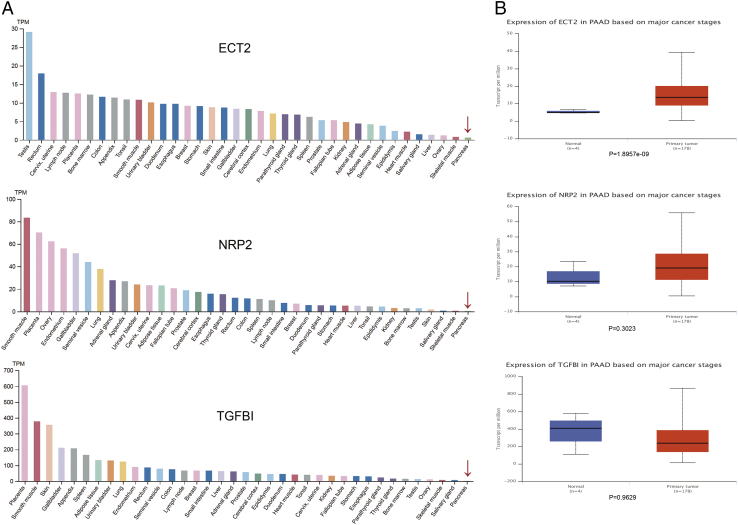
We used “ The Human Protein Atlas ” to perform the basic expression level of DEGs in different human organs. We found that the basic expression of ECT2, NRP2, and TGFBI was at lower level than other organs with HPA dataset, GTEx dataset, and FANTOM5 dataset (Figure 5A, GTEx and FANTOM% not shown). We also analyzed the basic expression of DEGs in pancreas and cancer tissues with TCGA dataset. Those results showed that the expression of ECT2 and NRP2 was significant higher in pancreatic cancer tissues, but there was no significant difference in expression on TGFBI (Figure 5B). Furthermore, we found that NRP2 expression in pancreatic cancer is a big difference.
Figure 5.
The basic expression of DEGs in different organs and cancer. The basic expression of DEGs in pancreas stood lower level in 37 kinds of organs (A). The expressions of ECT2 and NRP2 were significant higher in pancreatic cancer tissues (B).
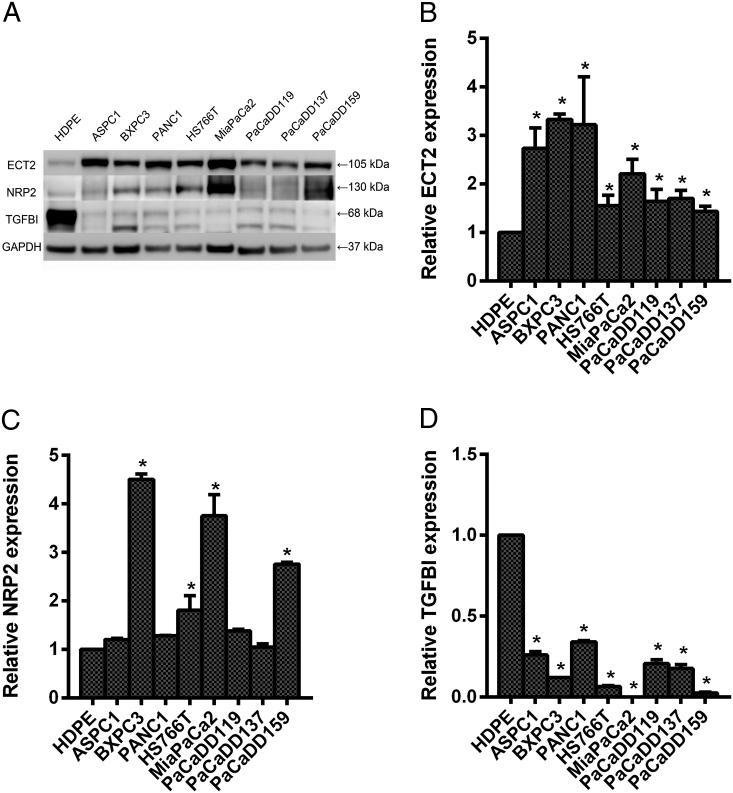
Expression of DEGs in Normal Pancreas and Pancreatic Cancer Cell Lines
For further research of the different expression of DEGs in pancreatic cancer, we detected the protein expression between normal pancreas HDPE cell and different pancreatic cancer cell lines by Western blot. Those results showed that the protein expression of ECT2 in pancreatic cancer cell lines was higher than that in normal pancreas cell line. The expression of NRP2 was higher in some kinds of pancreatic cancer cell lines, such as ASPC1, PaCaDD119, PaCaDD137, and PaCaDD159, than normal pancreas cell line. There was higher expression level of TGFBI in normal pancreas cell line (Figure 6A). The q-PCR showed the same results with Western blot (Figure 6B, C, D). ECT2 was completely overexpressed in pancreatic cancer cell lines. NRP2 and TGFBI were partly overexpressed and obviously not overexpressed, respectively, in pancreatic cancer cell lines.
