Abstract
Background
Aberrant DNA methylation is considered to be a potential cause of recurrent pregnancy loss (RPL), while potential mechanism has not yet been elucidated.
Methods
In order to uncover the contribution of the perturbation of DNA methylation in RPL, we performed genome-wide DNA methylation analysis combined with genome-wide gene expression in decidua tissue.
Findings
Totally, 539 differentially methylated regions (DMRs) were identified and significantly correlated with gene expressions. We observed that hypo-methylated DMR near CREB5 recruited transcription factors binding, such as P53 and SP1, and in turn upregulated CREB5. Compromised cell migration and apoptosis were observed in human CREB5 overexpression trophoblast cell lines, indicating dysfunctional trophoblast cells might contribute to RPL after hypo-methylation of CREB5. In addition, overexpression of CREB5 altered cell cycle.
Interpretation
Our data highlights a role of CREB5 involved in the pathogenesis of RPL, and CREB5 maybe a potential diagnostic biomarker for RPL.
Keywords: Recurrent pregnancy loss, DNA methylation, Gene expression, CREB5
Research in context.
Evidence before this study
The underlying mechanisms of 50% of cases with recurrent pregnancy loss (RPL) are unknown, it is an emerging area of interest to identify the risk factors contributing to RPL. We searched PubMed with the terms “recurrent pregnancy loss” and “DNA methylation” for original research articles up to March 1, 2016. There were no language restrictions. In these studies, DNA methylation was measured at either global level or small set of genes on the basis of existing hypotheses. Few studies focused on the genome wide DNA methylation and the regulatory role of DNA methylation on gene expression in RPL.
Added value of this study
In the present study, we identified CREB5 as a novel risk gene contributing to recurrent pregnancy loss by performing genome-wide DNA methylation analysis combined with RNA-seq analysis in decidua tissue. In mechanism, hypomethylation of CREB5 upregulated its expression and caused dysfunction of trophoblast cells, which might contribute to RPL.
Implications of all the available evidence
To our knowledge, this is the first study to explore the alternations of decidua DNA methylome on gene regulations in RPL. Our research provides new insights into CREB5 dependent regulatory effects during RPL and suggests DNA methylation of CREB5 may be a potential biomarker for RPL diagnosis.
Alt-text: Unlabelled Box
1. Introduction
Recurrent pregnancy loss (RPL), defined as the failure of two or more consecutive clinical pregnancies before 20 weeks of gestation, affects approximately 1% of couples attempting to conceive [1]. The known causes of RPL include chromosomal abnormalities, maternal reproductive tract abnormalities, maternal endocrine abnormalities, immune dysfunction, genital tract infections, cervical insufficiency and environmental exposures [2]. However, the underlying mechanisms of 50% of cases are unknown [3].
The placenta plays a crucial role during pregnancy, which allows oxygen exchange, nutrients uptake and waste removal. The maternal portion of the placenta is known as the decidua, which can provide a delicate balance between immune tolerance and defense to maintain the pregnancy [4]. During early pregnancy, failure of cell invasion, immune tolerance and arterial remodeling in the developing decidua, which are controlled by complicated epigenetic modification network, may lead to poor implantation, inflammation and impaired placental development [5].
DNA methylation, one of major epigenetic modifications, plays important roles in the regulation of gene expression, X-inactivation, genomic imprinting, genome stabilization and chromatin modification, which are crucial for normal embryonic development [[6], [7], [8]]. During the early stages of embryogenesis, extensive epigenetic modifications occur, and the total demethylation phase alternates with the remethylation phase to ensure the pluripotency of the developing embryo [9]. However, some parental epigenetic modifications may escape the second wave of demethylation, indicating a potential inheritance of epigenetic modifications set during gametogenesis [10, 11]. Aberrant DNA methylation has been reported to be involved in human miscarriage, pre-eclampsia, abnormal embryonic development and birth malformations [[12], [13], [14], [15]]. Epigenetic mechanism in early life altered function and structure of organs, which played an important role for diseases in later life [16]. It has been shown that abnormal DNA methylation in the chorionic villus, especially at the loci of the imprinting genes, resulted in RPL in patients with normal karyotype [17, 18]. In addition, epigenetic alterations in placenta have been reported to be associated with gestational diabetes mellitus and hypertension, which are also involved in pregnancy loss [19, 20]. Moreover, hyper-methylation at the promoter of FOXP3, which is indispensable for the differentiation of Treg cells, led to down-regulate FOXP3 and broke the immune balance in maternal-fetal interface [21]. Several studies suggested that abnormal methylation existed in semen samples from couples with RPL [22, 23]. However, most of these studies only measured small set of genes on the basis of existing hypotheses about the etiology of RPL, which might bias interpretation. Additionally, few studies reported the DNA methylation level of decidua and clarified the contribution of decidua methylation change on gene regulations involved in RPL.
Thus, we assessed alterations in DNA methylation by Infinium Human Methylation 850 K BeadChips and gene expression by RNA-sequencing analysis (RNA-seq) in decidua samples from RPL patients and controls with induced abortions. Eventually, CREB5 gene (cAMP responsive element binding protein 5), up-regulation in RPL through hypo-methylation in promoters, was found to compromise the trophoblast cellular functions, which might in turn lead to RPL.
2. Materials and methods
2.1. Ethics statement
This study was approved by the Institutional Ethics Committee of Nanjing Medical University. All experiment protocols were approved by the Institutional Review Board of Nanjing Medical University prior to the study. All activities involved in this study were done under full compliance with government policies and the Helsinki Declaration.
2.2. Study population and sample collection
20 normal pregnant women (non-medical reasons for abortion) and 80 RPL patients were recruited from the affiliated hospitals of Nanjing Medical University between 2012 and 2015. A questionnaire was used to collect recruited couples' characteristic information including personal information, lifestyle factors and medical history. Females with infection, endocrine abnormalities, antiphospholipid syndrome or other known risk factors for RPL were excluded from the study. Through array-based Comparative Genomic Hybridization (array-CGH), a genetic diagnosis method, we excluded cases with chromosomal abnormality or large fragment structure variation. Finally, 20 RPL patients and 20 normal pregnant women were enrolled (Table 1). At the time of dilation and curettage, we separated decidua tissues from the products of conception under a dissecting microscope. Firstly, we washed tissue samples thoroughly with sterile normal saline to remove excess blood, mucous and fetal tissues. Then, we transferred fragments of decidua and deciduas with attached chorionic villi to a petri dish with sterile normal saline and examined under a dissecting microscope. The deciduas were carefully dissected out from the branching chorionic villi, flashed frozen in liquid nitrogen and stored at −80 °C for following studies.
Table 1.
