Abstract
Quorum sensing (QS), where bacteria secrete and respond to chemical signals to coordinate population-wide behaviors, has revealed that bacteria are highly social. Here, we investigate how diversity in QS signals and receptors can modify social interactions controlled by the QS system regulating bacteriocin secretion in Streptococcus pneumoniae, encoded by the blp operon (bacteriocin-like peptide). Analysis of 4096 pneumococcal genomes detected nine blp QS signals (BlpC) and five QS receptor groups (BlpH). Imperfect concordance between signals and receptors suggested widespread social interactions between cells, specifically eavesdropping (where cells respond to signals that they do not produce) and crosstalk (where cells produce signals that non-clones detect). This was confirmed in vitro by measuring the response of reporter strains containing six different blp QS receptors to cognate and non-cognate peptides. Assays between pneumococcal colonies grown adjacent to one another provided further evidence that crosstalk and eavesdropping occur at endogenous levels of signal secretion. Finally, simulations of QS strains producing bacteriocins revealed that eavesdropping can be evolutionarily beneficial even when the affinity for non-cognate signals is very weak. Our results highlight that social interactions can mediate intraspecific competition among bacteria and reveal that competitive interactions can be modified by polymorphic QS systems.
Introduction
Quorum sensing (QS) is a mechanism of intercellular communication that allows bacterial populations to coordinately regulate gene expression in response to changes in population density. QS is controlled by the secretion and detection of diffusible signaling molecules that, at threshold concentrations, lead to increased signal secretion and the induction of coupled downstream pathways [1, 2]. By this process, QS ensures that metabolically costly products are only produced when this would benefit the bacterial population, i.e., when they are at high concentrations [2]. QS systems are coordinated by the fact that cells simultaneously send and detect a specific signal [2–4], a characteristic that increases the likelihood that QS functions as a private message between clonemates that share evolutionary interests [5–7]. However, although QS works as an effective means of gene regulation in the laboratory in single strain cultures, QS in nature may be less reliable because it is susceptible to signal eavesdropping (i.e., where a promiscuous QS receptor can detect a QS signal not produced by that genotype) and signal crosstalk (i.e., where a non-specific QS signal can activate QS receptors in genotypes that produce other QS signals) [4, 8]. This variation in QS signals and QS signal detection is widespread in nature [9–12] and distinct from well-studied cheater/cooperator dynamics (e.g., refs. [13, 14]). For example, signal-blind bacteria that produce, but are incapable of responding to, QS signals can engage in signal crosstalk to manipulate the behavior of other cells, e.g., by inducing them to produce expensive public goods [15]. Crosstalk and eavesdropping can occur even if all cells within a population are otherwise phenotypically wild-type if (i) QS signals and receptors are polymorphic and (ii) signals can bind and activate more than one receptor variant. Here we examine these issues using the polymorphic QS system regulating bacteriocin production in the Gram-positive opportunistic pathogen Streptococcus pneumoniae, where QS is integral for mediating intraspecific competition.
To initiate infection, S. pneumoniae must successfully colonize the nasopharynx and then persist during subsequent colonization attempts from other strains. Commensal carriage of S. pneumoniae is widespread, affecting up to 88% of children worldwide [16, 17], and between 5–52% of individuals are co-colonized with multiple strains [17–20]. Interactions between different strains during colonization are common and dynamic, and the rate of clonal turnover—where one strain displaces another—occurs on a timescale of days to months [21, 22]. Among the factors thought to mediate intraspecific competition among pneumococcal strains are small anti-microbial peptides with narrow target ranges called bacteriocins [23], which are regulated by QS. The most diverse bacteriocins in S. pneumoniae are encoded by the blp (bacteriocin-like peptides) locus [23, 24]. We recently showed that the number of possible combinations of bacteriocins and immunity genes at this locus can extend into the trillions, although only several hundred combinations are actually observed [25]. As with other Gram-positive peptide signals, the QS signal peptide (BlpC) regulating the blp operon is constitutively produced at low levels, but is auto-induced at high levels once a threshold concentration has been reached [24]. Secreted BlpC binds to the extracellular domain of the membrane-bound histidine kinase BlpH, and upon binding the kinase phosphorylates the response regulator BlpR (Fig. 1b; [26, 27]) which initiates production of the blp bacteriocin and immunity genes and increases production of the BlpC signal [26]. blpC expression is also enhanced by the response regulator ComE during the induction of pneumococcal competence, which is regulated by the paralogous com QS signaling system [28]. Both ABC transporters BlpAB [29] and ComAB [28, 30] cleave the N-terminal, double-glycine leader sequence of BlpC before export of the mature peptide signal by the same transporters (Fig. 1b). Using QS to regulate secretion presumably ensures that Blp bacteriocins are only produced when there is a sufficiently high cell number to allow these anti-competitor toxins to reach effective concentrations.
Fig. 1.

QS eavesdropping, crosstalk, and regulation. a Eavesdropping occurs when a QS receptor of a cell is activated by a QS signal that the cell does not produce, such as activation of the blue QS receptor by both the cognate blue square signal and non-cognate green triangle signal. Crosstalk occurs when a QS signal activates more than one receptor, such as the green triangle signal activating both the cognate green QS receptor and the non-cognate blue QS receptor. b blp QS regulation. External BlpC signal binds to histidine kinase receptor BlpH. This activates response regulator BlpR through phosphorylation, which increases transcription of blpABC, blpT, the blp bacteriocins (including blpK), and immunity genes. Pre-BlpC is processed and transported out of the cell by ABC transporters ComAB and BlpAB. Similarly, QS signal CSP binds to the histidine kinase receptor ComD, thereby phosphorylating the response regulator ComE, which increases transcription of blpC and the blp operon (although to a lower level than BlpR) as well as com-specific genes
Both the BlpC signal and its receptor, BlpH, are highly polymorphic [25]. What are the effects of this variation, and how does this diversity influence the competitive interactions between strains that are mediated by blp bacteriocins? One possibility is that each unique BlpC signal corresponds to a distinct BlpH receptor to which it specifically and exclusively binds. By this explanation, strains detect and respond only to their own signal to determine the threshold at which they induce the blp operon. Such exclusivity is found in the S. pneumoniae competence signaling system where the two dominant peptide signals, CSP1 and CSP2, only induce cells expressing the cognate receptor [31]. Similarly, there is near perfect concordance between the signal and receptor carried by any single genome, suggesting that tight coupling of these loci is crucial for the activation of competence [32]. An alternative possibility, considered in a recent experimental study [33], is that BlpC peptides cross-react via crosstalk or eavesdropping with different BlpH receptors, thereby leading to a scenario where competing strains interact socially to induce the production of either immunity or bacteriocins at densities that would be insufficient for activation by auto-induction. Bacterial strains may benefit from this cross-reactivity if they are forewarned of the threats from others, allowing them to induce their own bacteriocins or immunity. Alternatively, eavesdropping may be costly if strains with promiscuous receptors are induced to secrete bacteriocins at densities that are too low to provide sufficient benefits to offset the costs of their production. S. pneumoniae presents an ideal opportunity to study the evolution of QS systems beyond cheater/cooperator dynamics [34–36] in an easily manipulated, highly relevant study system in which much is already known about signal/receptor dynamics [33]. Our results extend previous work by highlighting the extensive diversity in both Blp signals and receptors and by demonstrating how this diversity can influence blp operon regulation in both theory and in vitro.