Figure 6.
The basic expression of DEGs in normal pancreas and pancreatic cancer cell lines. The protein expression of DEGs in normal pancreas and pancreatic cancer cell lines (A). The mRNA expression of DEGs in normal pancreas and pancreatic cancer cell lines (B, C, D). *P value < .05.
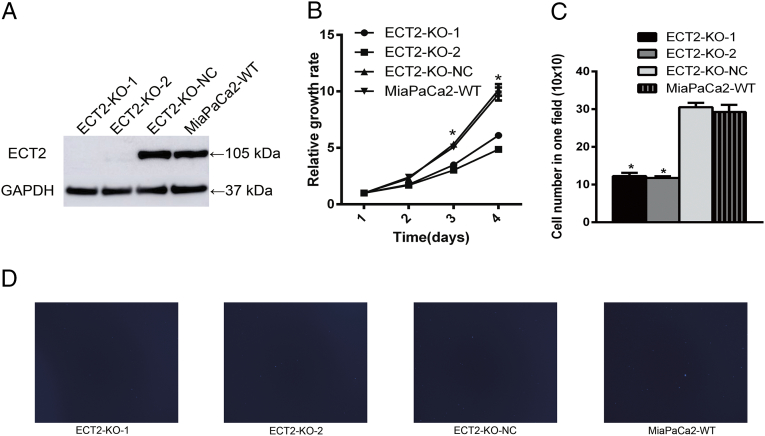
CRISPR/Cas9 Gene Editing
We detected ECT2 function to verify the data mining results. From the basic expression of different pancreatic cancer cell lines and normal pancreas cell line, we found that there was higher expression of ECT2 in MiaPaCa2 cell line. Then, we used Crispr Cas9 gene editing system to knockout ECT2 in MiaPaCa2 cells (Figure 7A). We detected the proliferation ability with ECT2 knocked out cells, negative control cells, and wild-type cells. Those results showed that ECT2 knocked out cells grew slower than negative control and wild-type cells (Figure 7B). Furthermore, we also detected the migration ability with ECT2 knocked out cells. Those results showed that the migration ability of ECT2 knocked out cells was lower than negative control and wild-type cells (Figure 7, C and D).
Figure 7.
Knockout of ECT2 decreased proliferation and migration ability in MiaPaCa2 cells. Knockout of ECT2 by Crispr Cas9 gene editing system inhibited protein expression of ECT2 in MiaPaCa2 cells (A). ECT2 knocked out cells grew slower than control groups (B). ECT2 knocked out cells migration slower than control groups (C and D). *P value < .05.
Discussion
Pancreatic cancer is a nearly universally fatal disease and is one of the few cancer types that continue to increase in incidence [33]. It is expected to become the second leading cause of cancer-related deaths in the United States in the next few years. Peak incidence of pancreatic cancer has been linked to few predisposing factors, including cigarette smoking, red meat, obesity, and heavy alcohol consumption along with pancreatic disease [34], [35], [36]. Because of its aggressive behavior and frequently late diagnosis, research aiming at early diagnosis of pancreatic cancer or its precursors is crucial. Molecular biomarkers play an important role in early diagnosis, and the rapid development of high-throughput technologies has resulted in an increasing number of biomarkers being screened for disease progression [37], [38]. Biomarkers enable the identification of genes, miRNAs, LncRNAs, and so on for early diagnosis, treatment, and prognosis of the disease, including pancreatic cancer. In particular, microarray and follow-up experiments performed in our laboratory over the last few years [39], [40] have shown that we can identify biomarkers and key genes in pancreatic cancer [39], [41].
