Genetically matching tusks from the same elephant in consecutive shipments links the same trafficker to multiple ivory seizures.
Abstract
Rapid growth in world trade has enabled transnational criminal networks to conceal their contraband among the 1 billion containers shipped worldwide annually. Forensic methods are needed to identify the major cartels moving the contraband into transit. We combine DNA-based sample matching and geographic assignment of tusks to show that the two tusks from the same elephant are often shipped by the same trafficker in separate large consignments of ivory. The paired shipments occur close in time from the same initial place of export and have high overlap in the geographic origins of their tusks. Collectively, these paired shipments form a linked chain that reflects the sizes, interconnectedness, and places of operation of Africa’s largest ivory smuggling cartels.
INTRODUCTION
Major transnational organized crimes (1) have increased dramatically since 2006, coincident with large increases in legal containerized cargo shipped worldwide (2). The illegal trade in African elephant ivory is no exception. That trade has grown into a multibillion dollar industry responsible for the deaths of up to 40,000 elephants annually. Seventy percent of all ivory seizures are shipped in large (≥0.5 metric tons) containerized consignments. Increases in ivory seizure rates have been dominated by shipments ≥100 kg (3), and the amount of ivory being trafficked continues to increase (4). Intelligence-led forensic tools are urgently needed to help stop this illegal wildlife trade from decimating the world’s largest land mammal.
Genetics offer a unique solution. Wasser et al. (5) developed DNA-based tools to identify the major poaching hotspots in Africa, hoping to prevent contraband from entering transit by focusing law enforcement at the poaching source. These methods were able to statistically assign geographic origin of ivory samples to within 300 km of the poaching source by comparing their genotypes at 16 microsatellite DNA loci to a comprehensive DNA reference map of elephants assembled from across Africa. Applying these methods to representative samples from numerous large ivory seizures (≥500 kg) enabled Wasser et al. to identify the two largest ivory poaching hotspots in Africa over the past decade. Nevertheless, containing large-scale poaching and ivory trafficking has remained a challenge (4). Presumably, poachers are hard to catch because they typically operate in large protected areas they know well. Even when apprehended in the field, poachers rarely have more ivory than they can carry. They are seldom prosecuted, and many other potential poachers are ready to take their place.
The above challenges led us to expand the genetic methods of Wasser et al. (5) to identify and focus law enforcement on the export cartels consolidating and shipping large volumes of ivory out of Africa. Most poachers appear to sell their ivory to a pyramid-shaped hierarchy of middlemen who progressively consolidate the ivory as it moves up the crime chain to the export cartels. These cartels are the final consolidators, exporting individual containers filled with multiple metric tons of ivory out of Africa, many times each year (2, 6). The high cost of munitions needed to kill elephants on such a large scale further suggests that these same cartels are directly or indirectly funding poaching operations on the ground. Targeting the major export cartels could thus provide some of the most direct ways to police this illegal trade and stop the killing. To this end, we use genetic methods to determine the number, scale, and location of Africa’s major ivory export cartels as well as their connection to in-country poaching hotspots.
Our approach stemmed from our discovery that over half the tusks in the large ivory seizures we have sampled appeared to be unpaired (that is, only one of the two tusks from the same animal could be identified in the shipment). We arrived at that conclusion after devising methods to reduce DNA analysis costs by physically identifying and excluding one of the two tusks from same elephant. Tusks were paired on the basis of similarities in their color, diameters at the base (where the tusks exit the jaw), and, most importantly, the distance from the base of the tusk to gum line (a highly visible line marking where the tusk protrudes from the lip of the elephant) (5). We hypothesized that tusk pairs from the same animal often become separated en route from the kill site to export location, resulting in the two tusks being shipped in separate consignments. However, assuming that cartels compete for increasing large shares of the trade, both tusks should still have a high likelihood of being acquired and exported by the same cartel. Assuming also that traffickers tend to repeatedly use the same ports to ship their containerized contraband out of Africa, separate seizures containing genetically matched tusks should tend to be shipped from the same port, close in time, with high overlap in the geographic origin of tusks comprising the two linked seizures. Overlap in geographic origin would additionally reflect a connection between the export cartel and poaching operations on the ground. Finally, the size of the cartel should be reflected by the number of links in the chain of genetically matched tusks connecting multiple shipments.