Characteristics of subjects.
| Variable | Control group (N = 20) | RPL group (N = 20) | t | P value |
|---|---|---|---|---|
| Materal age (year)⁎ | 24·333 ± 5·245 | 25·000 ± 2·563 | −0·332 | 0·744 |
| Body mass index (BMI) (kg/m2) ⁎ |
19·962 ± 2·210 | 21·895 ± 3·324 | −0·820 | 0·423 |
| Gestational age (weeks) ⁎ | 7·083 ± 1·567 | 7·715 ± 0·572 | −1·084 | 0·293 |
| Pregnant frequency⁎ | 2·000 ± 1·348 | 1·500 ± 1·069 | 0·878 | 0·391 |
| Childbirth frequency⁎ | 0·583 ± 0·793 | 0·444 ± 0·727 | 1·068 | 0·300 |
| NO. of pregnancy loss⁎ | – | 2·63 ± 0·518 | – |
The Student's two-tailed t-test was used to statistically compare groups for each variable, unless otherwise denoted.
Data are given as mean ± standard deviation.
2.3. DNA isolation and Infinium human methylation 850 K BeadChip
Genome-wide DNA methylation patterns were performed by Infinium Human Methylation 850 K BeadChips (Illumina), which determine the methylation levels of 853,307 CpG sites. After performing bisulfite treatment with EZ DNA Methylation Kit (Zymo Research) following the manufacturer's procedure, processed DNA samples were then hybridized to the BeadChip (Illumina) according to the Illumina Infinium HD Methylation Protocol.
Illumina intensity data (IDAT) files from the chip were further processed by the R/Bioconductor (version 3·3·3) package ChAMP. DNA methylation level was reported as β value, ranging from zero to one, where zero represents non-methylated probe and one represents fully methylated probe, for every CpG site. Probes that had a detection P value of >0·01 and those located on the X and Y chromosome were filtered away. We also removed probes with <three beads in at least 5% of samples per probe, SNP-related probes and all multi-hit probes. BMIQ algorithm was used to correct for the Infinium type I and type II probe bias.
Additionally, we identified differently methylated probes (DMPs) at significance of a Benjamini-Hochberg adjusted P < 0·05 and conducted gene set enrichment analysis to know if genes involved in these significant DMPs are enriched for specific biological terms or pathways. Differentially Methylated Regions (DMRs) were identified by Bumphunter with default settings.
Methylation array data were deposited into Gene Expression Omnibus (GEO) under accession GSE113600.
2.4. RNA isolation, qRT-PCR and RNA-seq
Total RNA from tissues and cells were extracted using TRIzol reagent (Life Technologies). For mRNA detection, 0·5 μg total RNA were reverse transcribed to generate cDNA using the reverse transcription kit (Takara). To measure the expression of mRNA, we employed qRT-PCR on the LightCycler 480 (Roche) along with the GAPDH as the endogenous control. All primers for PCR were listed in Supplementary Table S1. For RNA-seq, libraries were prepared from three biological replicates using the TruSeq RNA Sample Prep Kit and sequenced on the NextSeq (Illumina). After generating read alignments with hg19 by being processed with STAR [24], RNA-seq data was then normalized and analyzed using DEseq2 [25]. RNA-seq data has been submitted to the GEO repository (GSE113790).
2.5. Bisulfite sequencing PCR
Genomic DNA (gDNA) was isolated from the decidua tissue of pregnant women with DNeasy Blood and Tissue kit (Qiagen). 500 ng of gDNA was bisulfite-treated by EZ DNA Methylation Kit (Zymo Research). The sequences of primers used for Bisulfite Sequencing PCR are listed in Supplementary Table S2. Bisulfite-PCR products were amplified by GoTaq Hot Start Polymerase (Promega) according to the manufacturer's instructions. The PCR products were purified and ligated into pMD 19-T Vector (Takara). Then, the plasmids were transformed into E.coli DH5α Competent Cells (Takara). Sequencing reactions for single amplified clones were performed by Jie Li Biology Co (Shanghai). Sequence data were analyzed by Vector NTI software (Thermo Fisher SCIENTIFIC) and the QUMA website (http://quma.cdb.riken.jp/).
2.6. Dual-luciferase reporter assay
Functional analysis of CREB5 DMR was conducted with a dual-luciferase assay. A 1166 bp DNA fragment containing the CREB5 DMR was amplified by forward primer 5′-GAAGATCTTCTTGCTGGCATCGTTACCT- 3′ and reverse primer 5′-CGGGGTACCGTATTCGACTTCCTCTTTCAACATA-3′. The forward primer contained a BglII and the reverse primer contained a KpnI recognition site. The PCR product was purified, cloned into pGL3-basic vector and Sanger sequencing was used to check the cloned PCR product. Plasmid DNA was purified by EndoFree Plasmid Kit (QIAGEN). The pGL3-basic vector with CREB5 DMR insert was in vitro methylated by M.SssI (New England Biolabs) and the methylation sensitive enzyme HpaII (New England Biolabs) was used to confirm in vitro methylation. Finally, either methylated or unmethylated plasmid was co-transfected with pGL3-SV40 vector (in a ratio of 400:1) in three different cell lines, including HTR8-S/Vneo, JEG-3 and 293 T, using Lipofectamine 2000 (Invitrogen). Luciferase activity in cells was analyzed using the Dual-luciferase Assay System (Promega), luciferase normalized to renilla luciferase activity and the fold increase relative to that of empty pGL3-basic vector as recommended by the manufacturer.
2.7. Chromatin immunoprecipitation (ChIP)
ChIP was performed with the EZ-Magna ChIP A/G Chromatin Immunoprecipitation Kit (Millipore) following the instructions provided by the manufacturer. In brief, solubilized chromatin was sonicated to an average size between 300 and 500 bp and immunoprecipitated with the antibody against IgG (Millipore), P53 (Cell Signaling Technology Cat# 2527S, RRID:AB_10695803) and SP1 (Cell Signaling Technology Cat# 9389S, RRID:AB_11220235). Antibody–chromatin complexes were pulled-down using magnetic A/G beads, washed and eluted. Purified DNA was quantified with standard qPCR methods using SYBR Green PCR Master Mix (Applied Biosystems) on a 7900HT Fast-Real-Time PCR (ABI) and normalized to values obtained after amplification of unprecipitated (input) DNA. Primer sequences are listed as followed: forward primer 5′- GGGTAAAAGCTTCTGGGGTC- 3′ and reverse primer 5′-CTAGAGGAGAAGGCTTACTGGTGC-3′.
2.8. Western blot
Proteins from decidua tissues and cultured cells were extracted using RIPA buffer containing PMSF, and quantified by bicinchoninic acid (BCA) solution (Beyotime, Nantong, China). Protein samples were separated in a 10% SDS-PAGE gel and transferred to PVDF membranes (Millipore). The membrane was blocked with Tris Buffered Saline Tween (TBST), added with 5% non-fat dry milk, and incubated overnight at 4 °C with a primary antibody CREB5 (1:1000, Sigma, St Louis, MO, USA) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH, 1:1000, Beyotime, Nantong, China). After washing three times in TBST buffer, the secondary antibodies anti-mouse or anti-rabbit HRP-linked (Beyotime, Nantong, China) were used at appropriate dilution (1:1000) for 1 h at room temperature. The signals were detected by the addition of Immobilon Western Chemiluminescent HRP Substrate (Millipore).
