Abstract
The Deepwater Horizon (DWH) blowout resulted in the deposition to the seafloor of up to 4.9% of 200 million gallons of oil released into the Gulf of Mexico. The petroleum hydrocarbon concentrations near the wellhead were high immediately after the spill, but returned to background levels a few years after the spill. Microbial communities in the seafloor are thought to be responsible for the degradation of hydrocarbons, however, our knowledge is primarily based upon gene diversity surveys and hydrocarbon concentration in field sediment samples. Here, we investigated the oil degradation potential and changes in bacterial community by amending seafloor sediment collected near the DWH site with crude oil and both oil and Corexit dispersant. Polycyclic aromatic hydrocarbons were rapidly degraded during the first 30 days of incubation, while alkanes were degraded more slowly. With the degradation of hydrocarbons, the relative abundances of Colwelliaceae, Alteromonadaceae, Methylococales, Alcanivorax, Bacteriovorax, and Phaeobacter increased remarkably. However, the abundances of oil-degrading bacteria changed with oil chemistry. Colwelliaceae decreased with increasing oil degradation, whereas Alcanivorax and Methylococcales increased considerably. We assembled seven genomes from the metagenome, including ones belonging to Colwellia, Alteromonadaceae, Rhodobacteraceae, the newly reported genus Woeseia, and candidate phylum NC10, all of which possess a repertoire of genes for hydrocarbon degradation. Moreover, genes related to hydrocarbon degradation were highly enriched in the oiled treatment, suggesting that the hydrocarbons were biodegraded, and that the indigenous microflora have a remarkable potential for the natural attenuation of spilled oil in the deep-sea surface sediment.
Introduction
The Deepwater Horizon (DWH) oil spill is the largest accidental oil spill in the history of the petroleum industry, releasing about 780 million liters of light Louisiana crude oil into the Gulf of Mexico between April and July 2010 [1]. Discharged from the wellhead at 1500 m, the oil impacted large areas of the deep-sea and surface environments, marshes, and sandy beach shorelines. In addition, 7 million liters of Corexit dispersants were applied on the surface and subsurface environments, constituting the largest ever remediation response to an oil spill [2]. However, the fate of the oil remnants and the impact of the spilled oil are still being determined.
Estimates indicate that 11–30% of the spilled oil was unaccounted for, and a large fraction of this might have been deposited in the seafloor [3]. Chanton et al. [4] calculated that 3.0–4.9% of the oil released from the DWH was deposited in a 2.4 × 1010 m2 region of sediments surrounding the wellhead, based on the absence of 14C tracer; Valentine et al. [5] estimated 4–31% of the oil was deposited over a 3200 km2 area based on chemical tracers (hopane). High levels of polycyclic aromatic hydrocarbons (PAHs) (>24,000 μg kg−1) were detected in sediments near the wellhead following the spill, compared to those in distant cores (50 μg kg−1) [6]. Moreover, hydrocarbon concentrations were higher during 2010–2011 compared to years prior to 2010 [7, 8]. Oil deposition was likely facilitated by contact of the deep plume with continental slope surface sediments, and sinking of oil-rich marine snow [9, 10]. However, 1–3 years after the spill, PAH concentrations in many of these surface sediments were similar to pre-DWH conditions [11]. This depletion can be attributed to the combined effects of dissolution and biodegradation [12, 13]. In fact, microbially sourced radiocarbon-depleted organic molecular species were observed in the water column 4 years after the spill event, potentially accounting for a large proportion of “missing” spilled hydrocarbon [14].
Seafloor sediments harbor diverse and abundant microbial communities comparable to those in seawater [15]. In the northern Gulf of Mexico (nGOM) these bacteria are commonly influenced by hydrocarbon-rich cold-seeps and likely have developed the abilities to metabolize hydrocarbons [16, 17]. Oil-impacted seafloor sediments sampled 5–6 months after the DWH were enriched in uncultured Gammaproteobacterium and Colwellia species, and genes involved in degrading aliphatic and aromatic hydrocarbons [18, 19]. Genes associated with denitrification also increased significantly in highly contaminated sediments. Other sediments collected at similar times found aerobic microbial communities and associated functional genes present in all samples [20]. However, Deltaproteobacteria and anaerobic hydrocarbon-degradation genes were enriched only in sediments closer to the DWH site.
The hydrocarbon degradation abilities of bacterial communities have been widely studied in the deep-sea oil plume, but the deep-sea sediments have received much less attention. Previous studies sampled sediments, analyzed bacterial communities or metagenomes, and correlated the bacterial community to their degradation potential or residual oil [7, 18–22]. However, there is no systematic study reporting the abilities of the seafloor microbial communities to degrade different groups of hydrocarbons. Here, we amended deep-sea surface sediment collected near the DWH site with light Louisiana sweet crude oil and/or Corexit dispersant, and incubated the samples at in situ temperature for 180 days. We analyzed the residual hydrocarbons, bacterial community, and determined the functional genes to address the following questions: (1) How quickly does the crude oil biodegrade in deep-sea sediment? (2) How does the bacterial community composition change when exposed to crude oil? (3) Are functional genes associated with hydrocarbon degradation enriched in the presence of oil in deep-sea sediment? Our findings revealed that PAHs were more rapidly degraded than alkanes within 30 days, whereas alkane degradation continued until 180 days. Furthermore, increased hydrocarbon degradation corresponds to increased abundances of several members of Alteromonadales (Colwelliaceae and Alteromonadaceae), Methylococcales, Oceanospirillales (Alcanivorax, Olephilaceae, and Oceanospirillaceae), Rhodanobacterales (Phaeobacter and Octadecabacter), Bacteriovorax, and a myriad of hydrocarbon degradation genes.
Materials and methods
Sediment sampling
Sediment was collected by multicore aboard the R/V Pelican on June 25, 2013, from Station C12 (28.6381°N, 88.5192 °W, 1600 m deep) in the nGOM about 17 km away from the DWH Site [11]. The temperature, salinity, and dissolved oxygen of the overlying water were 4.4 °C, 35 ppt, and 4.6 mg L−1, respectively. The background PAHs concentration (ΣPAH43) at this site was ca. 110 ng g−1 sediment [11].