Materials and methods
Phylogenetic and sequence analysis
We analyzed S. pneumoniae genomes from eight publicly available sets, six of which contain strains that were randomly sampled from cases of disease or asymptomatic carriage: 3017 genomes from refugees in Maela, Thailand [37]; 616 genomes from Massachusetts carriage strains [38]; 295 genomes from GenBank, which include 121 genomes from Atlanta, Georgia, The United States [39]; 142 genomes from Rotterdam, the Netherlands (Hermans data set) [25, 40]; and 26 PMEN (Pneumococcal Molecular Epidemiology Network) genomes [25, 41]. The PMEN-1 [42] and Clonal Complex 3 [43] data sets, containing 240 and 82 genomes, respectively, were a result of targeted sampling for specific clonal complexes of S. pneumoniae; as such, these strains were excluded from analyses that assumed random sampling. Contigs for the 142 Rotterdam strains have been deposited the European Nucleotide Archive as Study ID PRJEB10892, and contigs for the 26 PMEN strains have been deposited as Study ID PRJEB10893. Sequences for BlpH for strains manipulated in this paper have been deposited as MG675558—MG675563 in NCIB. Details of the phylogenetic and sequence analysis are provided in the Supplemental Materials.
Bacterial strains and growth conditions
S. pneumoniae strains were grown as liquid cultures in C+Y medium [44] at 37 °C and transformed as described previously [28]. For selection, S. pneumoniae was plated on Columbia agar supplemented with 2% defibrinated sheep blood (Johnny Rottier, Kloosterzande, Netherlands) and 1 µg/ml tetracycline, 100 µg/ml spectinomycin or 0.25 µg/ml erythromycin, when appropriate. E. coli was grown in LB medium with shaking at 37 °C or plated on LA containing 100 µg/ml ampicillin.
Strain construction
Strains and plasmids used in this study are listed in Table S3. Full descriptions of strain construction for the expression of blpSRH alleles, the deletion of blpSRHC, and gene reporter constructs are given in the Supplemental Materials and Methods.
Luciferase assays
Luciferase assays were performed as described [28, 45]. Briefly, S. pneumoniae cultures grown to OD600 0.4 were diluted 100-fold in C+Y medium (pH 6.8) with 340 µg/ml luciferin. Luc-activity was measured in 96-well plates at 37 °C, and OD600 and luminescence (as relative luminescence units, RLU) were recorded every 10 min using Tecan Infinite 200 PRO. Synthetic peptides (BlpCs) were purchased from Genscript (Piscataway, NJ). Different concentrations of BlpCs were added to the culture wells after 100 min or in the beginning of the experiment, depending on the experiment. The data were plotted as RLU/OD over time to analyze induction of blp expression. Activation of a receptor from a signal was assigned when the luc-expression values (RLU/OD) were above the baseline in each experiment after the peptides were added, with the baseline defined as RLU/OD when no BlpC was added.
LacZ assays on agar plates
LacZ assays for testing induction by neighboring colonies on plates were performed on C+Y agar (pH 8) covered with 40 µl of 40 mg/ml solution X-gal (spread on top of the plates). All strains were pre-grown to OD600 0.4, before 2 μl of the wild-type strains (BlpC producers) were spotted and allowed to dry. Then 2 μl of the different reporter strains were spotted next to the dried spot. The plates were incubated at 37 °C overnight.
For induction with synthetic BlpC, C+Y agar plates (pH 7.2) were covered with 40 µl of 40 mg/ml solution X-gal and 5 µl 1 mg/ml BlpC (spread on top of the plates), and different reporter strains were spotted on top. The plates were incubated at 37 °C overnight.
Stochastic model
We built an individual-based spatial, stochastic model in which cells interact on a grid. We modeled four genotypes, which differ in the signaling molecule and bacteriocins that they produce as well as in the number and identity of signals that they respond to (Table S2). Bacteriocins produced by genotypes 1 and 2 specifically could kill genotypes 3 and 4 and vice versa. Signals produced by genotype 1 could induce genotypes 1 and 2 and similarly, signals produced by genotype 3 could induce genotypes 3 and 4; we therefore classify genotypes 2 and 4 as “eavesdropping genotypes”. Genotypes 1 and 3 can only respond to their own signal, as “signal-faithful genotypes”. All four genotypes have equivalent growth rates, which are only variable depending on if a cell is induced or uninduced. Eavesdropping cells respond to signals that they do not produce with variable degrees of affinity. We consider the affinity of a cell to its own signal as 100%, and ranged the affinity to the other signals in the case of eavesdropping genotypes to 0–90% for different simulations. Full model details are given in the Supplemental Materials and Methods.
Results
Molecular diversity of blpH and blpC
We examined 4096 S. pneumoniae genomes taken from six data sets of strains (Maela, Massachusetts Asymptomatic, GenBank, Hermans, Georgia GenBank, and PMEN: 4096 genomes in total) alongside two additional data sets that are intentionally biased to specific clonal sub-groups (Complex 3 and PMEN-1: 322 genomes in total). We identified blpC in 99% and blpH in 99% of the combined 4418 genomes using a DNA reciprocal BLAST algorithm [25]; 98.2% of these strains contained both blpC and blpH. We note that the few genomes apparently lacking a blp gene may still contain these genes, as the data sets contain incomplete draft genomes. Consistent with earlier work [25], we found extensive allelic variation within blpC, which contains 37 alleles at the nucleotide level, 29 protein precursors, and 20 different BlpC signal peptides (range of 18–51 residue predicted mature signals), including signal peptides lacking a canonical double-glycine cleavage site. Nine of these peptide signal sequences were found in more than 0.5% of genomes (i.e., over 20 genomes; Table 1), and together these nine comprise ~98% of all signal variants. All signals under this 0.5% threshold were each confined to a single clade in the whole-genome phylogeny (Fig. S1). Each unique BlpC signal was designated with a letter from the NATO phonetic alphabet (Table 1). As expected for the genomes from intentionally biased samples, the PMEN-1 data set almost exclusively carried the Golf signal (93.8%; Table S1), whereas the Clonal Complex 3 data set almost exclusively carried the Kilo signal (97.6%; Table 1). The Bravo and Hotel signal peptides were exclusively found in strains collected as part of the Maela data set possibly indicating limited admixture between these strains and those from the other collections.