In this study, we used three mRNA and one miRNA expression microarray datasets to identify biomarkers in pancreatic cancer. The four expression microarrays correspond to two different platforms, making the screen result more robust. We performed a comprehensive analysis of the DEMs, including functional and pathway enrichment analysis, and PPI analysis. Next, we screened the DEGs among the putative targets of the DEMs. Compared to previous studies [15], [42], [43], the samples included were more comprehensive and random. We identified a total of 236 DEGs among the three mRNA expression microarray datasets, consisting of 182 up-and 54 downregulated genes. Upregulated DEGs were mainly associated with the integrin-mediated signaling pathway, proteolysis, and the PI3K-Akt signaling pathway, which have been proved to have a close relationship with pancreatic cancer [44]. Twelve of the 236 DEGs were highly connected in the PPI network and were in the core of the PPI network. Such hub genes are usually the focus of microarray-based biomarker research. Indeed, an overall survival analysis with the clinical data from TCGA showed the expected trend, with 10 of these genes being strongly associated with overall survival in pancreatic adenocarcinoma. However, the correlation between increased overall survival and low expression levels is only strong for the bottom 10%, and the number of hub genes displaying a significant correlation decreases to 5 or 4 when then 20th and 40th percentiles are considered. In addition, we identified 21 DEMs and their more than 500 predicted target genes. In order to improve the generalizability and robustness of the predictions, DEM target predictions were obtained with an ensemble approach [30]. Four DEM targets were also DEGs: ECT2 (miR302c), NR5A2 (miR27a), NRP2 (miR27a and miR331-3p), and TGFBI (miR21). All four DEGs were upregulated in pancreatic cancer tissue. miRNA including miR27a, miR331-3p, and miR21 were upregulated; miR302c was downregulated. NR5A2 has been reported to be upregulated in pancreatic cancer [45]; ECT2, NRP2, and TGFBI are novel candidates. Overall survival analysis of these genes showed that upregulation of ECT2, NRP2, or TGFBI is significantly correlated with poor survival of pancreatic cancer patients. Interestingly, 7 patients had low expression levels for ECT2, NRP2, and TGFBI, and 13 patients had low expression levels for 2 of these 3 genes. However, this finding warrants further investigation.
ECT2 locates at 3q26.31, is a BRCT-containing guanidine exchange factor for Rho GTPases, and was found to be a responder to DNA damage [46]. It controls cell division and exerts oncogenic functions in multiple cancers [47], [48]. Patients with high expression of ECT2 also have poor overall survivals in lung (log-rank P value = .04) and liver (log-rank P value = .003) cancer. Additionally, ECT2 has been reported to play an important role in the Wingless/Wnt and KRAS signaling pathways [49], [50]. The KRAS is a key signaling pathway in pancreatic cancer. Typical alterations in the molecular signaling include mutations in KRAS in 95% of pancreatic cancer [41], [51]. Consistent with our results, previous studies have also shown that ECT2 is an oncogene which is overexpressed in pancreatic tumor tissues [52]. However, the oncogenic mechanism for pancreatic cancer has rarely been explored. NRP2 is located at 2q33.3; is a nontyrosine kinase receptor frequently overexpressed in various malignancies; and may play a role in cardiovascular development, axon guidance, and tumorigenesis [53]. Some studies have shown that it is required for maintaining endocytic activity in cancer cells, which supports their oncogenic activities and confers drug resistance [54]. Further, it has been suggested that NRP2 works its biological function via the GSK3β and VEGFC/VEGFR3 signaling pathway [55], [56], and GSK3β plays a key role in resistance to pancreatic cancer therapy [57]. We performed overall survival analysis of NRP2 in some common tumors and found that upregulated NRP2 is significantly correlated with poor overall survival of liver (log-rank P value = .008) tumor patients. In our study, NRP2 we could not confirm a significant correlation of NRP2 expression and survival in breast and gastric cancer, but other studies have shown a function in breast and gastric tumor, proving that it is an important gene in solid tumors [58], [59]. However, there is no study focused on NRP2 function in pancreatic