Incubation
On board, the top three centimeters of the sediment core were extruded immediately after collection then homogenized in a sterile beaker using a sterile metal spoon. Equal volumes of homogenized sediment (6 cm3) were transferred to 60-mL wide-mouth amber glass jars. Three treatments were prepared (Table 1): Sediment amended with light Louisiana sweet (LLS) crude oil and Corexit (SOC), sediment plus overlying water amended with both oil and Corexit (SWOC), and sediment plus overlying water amended with oil only (SWO). Before addition, the overlying water was serially filtered through 5 and 0.20 µm membranes to remove bacteria. The crude oil was added from a stock of crude oil in seawater [23, 24], and stirred to mix. Corexit 9500A dispersant was added at a 1:20 dispersant:oil ratio. Controls without added substrate were prepared for both the sediment only (S) and sediment plus overlying water (SW). All bottles were tightly sealed using Teflon-lined caps to minimize losses due to volatilization.
Table 1.
Description of experimental treatments
| Treatment | Description |
|---|---|
| S | Sediment only (Control) |
| SOC | Sediment + oil + Corexit |
| SW | Sediment + water (Control) |
| SWOC | Sediment + water + oil + Corexit |
| SWO | Sediment + water + oil |
Incubation was performed in sealed 60-mL wide-mouth amber glass jars containing 6 cm3 of sediment. Crude oil was added at a final concentration of 200 ppm (mass/volume).
The jars were incubated in the dark at 4–4.5 °C without shaking. Three replicate jars for each treatment were sampled for hydrocarbon and bacterial analysis after 15, 30, 60, 120, and 180 days. Only one replicate for controls was prepared for each sampling period, solely for bacterial analysis. Abiotic controls were not included, primarily because sediment quantities from the cores were limited, and we prioritized the experimental treatments. Our previous work showed that abiotic losses are negligible under our standard incubation conditions [24–28]. Our incubation bottles are tightly sealed with Teflon-lined caps to prevent evaporation. We did not observe appreciable losses in previous incubation experiments at 25–30 °C, and the ~4 °C incubation temperature in this study should further reduce any evaporative loss. The other potential abiotic loss of hydrocarbons is through sorption, as hydrocarbons are hydrophobic and can be sorbed to sediments irreversibly. However, deuterated standards added before the extraction of the samples were all recovered in excellent rates (85–97%), consistent with our previous work [7, 25, 29, 30], suggesting minimal loss due to sorption. Overall, we do not think abiotic loss was significant in our incubations.
Oil extraction and analysis
The entire volume of sample was freeze-dried and extracted using dichloromethane in a Soxhlet extractor for 24 h. The extract was filtered through a chromatographic column packed with anhydrous sodium sulfate [7], concentrated using Rotovap, and exchanged with hexane. Sample clean-up and fractionation were performed according to Wang et al. [31]. The saturated fraction was eluted with hexane while the aromatic fraction was eluted with hexane in benzene (1:1). Aliphatic hydrocarbons (C11–C35), 17α(H), 21β(H)-hopane (C30-hopane), and PAHs were analyzed using GC–MS (Shimadzu QP2010 plus) operated in selected ion mode (SIM) [24, 29]. Hydrocarbons were quantified by a five-point calibration curve of known mixed standards of alkanes and PAHs. The concentration of each compound was normalized to C30-hopane [32, 33]. The deuterated surrogate standards (hexadecane-d34, acenapthene-d10, phenanthrene-d10, pyrelene-d12, benzo[e]pyrene- d12) added prior to extraction had a recovery rate of 85–97%.
DNA extraction, sequencing, and analysis
Total genomic DNA was extracted from 250 mg of freeze-dried samples using the MO BIO Powersoil DNA Isolation Kit. The 400 bp V4/V5 region of the bacterial 16S rRNA gene was amplified using the primers Hyb515F_rRNA and Hyb909R_rRNA, and sequenced using the Illumina MiSeq platform at the University of Texas Genomic Sequencing and Analysis Facility (UTGSAF) [34].
At least 10,000 2 × 250 bp paired-end sequences were generated from each sample. After trimming primer sequences, Pandaseq (v2.8) was used to assemble forward and reverse reads into one contiguous sequence [35]. The contiguous sequences were assigned to operational taxonomic units (OTUs) via the closed reference-based OTU picking method in QIIME (v1.7.0) [36], using the Greengenes database (version 13_5) at 97% similarity [37]. Taxonomic classification was carried out using the RDP classifier in QIIME and the Greengenes database.
Genomic assembly, binning, and annotation
DNA samples of control (S-30) and oil treatment (SOC-30) collected at 30 days of incubation were sequenced by Illumina HiSeq 4000 at UTGSAF, resulting in about 120 million reads of 2 × 150 paired-end sequences per sample. Assembly, binning, and annotation was performed following an established protocol [38] with some modifications. Briefly, shotgun genomic reads were screened for adapter using Cutadapt [39], interleaved using interleave_fasta.py, and trimmed at both ends using Sickle (https://github.com/najoshi/sickle). Paired-end reads were assembled using IDBA-UD [40] with the following parameters (–pre_correction –mink 55 –maxk 95 –step 10 –seed_kmer 55). Assembled 1 kb fragments were submitted to JGI IMG/MER system [41] for annotation. The predicted proteins were further compared to proteins involved in hydrocarbon degradation in a custom database using blastp [42]. The resulting output was used to determine the relative abundances of genes in the control (S-30) and treatment (SOC-30).
Binning of the assembled fragments was performed using tetra-nucleotide frequency signatures using 4.5 kb fragments of the contigs [43]. Emergent Self-Organizing Maps were manually delineated and curated based on clusters within the map. Binning was enhanced by incorporating reference genomes for all assembled contigs into the Emergent Self-Organizing Maps [43]. The completeness, contamination, and strain heterogeneity of the genomes within bins were estimated using CheckM [44]. The distribution of the genes for hydrocarbon degradation was determined by comparing the predicted proteins to a custom database. The genomes are available in NCBI GenBank under BioProject ID PRJNA434806 and accession numbers SAMN08581018–SAMN08581024.
Statistical analysis
Non-metric multidimensional scaling (NMDS) was used to visualize the similarity of the bacterial community structures in control and treatments. NMDS was performed using Bray–Curtis distances in the PAST software package, V2.17 [45]. Treatments were then compared using one-way analysis of similarity (ANOSIM) to verify the significance of the clustering [30]. The hydrocarbon biodegradation rate constant was computed based on first-order kinetic model [25, 30].