Table 1.
Predicted blpC mature peptide signals in 4,096 genomes from six unbiased genome sets.
| Signal | Signal group | Mature AA sequencea | Frequency in unbiased sets | Genomes used in this paper with mature signal | Reference genomes with mature signal | Alternative name for signal |
|---|---|---|---|---|---|---|
| Alpha | Alpha/Bravo/Kilo | GWWEELLHETILSKFKITKALELPIQL | 0.125 | D39 | R6 | R6 |
| Bravo | Alpha/Bravo/Kilo | GLWEELLHETILSKFKITKALELPIQL | 0.028 | |||
| Kilo | Alpha/Bravo/Kilo | EWWEELLHETILSKFKITKALELPIQL | 0.007 | OXC141 | 6A | |
| Charlie | Charlie | GLWEDILYSLNIIKHNNTKGLHHPIQL | 0.291 | Hermans-1012, PMEN-14 | SPNA45, INV200 | |
| Delta | Delta | GWWKDLLHRFNVIEQNNTKGFNQPIQL | 0.035 | Hermans-33 | ECC_3510 | |
| Echo | Echo/Foxtrot | GWWEDFLYRFNIIEQKNTKGFHQPIQL | 0.087 | |||
| Foxtrot | Echo/Foxtrot | GWWEDFLYRFNIIEQKNTKGFYQPIQL | 0.125 | PMEN-2 | P164 | P164 |
| Golf | Golf/Hotel | GLWEDLLYNINRYAHYIT | 0.223 | PMEN-18 | TIGR4, INV104 | T4 |
| Hotel | Golf/Hotel | GLWEDLLYNINRYAHYITQELHHPIQL | 0.06 | |||
| Other, or no ‘GG’ cleavage site | 0.009 | |||||
| No blpC | 0.011 | |||||
a Conserved residues are in bold. Residues that differentiate signals within a signal group are underlined
blpC/blpH intragenomic pairing is highly biased but not exclusive
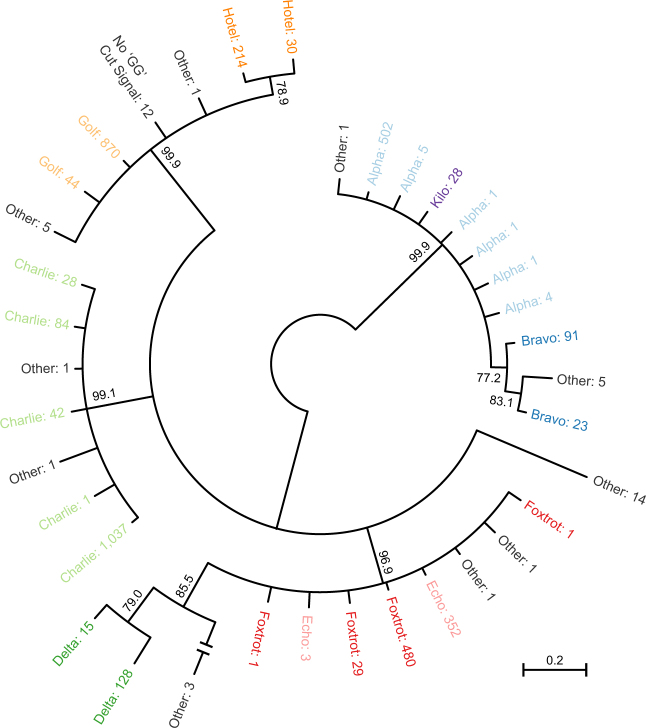
Phylogenetic analysis of blpC revealed four well-supported clades (Fig. 2) containing the following signals: (1) Alpha, Bravo, and Kilo; (2) Golf and Hotel; (3) Charlie; and (4) Delta, Echo, and Foxtrot. With the exception of the Delta signal, within-group signals are differentiated by a single amino acid or stop codon. The relationships between signaling groups within these major clades are uncertain, although there is evidence (0.75 < posterior probability < 0.95) that the Hotel, Bravo, and Delta signals are each monophyletic within their respective larger clades.
Fig. 2.
Bayesian unrooted phylogenetic tree of blpC. Taxa are colored by mature BlpC signal with the signal designation followed by the number of genomes containing the allele. Internal nodes show the posterior probabilities of clades; we collapsed clades with less than 0.75 posterior probability
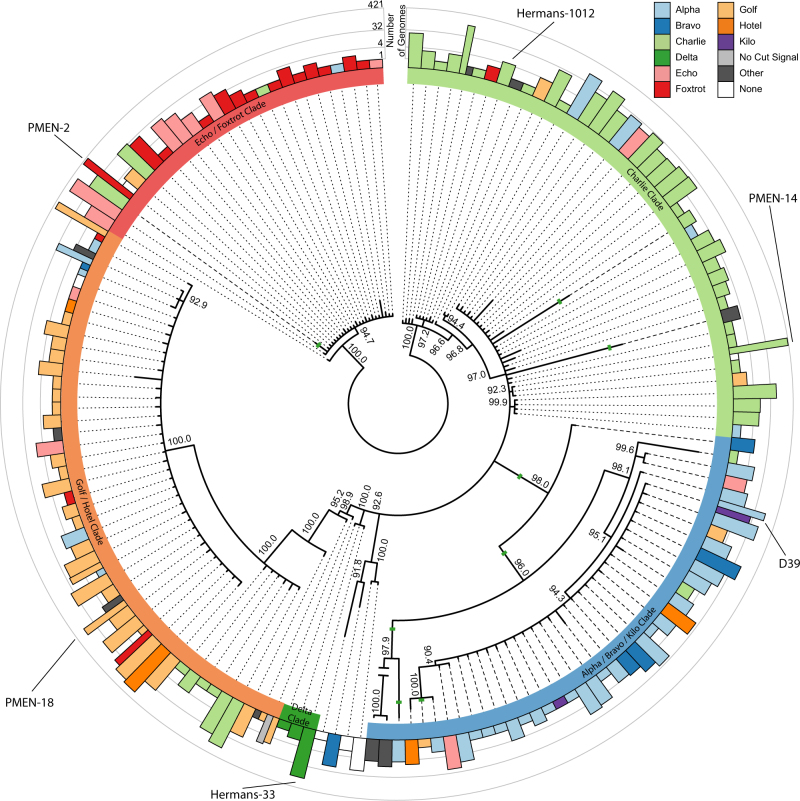
After accounting for recombination, phylogenetic analysis of the receptor domain of blpH (residues 1–229) identified five paraphyletic clades that are broadly concordant with the divisions observed for BlpC signals (Fig. 3), although there are many exceptions to this correspondence. Across the five clades, the classification of blpH alleles correlated with the BlpC signal in at least 75% of cases: (Alpha/Bravo/Kilo Clade: 86.6%; Echo/Foxtrot Clade: 90%; Delta Clade: 100%: Charlie Clade: 86.2%; Golf/Hotel Clade: 75%). Evidence of extensive recombination affecting the blpH kinase, intergenic region, and blpC signal (Fig. S2) suggests that recombination has caused some of these mismatches. Overall, from the 4002 genomes with full-length blpH genes, 16.7% (667 genomes) show a lack of correspondence between signal and peptide, suggesting either that these strains are deficient in blp signaling or that these BlpH histidine kinase receptors can be cross-induced by non-cognate BlpC signals. Overall frequencies by signal and receptor class are summarized in Fig. 4a.