cancer. Finally, TGFBI, also known as βig-H3, is a protein inducible by TGFβ1 and secreted by many cell types [60]. Furthermore, TGFBI is an important component of the Wnt signaling pathway [61]. It is located at 5q31.1. In our study, the patients with upregulated TGFBI have poor survival rate in gastric (log-rank P value = .004) and pancreatic (log-rank P value = 2.0 × 10−2) cancer. TGFBI has been studied in the context of many different diseases, but no study has revealed its function and mechanism in pancreatic cancer. Interestingly, all ECT2, NRP2, and TGFß1 are involved in the Wnt signaling pathway, which has been reported to be an important regulated part and potential treatment target in pancreatic cancer in multiple studies. These observations support our hypothesis that ECT2, NRP2, and TGFBI are potential regulated components of pancreatic cancer and that there may be more interactions among them. Together with NR5A2, these four genes grant further in vivo and in vitro study.
Among the 21 DEMs in pancreatic cancer, we identified 19 upregulated and 2 downregulated miRNAs. miR27a was the strongest upregulated miRNA, and miR630 was the strongest downregulated miRNA. Some studies have reported that miR27a is a cooperative repressor of a network of tumor suppressor genes, and inhibition of miR27a has been shown to have synergistic effects in reducing proliferation of PDAC cells in culture and growth of xenograft tumors in mice [22]. NR5A2 and NRP2 are the predicted targets of miR27a, and ECT2, NR5A2, NRP2, and TGFBI were DEGs in the three mRNA expression datasets. miR21, miR302c, and miR331-3p potentially play a role in pancreatic cancer.
We also checked the basic expression of DEGs in different organ. We found that the basic expression of ECT2, NRP2, and TGFBI was at a lower level than other organs with HPA dataset, GTEx dataset, and FANTOM5 dataset. In HPA dataset, those 3 DEGs were the last one of 37 organs. However, in the comparison of expression between pancreas and pancreatic cancers from TCGA, we can that found ECT2 and NRP2 were statistically significantly higher in cancer tissues. It predicted that the higher expression of ECT2 and NRP2 may play a role in tumorigenicity of pancreatic cancer.
For further verification of above data-mining results, we used basic biology experiment to detect the basic expression of ECT2, NRP2, and TGFBI in normal pancreas cell line and pancreatic cancer cell lines. Those results proved that ECT2 and NRP2 had higher expression level in pancreatic cancer. However, there was higher expression level of TGFBI in normal pancreas cell line. Globally expressed ECT2 was associated with worse overall survival stably. Mixed expressed NRP2 was associated with worse overall survival based on the percentiles of expression values and cancer kinds. Outlier expressed TGFBI was associated with worse overall survival occasionally (Table 3, Table 5). We think ECT2 and NRP2 may play an important role in pancreatic cancer.
Furthermore, we also did gene function analysis by experiment. We made MiaPaCa2 ECT2 knockout stable cell lines to verify the above results with Crispr Cas9 system. As those results showed, knockout of ECT2 decreased the proliferation and migration ability of MiaPaCa2 cells. It predicted that ECT2 was an important gene in pancreatic cancer. This is in line with expectations based on above results.
This study is a next step in identifying relevant biomarkers for early diagnosis and progression of pancreatic cancer. In this study, we combined high-throughput mRNA and miRNA expression datasets with functional and regulatory network analyses to screen for biomarkers. miR21, miR27a, miR302c, and miR331-3p and their predicted targets genes ECT2, NRP2, and TGFBI were selected as promising candidates. Then, we detected the basic expression level of DEGs in various organ tissues and different pancreatic cancer with data mining and experiment methods. Those showed that ECT2 and NRP2 were in line with data mining results. We also used Cripsr Cas9 gene editing system to knockout ECT2 and proved its important function in pancreatic cancer. Meanwhile, we will continue to research ECT2 and NRP2 functions and mechanisms in pancreatic cancer and assess their clinical application prospects. We believe that ECT2 and NRP2 can play as potential biomarker in pancreatic cancer patients, and they may be potential target therapy site with Crispr Cas9 gene editing system for pancreatic cancer.