Results
Hydrocarbon degradation
PAHs were degraded faster than alkanes in all treatments during the first 30 days of incubation (Fig. 1a–c). In SOC, only 30 and 21% of PAHs and alkylated PAHs remained at day 20, while 60% of alkanes were not transformed. Moreover, the percent degradation of alkanes and PAHs was higher at all time points in SOC compared to both slurry treatments (SWO and SWOC). Corexit enhanced the degradation of alkanes at 120 days, but did not affect PAH degradation. While alkane degradation continued until 180 days, PAH degradation was negligible after 60 days. The degradation rate constant (k) of alkanes was higher in SOC than SWOC, which was higher than SWO (Figure S1A). Whereas the rate constants of the PAHs and alkylated PAHs varied, SOC generally showed the highest degradation rate for most PAHs, and the rate of SWO was slightly higher than that of SWOC (Figure S1B). Note also that rate constants of PAHs and alkanes with lower molecular weights were generally higher than those with higher molecular weights in all treatments.
Fig. 1.
Biodegradation of (a) total alkanes, (b) total PAHs, and (c) total alkalyted PAHs in deep-sea sediment amended with crude oil. SOC sediment + oil + Corexit, SWOC sediment + water + oil + Corexit, SWO sediment + water + oil
Bacterial community structure and dynamics
Incubations in the presence or absence of oil showed shifts in bacterial community structure, but the effect of oil addition was much greater (Fig. 2). Control communities (S and SW) do not vary from each other (P = 0.1212), but differ significantly from oiled treatments (P = 0.0001) (Table S1). There was no significant difference between the slurry treatments with oil alone or with both oil and dispersant (P = 0.8865). Moreover, sediment alone with both oil and Corexit was not different from the sediment–water slurry (P = 0.8272).
Fig. 2.
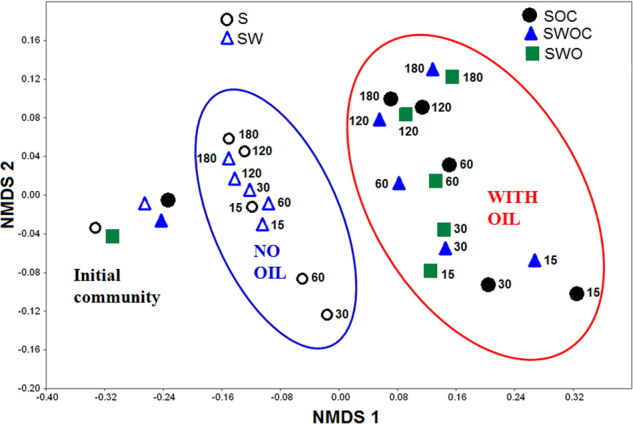
Non-metric multidimensional scaling (NMDS) of bacterial community structures in deep-sea sediment incubated with or without crude oil. The ordination is based on Bray–Curtis coefficient matrix of relative abundances. S sediment only, SOC sediment + oil + Corexit, SW sediment + water, SWOC sediment + water + oil + Corexit, SWO sediment + water + oil. The numbers indicate days of incubation
The initial bacterial community was comprised mainly of Gammaproteobacteria, Deltaproteobacteria, Alphaproteobacteria, Planctomycetes, Acidobacteria, and Actinobacteria (Fig. 3). Changes in bacterial community structure, most notably the Gammaproteobacteria, were observed both in control and oil-amended treatments. On average, Gammaproteobacteria abundance in the controls increased by 1.2-fold, whereas their abundances in the oil treatments increased by about two-fold. At the order level, the increase of Gammaproteobacteria was mainly due to Alteromonadales, Oceanospirillales, and Methylococcales (Figure S2). Besides the Gammaproteobacteria, Rhodobacterales (Alphaproteobacteria), and Bdellovibrionales (Deltaproteobacteria) increased in oil treatments.
Fig. 3.
Bacterial community dynamics at class level in deep-sea sediment incubated with or without crude oil. S sediment only, SOC sediment + oil + Corexit, SW sediment + water, SWOC sediment + water + oil + Corexit, SWO sediment + water + oil. The numbers in the horizontal axis indicate the days of incubation
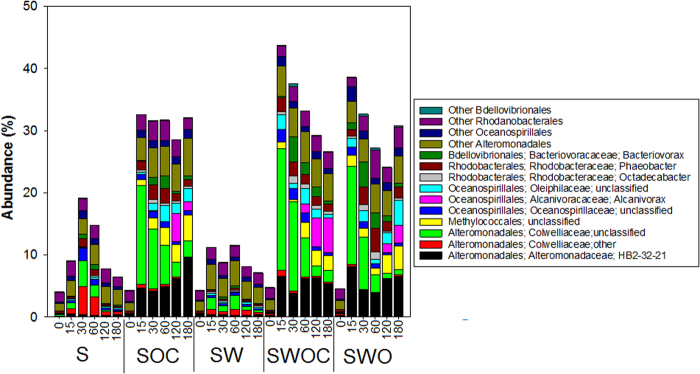
Alteromonadales sequences included Altermonadaceae closely affiliated to HB2-32-21, which increased 2–3-fold in control and 20–34-fold in treatment, and two genera belonging to Colwelliaceae (Fig. 4). The unclassified Colwelliaceae responded greatly to oil amendment during the initial 15 days, increasing up to 100-fold, and then decreased steadily. This gradual decrease of Colwelliaceae correlated with an increase in the abundances of unclassified Methylococcales (50-fold at 15 days to 180-fold at 180 days), Alcanivorax (10-fold at 30 days to 700 to 900-fold at 180 days), and Oleiphilaceae. The increase of other Colwelliaceae in S-control at 30–60 days of incubation comprised more than 50% of the total Colwelliaceae, but this OTU did not seem to metabolize oil as they showed a negligible increase in the oil treatment. Bacteriovorax increased greatly at 30 and 60 days by 40-fold to 300-fold, respectively, in the treatments, yet only about 10–20-fold in control. Although present in relatively low abundance, Phaeobacter and Octadecabacter increased by 10-fold with oil addition.
Fig. 4.