2.9. Cell culture and transfection
We obtained human HTR8-S/Vneo cell, JEG-3 cell and 293 T cell from American Type Culture Collection (ATCC). HTR8-S/Vneo cell was cultured in 1640 medium (Gibco), JEG-3 cell was cultured in MEM medium (Gibco) and 293 T cell was cultured in DMEM medium (Gibco), all of which were mixed with 10% fetal bovine serum (FBS) (Gibco), 100 U /ml penicillin (Gibco), and 100 μg/ml streptomycin (Gibco) at 37 °C, 5% CO2. 4 μg of CREB5 overexpression plasmid (pcDNA-CREB5) or negative control (pcDNA-NC) were used in transfection experiments with Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions.
2.10. Analysis of CREB5 overexpression and knockdown on cell function
5*105 cells transfected with pcDNA-CREB5, pcDNA-NC, si-NC or si-CREB5 about 24 h were incubated in 100 μl serum-free medium inside upper chamber (Millipore) while 600 μl DMEM with 10%FBS were added into the lower chamber. After 24 h, cells were dyed with crystal violet staining solution (Beyotime) and photographed under 20× and 10× magnification. For cell cycle assay, transfected cells were collected and detected by FACS (BD Biasciences). As for cell apoptosis, cells were stained with annexin-V/PI (BD Biopharmingen), and then analyzed by FlowJo V7 software (Tree Star). All experiments were repeated three times independently. CREB5 siRNA were purchased from Gene Pharma (Shanghai, China). The siRNA sequence is as follows: sense: 5′- GCGGAAUAUCUCGAUGCAUTT- 3′, antisense: 5′- AUGCAUCGAGAUAUUCCGCTT- 3′.
2.11. Statistical analyses
Statistical analysis was performed using R software (version 3·3·3). Pearson's chi-squared test and Student's t-test was used to assess statistical differences in demographic and clinical characteristics, including drinking or smoking status, body mass index (BMI), age, abortion frequency and gestational weeks between cases and controls. Each experiment was performed in triplicate and results were considered statistically significant at P value <0·05.
3. Results
3.1. Genome-wide DNA methylation analysis of decidua tissues in RPL and controls
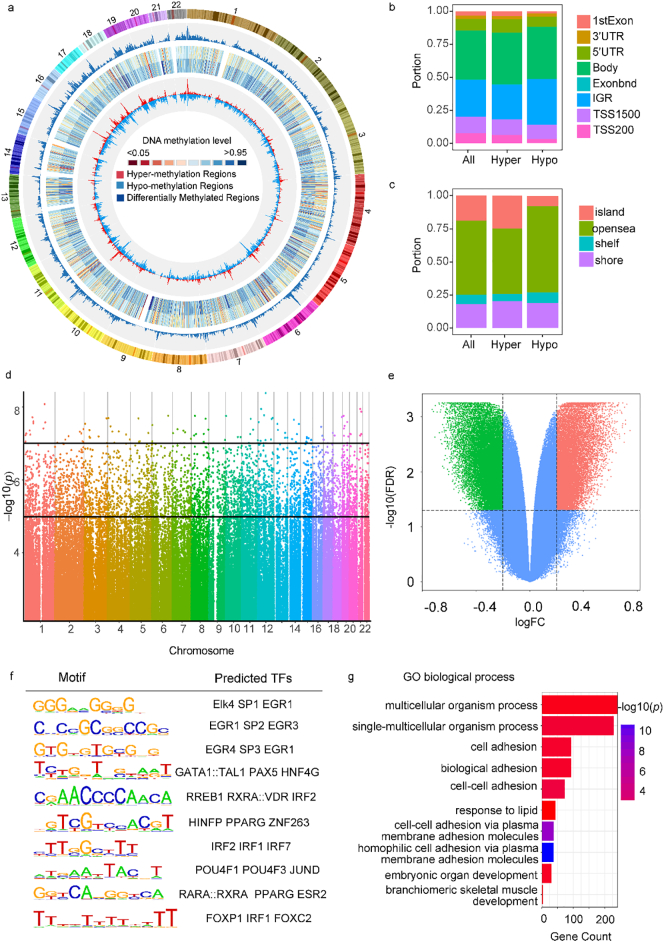
To better understand the role of DNA methylation in RPL, we performed the Infinium Human Methylation 850 K BeadChip arrays in decidua samples from the patients with RPL (n = 4) and controls (n = 2). The density plot of methylation level between RPL and controls was largely bimodal, indicating that there were no apparent global DNA methylation differences between these two groups (Supplementary Fig. S1a, b). By averaging β-value per Mb in the genome, we described global DNA methylation patterns between RPL and controls using circos plot (Fig. 1a). The distribution of differentially methylated CpG sites was largely overlapped between these two groups with majority in gene body and intergenic gene regions (IGR) (Fig. 1b, c). Additionally, differentially methylated probes (DMPs) are evenly distributed on autosomes (Fig. 1d). Using β-value as DNA methylation level, we identified 107,550 differentially methylated CpG sites between RPL and controls with FDR < 0·05, among which 45,245 sites showed >20% β-value difference (Fig. 1e).
Fig. 1.
DNA methylation patterns in RPL and controls. (a) Circular representation of whole genome DNA methylation level for RPL and controls. CpG methylation level were averaged per Mb in the genome and represent as histograms and heatmaps. (b) Distributions of hyper-methylated and hypo-methylated DMPs were investigated throughout each genomic region, FDR < 0·05. (c) Genomic locations divided into island, opensea, shelf and shore according to the distance between CpG island and DMPs, FDR < 0·05. (d) Manhattan plot showing P value based on DMPs analyses. The two horizontal lines is the suggestive DNA methylation chip significance threshold cutoff. (e) A volcano plot of the distribution between FDR (adjusted P value) and difference in β value (logFC). Lines represent used cutoff values to identify the most hyper-methylated DMPs (red) and hypo-methylated DMPs (green). (f) DNA sequences motifs identified to be enriched in DMRs by CisGenome Browser software. The right column contains the top three transcription factor similar to these motifs using JASPAR database. (g) Gene enrichment analysis of gens with DMPs in RPL.
Given that at CpG island might have stronger function than that single site, we then analyzed differentially methylated regions (DMRs) between RPL and controls. By using the algorithm of Bumphunter, we identified 539 DMRs in autosomal between these two groups including 129 hypo-DMRs and 410 hyper-DMRs (Supplementary Table S3). Functional genomic elements were usually associated with DNA methylation signatures [26], thus we hypothesized that RPL-related DMRs may be enriched with transcription factors binding motif. To explore potential transcription factors occupancy in these DMRs, we searched for enriched DNA motifs in DMRs using CisGenome Browser software [27], and identified ten new motifs which might regulate the transcription of DMR-related genes through JASPAR database (Fig. 1f) [28]. We next performed gene enrichment analysis using ChAMP to assess whether the DMRs related genes were functionally linked to pregnancy loss. GO analysis indicated an enrichment of genes that are implicated in developmental processes, such us embryonic organ development and branchiomeric skeletal muscle development (Fig. 1g).