Fig. 3.
Bayesian unrooted phylogenetic tree of blpH alleles. The outer ring shows the number of 4096 genomes with each blpH allele, color-coded by their co-occurring BlpC signal and on a log scale. The inner ring denotes the blpH clade type, and recombination events within blpH are shown as solid green lines. Internal nodes show the posterior probabilities of clades; we collapsed clades with <90% posterior probability
Fig. 4.
a Proportion of each BlpC signal within genomes containing each blpH clade. The phylograms are simplified versions of Figs. 2 and 3. b The relative maximal expression levels of luc following addition of 1 μg/ml of synthesized BlpC signal peptide. The maximum expression level for each reporter strain was set to 1. Raw data are found in Fig. S5. c The minimum concentration of synthetic BlpC signal peptide required for luc induction in reporter strains with different BlpH. Asterisks indicate receptor/signal pair blp activation reported in [33]. Examples of raw data are provided in Fig. S6. The Bravo, Kilo, and Hotel signal peptides were not synthesized and are denoted with slashes
Crosstalk and eavesdropping between BlpC signals and BlpH receptors
To examine the incidence of crosstalk and eavesdropping between signals and receptors experimentally, we measured the responsiveness of each of the major BlpH clades to synthetic peptides from each BlpC class. We transformed a S. pneumoniae D39 strain lacking the native blp regulatory genes (blpSRHC) with constructs expressing one of six different BlpH histidine kinases alleles: blpSRHD39 from the Alpha/Bravo/Kilo clade, blpSRHPMEN-2 from the Echo/Foxtrot clade, blpSRHHermans-33 from the Delta clade, blpSRHHermans-1012 and blpSRHPMEN-14 from the Charlie clade, and blpSRHPMEN-18 from the Golf/Hotel clade. These strains also contained a reporter cassette, in which the blp-promoter from either PblpK or PblpT controlled expression of firefly luciferase (luc), GFP (sf)gfp), and β-galactosidase (lacZ) [28]. Deletion of the blpC signal gene and the native blpSRH genes from the D39 ancestor ensured that the reporter strains would only be induced in response to exogenously added signal via the introduced blpSRH systems. By exposing cells to a concentration gradient of exogenous peptide, we could estimate the peptide concentration that induced the maximum response as well the minimum concentration required to elicit a response. Although the maximum response indicates the overall influence of a given peptide on each receptor, the minimal concentration required to induce a response provides an indication of the sensitivity of each receptor to every potential peptide partner.
Figure 4a and b shows that five of six PblpK reporter strains were maximally induced by the BlpC signal carried by a significant majority of their wild-type counterparts. However, we also see extensive evidence for crosstalk and eavesdropping between mismatched peptide:receptor pairs, demonstrating that some BlpH receptors are highly promiscuous while equally, several BlpC peptides can induce the blp operon in strains carrying non-cognate BlpH receptors. For example, blpSRHPMEN-2 (Echo/Foxtrot BlpH clade) could be induced by 4 out of 6 synthetic peptides, and the strain with blpSRHHermans-1012 (Charlie BlpH clade) was strongly induced by the Echo and Foxtrot signals at 65 and 71% expression of its cognate signal. Although there is clear evidence for cross-induction, these responses tended to be less sensitive to non-cognate peptides, with a minimum concentration required for induction of between 2 and 500-fold greater than with the cognate signal (Fig. 4c). By contrast, the strain with blpSRHHermans-1012 (Charlie BlpH clade) was more sensitive to the non-cognate Echo and Foxtrot signals (1 and 3.9 ng/ml) than to its cognate Charlie signal (7.8 ng/ml; Fig. 4c). The reporter strain carrying blpSRHHermans-33 did not respond to any of the BlpC peptides, including its cognate Delta BlpC (Fig. 4b, c). Interestingly, blpSRHHermans-33, as well as all other strains with blpH alleles in the Delta clade, contains a frameshift in the blpR gene, encoding the response regulator, thus preventing expression of the full-length blpR. This probably renders the QS systems non-functional and therefore not responsive to added peptide. All results with PblpK were mirrored with a different set of reporter strains that used the blpT promoter for the reporter cassette (Fig. S3).
We conclude from these results that crosstalk among quorum-dependent peptide BlpC signals is common and concentration dependent, with strains able to eavesdrop on multiple signals using cross-responsive receptors. Furthermore, these results are concordant with the patterns of co-association observed in our bioinformatics survey of pneumococcal strains. When only considering genomes carrying blpC and blpH alleles potentially capable of blp activation (as determined in Fig. 4b, c), 88% of the strains are predicted to autoinduce blp expression under appropriate conditions, i.e., their genomes contain functionally active blpC/blpH pairs. Notably, however, this also indicates that a substantial proportion of strains (12%, 364 of 3046 genomes) may not be able to autoinduce blp expression since they carry blpC/blpH pairs that were inactive in our experimental assay; this is in addition to strains carrying Delta blpC/blpH, which was also unable to autoinduce blp expression in our assay.
Cross-induction between colonies
Pneumococci in the nasopharynx live in spatially structured colonies or biofilms. In order to determine if cross-induction between signaling cells could occur under these conditions where QS efficiency may be limited by signal diffusion [4, 46, 47], we examined interactions between neighboring colonies endogenously secreting either cognate or non-cognate signals (Fig. 5). In control assays, we first demonstrated that colonies were induced by exogenous addition of peptide to the plate surface; these results were concordant with those in Fig. 4b in 14 of 15 combinations (Fig. S4). Next, we measured expression of reporter strains when grown adjacent to wild-type colonies that secreted BlpC peptides at endogenous levels (Fig. 5a). We observed a response in the reporter strains as estimated by increased LacZ activity in 3 out of 6 strains, with 2 examples of induction by non-cognate BlpC signals. Interestingly, when the reporter strain expressing the BlpH from Hermans-1012 was grown adjacent to its wild-type counterpart, there was no induction; instead this strain was induced by PMEN-14, which also produced the Charlie signal. The same strain was also induced by PMEN-2, which produced the Foxtrot signal (which induces Hermans-1012 at a lower concentration than with its cognate signal; Fig. 4c), and strain PMEN-18 (Golf/Hotel BlpH clade) was induced by PMEN-14, which produced the Charlie signal (Fig. 5). This may suggest that in addition to differences in the binding sensitivity of BlpC and BlpH, strains may also vary in the concentration of the diffusible signals that they secrete, at least under these experimental conditions. Consistent with our in vitro assays with synthetic peptides, these results show that blp operon expression can be activated by crosstalk between neighboring competing colonies secreting peptides at wild-type concentrations.