RESULTS
Genetically matched samples in separate seizures
We sampled 38 large ivory seizures (≥0.5 metric tons) made around the world between 2006 and 2015. An average of 36% of the total number of tusks were sampled per seizure, using methods detailed in Wasser et al. (5). Briefly, after first excluding one tusk from each visually identified pair, all remaining tusks were divided into groups based on similar physical characteristics (for example, presence versus absence of burn marks caused by poachers to remove tissue, writing on tusks, color). Tusks were then randomly sampled proportionately from each group until the desired sample size was obtained. All sampled tusks were genotyped at 16 microsatellite DNA loci (5). Any additional samples in the same seizure found to have matching genotypes were excluded. Samples that failed to amplify both alleles at a minimum of 10 of 16 loci were also excluded from further analysis. This yielded 3315 samples with unique genotypes within seizures to be examined for potential genetic matches between seizures. Bone samples from 10 unique elephants killed in a high-profile poaching incident involving a helicopter in northeastern (NE) Democratic Republic of the Congo (DRC) (6) were similarly examined for matches to tusks in these seizures.
Samples between seizures were considered a match if they had identical genotypes, confirmed at 10 or more loci for both alleles with any discrepancies beyond the 10 loci explained by allelic dropout (Materials and Methods). Twenty-eight sample pairs met these matching criteria among 5,336,302 multilocus genotype comparisons. We then computed forensic genetic likelihood ratios (7) for pairs that met the criteria to quantify the evidentiary strength of the match, taking into account repeat measurement (8), the effect of population structure on allele frequencies (9), and the occurrence of allelic dropout/in (Table 1) (10). We considered multiple defense hypotheses, allowing for the genotypes to come from either related or unrelated individuals in the same subpopulation. Only samples with a likelihood ratio for unrelated alternates ≥107 were considered a match, indicating that the observed genotypes are at least 10 million times more likely if they came from the same individual than if they came from unrelated individuals in the same subpopulation. Twenty-six of the 28 matches met that criterion.
Table 1. Likelihood ratios for matched pairs of ivory samples between seizures.
| Sample 1 | Sample 2 | Species | LR (unrelated) | LR (half sib) | LR (parent-offspring) | LR (full sib) |
| AVP.EBB.154 | KKP015 | Savannah | 5.92 × 1011 | 4.57 × 108 | 6.19 × 106 | 3.54 × 104 |
| Uganda Oct 2013 2.9t | Kenya Jun 2013 1.5t | |||||
| AVP.EBB.239 | KKP009 | Savannah | 4.60 × 1012 | 1.82 × 109 | 1.27 × 107 | 1.02 × 105 |
| Uganda Oct 2013 2.9t | Kenya Jun 2013 1.5t | |||||
| AVP.EBB.391 | KKP347 | Savannah | 1.04 × 1011 | 1.37 × 108 | 1.86 × 106 | 3.72 × 104 |
| Uganda Oct 2013 2.9t | Kenya Jun 2013 1.5t | |||||
| DUB030 | KSP046 | Savannah | 6.05 × 109 | 1.38 × 106 | 9.94 × 103 | 5.24 × 101 |
| Dubai May 2013 1.5t | Kenya Jan 2013 3.8t | |||||
| DUB060 | KCM045 | Savannah | 1.14 × 1011 | 8.65 × 107 | 2.08 × 106 | 8.36 × 103 |
| Dubai May 2013 1.5t | Kenya Oct 2013 2.9t | |||||
| DUB065 | KSP139 | Savannah | 1.23 × 1011 | 2.61 × 108 | 4.51 × 106 | 4.40 × 104 |
| Dubai May 2013 1.5t | Kenya Jan 2013 3.8t | |||||
| DUB141 | KCM026 | Savannah | 1.07 × 1012 | 6.61 × 108 | 8.73 × 106 | 5.14 × 104 |
| Dubai May 2013 1.5t | Kenya Oct 2013 2.9t | |||||