Abundances of genera under orders Alteromonadales, Methylococcales, Oceanospirillales, Rhodanobacterales, and Bdellovibrionales that showed increase in deep-sea sediment incubated with or without crude oil. S sediment only, SOC sediment + oil + Corexit, SW sediment + water, SWOC sediment + water + oil + incubation with or without crude Corexit, SWO sediment + water + oil. The numbers in the horizontal axis indicate the days of incubation
Abundance of hydrocarbon-degradation genes
Metagenomic analysis showed that a variety of genes involved in hydrocarbon degradation were enriched in the oil treatment (SOC-30) relative to the control (S-30) (Fig. 5). Genes associated with initial oxidation of aromatic rings (benzene dioxygenase to aromatic ring hydroxylating dioxygenase) were generally more abundant than alkane monooxygenase in the control. However, while these aromatic genes increased on average by 2.5-fold, alkane monooxygenase increased by 4.5-fold in the treatment. Undetected in the control, toluene, methan, and butane dioxygenases emerged in the presence of oil. Moreover, genes involved in metabolism of the by-products of hydrocarbon degradation, including alcohol and cyclohexanol dehydrogenases, and those relating to fatty acid metabolism are the most abundant. A set of genes involved in the complete oxidation of alkanes was abundant in the deep-sea sediment and increased after oil addition. Similarly, we found a nearly complete set of genes for the degradation of cyclohexane (alkane monooxygenase, cyclohexanol dehydrogenase, cyclohexanone monoxygenase, and fatty acid metabolism). Catechol and benzoate dioxygenases, two genes commonly responsible for the total oxidation of aromatic rings, were detected at lower abundance.
Fig. 5.
Abundance of genes involves in hydrocarbon degradation in the metagenome data of sediment + oil + Corexit (SOC-30) and sediment only (S-30) sampled at 30 days of incubation
Genes associated with anaerobic metabolism were also enriched. Abundances of denitrifying genes, such as nitrate reductase, nitrite reductase, nitric oxide, and nitrous oxide reductases increased 4-fold in the presence of oil compared to the control (Figure S3). Notably, the genetic potential for anaerobic alkane and aromatic degradation was indicated by a slight increase in alkysuccinate synthase (assA) and benzylsuccinate synthase (bssA), respectively. Moreover, benzoyl-CoA reductase, another key enzyme of anaerobic aromatic metabolism, increased when oil was added [46].
Reconstructed genomes
Assembly and binning of metagenomic data resulted in reconstruction of seven genomic bins (designated Bin 6, 8, 17, 18, 19, 20, and 21) from the oiled treatment (Table 2). These bins have an estimated completeness from 17 to 50%, and low contamination (<10%). Phylogenetic analyses of 15 concatenated ribosomal proteins identified Bin 20 as a Gammaproteobacterium belonging to genus Colwellia and closely related to Bin 37 of Dombrowski et al. [42] (Fig. 6). The 46% complete Bin 21 was also closely affiliated to Colwellia, but distinct from Bin 20. Both genomes possess naphthalene dioxygenase, but lack genes for the degradation of natural gas and benzene, toluene, ethylbenzene, and xylene (BTEX). Bin 8 (50% complete) was similar to Woeseia oceani XK5, a new genus in the order Chromatiales identified from coastal sediment [47]. This is the first study to report the presence of this genus in the deep-sea sediment. Bin 8 harbors a repertoire of genes for complete degradation of naphthalene and initial oxidation of BTEX compounds. Bin 17 belongs to Candidate Phylum NC10 whose members are linked to anaerobic oxidation of methane coupled to denitrification [48, 49]. Genomes of Bin 6 (Alteromonadales), Bin 18 (unidentified), and Bin 19 (Rhodobacteraceae) possess a myriad of aromatic degradation genes, which are thought to be involved in PAH degradation. Alkane monooxygenase, the key enzyme in alkane degradation [50], was only present in Bins 6 and 21. Notably, Bin 21 has a complete set of pathway genes for total oxidation of alkanes. The denitrification genes narG and nirK were found in 5 of 7 reconstructed genomes.
Table 2.
Distribution of genes involved in hydrocarbon degradation in seven reconstructed genomes
Fig. 6.
Phylogenetic characterization of genomic Bin 8, Bin 17, and Bin 20
Discussion
To date, research documenting the bacterial community and potential hydrocarbon degradation of deep-sea sediment in the nGoM is relatively scarce [18–22]. In each of these studies, sediment cores were collected from different distances around the wellhead, analyzed for microbial community or metagenomes, and examined for possible metabolic or hydrocarbon degradation potential. Succession of oil-degrading bacteria and abundance of functional genes, together with the change in oil chemistry at a molecular level, were not documented in these, or, to the best of our knowledge, any other studies.
PAHs and their alkylated homologs were degraded faster than alkanes within the first 30 days in the sediment. These data agree with the results of Yergeau et al. [22] who reported high concentrations of alkanes in the 0-cm-depth sediments around the DWH wellhead, while both PAHs and methylated PAHs were below the detection limit. Using 13C-labeled dodecane, toluene, and phenanthrene, Mason et al. [19] found that dodecane was degraded faster than both phenanthrene and toluene, which is contrary to our findings. Using single substrates may alter the preferential degradation of hydrocarbons by the bacteria, thus crude oil may be more representative of oil spill situations [27]. Our data follow the typical pattern that short chain (C11–C14) alkanes were metabolized faster than mid-chain (C15–C22), and long chain >C30 were the least degraded [7, 24, 25, 29]. After the 30-day period of rapid loss, PAH degradation slowed, as the remaining 4–5 ring PAHs are more recalcitrant. However, many alkylated PAHs disappeared faster than the parent PAHs, particularly monoalkylated naphthalenes, which represent a large fraction of alkylated PAHs in LLS oil [24]. Biodegradation of all hydrocarbon groups was also faster in sediment alone, suggesting that overlying water reduced hydrocarbon degradation likely by decreasing bioavailability, as floating oil sheen was observed on the water. Using seawater alone, Kleindienst et al. [51] showed that the addition of Corexit negatively affected the degradation of both n-alkanes and PAHs. However, our results revealed that Corexit enhanced the degradation of n-alkanes, but not PAHs and alkylated PAHs, in slurry sediments.