Fig. 5.
LacZ induction by neighboring colonies on agar plates. a The wild-type strains were spotted next to the reporter strains (see box), and induction of blp expression by the wild-type produced BlpC is shown as faint blue colonies (indicated by brackets). The experiment was repeated three times with the same results, and a representative photo of the plates is shown. b Summary of the results from (a). Squares in white indicate no induction of the reporter strain for colony pairs, whereas black and gray indicate induction by cognate and non-cognate BlpCs, respectively
Evolutionary consequences of eavesdropping genotypes
Because the blp operon is auto-induced via a quorum-dependent process, cross-induction can potentially influence other strains by lowering the population density required for auto-induction. To examine the possible effects of cross-induction on bacteriocins, we developed a spatially explicit stochastic model to investigate conditions where genotypes with eavesdropping receptors may be favored over strains only able to respond to a single peptide signal. We further varied the signal affinity to eavesdropping receptors to determine how this altered the selective benefits of cross-induction. Simulations are initiated with cells of four genotypes randomly spaced upon a plane. The four genotypes each release their own QS signal at equal concentrations (Table S2). Cells bind these secreted signals in a concentration dependent manner, at which point they are induced to produce bacteriocins that kill susceptible neighbor cells at the cost of reduced growth for the producer [48]. Although two faithful-signaling genotypes are only able to respond to their own signals, the two other eavesdropping genotypes can respond to multiple signals. The model does not take into account the complex regulatory interplay between QS systems (i.e., blp and com systems; [28, 30]), but instead serves as a general model for studying eavesdropping in social bacteria. Our results shown in Fig. 6 lead to two conclusions. First, we observe strong benefits to eavesdropping cells that depends on the degree of cross-sensitivity, or affinity, to non-cognate signals. Specifically, we found that higher affinity to non-cognate signal provides stronger ecological benefits. This results from earlier potential activation (Fig. S3) and secretion of bacteriocins in these cells, an effect that increases with greater affinity to non-cognate signals. Second, we find that the benefits to eavesdropping are strongly negative frequency-dependent, i.e., eavesdropping cells only gain benefits (in the form of earlier bacteriocin induction) when surrounded by faithful-signaling cells. When eavesdropping cells are rare, they benefit through maximum exposure to the alternative peptide, whereas after they increase in frequency they must rely solely on auto-induction. Because the benefits of eavesdropping are frequency-dependent, these simple simulations thus suggest that promiscuous receptor mutants with increased affinity to non-cognate signals will be able to rapidly invade populations of cells that can only respond to a single signal. Interestingly, the simulations also clarify that the affinity to non-cognate signals can be extremely low—even at 10% of the affinity to cognate signals—to provide benefits (Fig. 6).
Fig. 6.
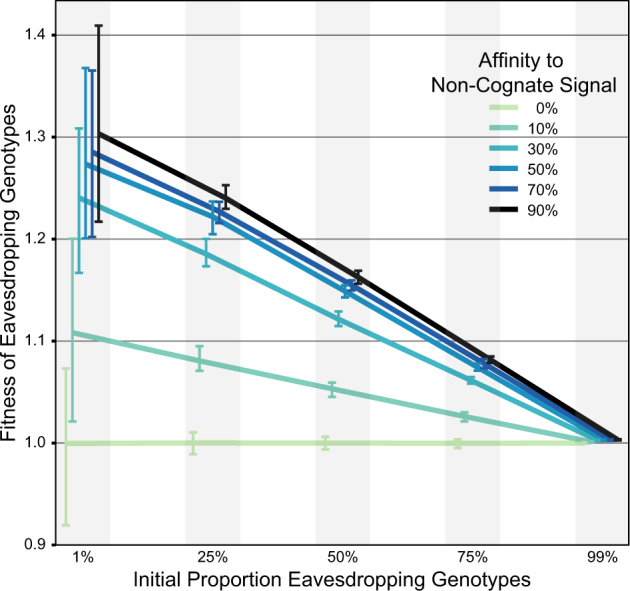
Average fitness of eavesdropping genotypes that produce bacteriocins in response to multiple signals in a spatially explicit, stochastic model. Simulations were started with five proportions of eavesdropping genotypes mixed with signal-faithful genotypes, as indicated on the x-axis. Absolute fitness values on the y-axis above 1 indicate that the genotype can increase in frequency in the population. Affinity to other genotypes’ signals are a percentage of affinity to a genotype’s own signal for eavesdropping genotypes. Error bars link the 25 and 75% quantiles for the final eavesdropping genotypes’ fitness across 100 simulations
Discussion
Pneumococcal bacteriocins are believed to have an important role in mediating intraspecific competitive interactions [23], and bacteriocins are found in nearly all strains [25]. However, individual strain blp composition shows no patterns with which strains are found to co-colonize the nasopharynx [49]. One possible explanation is that these bacteriocin dynamics are influenced by the social dynamics of their respective signaling systems, although this remains to be tested. Here, we show that the QS system regulating blp bacteriocins is highly polymorphic, that QS signals are frequently cross-reactive (crosstalk), and that promiscuous receptors can detect and respond to non-cognate signals (eavesdropping). Assays between adjacent colonies revealed that both behaviors occur at endogenous concentrations of secreted peptides, and simulations showed ecological benefits to strains that express promiscuous receptors. Together, these results suggest that social interactions influenced by QS signaling may strongly influence pneumococcal competition.
Previous surveys [26, 27] of BlpC and BlpH identified four BlpC signals: the Alpha, Charlie, Foxtrot and Golf signals in our nomenclature, which together represent ~75% of the strains in our sample (Table 1). By expanding our survey to thousands of strains, we identified several additional signal peptide families beyond those signals previously investigated [33]: the Echo, Hotel, Delta, Bravo, and Kilo signals. The concordance between the phylogenies of blpC and blpH and the extensive co-occurrence in individual genomes suggest that these genes are co-evolving (Figs. 2 and 3).
Previous work suggested that differences in the electric charge of residue 14 in BlpC is crucial for specificity [33]. This residue is undoubtedly important because it differentiates the Alpha/Bravo/Kilo, Echo/Foxtrot, and Golf/Hotel signal groups; however, other BlpC residues are also likely to be important for BlpH binding, as signals that are identical at position 14 (e.g., Golf/Hotel, and Alpha/Bravo/Kilo/Charlie) differentially activate BlpH (Fig. 4). The co-variation between BlpC and BlpH also allows us to identify residues in BlpH that are correlated with the QS signal. These results can serve as a guide for future experiments to unravel the specificity of the BlpC/BlpH interaction (Figs. S7, S8). Notably, our analysis of the BlpH receptor support previous findings that residues 17 and 119–124 are important for activation by BlpC signals [33], although additional residues also co-vary with specific BlpC signals (Figs. S7, S8).