| DUB142 | KCM019 | Savannah | 2.34 × 1011 | 1.12 × 107 | 5.42 × 104 | 1.81 × 102 |
| Dubai May 2013 1.5t | Kenya Oct 2013 2.9t | |||||
| KAD06 | KSP001 | Savannah | 3.01 × 107 | 5.39 × 104 | 8.26 × 102 | 2.64 × 101 |
| Kenya Sep 2011 0.3t | Kenya Jan 2013 3.8t | |||||
| KCM054 | KX093 | Savannah | 6.91 × 102 | 5.56 × 100 | 2.46 × 10−1 | 7.73 × 10−2 |
| Kenya Oct 2013 2.9t | Kenya Oct 2013 2.0t | |||||
| KCM111 | KX038 | Savannah | 3.14 × 1012 | 1.50 × 109 | 1.49 × 107 | 6.96 × 104 |
| Kenya Oct 2013 2.9t | Kenya Oct 2013 2.0t | |||||
| KCM118 | KX100 | Savannah | 1.71 × 1011 | 2.01 × 108 | 3.77 × 106 | 2.69 × 104 |
| Kenya Oct 2013 2.9t | Kenya Oct 2013 2.0t | |||||
| KCM140 | MYS1916 | Savannah | 2.32 × 1013 | 4.75 × 109 | 4.18 × 107 | 9.87 × 104 |
| Kenya Oct 2013 2.9t | Malaysia Dec 2012 6.0t | |||||
| KCM160 | UG.B137 | Savannah | 3.80 × 106 | 1.37 × 103 | 1.43 × 101 | 2.38 × 101 |
| Kenya Oct 2013 2.9t | Uganda May 2014 1.8t | |||||
| KCM168 | UG.B131 | Savannah | 1.07 × 1014 | 7.15 × 109 | 4.20 × 107 | 7.64 × 104 |
| Kenya Oct 2013 2.9t | Uganda May 2014 1.8t | |||||
| KCM170 | KX130 | Savannah | 3.43 × 1012 | 1.45 × 109 | 1.54 × 107 | 5.59 × 104 |
| Kenya Oct 2013 2.9t | Kenya Oct 2013 2.0t | |||||
| KKP147 | UWA.A.040 | Savannah | 4.61 × 1010 | 6.23 × 107 | 8.83 × 105 | 1.29 × 104 |
| Kenya Jun 2013 1.5t | Uganda Dec 2013 1.4t | |||||
| KKP323 | UWA.B.100 | Savannah | 2.56 × 1013 | 4.42 × 109 | 2.46 × 107 | 1.43 × 105 |
| Kenya Jun 2013 1.5t | Uganda Dec 2013 1.4t | |||||
| KMN004 | SL324 | Savannah | 1.48 × 1013 | 2.96 × 109 | 1.86 × 107 | 1.16 × 105 |
| Kenya Dec 2011 1.5t | Sri Lanka May 2012 1.5t | |||||
| MYS0012 | TOGI030 | Savannah | 1.20 × 1014 | 8.30 × 109 | 3.80 × 107 | 1.34 × 105 |
| Malaysia Dec 2012 6.0t | Togo Jan 2014 3.9t | |||||
| MYS0062 | TOGI176 | Savannah | 1.25 × 108 | 8.39 × 105 | 4.37 × 104 | 1.34 × 103 |
| Malaysia Dec 2012 6.0t | Togo Jan 2014 3.9t | |||||
| MYS1119 | SL129 | Savannah | 2.76 × 1016 | 2.28 × 1011 | 4.49 × 108 | 4.87 × 105 |
| Malaysia Dec 2012 6.0t | Sri Lanka May 2012 1.5t | |||||
| G04 | KKP021 | Forest | 3.25 × 1015 | 6.98 × 109 | 9.74 × 106 | 8.89 × 101 |
| Garamba incident | Kenya Jun 2013 1.5t | |||||
| G04 | KKP123 | Forest | 3.25 × 1015 | 6.98 × 109 | 9.74 × 106 | 8.89 × 101 |
| Garamba incident | Kenya Jun 2013 1.5t | |||||
| G05 | KKP020 | Forest | 2.05 × 1020 | 2.32 × 1014 | 2.19 × 1011 | 5.71 × 106 |
| Garamba incident | Kenya Jun 2013 1.5t | |||||
| G09 | KKP140 | Hybrid | 1.70 × 1019 | 4.17 × 1013 | 5.33 × 1010 | 3.20 × 106 |
| Garamba incident | Kenya Jun 2013 1.5t | |||||
| G15 | KKP118 | Forest | 8.46 × 1017 | 1.02 × 1013 | 2.43 × 1010 | 1.48 × 106 |
| Garamba incident | Kenya Jun 2013 1.5t | |||||
| MYS0648 | TG59 | Forest | 2.33 × 1017 | 2.87 × 1012 | 8.71 × 109 | 7.37 × 105 |
| Malaysia Dec 2012 6.0t | Aug 2013 0.7t |
The 26 matched samples found in separate seizures (termed matched seizures) are shown in Fig. 1A. Matched seizures are connected by double-headed arrows. Arrow thickness denotes the number of matching samples among the two seizures, and arrow color corresponds to the common port that both seizures passed through. Four of the 26 sample matches, indicated by a single-headed arrow, were bone samples from the carcasses shot by helicopter in NE DRC in May 2012. Those samples were matched to tusks in a seizure being exported out of Mombasa [Kenya Jun 2013 1.5 metric tons (t)] after transiting Entebbe, Uganda. That same seizure also matched two other seizures made in Entebbe (Fig. 1A).