Funding
B. L. is funded by the China Scholarship Council (CSC) scholarship (No. 201406210072).
Availability of Data and Materials
All source data are available on the appropriate Web sites.
Author’s Contributions
Conceptualization: B. L., C. P.
Data curation: B. L.
Formal analysis: B. L.
Funding acquisition: B. L C. P., R. G.
Supervision: C. P.
Writing, original draft: B. L.
Writing, review & editing: B. L., H. Y., L. T., A. D., R. G., C. P., G.W.
All authors participated in the process of revision and finalizing the manuscript. All authors read and approved the final manuscript.
Competing Interests
All authors declare that they have no competing interest.
Consent for Publication
Not applicable.
Ethics Approval and Consent to Participate
Not applicable.
Previous Presentation as Congress Abstract
Not applicable.
Acknowledgements
We like to thank all the scientists who were able to put their software and data online. Without the efforts of the research community, this study would have been impossible.
References
- 1.New M, Van Acker T, Long JS, Sakamaki JI, Ryan KM, Tooze SA. Molecular pathways controlling autophagy in pancreatic Cancer. Front Oncol. 2017;7 doi: 10.3389/fonc.2017.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.De Angelis R, Sant M, Coleman MP, Francisci S, Baili P, Pierannunzio D, Trama A, Visser O, Brenner H, Ardanaz E. Cancer survival in Europe 1999-2007 by country and age: results of EUROCARE--5-a population-based study. Lancet Oncol. 2014;15:23–34. doi: 10.1016/S1470-2045(13)70546-1. [DOI] [PubMed] [Google Scholar]
- 5.Lianos GD, Christodoulou DK, Katsanos KH, Katsios C, Glantzounis GK. Minimally invasive surgical approaches for pancreatic adenocarcinoma: recent trends. Int J Gastrointest Cancer. 2017;48:129–134. doi: 10.1007/s12029-017-9934-9. [DOI] [PubMed] [Google Scholar]
- 6.Griffin JF, Poruk KE, Wolfgang CL. Pancreatic cancer surgery: past, present, and future. Chin J Cancer Res. 2015;27:332–348. doi: 10.3978/j.issn.1000-9604.2015.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou B, Xu JW, Cheng YG, Gao JY, Hu SY, Wang L, Zhan HX. Early detection of pancreatic cancer: Where are we now and where are we going? Int J Cancer. 2017;141:231–241. doi: 10.1002/ijc.30670. [DOI] [PubMed] [Google Scholar]
- 8.Halbrook CJ, Lyssiotis CA. Employing metabolism to improve the diagnosis and treatment of pancreatic cancer. Cancer Cell. 2017;31:5–19. doi: 10.1016/j.ccell.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Chang JC, Kundranda M. Novel diagnostic and predictive biomarkers in pancreatic adenocarcinoma. Int J Mol Sci. 2017;18:E667. doi: 10.3390/ijms18030667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stojkovic Lalosevic M, Stankovic S, Stojkovic M, Markovic V, Dimitrijevic I, Lalosevic J, Petrovic J, Brankovic M, Pavlovic Markovic A, Krivokapic Z. Can preoperative CEA and CA19-9 serum concentrations suggest metastatic disease in colorectal cancer patients? Hell J Nucl Med. 2017;20:41–45. doi: 10.1967/s002449910505. [DOI] [PubMed] [Google Scholar]
- 11.Zhou G, Liu X, Wang X, Jin D, Chen Y, Li G, Li C, Fu D, Xu W, Wang X. Combination of preoperative CEA and CA19-9 improves prediction outcomes in patients with resectable pancreatic adenocarcinoma: results from a large follow-up cohort. Onco Targets Ther. 2017;10:1199–1206. doi: 10.2147/OTT.S116136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Distler M, Pilarsky E, Kersting S, Grutzmann R. Preoperative CEA and CA 19-9 are prognostic markers for survival after curative resection for ductal adenocarcinoma of the pancreas - a retrospective tumor marker prognostic study. Int J Surg. 2013;11:1067–1072. doi: 10.1016/j.ijsu.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Zhu L, Xue HD, Liu W, Wang X, Sui X, Wang Q, Zhang D, Li P, Jin ZY. Enhancing pancreatic mass with normal serum CA19-9: key MDCT features to characterize pancreatic neuroendocrine tumours from its mimics. Radiol Med. 2017;122:337–344. doi: 10.1007/s11547-017-0734-x. [DOI] [PubMed] [Google Scholar]