3.2. The correlation of DMRs with genome-wide gene expressions
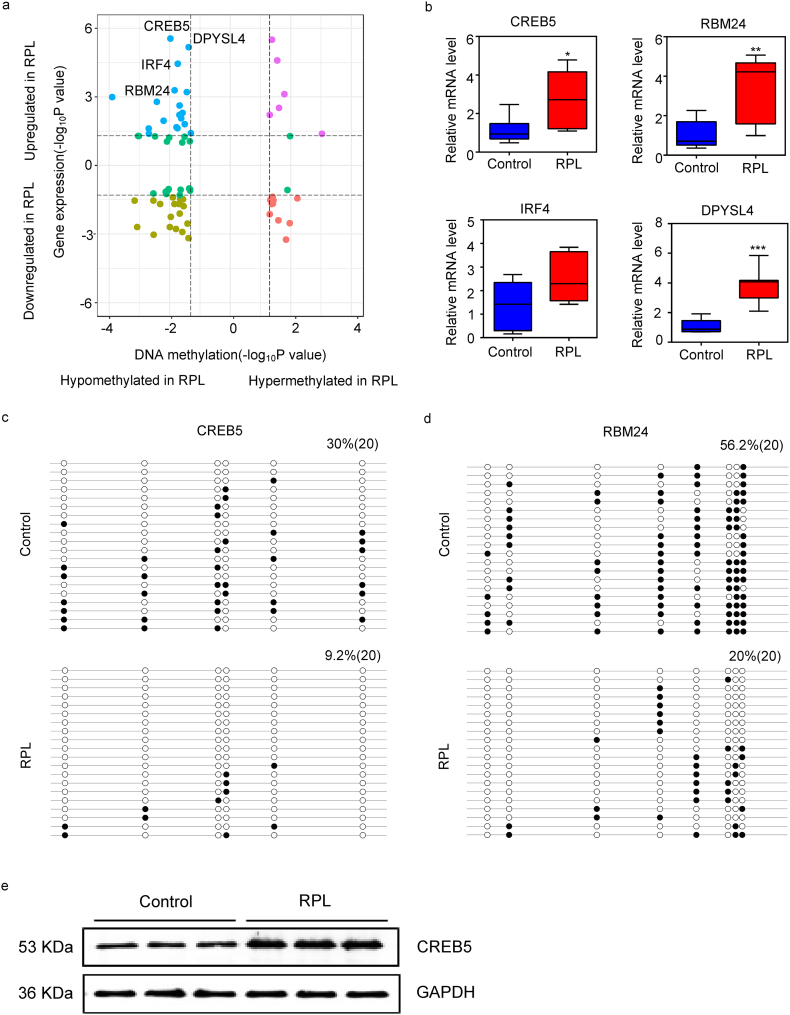
In order to investigate how DMRs affected the gene transcriptions, we made an unbiased assessment of genome-wide mRNA levels by RNA-seq in decidua samples from the patients with RPL (n = 3) and controls (n = 3). With FDR < 0·05 and fold change>2, we identified 225 differential expressed genes (DEGs) between RPL and controls (Supplementary Table S4). To verify the RNA-seq results, 20 DEGs were randomly selected and validated in an independent group of RPL and controls. All the results were highly consistent with RNA-seq results (Supplementary Fig. S2a, b). Potential relationships between alterations in DNA methylation and related-genes expression were carried out by comparing the P values of DMRs and P values of DEGs (Fig. 2a). By setting a threshold of 5% FDR, we selected four genes, including CREB5, RBM24, IRF4, DPYSL4, in the further study. Interestingly, all of these genes had hypo-methylated regions or up-regulated gene expression in RPL. Next, we validated these gene expressions by qRT-PCR and they all significantly higher expressed in RPL group except for IRF4 (Fig. 2b).
Fig. 2.
Filtration of genes with synergetic alterations in DNA methylation and gene expression. (a) Quadrant plot showing DMRs and expression of corresponding genes. The dashed lines indicate a threshold of P value below 0·05. The blue dots signify hypo-methylated and up-regulated genes in RPL, the brown bots signify hypo-methylated and down-regulated genes in RPL, the purple bots signify hyper-methylated and up-regulated genes in RPL and the red bots signify hyper-methylated and down-regulated genes in RPL. (b) Box plots of relative expression level of four genes selected by a threshold of 5% FDR. Error bars, SEM. *P < 0·05, **P < 0·01, ***P < 0·001. Bisulfite sequencing of CREB5 (c) and RBM24 (d). Open and closed circles represent unmethylated and methylated CpG sites and each row corresponds to a single clone sequenced. All loci shown in picture were found to be hypo-methylated in RPL according to the analysis of Infinium Human Methylation 850 K BeadChip data. (e) The expression of CREB5 protein in control group and RPL group.
Since the DMRs of CREB5 and RBM24 located in the promoters, which might affect gene transcription, bisulfite sequencing was used to determine the DNA methylation in promoter regions of these two genes (Table 2, Fig. 2c, d). Our results demonstrated that these two genes' promoters were hypo-methylated in the RPL group compared with the control group. In addition, CREB5 DMRs in the promoter regions de-methylated more completely than RBM24. Then, we chose CREB5 as a potentially functional gene for further study.
Table 2.
Significant RPL-associated differentially methylated regions (DMRs).
| Gene | Region | Probes | Locationa | βb (RPL) | βb (Controls) | FDRc |
|---|---|---|---|---|---|---|
| CREB5 | Chr7:28450001-28,451,011 | cg11093548 | 5'UTR | 0·07 | 0·36 | 0·01 |
| cg22396147 | 5'UTR | 0·09 | 0·28 | 0·001 | ||
| cg14803765 | TSS1500 | 0·08 | 0·31 | 0·008 | ||
| cg14927519 | TSS1500 | 0·17 | 0·33 | 0·03 | ||
| cg01332259 | TSS1500 | 0·06 | 0·25 | 0·003 | ||
| cg17067116 | TSS1500 | 0·04 | 0·27 | 0·03 | ||
| RBM24 | Chr6:17282284–147,282,863 | cg25302957 | TSS1500 | 0·05 | 0·32 | 0·002 |
| cg02685016 | 1stExon | 0·11 | 0·45 | 0·01 | ||
| cg15168816 | TSS1500 | 0·13 | 0·52 | 0·006 | ||
| cg14466942 | TSS1500 | 0·42 | 0·70 | 0·006 | ||
| cg02066331 | TSS200 | 0·48 | 0·69 | 0·02 | ||
| cg02311932 | TSS200 | 0·38 | 0·68 | 0·01 | ||
| cg23146346 | TSS200 | 0·65 | 0·91 | 0·002 | ||
| cg01729060 | 5'UTR | 0·66 | 0·89 | 0·007 | ||
Chr, chromosome;FDR, false discovery rate.