Although the correlation between blpH clade and co-occurring BlpC signal is high, in some clades the correlation drops to 75%, and BlpH/BlpC mismatches (Fig. 3) are common across the pneumococcal phylogeny. This can be compared to the exceptionally tight, >99% correlation between the ComD QS receptor and CSP signal also in S. pneumoniae [32]. There are at least two explanations for this difference. First, we do not know if different BlpH variants are functionally distinct. All blpH alleles could, in principle, be most responsive to their co-occurring BlpC. This seems unlikely, given the high frequency (up to 45 signal:receptor pairs) of blpH clade/BlpC mismatches (Fig. 3). Second, weaker selection for a highly auto-inducing blp QS could explain the difference between the blp and com QS systems. After a recombination event that results in a sub-optimal BlpH/BlpC pair for auto-induction, the BlpC signal may still be able to activate the co-occurring BlpH variant through crosstalk, albeit at a higher concentration of BlpC (Fig. 4c). Although auto-induction may be decreased, such a genotype would gain an eavesdropping receptor that can potentially detect signals of surrounding genotypes. For comparison, there is no eavesdropping between CSP pherotypes in the com QS system, and very rare signal/receptor mismatches [31, 32].
Signal/receptor mismatches can result in two outcomes for cell:cell communication. First, cells may be unable to detect the signal that they produce, rendering them unable to autoinduce. The lack of QS activation in strains producing the Delta signal (Hermans-33; Fig. 4) seemingly fits this description; however, interestingly, this is not caused by signal/receptor mismatch because there is perfect concordance between the Delta signal and the Delta blpH clade, and no tested signal activated strains with Delta blpH. Instead, all 143 strains carrying the Delta signal have a frameshift in blpR, which suggests functional deterioration of the QS system in these strains, which has not yet led to deterioration of blpH and blpC. At present, it is unclear if these Delta BlpC strains benefit in some way from the inactivation of blpR; however, such strains may continue to pay the cost of synthesizing BlpC if blpC is actively transcribed through another mechanism, such as via phosphorylated ComE from an activated competence system. This possibility suggests there may be weakened selection for functional blp QS.
The second outcome of signal/receptor mismatches for cell-to-cell communication is crosstalk and eavesdropping. We have ample evidence for crosstalk in the blp QS system, as all signal peptides except for the Alpha signal activated QS receptors in genotypes that produce other QS signals (Fig. 4b, c). Similarly, BlpH receptors (aside from the Alpha clade) were eavesdropping QS receptors able to detect more than one QS peptide signal (Fig. 4b, c). Each of the receptors we tested (except for the signal-blind BlpH Delta clade) was maximally induced with a single set of related signals and decreased to 3–71% with signals that the receptors were eavesdropping upon (Fig. 4b). This suggests that there are no ‘generalist’ receptors that are able to listen to multiple signals with equal responsiveness. Crosstalk was observed in previous research on the blp system ([33]; see asterisks in Fig. 4c and Table 1 alternative signal names), and results from this study indicated that blpH alleles with more crosstalk were less sensitive to BlpC [33]. However, the results reported here show that receptors from strains PMEN-2 and PMEN-14 were highly sensitive to their cognate signal (≤1 ng/ml) despite showing crosstalk at low (≤15.6 ng/ml; Fig. 4c) signal concentrations of non-cognate peptides, thereby suggesting that the trade-off between crosstalk and sensitivity of blpH alleles is not universal. Building upon the results of previous studies [33], we demonstrate that eavesdropping can activate blp expression patterns in neighboring colonies.
What are the potential consequences of crosstalk and eavesdropping? Crosstalk may enable one strain to manipulate competing strains into inducing their QS system at lower densities, thereby causing them to secrete bacteriocins and induced immunity proteins earlier. At present, it is unclear how such crosstalk would be beneficial to cells producing cross-reactive signals, unless premature production of bacteriocins or immunity introduces energetic or other costs to cells responding at sub-quorum densities. Similar benefits have been shown to exist for public goods in Gram-negative bacteria [15, 50] and specifically for public goods that offer higher benefits to QS responding cells in Bacillus subtilus [51]. By contrast, it is easier to envision the potential benefits of eavesdropping, which can both lead to earlier activation of bacteriocins (although this may also have attendant costs) and earlier induction of cross-reactive immunity. Our simulations suggest that this could be beneficial even if the affinity of promiscuous receptors is only 10% of the affinity for their cognate signal (Fig. 6). This value falls within the range of responses we measured experimentally (Fig. 4c). This level of responsiveness is also sufficient to induce the blp operon among adjacent colonies secreting peptides at endogenous levels (Fig. 5).
How does this amount of crosstalk specifically affect bacteriocin-mediated competition between S. pneumoniae strains? This is challenging to answer conclusively. First, extensive variation in the kinase domain of BlpH, the response regulator BlpR, and the leader sequences of the blp bacteriocins [25] prevents a full understanding of how signal concentrations translate into increased bacteriocin export. A systematic approach to investigate each of these molecules and their variants in the laboratory will be required to address this question. Second, a bioinformatics approach to examine evidence of selection in coordination with the BlpH receptor or BlpC signal is not possible due to the inability to align the entire blp operon and because recombination breaks up potential associations that are otherwise selected for. Third, the effects of crosstalk and eavesdropping will also depend on the activation of the com QS system, which promotes the expression and export of blpC at a low level (Fig. 1), even when the ABC-transporter genes blpAB are disrupted by early termination mutations [28, 30]. For example, we found that both wild-type strains D39 and PMEN-14 could activate blp expression in neighboring colonies (Fig. 5) despite having disrupted blpA (for PMEN-14) or disrupted blpA and blpB (for D39).
Signaling interactions in vitro can lead to complex ecological outcomes that may influence competitive interactions between strains. As yet, however, it is unclear how these interactions will play out in the complex within-host environment of the human nasopharynx [49]. In addition, it remains unclear how these interactions directly influence bacteriocin-mediated killing and immunity. Clearly, the heterogeneous conditions in vivo differ markedly between liquid cultures or agar surfaces. Diffusion is more limited, population densities may be strongly constrained overall and spatially, and unknown host factors may further modify intra-strain competitive dynamics. These factors, among others, may alter the level and dispersion of signal peptides as well as the sensitivity of individual strains to these signals. More generally, our results reinforce the importance of social interactions among bacteria for mediating competitive dynamics. Many ecologically relevant bacterial traits are regulated by QS, and many of these systems, especially in Gram-positive peptide signaling systems, are polymorphic. Although some of these systems (e.g., pneumococcal competence regulated by the com QS system) have only few signal types and show no cross-reactivity, many others signal are polymorphic with substantial cross-reactivity (e.g., Agr in S. aureus [10] and ComX in B. subtilis [52]). It remains to be investigated which of these polymorphic QS signals have ecological effects and which factors (such as co-colonization or extensive intraspecific competition) result in the evolution of crosstalk and eavesdropping.