We observed greater shifts in microbial community composition with oil as compared to controls. Although the top centimeters of the core were oxic to suboxic [22], changes in the controls were expected as oxygen was introduced during experiment preparation. Moreover, we observed changes in bacterial community in seawater only during previous incubation studies [52]. Regardless, the difference in the bacterial composition between the control and the treatment should be attributed to oil as the key driver. For example, members of Alteromonadales, Methylococcales, Oceanospirillales, Rhodobacterales, and Bdellovibrionales increased with reference to the control in the presence of oil or both oil and dispersant. The bacterial communities in our incubations share some features with communities from sediments collected following the DWH spill. Contaminated sediments near the DWH site contained abundant Gammaproteobacteria, particularly uncultured Gammaproteobacterium, Colwellia, and Rhodobacteraceae [18, 19]. These studies represent single time point collections. Here, we showed that Colwellia initially dominated, but decreased remarkably with oil degradation and was succeeded by other taxa including Alcanivorax and Methylococcales. Surface sediment collected 19 months after DWH by Yergeau et al. [22] included low abundances of Oceanospirillales, Alcanivorax, Colwellia, and Methylococcaceae, comparable to our initial sediment communities. In contrast, Kimes et al. [20] found higher levels of anaerobic Deltaproteobacteria, which were not abundant in this study or Mason et al. [19]. Our previous report on surface bacterial communities of deep-sea sediment samples found mainly Methylococcus, Vibrio, Pseudomonas, Methylobacter, Flavobacteria, and Acidobacteria [21], whereas Vibrio and Pseudomonas were not dominant in our cores. This variation in initial bacterial composition may be due to differences in sampling site or time, and degree of oil contamination and weathering.
The dynamics of several bacterial taxa correlated with degradation of oil components. Colwelliaceae and Altermonadaceae were barely detectable in the initial community, yet comprised 15–20% and 4–8% of the community after 15 days. This suggests the key roles of these bacteria played in the early stages of degradation, as 25–50% of n-alkanes C11–C15 and 60–80% of naphthalene were degraded during this period. Alteromonas is a known naphthalene degrader [53]. Colwellia in the DWH plume incorporated ethane, propane, and benzene [54], suggesting its flexibility to degrade both light alkanes and aromatics. Our results support these findings, because Colwellia abundance decreased dramatically as lower molecular weight hydrocarbons disappeared (Fig. 4; Fig. S4). Another Colwelliaceae representative was abundant in sediment controls, indicating a diversity of Colwelliaceae phylotypes in the nGoM with functions other than hydrocarbon degradation. For example, Colwellia RC25 isolated from the GoM degraded components of Corexit when incubated at 4 °C [53]. After 15 days, Alteromonas remained abundant while Colwelliaceae numbers declined, with a concurrent increase of Alcanivorax and Oleiphilaceae, which can degrade long chain alkanes and are abundant in weathered oil [54, 56, 57]. This is unsurprising as by that point the treatments were already depleted of short alkanes, leaving the mid-chain to long-chain alkanes.
The metagenomes of the control and oil treatments provide further evidence of oil degradation abilities in deep-sea sediment communities. Functional gene categories related to aliphatic and aromatic hydrocarbon metabolism were present in the sediment and increased in abundance with oil addition (Fig. 5). Genes involved in initial oxidation of aromatic compounds were more abundant than alkane oxygenase, which supports the observation that PAHs were readily degraded during the first 30 days of incubation, similar to Lu et al. [58]. The most abundant genes were alcohol dehydrogenase, cyclohexanol dehydrogenase, and those involved in fatty acid oxidation, consistent with previous observations in deep-sea sediments [19]. Like Mason et al. [19], we also found a complete set of genes for cyclohexane degradation. They report that these genes were abundant in sediments with high hydrocarbon concentrations, and here we demonstrated that these genes increased with oil addition. We observed a slight increase in genes for anaerobic hydrocarbon degradation (assA, bssA), a process that occurs very slowly [20]. This is not surprising, as while our initial conditions were oxic, areas within the sediment might become suboxic to anoxic. The 60-ml sealed jars contain about 6 cm3 of sediment, and a headspace with sufficient O2 for the complete oxidation of the quantifiable amount of hydrocarbons. The surface might be oxic throughout the incubation period, but extensive hydrocarbon degradation could cause suboxic conditions below, reflective of in situ conditions in the seafloor. Oil exposure in deep-sea sediment can also alter other metabolic pathways, such as nitrogen cycling, evidenced by the tremendous increase in denitrifying genes observed here and by Mason et al. [19]. Thus, degradation of large amounts of labile hydrocarbons could create microenvironments that enhanced denitrification under the experimental conditions.
The partial genomes of Colwelliaceae (Bin 20 and Bin 21) contained genes for denitrification similar to Colwellia psychrerythraea 34H, which is a facultative anaerobe, and to the Colwellia described by Mason et al. [59], suggesting that both aerobic and anaerobic processes occurred during incubation. This is not surprising as the surface sediment and overlying water in this area are enriched in nitrate, 7.5–10.5 µM [19, 30]. While PAH dioxygenase and naphthalene dioxygenase were abundant in four genomes (Bins 6, 8, 18, 19), alkane monoxygenase was only detected in two genomes (Bins 6 and 21). Nonetheless, a repertoire of alkane metabolite degradation genes was found in four genomes, indicating a strong likelihood that many alkanes could have been totally oxidized. Colwelliaceae Bin 21 had a broad spectrum of genes for substrate utilization that may explain its ability to degrade a variety of hydrocarbons. Notably, Bin 8 closely related to Woeseia has a nearly complete repertoire of genes for degrading BTEX compounds, aliphatic and aromatic hydrocarbons. Woeseia is a facultative, chemoheterotrophic anaerobe recently classified as a novel genus [47]; this is the first report of its ability to degrade hydrocarbons. Bin 17 is closely affiliated to candidate phylum NC10 that links anaerobic methane oxidation to denitrification [48, 49], another indication that anoxic conditions were present during incubation. The presence of nitrite and nitrate reductases and naphthalene dioxygenase in the Bin 17 genome suggests a potential for hydrocarbon degradation coupled to denitrification. NC10-related bacteria have been found in oxygen-limited freshwater habitats, lake sediments, rice paddy soils, wastewater sludge, and marine water column oxygen minimum zones [60–63], but this is the first report on NC10 bacterium in GOM deep-sea sediments, particularly related to hydrocarbon degradation. Our mass-balance calculation revealed that the oxygen in the headspace was sufficient for the complete oxidation of hydrocarbons to CO2 and H2O. However, only 20–60% of biodegraded oil could be converted to CO2, and the rest could be transformed to metabolites or be converted into biomass [27, 64]. The degradation of other organic compounds in the sediment might have also contributed to the depletion of oxygen in the incubation bottles at the later part of the experiment, resulting in the oxic surface layer and suboxic/anoxic bottom layer. The bacterial communities here are consistent with oxic to suboxic environments, corresponding to the top few centimeters of the core used in this study. Our incubations represent a model for the fate of oil deposited on surface sediments, which could undergo biodegradation due to aerobes and facultative anaerobes. This study does not represent the anoxic subsurface environment, primarily inhabited by obligate anaerobes.