Fig. 1. Genetically matched tusks between large ivory seizures.
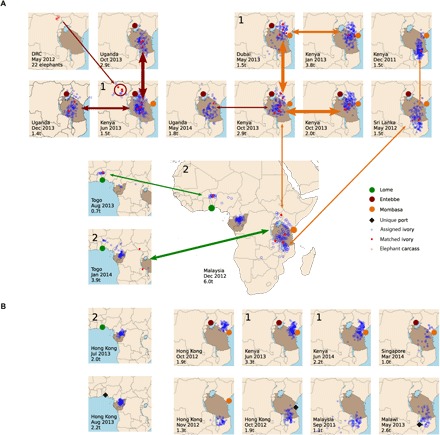
Large ivory seizures made between December 2011 and May 2014, with (A) and without (B) genetically matched tusks between the seizures. The location, date, and weight [in metric tons (t)] of each seizure are written in the lower left corner of each map. The locations where each seizure transited out of Africa are indicated by large colored circles (maroon, orange, and green), with the exception of unique ports (black diamonds) that appear only once. Countries colored dark brown indicate poaching centers (5). The genetically assigned geographic origin of each tusk within the seizure is illustrated by blue circles. Matched tusks between seizures are indicated by solid red circles. Double headed arrows connect matched seizures; arrow thickness denotes the number of matching samples between the two seizures; and arrow color corresponds to the last common port of transit between matched seizures. Seizures linked by one or more types of nongenetic forensic evidence are indicated by the same number in the upper left-hand corner of their respective maps. 1 = same clearing agent, shared cell phone number, vehicle or owner of vehicle transporting ivory, owner of shipment, law enforcement impressions; 2 = ivory concealed under sawn mahogany planks.
The three different arrow colors connecting multiple linked seizures in Fig. 1A indicate three major export networks moving some of the greatest quantities of ivory out of Africa. All matched seizures in a network passed through a common port, and, in all but one case, matched seizures were always shipped ≤10 months apart. The one exceptional case involved a Togo (Jan 2014 3.9t) seizure that matched the unusually large Malaysia (Dec 2012 6.0t) seizure that transited Togo 14 months earlier. That match had multiple other unique features, as discussed in greater detail below.
Predictors of between-seizure sample matches
The 38 seizures make 703 seizure pairs (37 × 38/2). A Poisson regression (see Materials and Methods) was run for all 703 possible seizure pairs. The Poisson regression was also run separately for just the 630 possible pairs (35 × 36/2), consisting only of savannah elephant sample matches since all but one matching tusk pair occurred among savannah elephant tusks. Both models agree that the number of sample matches between two seizures increases when they share a port (P: P < 0.0001 overall, P < 0.0003 savannah only), the time separating the two seizures decreases (T: P < 0.0001 for both models), and the overlap in the assigned geographic origins of the tusks in the two seizures increases (O: P < 0.03 overall, P < 0.003 savannah only).