- 14.Swords DS, Firpo MA, Scaife CL, Mulvihill SJ. Biomarkers in pancreatic adenocarcinoma: current perspectives. Onco Targets Ther. 2016;9:7459–7467. doi: 10.2147/OTT.S100510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grutzmann R, Boriss H, Ammerpohl O, Luttges J, Kalthoff H, Schackert HK, Kloppel G, Saeger HD, Pilarsky C. Meta-analysis of microarray data on pancreatic cancer defines a set of commonly dysregulated genes. Oncogene. 2005;24:5079–5088. doi: 10.1038/sj.onc.1208696. [DOI] [PubMed] [Google Scholar]
- 16.Krizkova S, Kepinska M, Emri G, Rodrigo MA, Tmejova K, Nerudova D, Kizek R, Adam V. Microarray analysis of metallothioneins in human diseases—a review. J Pharm Biomed Anal. 2016;117:464–473. doi: 10.1016/j.jpba.2015.09.031. [DOI] [PubMed] [Google Scholar]
- 17.Stewart JP, Richman S, Maughan T, Lawler M, Dunne PD, Salto-Tellez M. Standardising RNA profiling based biomarker application in cancer-The need for robust control of technical variables. Biochim Biophys Acta. 2017;1868:258–272. doi: 10.1016/j.bbcan.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepato-Gastroenterology. 2008;55:2016–2027. [PubMed] [Google Scholar]
- 19.Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, Petersen G, Lou Z, Wang L. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16:259–266. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang G, He P, Tan H, Budhu A, Gaedcke J, Ghadimi BM, Ried T, Yfantis HG, Lee DH, Maitra A. Integration of metabolomics and transcriptomics revealed a fatty acid network exerting growth inhibitory effects in human pancreatic cancer. Clin Cancer Res. 2013;19:4983–4993. doi: 10.1158/1078-0432.CCR-13-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang G, Schetter A, He P, Funamizu N, Gaedcke J, Ghadimi BM, Ried T, Hassan R, Yfantis HG, Lee DH. DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity and predicts clinical outcome in pancreatic ductal adenocarcinoma. PLoS One. 2012;7:e31507. doi: 10.1371/journal.pone.0031507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frampton AE, Castellano L, Colombo T, Giovannetti E, Krell J, Jacob J, Pellegrino L, Roca-Alonso L, Funel N, Gall TM. MicroRNAs cooperatively inhibit a network of tumor suppressor genes to promote pancreatic tumor growth and progression. Gastroenterology. 2014;146:268–277 e218. doi: 10.1053/j.gastro.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 23.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gan Z, Wang J, Salomonis N, Stowe JC, Haddad GG, McCulloch AD, Altintas I, Zambon AC. MAAMD: a workflow to standardize meta-analyses and comparison of affymetrix microarray data. BMC Bioinformatics. 2014;15:69. doi: 10.1186/1471-2105-15-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vilming Elgaaen B, Olstad OK, Haug KB, Brusletto B, Sandvik L, Staff AC, Gautvik KM, Davidson B. Enhancing pancreatic mass with normal serum CA19-9: key MDCT features to characterize pancreatic neuroendocrine tumours from its mimics. BMC Cancer. 2014;14:80. [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the false discovery rate — a practical and powerful approach to multiple testing. J R Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 27.Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 28.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. Peer J Comput Sci. 2016;2 [Google Scholar]