In this study, we demonstrated the response of deep-sea sediment bacteria in the nGoM to light Louisiana sweet crude oil, in terms of community composition and hydrocarbon metabolism. We showed that PAHs were readily degraded when oil was added, but alkane degradation occurred more slowly. We calculated degradation rate constants based on the first-order model for alkanes, PAHs, and alkylated PAHs that can be used for modeling the fate of oil in the deep-sea surface sediment. Using 16S rRNA gene sequencing and metagenomics we showed that Colwelliacea, Altermonadaceae, Methylococcales, Rhodobacteraceae, and Alcanivorax increased in abundance with oil addition. Colwelliacea later decreased as oil degradation continued, with corresponding increases in Alcanivorax and Methylococcales. We correlated the putative roles of these bacteria in the degradation of hydrocarbons, based on changes in oil composition. Lastly, assembled metagenomes assign the degradation potential to individual bacteria within the microbial community in the sediment. Our findings are crucial in understanding the role of bacteria taxa in the fate of oil in deep-sea sediment and can inform remediation efforts in future oil spills in the region.
Electronic supplementary material
Acknowledgements
This research was made possible by grants from The Gulf of Mexico Research Initiative (GoMRI) through the research consortium on Dispersion Research on Oil: Physics and Plankton Studies (DROPPS I and II), and the Consortium on the Advanced Research of Transport of Hydrocarbon in the Environment (CARTHE). Data are publicly available through the Gulf of Mexico Research Initiative Information & Data Cooperative (GRIIDC) at https://data.gulfresearchinitiative.org (10.7266/N7Q81B3J and 10.7266/N70G3H4C). We thank Nina Dombrowski for assistance in metagenomic analysis, and Puspa Adhikari and Somiddho Bosu for help in sampling.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0190-1) contains supplementary material, which is available to authorized users.
References
- 1.Atlas RM, Hazen TC. Oil biodegradation and bioremediation: a tale of the two worst spills in U.S. history. Environ Sci Technol. 2011;45:6709–15. doi: 10.1021/es2013227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Federal Integracy Solutions Group (TFISG). Oil Budget Calculator Science and Engineering Team. Oil budget calculator deepwater horizon. 2010.
- 3.Lehr W, Bristol S, Possolo A. Federal Interagency Solutions Group, Oil budget calculator science and engineering team. Oil budget calculator. Technical document http://www.restorethegulf.gov/sites/default/files/documents/pdf/OilBudgetCalc_Full_HQ-Print_111110. pdf. 2010. Last accessed, 24 July 2014.
- 4.Chanton J, Zhao T, Rosenheim BE, Joye S, Bosman S, Brunner C. Using natural abundance radiocarbon to trace the flux of petrocarbon to the seafloor following the deepwater horizon oil spill. Environ Sci Technol. 2015;49:847–54. doi: 10.1021/es5046524. [DOI] [PubMed] [Google Scholar]
- 5.Valentine DL, Fisher GB, Bagby SC, Nelson RK, Reddy CM, Sylva SP, et al. Fallout plume of submerged oil from Deepwater Horizon. Proc Natl Acad Sci USA. 2014;111:15906–11. doi: 10.1073/pnas.1414873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.OSAT-I. Summary report for sub-sea and sub-surface oi land dispersant detection: sampling and monitoring. Paul F. Zukunft, RADM, US CoastGuard, Federal On-Scene Coordinator, Deepwater Horizon MC252. 2010.
- 7.Liu Z, Liu J, Zhu Q, Wu W. The weathering of oil after the Deepwater Horizon oil spill: insights from the chemical composition of the oil from the sea surface, salt marshes and sediments. Environ Res Lett. 2012;7:035302. doi: 10.1088/1748-9326/7/3/035302. [DOI] [Google Scholar]
- 8.Romero IC, Schwing PT, Brooks GR, Larson RA, Hastings DW, Ellis G, et al. Hydrocarbons in deep-sea sediments following the 2010 Deepwater Horizon blowout in the northeast Gulf of Mexico. PLoS ONE. 2015;10:e0128371. doi: 10.1371/journal.pone.0128371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan B, Passow U, Chanton JP, Nöthig EM, Asper V, Sweet J, et al. Sustained deposition of contaminants from the Deepwater Horizon spill. Proc Natl Acad Sci USA. 2016;113:e3332–e3340. doi: 10.1073/pnas.1513156113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ziervogel K, Joye SB, Arnosti C. Microbial enzymatic activity and secondary production in sediments affected by the sedimentation pulse following the Deepwater Horizon oil spill. Deep-Sea Res II. 2014;129:241–8. doi: 10.1016/j.dsr2.2014.04.003. [DOI] [Google Scholar]
- 11.Adhikari PL, Maiti K, Overton EB, Rosenheim BE, Marx BD. Distributions and accumulation rates of polycyclic aromatic hydrocarbons in the northern Gulf of Mexico sediments. Environ Pollut. 2016;212:413–23. doi: 10.1016/j.envpol.2016.01.064. [DOI] [PubMed] [Google Scholar]
- 12.Stout SA, Payne JR. Macondo oil in deep-sea sediments: Part 1—sub-sea weathering of oil deposited on the seafloor. Mar Pollut Bull. 2016;111:365–80. doi: 10.1016/j.marpolbul.2016.07.036. [DOI] [PubMed] [Google Scholar]
- 13.Stout SA, Payne JR, Ricker RW, Baker G, Lewis C. Macondo oil in deepsea sediments: Part 2 d distribution and distinction from background and natural oil seeps. Mar Pollut Bull. 2016;111:381–401. doi: 10.1016/j.marpolbul.2016.07.041. [DOI] [PubMed] [Google Scholar]
- 14.Walker BD, Druffel ERM, Kolasinski J, Roberts BJ, Xu X, Rosenheim BE. Stable and radiocarbon isotopic composition of dissolved organic matter in the Gulf of Mexico. Geophys Res Lett. 2017;44:8424–34. doi: 10.1002/2017GL074155. [DOI] [Google Scholar]