Occurrence of tusk pairs shipped in separate seizures
Of the 38 seizures analyzed, all 11 seizures containing the 26 pairs of matching samples occurred among the 23 seizures made between December 2011 and May 2014 (Fig. 1A). Those 23 seizures represent 60% of all reported large ivory seizures during that period (3). A much smaller percentage of total seizures were obtained before and after those dates, highlighting the importance of good seizure access to the success of this method. It is similarly important to know how often sample pairs are actually shipped in separate seizures and the sampling intensity needed to detect them. To address this issue, we used the matched seizures examined during 2011–2014 to estimate the actual number of matching samples (Mij) needed to maximize the probability of observing mij matching samples between seizures i and j, taking into account the number of tusks we genotyped relative to the total number of tusks in the matching seizures (see Materials and Methods). On average, we estimated that 2.25% of samples per seizure [Mij/min(Ni,Nj)] matched tusks shipped in a separate seizure (table S5). Since our sampling protocol was sufficient to identify these matches, we recommend this protocol be used in future sampling efforts: randomly sampling 35% of the total number of tusks per seizure, after excluding one tusk from all visually identified pairs (5). However, for seizures in excess of 600 samples, we recommend capping the number of samples at 200 should analysis cost become an issue.
Connecting multiple ivory cartels
Some seizures matched multiple other seizures (Fig. 1A). Although not reflected in the 2.25% estimate above, seizures that matched multiple others may be particularly strategic for understanding the criminal networks being investigated. For example, the Malaysia seizure (Dec 2016 6.0t) passed through the Mombasa and Togo ports and included tusks with matches in two other seizures at each of those ports. Those combined matches also link together the major export cartels in Mombasa and Togo (respectively shown by the orange and green arrows pointing to the Malaysia seizure in Fig. 1A). One of the Mombasa seizures with matches to the Malaysia seizure (Kenya Oct 2013 2.9t) also included tusks that matched three other seizures, and one of those matches (Uganda May 2014 1.8t) potentially connects the Mombasa cartel to the Entebbe cartel (see also below).
The 6.0–metric ton Malaysia seizure is also noteworthy for including nearly 4 metric tons of savannah and 2 metric tons of forest elephant ivory, respectively, originating from Africa’s two largest poaching hotspots (5). However, for the following reasons, we concluded that the Malaysia seizure included only savannah tusks when it left Mombasa and was offloaded in Togo where the Central and West African forest elephant tusks were subsequently added to the Malaysia shipment before the final export of that seizure out of Togo: The large quantities of forest and savannah elephant tusks in the Malaysia seizure, respectively, came from opposite sides of Africa, and samples in the Malaysia seizure were matched to tusks from two East African and two West African seizures. Two additional observations further suggest that the repacking and export of the Malaysia seizure from Togo involved the same West African trafficker(s) implicated in the two Togo seizures it matched. First, one of the West African tusks in the Malaysia seizure matched a tusk from a seizure made in the warehouse of an individual alleged to be a major ivory trafficker in West Africa (Togo Aug 2013 0.7t). That finding strongly suggests that the matching tusk in the Malaysia seizure was taken from the West African trafficker’s warehouse and added to the Malaysia shipment by the West African trafficker (5). Second, the two tusks in the other Togo seizure (Jan 2014 3.9t) that had matches in the Malaysia seizure originated from East African savannah elephants (Fig. 1A). The likelihood of those two savannah elephant tusks being in the 3.9–metric ton Togo seizure is extremely low since 92% of the tusks in that seizure were from forest elephants (5). In fact, we estimated that the Togo (Jan 2014 3.9t) seizure would have had to include 500 savannah elephant tusk matches among its 1500 total tusks to maximize the likelihood of observing the two savannah tusk matches among the Togo and Malaysia seizures (table S5). (That is also why we excluded that case from the 2.25% estimate mentioned above.) Undoubtedly, the savannah tusks in the second Togo seizure were removed from the Malaysia seizure prior to its export out of Togo (presumably by the aforementioned West African trafficker), only to be reexported in that 3.9–metric ton Togo seizure 14 months later. The trafficker convicted for the first Togo seizure (Aug 2013 0.7t) was already in custody by this time, and the second seizure (Jan 2014 3.9t) may have been a failed attempt by his associates to dump additional evidence that could be used against him.