- 32.Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F. A pathology atlas of the human cancer transcriptome. Science. 2017;357 doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 33.Malvezzi M, Carioli G, Bertuccio P, Boffetta P, Levi F, La Vecchia C, Negri E. European cancer mortality predictions for the year 2017, with focus on lung cancer. Ann Oncol. 2017;28:1117–1123. doi: 10.1093/annonc/mdx033. [DOI] [PubMed] [Google Scholar]
- 34.Larson A, Kwon RS. Natural history of pancreatic cysts. Dig Dis Sci. 2017;62:1770–1777. doi: 10.1007/s10620-017-4542-x. [DOI] [PubMed] [Google Scholar]
- 35.Maisonneuve P, Amar S, Lowenfels AB. Periodontal disease, edentulism and pancreatic cancer: a meta analysis. Ann Oncol. 2017;28:985–995. doi: 10.1093/annonc/mdx019. [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Useros J, Li W, Cabeza-Morales M, Garcia-Foncillas J. Oxidative stress: a new target for pancreatic cancer prognosis and treatment. Nippon Rinsho. 2017:6. doi: 10.3390/jcm6030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boccardi M, Gallo V, Yasui Y, Vineis P, Padovani A, Mosimann U, Giannakopoulos P, Gold G, Dubois B, Jack CR., Jr. The biomarker-based diagnosis of Alzheimer's disease. 2-lessons from oncology. Neurobiol Aging. 2017;52:141–152. doi: 10.1016/j.neurobiolaging.2017.01.021. [DOI] [PubMed] [Google Scholar]
- 38.Kramer F, Sabbah HN, Januzzi JJ, Zannad F, Peter van Tintelen J, Schelbert EB, Kim RJ, Milting H, Vonk R, Neudeck B. Redefining the role of biomarkers in heart failure trials: expert consensus document. Heart Fail Rev. 2017;22:263–277. doi: 10.1007/s10741-017-9608-5. [DOI] [PubMed] [Google Scholar]
- 39.Pilarsky C, Ammerpohl O, Sipos B, Dahl E, Hartmann A, Wellmann A, Braunschweig T, Lohr M, Jesenofsky R, Friess H. Activation of Wnt signalling in stroma from pancreatic cancer identified by gene expression profiling. J Cell Mol Med. 2008;12:2823–2835. doi: 10.1111/j.1582-4934.2008.00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winter C, Kristiansen G, Kersting S, Roy J, Aust D, Knosel T, Rummele P, Jahnke B, Hentrich V, Ruckert F. Google goes cancer: improving outcome prediction for cancer patients by network-based ranking of marker genes. PLoS Comput Biol. 2012;8 doi: 10.1371/journal.pcbi.1002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jahny E, Yang H, Liu B, Jahnke B, Lademann F, Knosel T, Rummele P, Grutzmann R, Aust DE, Pilarsky C. The G protein-coupled receptor RAI3 is an independent prognostic factor for pancreatic cancer survival and regulates proliferation via STAT3 phosphorylation. PLoS One. 2017;12:e0170390. doi: 10.1371/journal.pone.0170390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, Cooc J, Weinkle J, Kim GE, Jakkula L. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newhook TE, Blais EM, Lindberg JM, Adair SJ, Xin W, Lee JK, Papin JA, Parsons JT, Bauer TW. A thirteen-gene expression signature predicts survival of patients with pancreatic cancer and identifies new genes of interest. PLoS One. 2014;9 doi: 10.1371/journal.pone.0105631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pihlak R, Valle JW, McNamara MG. Germline mutations in pancreatic cancer and potential new therapeutic options. Oncotarget. 2017;8:73240–73257. doi: 10.18632/oncotarget.17291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 46.He D, Xiang J, Li B, Liu H. The dynamic behavior of Ect2 in response to DNA damage. Sci Rep. 2016;6:24504. doi: 10.1038/srep24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cook DR, Rossman KL, Der CJ. Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease. Oncogene. 2014;33:4021–4035. doi: 10.1038/onc.2013.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mack NA, Georgiou M. The interdependence of the Rho GTPases and apicobasal cell polarity. Small GTPases. 