- 15.Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D’Hondt S. Global distribution of microbial abundance and biomass in sub- seafloor sediment. Proc Natl Acad Sci USA. 2012;109:16213–6. doi: 10.1073/pnas.1203849109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mills HJ, Martinez RJ, Story S, Sobecky PA. Identification of members of the metabolically active microbial populations associated with Beggiatoa species mat communities from Gulf of Mexico cold-seep sediments. Appl Environ Microbiol. 2004;70:5447–58. doi: 10.1128/AEM.70.9.5447-5458.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orcutt BN, Joye SB, Kleindienst S, Knittel K, Ramette A, Reitz A, et al. Impact of natural oil and higher hydrocarbons on microbial diversity, distribution, and activity in Gulf of Mexico cold-seep sediments. Deep-Sea Res Part II. 2010;57:2008–21. doi: 10.1016/j.dsr2.2010.05.014. [DOI] [Google Scholar]
- 18.Handley KM, Piceno YM, Hu P, Tom LM, Mason OU, Andersen GL, Jansson JK, Gilbert JA. Metabolic and spatio-taxonomic response of uncultivated seafloor bacteria following the Deepwater Horizon spill. ISME J. 2017;11:2569–83. doi: 10.1038/ismej.2017.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mason OU, Scott NM, Gonzalez A, Robbins-Pianka A, Bælum J, Kimbrel J, et al. Metagenomics reveals sediment microbial community response to Deepwater Horizon oil spill. ISME J. 2014;8:1464–75. doi: 10.1038/ismej.2013.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimes NE, Callaghan AV, Aktas DF, Smith WL, Sunner J, Golding BT, et al. Metagenomic analysis and metabolite profiling of deep-sea sediments from the Gulf of Mexico following the Deepwater Horizon oil spill. Front Microbiol. 2013;4:50. doi: 10.3389/fmicb.2013.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z, Liu J. Evaluating bacterial community structures in oil collected from the sea surface and sediment in the northern Gulf of Mexico after the Deepwater Horizon oil spill. Microbiol Open. 2013;2:492–504. doi: 10.1002/mbo3.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yergeau E, Maynard C, Sanschagrin S, et al. Microbial community composition, functions, and activities in the Gulf of Mexico 1 year after the Deepwater Horizon accident. Appl Environ Microb. 2015;81:5855–66. doi: 10.1128/AEM.01470-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Almeda R, Wambaugh Z, Wang Z, Hyatt C, Liu Z, Buskey EJ. Interactions between zooplankton and crude oil: Toxic effects and bioaccumulation of polycyclic aromatic hydrocarbons. PLoS ONE. 2013;8:e67212. doi: 10.1371/journal.pone.0067212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bacosa HP, Erdner DL, Liu Z. Differentiating the roles of photooxidation and biodegradation in the weathering of Light Louisiana Sweet crude oil in surface water from the Deepwater Horizon site. Mar Poll Bull. 2015;95:265–72. doi: 10.1016/j.marpolbul.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Bacosa H, Suto K, Inoue C. Preferential degradation of aromatic hydrocarbons in kerosene by a microbial consortium. Int Biodeterior Biodegrad. 2010;64:702–10. doi: 10.1016/j.ibiod.2010.03.008. [DOI] [Google Scholar]
- 26.Bacosa HP, Suto K, Inoue C. Preferential utilization of petroleum oil hydrocarbon components by microbial consortia reflects degradation pattern in aliphatic–aromatic hydrocarbon binary mixtures. World J Microbiol Biotechnol. 2011;27:1109–17. doi: 10.1007/s11274-010-0557-6. [DOI] [Google Scholar]
- 27.Bacosa HP, Suto K, Inoue C. Degradation potential and microbial community structure of heavy oil-enriched microbial consortia from mangrove sediments in Okinawa, Japan. J Environ Sci Health A. 2013;48:1–12. doi: 10.1080/10934529.2013.761476. [DOI] [PubMed] [Google Scholar]
- 28.Bacosa HP, Inoue C. Polycyclic aromatic hydrocarbons (PAHs) biodegradation potential and diversity of microbial consortia enriched from tsunami sediments in Miyagi, Japan. J Hazard Mater. 2015;283:689–97. doi: 10.1016/j.jhazmat.2014.09.068. [DOI] [PubMed] [Google Scholar]
- 29.Bacosa HP, Thyng K, Plunkett S, Erdner DL, Liu Z. The tarballs on Texas beaches following the 2014 Texas City “Y” Spill: Modeling, chemical, and microbiological studies. Mar Pollut Bull. 2016;109:236–44. doi: 10.1016/j.marpolbul.2016.05.076. [DOI] [PubMed] [Google Scholar]
- 30.Liu J, Bacosa HP, Liu Z. Potential environmental factors affecting oil-degrading bacterial populations in deep and surface waters of the northern Gulf of Mexico. Front Microbiol. 2017;7:2131. doi: 10.3389/fmicb.2016.02131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang ZD, Fingas M, Lambert P, Zeng G, Yang C, Hollebone B. Characterization and identification of the Detroit River mystery oil spill (2002) J Chromatogr A. 2004;1038:201–14. doi: 10.1016/j.chroma.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 32.Aeppli C, Nelson RK, Radović JR, Carmichael CA, Valentine DL, Reddy CM. Recalcitrance and degradation of petroleum biomarkers upon abiotic and biotic natural weathering of Deepwater Horizon oil. Environ Sci Technol. 2014;48:6726–34. doi: 10.1021/es500825q. [DOI] [PubMed] [Google Scholar]
- 33.Prince RC, Elmendorf DL, Lute JR, Hsu CS, Halth CE, Senlus JD, et al. 17a(H), 21b(H)-hopane as a conserved internal marker forestimating the biodegradation of crude oil. Environ Sci Technol. 1994;28:142–5. doi: 10.1021/es00050a019. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Qian PY. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE. 2009;4:e7401. doi: 10.1371/journal.pone.0007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for Illumina sequences. BMC Bioinforma. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seitz KW, Lazar CS, Hinrichs KU, Teske AP, Baker BJ. Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction. ISME J. 2016;10:1696–705. doi: 10.1038/ismej.2015.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–2. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 40.Peng Y, Leung HCM, Yiu SM, Chin FYL. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–8. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 41.Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, et al. IMG, the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dombrowski N, Donaho JA, Gutierrez T, Seitz KW, Teske AP, Baker BJ. Reconstructing metabolic pathways of hydrocarbon-degrading bacteria from the Deepwater Horizon oil spill. Nat Microbiol. 2016;1:16057. doi: 10.1038/nmicrobiol.2016.57. [DOI] [PubMed] [Google Scholar]