Locations of probes in 5’UTR, 1stExon, TSS200 and TSS 1500.
Mean CpGs methylation level in RPL and controls measured by Illumina's 850 K Methylation BeadChip.
Adjusted P-values by R package ChAMP.
Furthermore, the expression of CREB5 protein was consist with mRNA level by western blot (Fig. 2e).
3.3. Hypo-DMR of CREB5 up-regulated its expression by recruiting P53 and SP1
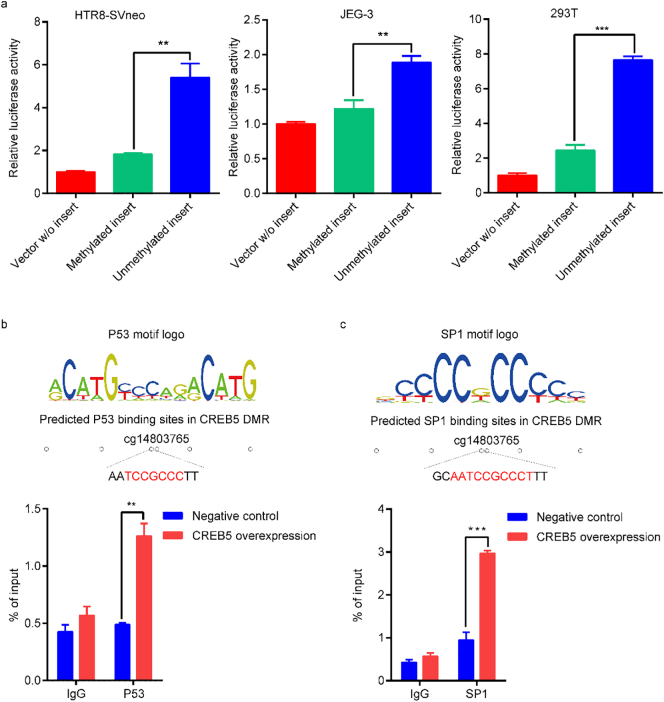
To evaluate the effect of CREB5 DMR on gene regulation, we cloned the CREB5 DMR into pGL3-basic luciferase vector. The pGL3-basic vector with methylated or unmethylated insert were co-transfected with an internal control (pGL3-SV40 vector) into the HTR8-S/Vneo cells and JEG-3 cells. Compared to the unmethylated insert vector group, we found that the luciferase activities of methylated insert vectors were significantly decreased in both HTR8-S/Vneo cells and JEG-3 cells (t = 5·42, P = 5·60*10−3 and t = 4·13, P = 4·410*10−3, respectively), suggesting that CREB5 DMR could regulate gene transcription (Fig. 3a). Given the low transfection efficiency of HTR8-S/Vneo cells and JEG-3 cells, we repeated the report assay in 293 T cells, and the results were consistent with the above two cell lines (t = 13·54, P = 1·72*10−4) (Fig. 3a).
Fig. 3.
CREB5 DMR regulates gene expression through recruiting P53 and SP1. (a) The pGL3-basic vector with methylated or unmethylated CREB5 DMR were co-transfected with pGL3-SV40 vector (negative control) in three different cell lines, HTR8-S/Vneo cell, JEG-3 cell and 293 T cell. Firefly/Renilla ratio was normalized to the empty vector. (b) ChIP-qPCR showing P53 enrichment over CREB5 DMR in HTR8-S/Vneo cells. (c) ChIP-qPCR showing SP1 enrichment over CREB5 DMR in HTR8-S/Vneo cells. **P < 0·01, ***P < 0·001. All histograms represent mean ± SEM.
To uncover CREB5 DMR's regulatory mechanisms on gene expression, PROMO was used to search the transcription factor consensus binding motifs within DMR, and P53 and SP1 binding sites were enriched and contained cg14927519 [29]. ChIP-qPCR was used to validate the P53 and SP1 binding in CREB5 DMR. In CREB5 overexpression cells, both P53 and SP1 were enriched in CREB5 DMR, indicating that P53 and SP1 might be recruited to hypo-DMR andup-regulated CREB5 (Fig. 3b, c).
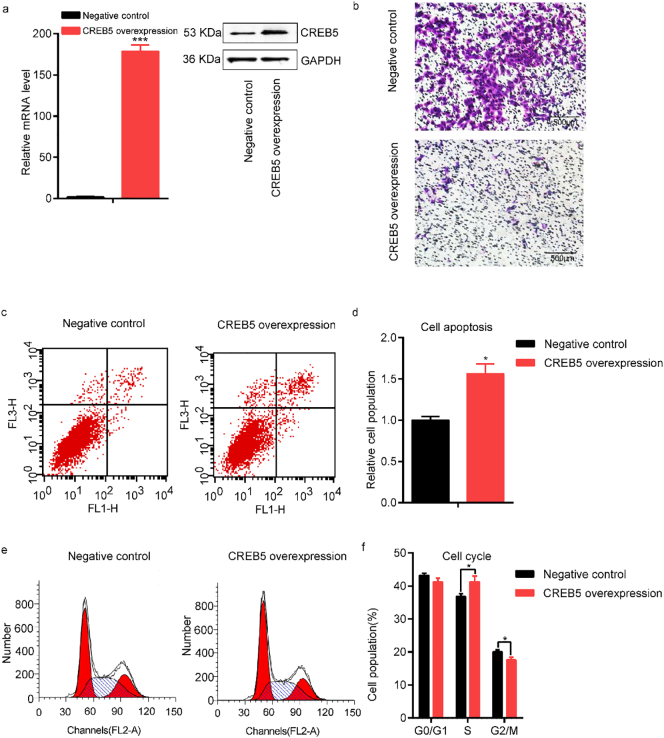
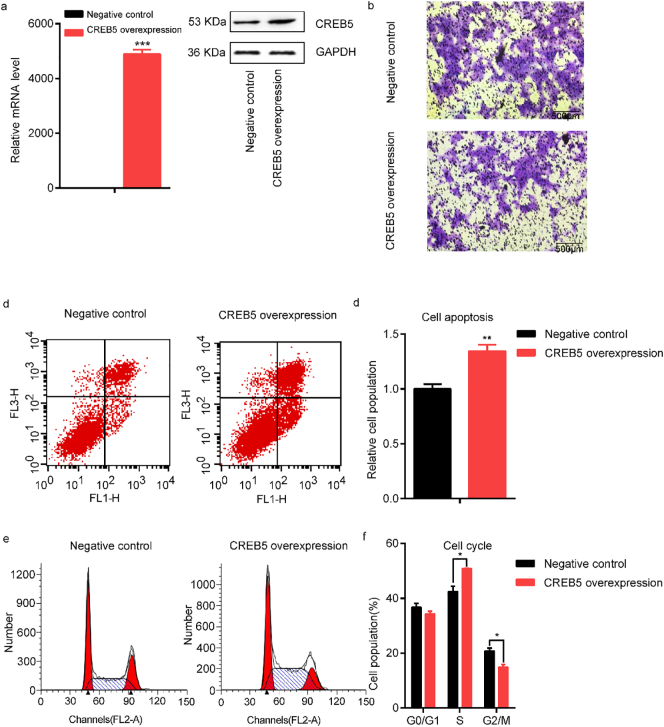
3.4. Effects of CREB5 overexpression on cell function
In an attempt to assure whether up-regulated CREB5 affects cell function, we overexpressed CREB5 in HTR8-S/Vneo and JEG-3 cell lines (Fig. 4a and Fig. 5a). Next, we assessed the effects of CREB5 overexpression on cell migration, and the number of migrated cells were significantly induced after overexpression of CREB5 (Fig. 4band Fig. 5b). Besides, by flow cytometer assays, we observed that cell apoptosis was significantly increased in HTR8-S/Vneo cells (t = 4·39, P = 1·18*10−2) (Fig. 4c, d) and JEG-3 cells (t = 4·87, P = 8·19*10−3) (Fig. 5c, d). In addition, CREB5 overexpression had effects on cell cycle with significantly increased S phase cells (t = 2·49, P = 2·83*10−2) and induced G2/M phase cells (t = 2·37, P = 3·51*10−2) in HTR8-S/Vneo cells (Fig. 4f, g), revealing that CREB5 blocked the cells in the S phase, similar results were also observed in JEG-3 cells (Fig. 5f, g).