Electronic supplementary material
Acknowledgements
We would like to thank Frank Lake for technical assistance. This work was supported by the Biotechnology and Biological Sciences Research Council (Grant Number BB/J006009/1) to DER and ISR and by the Wellcome Trust (105610/Z/14/Z) to the University of Manchester. MA is supported by the Biotechnology and Biological Sciences Research Council (Grant Number BB/M000281/1). Work in the Veening lab is supported by the EMBO Young Investigator Program, a VIDI fellowship (864.12.001) from the Netherlands Organisation for Scientific Research, Earth and Life Sciences (NWO-ALW) and ERC starting grant 337399-PneumoCell. MK is supported by a grant from The Research Council of Norway (250976/F20).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Ian S. Roberts, Email: i.s.roberts@manchester.ac.uk
Jan-Willem Veening, Email: Jan-Willem.Veening@unil.ch.
Daniel E. Rozen, Email: d.e.rozen@biology.leidenuniv.nl
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0178-x) contains supplementary material, which is available to authorized users.
References
- 1.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–99. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 2.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–46. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 3.Bassler BL, Greenberg EP, Stevens AM. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol. 1997;179:4043–5. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redfield RJ. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 2002;10:365–70. doi: 10.1016/S0966-842X(02)02400-9. [DOI] [PubMed] [Google Scholar]
- 5.Schluter J, Schoech AP, Foster KR, Mitri S. The evolution of quorum sensing as a mechanism to infer kinship. PLoS Comput Biol. 2016;12:e1004848. doi: 10.1371/journal.pcbi.1004848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 7.Crespi BJ. The evolution of social behavior in microorganisms. Trends Ecol Evol. 2001;16:178–83. doi: 10.1016/S0169-5347(01)02115-2. [DOI] [PubMed] [Google Scholar]
- 8.Atkinson S, Williams P. Quorum sensing and social networking in the microbial world. J R Soc. 2009;6:959–78. doi: 10.1098/rsif.2009.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ansaldi M, Dubnau D. Diversifying selection at the Bacillus quorum-sensing locus and determinants of modification specificity during synthesis of the ComX pheromone. J Bacteriol. 2004;186:15–21. doi: 10.1128/JB.186.1.15-21.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji G, Beavis R, Novick RP. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–30. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 11.Swem LR, Swem DL, Wingreen NS, Bassler BL. Deducing receptor signaling parameters from in vivo analysis: LuxN/AI-1 quorum sensing in Vibrio harveyi. Cell. 2008;134:461–73. doi: 10.1016/j.cell.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouillaut L, Perchat S, Arold S, Zorrilla S, Slamti L, Henry C, et al. Molecular basis for group-specific activation of the virulence regulator PlcR by PapR heptapeptides. Nucleic Acids Res. 2008;36:3791–801. doi: 10.1093/nar/gkn149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiricny N, Diggle SP, West SA, Evans BA, Ballantyne G, Ross-Gillespie A, et al. Fitness correlates with the extent of cheating in a bacterium. J Evol Biol. 2010;23:738–47. doi: 10.1111/j.1420-9101.2010.01939.x. [DOI] [PubMed] [Google Scholar]
- 14.Strassmann JE, Queller DC. Evolution of cooperation and control of cheating in a social microbe. Proc Natl Acad Sci USA. 2011;108:10855–62. doi: 10.1073/pnas.1102451108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diggle SP, Griffin AS, Campbell GS, West SA. Cooperation and conflict in quorum-sensing bacterial populations. Nature. 2007;450:411–4. doi: 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- 16.Regev-Yochay G, Raz M, Dagan R, Porat N, Shainberg B, Pinco E, et al. Nasopharyngeal carriage of Streptococcus pneumoniae by adults and children in community and family settings. Clin Infect Dis. 2004;38:632–9. doi: 10.1086/381547. [DOI] [PubMed] [Google Scholar]
- 17.Wyllie AL, Chu MLJN, Schellens MHB, Gastelaars JVE, Jansen MD, Van Der Ende A, et al. Streptococcus pneumoniae in saliva of Dutch primary school children. PLoS ONE. 2014;9:1–8. doi: 10.1371/journal.pone.0102045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Rodríguez JA, Fresnadillo Martínez MJ. Dynamics of nasopharyngeal colonization by potential respiratory pathogens. J Antimicrob Chemother. 2002;50:59–73. doi: 10.1093/jac/dkf506. [DOI] [PubMed] [Google Scholar]
- 19.Sauver JS, Marrs CF, Foxman B, Somsel P, Madera R, Gilsdorf JR. Risk factors for otitis media and carriage of multiple strains of Haemophilus influenzae and Streptococcus pneumoniae. Emerg Infect Dis. 2000;6:622–30. doi: 10.3201/eid0606.000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brugger SD, Frey P, Aebi S, Hinds J, Muhlemann K. Multiple colonization with S. pneumoniae before and after introduction of the seven-valent conjugated pneumococcal polysaccharide vaccine. PLoS ONE. 2010;5:e11638. doi: 10.1371/journal.pone.0011638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meats E, Brueggemann AB, Enright MC, Sleeman K, Griffiths DT, Crook DW, et al. Stability of serotypes during nasopharyngeal carriage of Streptococcus pneumoniae. J Clin Microbiol. 2003;41:386–92. doi: 10.1128/JCM.41.1.386-392.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner Paul, Turner Claudia, Jankhot Auscharee, Helen Naw, Lee Sue J., Day Nicholas P., White Nicholas J., Nosten Francois, Goldblatt David. A Longitudinal Study of Streptococcus pneumoniae Carriage in a Cohort of Infants and Their Mothers on the Thailand-Myanmar Border. PLoS ONE. 2012;7(5):e38271. doi: 10.1371/journal.pone.0038271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dawid S, Roche AM, Weiser JN. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect Immun. 2007;75:443–51. doi: 10.1128/IAI.01775-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lux T, Nuhn M, Hakenbeck R, Reichmann P. Diversity of bacteriocins and activity spectrum in Streptococcus pneumoniae. J Bacteriol. 2007;189:7741–51. doi: 10.1128/JB.00474-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller EL, Abrudan MI, Roberts IS, Rozen DE. Diverse ecological strategies are encoded by Streptococcus pneumoniae bacteriocin-like peptides. Genome Biol Evol. 2016;8:1072–90. doi: 10.1093/gbe/evw055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Saizieu A, Gardes C, Flint N, Wagner C, Kamber M, Mitchell TJ, et al. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J Bacteriol. 2000;182:4696–703. doi: 10.1128/JB.182.17.4696-4703.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reichmann P, Hakenbeck R. Allelic variation in a peptide-inducible two-component system of Streptococcus pneumoniae. FEMS Microbiol Lett. 2000;190:231–6. doi: 10.1111/j.1574-6968.2000.tb09291.x. [DOI] [PubMed] [Google Scholar]