- 43.Dick GJ, Andersson AF, Baker BJ, Simmons SL, Thomas BC, Yelton AP, et al. Community-wide analysis of genome sequence signatures. Gen Biol. 2009;10:R85. doi: 10.1186/gb-2009-10-8-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Gen Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hammer Oslash, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:9. [Google Scholar]
- 46.Boll M, Albracht SSP, Fuchs G. Benzoyl-CoA reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism—a study of adenosinetriphosphatase activity, ATP stoichiometry of the reaction and EPR properties of the enzyme. Eur J Biochem. 1997;244:840–51. doi: 10.1111/j.1432-1033.1997.00840.x. [DOI] [PubMed] [Google Scholar]
- 47.Du ZJ, Wang ZJ, Zhao JX, Chen GJ. Woeseia oceani gen. nov., sp. nov., a chemoheterotrophic member of the order Chromatiales, and proposal of Woeseiaceae fam. nov. Int J Syst Evol Microbiol. 2016;66:107–12. doi: 10.1099/ijsem.0.000683. [DOI] [PubMed] [Google Scholar]
- 48.Ettwig KF, van Alen T, van de Pas-Schoonen KT, Jetten MSM, Strous M. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 Phylum. Appl Environ Microbiol. 2009;75:3656–62. doi: 10.1128/AEM.00067-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu ML, Ettwig KF, Jetten MSM, Strous M, Keltjens JT, van Niftrik L. A new intra-aerobic metabolism in the nitrite-dependent anaerobic methane-oxidizing bacterium Candidatus ‘Methylomirabilis oxyfera’. Biochem Soc Trans. 2011;39:243–8. doi: 10.1042/BST0390243. [DOI] [PubMed] [Google Scholar]
- 50.Whyte LG, Schultz A, van Beilen JB, Luz AP, Pellizari V, Labbe D, Greer CW. Prevalence of alkane monooxygenase genes in Arctic and Antarctic hydrocarbon-contaminated and pristine soils. FEMS Microbiol Ecol. 2002;41:141–50. doi: 10.1111/j.1574-6941.2002.tb00975.x. [DOI] [PubMed] [Google Scholar]
- 51.Kleindienst S, Seidel M, Ziervogel K, Grim S, Loftis K, Harrison S, et al. Chemical dispersants can suppress the activity of natural oil-degrading microorganisms. Proc Natl Acad Sci. 2015;112:14900–5. doi: 10.1073/pnas.1507380112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bacosa HP, Liu Z, Erdner DL. Natural sunlight shapes crude oil-degrading bacterial communities in northern Gulf of Mexico surface waters. Front Microbiol. 2015;6:1325. doi: 10.3389/fmicb.2015.01325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chakraborty R, Borglin S.E, Dubinsky E.A, Andersen G.L, Hazen T.C. Microbial Response to the MC-252 Oil and Corexit 9500 in the Gulf of Mexico. Front. Microbiol. 2012;3:357–104. doi: 10.3389/fmicb.2012.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gutierrez T, Singleton DR, Berry D, Yang T, Aitken MD, Teske A. Hydrocarbon-degrading bacteria enriched by the Deepwater Horizon oil spill identified by cultivation and DNA-SIP. ISME J. 2013;7:2091–104. doi: 10.1038/ismej.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Redmond MC, Valentine DL. Natural gas and temperature structured a microbial community response to the Deepwater Horizon oil spill. Proc Natl Acad Sci USA. 2012;109:20292–7. doi: 10.1073/pnas.1108756108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Golyshin PN, Chernikova TN, Abraham WR, Lünsdorf H, Timmis KN, Yakimov MM. Oleiphilaceae fam. nov., to include Oleiphilus messinensis gen. nov., sp nov., a novel marine bacterium that obligately utilizes hydrocarbons. Int J Syst Evol Microbiol. 2002;52:901–11. doi: 10.1099/00207713-52-3-901. [DOI] [PubMed] [Google Scholar]
- 57.Kostka JE, Prakash O, Overholt WA, Green SJ, Freyer G, Canion A, et al. Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico Beach sands impacted by the deepwater horizon oil spill. Appl Environ Microbiol. 2011;77:7962–74. doi: 10.1128/AEM.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu Z, Deng Y, Van Nostrand JD, He Z, Voordeckers J, Zhou A, et al. Microbial gene functions enriched in the Deepwater Horizon deep-sea oil plume. ISME J. 2011;6:451–60. doi: 10.1038/ismej.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mason OU, Han J, Woyke T, Jansson JK. Single-cell genomics reveals features of a Colwellia species that was dominant during the Deepwater Horizon oil spill. Front Microbiol. 2014;5:332. doi: 10.3389/fmicb.2014.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deutzmann JS, Schink B. Anaerobic oxidation of methane in sediments of Lake Constance, an oligotrophic freshwater lake. Appl Environ Microbiol. 2011;77:4429–36. doi: 10.1128/AEM.00340-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He Z, Cai C, Wang J, Xu X, Zheng P, Jetten MSM, et al. A novel denitrifying methanotroph of the NC10 phylum and its microcolony. Sci Rep. 2016;6:32241. doi: 10.1038/srep32241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lueske FA, van Alen TA, van der Biezen E, Frijters C, Toonen G, Kampman C, et al. Diversity and enrichment of nitrite-dependent anaerobic methane oxidizing bacteria from wastewater sludge. Appl Microbiol Biotechnol. 2011;92:845–54. doi: 10.1007/s00253-011-3361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Padilla CC, Bristow LA, Sarode N, Garcia-Robledo E, Gómez-Ramírez E, Benson CR, et al. NC10 bacteria in marine oxygen minimum zones. ISME J. 2016;10:2067–71. doi: 10.1038/ismej.2015.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Solano-Serena F, Marchal R, Ropars M, Lebeault JM, Vandecasteele JP. Biodegradation of gasoline: kinetics, mass balance and fate of individual hydrocarbons. J Appl Microbiol. 1999;86:1008–16. doi: 10.1046/j.1365-2672.1999.00782.x. [DOI] [PubMed] [Google Scholar]
- 65.Garrett RM, Pickering IJ, Haith CE, Prince RC. Photooxidation of crude oils. Environ Sci Technol. 1998;32:3719–23. doi: 10.1021/es980201r. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.