Fig. 4.
Functional analysis of CREB5 overexpression in HTR8-S/Vneo cells. (a) Evaluation of the mRNA and protein level of CREB5 after transfection. (b) Transwell analysis of HTR8-S/Vneo cells transfected CREB5 overexpression plasmid and empty plasmid (negative control). Scale bars; 500 μm. (c and d) Assessment of cell apoptosis was detected by flow cytometer. (e and f) Analysis of the effects of CREB5 overexpression on cell cycle by flow cytometer. The data are presented as mean ± SEM. *P < 0·05, **P < 0·01, ***P < 0·001.
Fig. 5.
Functional analysis of CREB5 overexpression in JEG-3 cells. (a) qRT-PCR and western blot was used to detect the expression level of CREB5 mRNA and protein CREB5after transfection. (b) Transwell assay showed that CREB5 overexpression delayed cell migration. Scale bars; 500 μm. (c and d) The result of cell apoptosis indicated that CREB5 overexpression increased significantly the percentage of apoptotic cells. (e and f) Cell counts showed CREB5 overexpression increased significantly S-phase cells and reduced significantly G2/M-phase cells. The data are presented as mean ± SEM. *P < 0·05, **P < 0·01, ***P < 0·001.
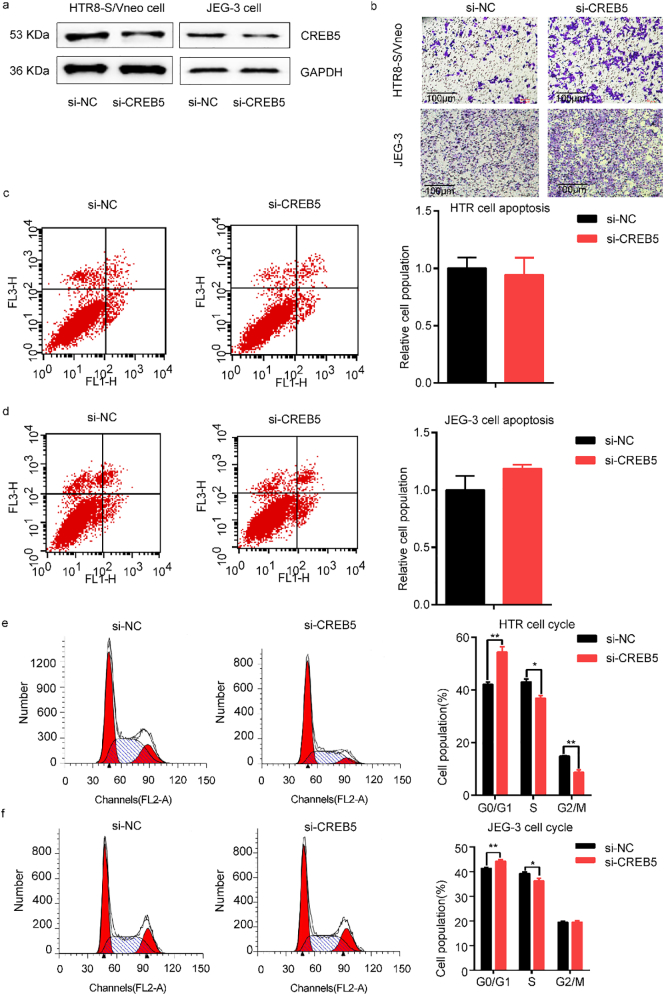
To further understand the function of CREB5, we knocked down CREB5 in HTR8-S/Vneo and JEG-3 cell lines (Fig. 6a). As shown in Fig. 6, cell migration was markedly increased after transfection of CREB5 siRNA both in HTR8-S/Vneo and JEG-3 cell lines (Fig. 6b). However, no significant difference of cell apoptosis was found in HTR8-S/Vneo and JEG-3 cells (Fig. 6c and d). Besides, CREB5 knockdown affected cell cycle both in HTR8-S/Vneo and JEG-3 cells (Fig. 6e and f), indicating that CREB5 delayed cell progression in G1 to S transition.
Fig. 6.
Functional analysis of CREB5 knockdown in HTR8-S/Vneo and JEG-3 cell lines. (a) The expression level of CREB5 protein after transfection. (b) Transwell analysis of HTR8-S/Vneo and JEG-3 cells transfected CREB5 siRNA and negative control. Scale bars; 100 μm. (c and d) The result of cell apoptosis HTR8-S/Vneo and JEG-3 cells. (e and f) Analysis of the effects of CREB5 overexpression on cell cycle by flow cytometer. The data are presented as mean ± SEM. *P < 0·05, **P < 0·01.
4. Discussion
RPL, the commonest complication of pregnancy, affecting about 1% of couples [30]. Several pregnancy complications like pre-eclampsia and intrauterine growth restriction (IUGR) are caused by defects in decidualization of the endometrium during early pregnancy, which is considered to be important in the pathogenesis of RPL. In order to explore the potential mechanism of DNA methylation involved in RPL, we carried out combined analysis of DNA methylation and gene expression, and identified CREB5 as contributor to RPL.