2014;5:10. doi: 10.4161/21541248.2014.973768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Justilien V, Ali SA, Jamieson L, Yin N, Cox AD, Der CJ, Murray NR, Fields AP. Ect2-dependent rRNA synthesis is required for KRAS-TRP53-driven lung adenocarcinoma. Cancer Cell. 2017;31:256–269. doi: 10.1016/j.ccell.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greer ER, Chao AT, Bejsovec A. Pebble/ECT2 RhoGEF negatively regulates the Wingless/Wnt signaling pathway. Development. 2013;140:4937–4946. doi: 10.1242/dev.101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Werner K, Lademann F, Thepkaysone ML, Jahnke B, Aust DE, Kahlert C, Weber G, Weitz J, Grutzmann R, Pilarsky C. Simultaneous gene silencing of KRAS and anti-apoptotic genes as a multitarget therapy. Oncotarget. 2016;7:3984–3992. doi: 10.18632/oncotarget.6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang ML, Lu S, Zhou L, Zheng SS. Correlation between ECT2 gene expression and methylation change of ECT2 promoter region in pancreatic cancer. Hepatobiliary Pancreat Dis Int. 2008;7:533–538. [PubMed] [Google Scholar]
- 53.Dutta S, Roy S, Polavaram NS, Baretton GB, Muders MH, Batra S, Datta K. NRP2 transcriptionally regulates its downstream effector WDFY1. Sci Rep. 2016;6:23588. doi: 10.1038/srep23588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fujii T, Shimada K, Asano A, Tatsumi Y, Yamaguchi N, Yamazaki M, Konishi N. MicroRNA-331-3p suppresses cervical cancer cell proliferation and E6/E7 expression by targeting NRP2. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17081351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ng T, Hor CH, Chew B, Zhao J, Zhong Z, Ryu JR, Goh EL. Neuropilin 2 signaling is involved in cell positioning of adult-born neurons through glycogen synthase kinase-3beta (GSK3beta) J Biol Chem. 2016;291:25088–25095. doi: 10.1074/jbc.M116.755215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ou JJ, Wei X, Peng Y, Zha L, Zhou RB, Shi H, Zhou Q, Liang HJ. Neuropilin-2 mediates lymphangiogenesis of colorectal carcinoma via a VEGFC/VEGFR3 independent signaling. Cancer Lett. 2015;358:200–209. doi: 10.1016/j.canlet.2014.12.046. [DOI] [PubMed] [Google Scholar]
- 57.Domoto T, Pyko IV, Furuta T, Miyashita K, Uehara M, Shimasaki T, Nakada M, Minamoto T. Glycogen synthase kinase-3beta is a pivotal mediator of cancer invasion and resistance to therapy. Cancer Sci. 2016;107:1363–1372. doi: 10.1111/cas.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yasuoka H, Kodama R, Tsujimoto M, Yoshidome K, Akamatsu H, Nakahara M, Inagaki M, Sanke T, Nakamura Y. Neuropilin-2 expression in breast cancer: correlation with lymph node metastasis, poor prognosis, and regulation of CXCR4 expression. BMC Cancer. 2009;9:220. doi: 10.1186/1471-2407-9-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uronis HE, Bendell JC, Altomare I, Blobe GC, Hsu SD, Morse MA, Pang H, Zafar SY, Conkling P, Favaro J. A phase II study of capecitabine, oxaliplatin, and bevacizumab in the treatment of metastatic esophagogastric adenocarcinomas. Oncologist. 2013;18:271–272. doi: 10.1634/theoncologist.2012-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qiu WY, Zheng LB, Pan F, Wang BB, Yao YF. New histopathologic and ultrastructural findings in Reis-Bucklers corneal dystrophy caused by the Arg124Leu mutation of TGFBI gene. BMC Ophthalmol. 2016;16:158. doi: 10.1186/s12886-016-0325-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang F, Hu W, Xian J, Ohnuma S, Brenton JD. The Xenopus Tgfbi is required for embryogenesis through regulation of canonical Wnt signalling. Dev Biol. 2013;379:16–27. doi: 10.1016/j.ydbio.2012.11.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All source data are available on the appropriate Web sites.