- 28.Kjos M, Miller E, Slager J, Lake FB, Gericke O, Roberts IS, et al. Expression of Streptococcus pneumoniae bacteriocins is induced by antibiotics via regulatory interplay with the competence system. PLoS Pathog. 2016;12:e1005422. doi: 10.1371/journal.ppat.1005422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Håvarstein LS, Diep DB, Nes IF. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol. 1995;16:229–40. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 30.Wholey WY, Kochan TJ, Storck DN, Dawid S. Coordinated bacteriocin expression and competence in Streptococcus pneumoniae contributes to genetic adaptation through neighbor predation. PLoS Pathog. 2016;12:e1005413. doi: 10.1371/journal.ppat.1005413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iannelli F, Oggioni MR, Pozzi G. Sensor domain of histidine kinase ComD confers competence pherotype specificity in Streptoccoccus pneumoniae. FEMS Microbiol Lett. 2005;252:321–6. doi: 10.1016/j.femsle.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Miller EL, Evans BA, Cornejo OE, Roberts IS, Rozen D. Pherotype polymorphism in Streptococcus pneumoniae has no obvious effects on population structure and recombination. Genome Biol Evol. 2017;9:2546–59. doi: 10.1093/gbe/evx188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinchas MD, LaCross NC, Dawid S. An electrostatic interaction between BlpC and BlpH dictates pheromone specificity in the control of bacteriocin production and immunity in Streptococcus pneumoniae. J Bacteriol. 2015;197:1236–48. doi: 10.1128/JB.02432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eldar A. Social conflict drives the evolutionary divergence of quorum sensing. Proc Natl Acad Sci USA. 2011;108:13635–40. doi: 10.1073/pnas.1102923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollak S, Omer-Bendori S, Even-Tov E, Lipsman V, Bareia T, Ben-Zion I, et al. Facultative cheating supports the coexistence of diverse quorum-sensing alleles. Proc Natl Acad Sci USA. 2016;113:2152–7. doi: 10.1073/pnas.1520615113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Son MR, Shchepetov M, Adrian P V, Madhi SA, de Gouveia L, von Gottberg A, et al. Conserved mutations in the pneumococcal bacteriocin transporter gene, blpA, result in a complex population consisting of producers and cheaters. MBio. 2012;2. 10.1128/mBio.00179-11. [DOI] [PMC free article] [PubMed]
- 37.Chewapreecha C, Harris SR, Croucher NJ, Turner C, Marttinen P, Cheng L, et al. Dense genomic sampling identifies highways of pneumococcal recombination. Nat Genet. 2014;46:305–9. doi: 10.1038/ng.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Croucher NJ, Finkelstein JA, Pelton SI, Mitchell PK, Lee GM, Parkhill J, et al. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat Genet. 2013;45:656–63. doi: 10.1038/ng.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chancey ST, Agrawal S, Schroeder MR, Farley MM, Tettelin HH, Stephens DS. Composite mobile genetic elements disseminating macrolide resistance in Streptococcus pneumoniae. Front Microbiol. 2015;6:1–14. doi: 10.3389/fmicb.2015.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bogaert D, Engelen MN, Timmers-Reker AJM, Elzenaar KP, Peerbooms PGH, Coutinho RA, et al. Pneumococcal carriage in children in the Netherlands: a molecular epidemiological study. J Clin Microbiol. 2001;39:3316–20. doi: 10.1128/JCM.39.9.3316-3320.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGee L, McDougal L, Zhou J, Spratt BG, Tenover FC, George R, et al. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J Clin Microbiol. 2001;39:2565–71. doi: 10.1128/JCM.39.7.2565-2571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–4. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Croucher Nicholas J., Mitchell Andrea M., Gould Katherine A., Inverarity Donald, Barquist Lars, Feltwell Theresa, Fookes Maria C., Harris Simon R., Dordel Janina, Salter Susannah J., Browall Sarah, Zemlickova Helena, Parkhill Julian, Normark Staffan, Henriques-Normark Birgitta, Hinds Jason, Mitchell Tim J., Bentley Stephen D. Dominant Role of Nucleotide Substitution in the Diversification of Serotype 3 Pneumococci over Decades and during a Single Infection. PLoS Genetics. 2013;9(10):e1003868. doi: 10.1371/journal.pgen.1003868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreno-Gamez S, Sorg R, Kjos M, Weissing F, van Doorn GS, Veening J. Quorum sensing integrates environmental cues, cell density and cell history to control bacterial competence. bioRxiv. 2016;31:75762. doi: 10.1038/s41467-017-00903-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slager J, Kjos M, Attaiech L, Veening JW. Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell. 2014;157:395–406. doi: 10.1016/j.cell.2014.01.068. [DOI] [PubMed] [Google Scholar]
- 46.Kümmerli R, Brown SP. Molecular and regulatory properties of a public good shape the evolution of cooperation. Proc Natl Acad Sci USA. 2010;107:18921–6. doi: 10.1073/pnas.1011154107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, Evans BA, Rozen DE. Signal diffusion and the mitigation of social exploitation in pneumococcal competence signalling. Proc R Soc B Biol Sci. 2010;277:2991–9. doi: 10.1098/rspb.2010.0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruparell A, Dubern JF, Ortori CA, Harrison F, Halliday NM, Emtage A, et al. The fitness burden imposed by synthesizing quorum sensing signals. Sci Rep. 2016;6:33101. doi: 10.1038/srep33101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valente C, Dawid S, Pinto FR, Hinds J, Simões AS, Gould KA, et al. The blp locus of Streptococcus pneumoniae plays a limited role in the selection of strains that can cocolonize the human nasopharynx. Appl Environ Microbiol. 2016;82:5206–15. doi: 10.1128/AEM.01048-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.West SA, Winzer K, Gardner A, Diggle SP. Quorum sensing and the confusion about diffusion. Trends Microbiol. 2012;20:586–94. doi: 10.1016/j.tim.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 51.Zhang F, Kwan A, Xu A, Süel GM. A synthetic quorum sensing system reveals a potential private benefit for public good production in a biofilm. PLoS ONE. 2015;10:15–19. doi: 10.1371/journal.pone.0132948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stefanic P, Decorosi F, Viti C, Petito J, Cohan FM, Mandic-Mulec I. The quorum sensing diversity within and between ecotypes of Bacillus subtilis. Environ Microbiol. 2012;14:1378–89. doi: 10.1111/j.1462-2920.2012.02717.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.