In current study, we performed 850 K assays to analyze the differential methylation of about 853,307 CpGs throughout the whole genome and used strict statistical criteria to define differential methylated sites between RPL and controls. Interestingly, we observed subtle concordant changes rather than all-or-none methylation changes in multiple CpGs distributed at specific gene loci. To further investigate the influence of DNA methylation on transcriptional regulation, we employed RNA-seq and found a set of genes that were hypo-methylated and up-regulated in RPL, including CERB5, RBM24, DPYSL4 and IRF4. Considering the levels of DMRs methylation and gene expressions, we finally selected CREB5 as a candidate gene for further study.
Transcriptome analysis showed that dysregulation of genes associated with cell invasion and immunity might contribute to idiopathic RPL [31]. CREB5 gene, belonging to the CRE (cAMP response element)-binding protein family, has crucial roles in regulating cell growth, proliferation, differentiation and cell cycle [32, 33]. Recently, relevant studies have suggested that the up-regulation of CREB5 is negatively correlated with the prognosis of epithelial ovarian cancer and non-small cell lung cancer and accelerate the metastasis of cancers [33, 34]. In addition, modifications of CREB5 methylation and expression were reported to be associated with plasma interleukin-6 (IL-6) levels and map to the network of inflammatory response genes [35]. Notably, knockdown CREB5 in monocytes could increase tumor necrosis factor alpha (TNF-α) level, decrease interleukin-10 (IL-10) level in cell-free plasma and enhance expression of phosphorylated–NF-κB p65 and NF-κBp65, thus causing immunosuppression [36]. Moreover, several investigations suggested the functions of CREB in immune responses, including macrophage survival, regulation of T and B lymphocytes and inducing transcription of immune-related genes, for example, IL-10 [37]. Thus, we speculate that CREB5 hypo-methylation may contribute to pregnancy immunity regulation and upregulation of CREB5 in the decidua tissue may be associated with the occurrence of RPL. However, the discussion is speculative because we do not demonstrate in which cell type CREB5 is expressed or deregulated at the feto-maternal interface.
DNA hypo-methylation, particularly in promoter and enhancer regions, is often accompanied with increased binding of transcription factors [38]. We found p53 and SP1 might bind to DMR near CREB5, which were validated by ChIP-qPCR. SP1 is a widely expressed transcription factor and plays critical roles in transcriptional regulation by binding to GC-rich cis-elements in many promoter regions [39]. SP1 can also interact with other transcription factors like methyl transferases, acetylases/de-acetylases, and other chromatin modifying molecules, which are involved in numerous cellular processes including cell growth, apoptosis, immune responses and so on [40]. P53, a transcription factor and tumor suppressor, plays a vital role in the cell cycle and functions as an important signaling hub for the cellular response to various stresses [41, 42]. Both of SP1 and P53 have effects on cell function, which are consist with our functional analysis of CREB5 overexpression. These findings support our speculation that hypo-DMR near CREB5 could up-regulate CREB5 gene expression by recruiting P53 and SP1, thus increasing cell migration and cell apoptosis, blocking cell cycle and eventually resulting in RPL by causing immune abnormalities.
In conclusion, in this study, we found a set of genes relevant to RPL by integrating analysis of DNA methylation and gene expression. Hypo methylation of CREB5 could recruit P53 and SP1 binding to CREB5 DMR and upregulate CREB5 in turn, which altered trophoblast cell functions. Our results highlight that CREB5 may play a critical role in the pathogenesis of RPL.
Conflict of Interest statement
None declared.
Author contributions
CL directed the study, obtained financial support and were responsible for study design. MY performed overall project management with GD, performed statistical analysis with QX, ZH and XH, and drafted the initial manuscript. YQ and LH were responsible for sample preparation. YF, YZ, XH, ZJ, YX and XW conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Funding
National Natural Science Foundation of China (81671461, 81402706 and 81502832), the Priority Academic Program for the Development of Jiangsu Higher Education Institutions.
Acknowledgements
This study was supported by grants from National Natural Science Foundation of China (81671461, 81402706 and 81502832) and the Priority Academic Program for the Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.07.042.
Appendix A. Supplementary data
Supplementary material table S1
Supplementary material table S2
Supplementary material table S3
Supplementary material table S4
Supplementary material fig. S1 and S2
References
- 1.Rai R., Regan L. Recurrent miscarriage. Lancet. 2006;368:601–611. doi: 10.1016/S0140-6736(06)69204-0. [DOI] [PubMed] [Google Scholar]
- 2.Li T.C., Makris M., Tomsu M., Tuckerman E., Laird S. Recurrent miscarriage: aetiology, management and prognosis. Hum Reprod Update. 2002;8:463–481. doi: 10.1093/humupd/8.5.463. [DOI] [PubMed] [Google Scholar]
- 3.Allison J.L., Schust D.J. Recurrent first trimester pregnancy loss: revised definitions and novel causes. Curr Opin Endocrinol Diabetes Obes. 2009;16:446–450. doi: 10.1097/MED.0b013e3283327fc5. [DOI] [PubMed] [Google Scholar]
- 4.Norwitz E.R., Schust D.J., Fisher S.J. Implantation and the survival of early pregnancy. N Engl J Med. 2001;345:1400–1408. doi: 10.1056/NEJMra000763. [DOI] [PubMed] [Google Scholar]
- 5.Krieg S.A., Fan X., Hong Y. Global alteration in gene expression profiles of deciduas from women with idiopathic recurrent pregnancy loss. Mol Hum Reprod. 2012;18:442–450. doi: 10.1093/molehr/gas017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 7.Fulka H., Mrazek M., Tepla O., Fulka J.J. DNA methylation pattern in human zygotes and developing embryos. Reproduction. 2004;128:703–708. doi: 10.1530/rep.1.00217. [DOI] [PubMed] [Google Scholar]
- 8.Wu H., Zhang Y. Early embryos reprogram DNA methylation in two steps. Cell Stem Cell. 2012;10:487–489. doi: 10.1016/j.stem.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Messerschmidt D.M., Knowles B.B., Solter D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 2014;28:812–828. doi: 10.1101/gad.234294.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cutfield W.S., Hofman P.L., Mitchell M., Morison I.M. Could epigenetics play a role in the developmental origins of health and disease? Pediatr Res. 2007;61:68R–75R. doi: 10.1203/pdr.0b013e318045764c. [DOI] [PubMed] [Google Scholar]
- 11.Schaefer S., Nadeau J.H. The genetics of epigenetic inheritance: modes, molecules, and mechanisms. Q Rev Biol. 2015;90:381–415. doi: 10.1086/683699. [DOI] [PubMed] [Google Scholar]
- 12.Blair J.D., Yuen R.K., Lim B.K., McFadden D.E., von Dadelszen P., Robinson W.P. Widespread DNA hypomethylation at gene enhancer regions in placentas associated with early–onset pre–eclampsia. Mol Hum Reprod. 2013;19:697–708. doi: 10.1093/molehr/gat044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feinberg A.P. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 14.Reamon Buettner S.M., Borlak J. A new paradigm in toxicology and teratology: altering gene activity in the absence of DNA sequence variation. Reprod Toxicol. 2007;24:20–30. doi: 10.1016/j.reprotox.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Hanna C.W., McFadden D.E., Robinson W.P. DNA methylation profiling of placental villi from karyotypically normal miscarriage and recurrent miscarriage. Am J Pathol. 2013;182:2276–2284. doi: 10.1016/j.ajpath.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 16.Reichetzeder C., Dwi P.S., Li J., Hocher B. Developmental origins of disease – crisis precipitates change. Cell Physiol Biochem. 2016;39:919–938. doi: 10.1159/000447801. [DOI] [PubMed] [Google Scholar]
- 17.Hanna C.W., McFadden D.E., Robinson W.P. DNA methylation profiling of placental villi from karyotypically normal miscarriage and recurrent miscarriage. Am J Pathol. 2013;182:2276–2284. doi: 10.1016/j.ajpath.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 18.Zheng H.Y., Tang Y., Niu J. Aberrant DNA methylation of imprinted loci in human spontaneous abortions after assisted reproduction techniques and natural conception. Hum Reprod. 2013;28:265–273. doi: 10.1093/humrep/des358. [DOI] [PubMed] [Google Scholar]
- 19.Reichetzeder C., Dwi P.S., Pfab T. Increased global placental DNA methylation levels are associated with gestational diabetes. Clin Epigenetics. 2016;8:82. doi: 10.1186/s13148-016-0247-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dwi P.S., Reichetzeder C., Meixner M., Liere K., Slowinski T., Hocher B. DNA methylation of the glucocorticoid receptor gene promoter in the placenta is associated with blood pressure regulation in human pregnancy. J Hypertens. 2017;35:2276–2286. doi: 10.1097/HJH.0000000000001450. [DOI] [PubMed] [Google Scholar]
- 21.Hou W., Li Z., Li Y. Correlation between protein expression of FOXP3 and level of FOXP3 promoter methylation in recurrent spontaneous abortion. J Obstet Gynaecol Res. 2016;42:1439–1444. doi: 10.1111/jog.13076. [DOI] [PubMed] [Google Scholar]
- 22.Lucas E.S., Dyer N.P., Murakami K. Loss of endometrial plasticity in recurrent pregnancy loss. Stem Cells. 2016;34:346–356. doi: 10.1002/stem.2222. [DOI] [PubMed] [Google Scholar]
- 23.Ankolkar M., Patil A., Warke H. Methylation analysis of idiopathic recurrent spontaneous miscarriage cases reveals aberrant imprinting at H19 ICR in normozoospermic individuals. Fertil Steril. 2012;98:1186–1192. doi: 10.1016/j.fertnstert.2012.07.1143. [DOI] [PubMed] [Google Scholar]
- 24.Dobin A., Davis C.A., Schlesinger F. STAR: ultrafast universal RNA–seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA–seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kellis M., Wold B., Snyder M.P. Defining functional DNA elements in the human genome. Proc Natl Acad Sci U S A. 2014;111:6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang H., Wang F., Dyer N.P., Wong W.H. CisGenome browser: a flexible tool for genomic data visualization. Bioinformatics. 2010;26:1781–1782. doi: 10.1093/bioinformatics/btq286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan A., Fornes O., Stigliani A. JASPAR 2018: update of the open–access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46:D260–D266. doi: 10.1093/nar/gkx1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Messeguer X., Escudero R., Farre D., Nunez O., Martinez J., Alba M.M. PROMO: detection of known transcription regulatory elements using species–tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 30.Menasha J., Levy B., Hirschhorn K., Kardon N.B. Incidence and spectrum of chromosome abnormalities in spontaneous abortions: new insights from a 12–year study. Genet Med. 2005;7:251–263. doi: 10.1097/01.gim.0000160075.96707.04. [DOI] [PubMed] [Google Scholar]
- 31.Krieg S.A., Fan X., Hong Y. Global alteration in gene expression profiles of deciduas from women with idiopathic recurrent pregnancy loss. Mol Hum Reprod. 2012;18:442–450. doi: 10.1093/molehr/gas017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zu Y.L., Maekawa T., Nomura N., Nakata T., Ishii S. Regulation of trans–activating capacity of CRE–BPa by phorbol ester tumor promoter TPA. Oncogene. 1993;8:2749–2758. [PubMed] [Google Scholar]
- 33.He S., Deng Y., Liao Y., Li X., Liu J., Yao S. CREB5 promotes tumor cell invasion and correlates with poor prognosis in epithelial ovarian cancer. Oncol Lett. 2017;14:8156–8161. doi: 10.3892/ol.2017.7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi L., Ding Y. Involvement of the CREB5 regulatory network in colorectal cancer metastasis. Yi Chuan. 2014;36:679–684. doi: 10.3724/SP.J.1005.2014.0679. [DOI] [PubMed] [Google Scholar]
- 35.Nevalainen T., Kananen L., Marttila S. Transcriptomic and epigenetic analyses reveal a gender difference in aging–associated inflammation: the Vitality 90+ study. Age (Dordr) 2015;37:9814. doi: 10.1007/s11357-015-9814-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long X., Li Y., Qiu S., Liu J., He L., Peng Y. MiR–582–5p/miR–590–5p targeted CREB1/CREB5–NF–kappaB signaling and caused opioid–induced immunosuppression in human monocytes. Transl Psychiatry. 2016;6 doi: 10.1038/tp.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lv X.F., Sun L.L., Cui C.L., Han J.S. NAc Shell Arc/Arg3.1 Protein Mediates Reconsolidation of Morphine CPP by Increased GluR1 Cell Surface Expression: Activation of ERK–Coupled CREB is Required. Int J Neuropsychopharmacol. 2015:18. doi: 10.1093/ijnp/pyv030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozanne S.E., Constancia M. Mechanisms of disease: the developmental origins of disease and the role of the epigenotype. Nat Clin Pract Endocrinol Metab. 2007;3:539–546. doi: 10.1038/ncpendmet0531. [DOI] [PubMed] [Google Scholar]
- 39.Koizume S., Miyagi Y. Diverse Mechanisms of Sp1–Dependent Transcriptional Regulation Potentially Involved in the Adaptive Response of Cancer Cells to Oxygen–Deficient Conditions. Cancers (Basel) 2015;8 doi: 10.3390/cancers8010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boumber Y.A., Kondo Y., Chen X. An Sp1/Sp3 binding polymorphism confers methylation protection. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greenblatt M.S., Bennett W.P., Hollstein M., Harris C.C. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 42.Soussi T., Legros Y., Lubin R., Ory K., Schlichtholz B. Multifactorial analysis of p53 alteration in human cancer: a review. Int J Cancer. 1994;57:1–9. doi: 10.1002/ijc.2910570102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material table S1
Supplementary material table S2
Supplementary material table S3
Supplementary material table S4
Supplementary material fig. S1 and S2