Nongenetic forensic evidence linking separate seizures
Ten seizures (Fig. 1B) made during the 2011–2014 period were not observed to match each other or any of the 13 matching seizures in Fig. 1A. However, 5 of those 10 (top row Fig. 1B) shared all three of the predictive matching criteria with one or more of the 13 seizures in Fig. 1A: shipped from a common port, close in time, and with high overlap in the assigned geographic origins of tusks in those seizures (Fig. 1, A and B). Nongenetic forensic evidence (for example, shared cell phone number, same clearing agent, same driver, similar methods used to hide the ivory) acquired from open source information and/or collaborating law enforcement authorities linked three of those five seizures to each other and/or to four seizures in Fig. 1A. Each of those nongenetic linkages is indicated by the same number in the upper left-hand corner of the seizures in Fig. 1 (A and B). One set of connections [designated by a 1 in the upper left corner of seizures from Kenya Jun 2013 3.3t and Kenya Jun 2013 2.2t (Fig. 1B) and Kenya Jun 2013 1.5t and Dubai May 2013 1.5t (Fig. 1A)] is consistent with the previously described genetically matched links between the Entebbe and Mombasa cartels (for example, Uganda May 2014 1.8t and Kenya Oct 2013 2.9t seizures). The Kenya Jun 2014 2.2t seizure in Fig. 1B is also the seizure that led to the conviction and 20-year prison sentence of one of the biggest ivory traffickers in Africa (11). The Kenya (Jun 2013 1.5t) seizure, allegedly connected to the Jun 2014 2.2t seizure, was the same one that had multiple matches to tusks in two large Uganda seizures and to the elephants killed by helicopter in DRC (May 2012).
DISCUSSION
We identified three major export cartels operating in Africa between 2011 and 2014. Each cartel was identified by a chain of multiple linked seizures exported or about to be exported out of Africa from the same port, with the two matched seizures in any given link occurring close in time and composed of tusks largely derived from the same poaching locale. Targeting such cartels could have a major direct impact on combating the illegal ivory trade by preventing contraband from transiting out of Africa before it becomes far more diffuse and expensive to trace. High overlap in the genetically identified geographic origins of the ivory in the matched seizures, along with the convergence of large quantities of ivory to the same cartels over time, also implies a strong link between the cartels and the poaching hotspots where those elephants are being killed. Thus, targeting these cartels could also directly reduce the rapid loss of (keystone) wildlife and the associated damage caused by that loss. The tools described in this paper may further provide information that can help law enforcement target other forms of contraband to the extent that these major ivory cartels are also involved in smuggling these other products (11).
Most ivory traffickers facing prosecution are charged for a single seizure. Methods that can connect individual traffickers to multiple large seizures have the potential to elevate their charges to major transnational crimes, simultaneously increasing the severity of their sentences. The trafficker implicated by our results in the two Togo seizures and the 6.0–metric ton Malaysia seizure was convicted only for the Aug 2013 0.7t Togo seizure, partly from genetic evidence we provided (12). He received the maximum sentence of 2 years in Togo and is already out of prison. His sentence could have been much stiffer had the additional findings in this report been available at the time of his prosecution. Another major trafficker was similarly convicted for one seizure (Kenya Jun 2014 2.2t), but this report potentially connects him to many others. His case is now under appeal. There is currently one suspect in custody, awaiting prosecution for one of the Uganda seizures in the Entebbe network. However, findings in this paper connect that network to at least two other seizures, one of which is linked to a major international incident involving poaching from a military helicopter belonging to a neighboring country (Fig. 1A).
The effectiveness of geographic origin assignment and sample matching methods as tools to target Africa’s major ivory traffickers depends on the willingness of countries making large seizures to submit samples for DNA analysis as close as possible to the time of the seizure. Previous work found that poaching hotspots are slow to change (5) and that most of this ivory is from recently killed elephants (13). Collectively, that makes recent seizures predictive of near future poaching events. Cartels may be even slower to change owing to the connections required to maintain their operations and dominance. Recognizing this, CITES (Convention on International Trade in Endangered Species) Resolution Conf. 10.10 urges all countries making large ivory seizures to provide representative samples for origin analysis as soon as possible. However, to date, compliance has been exceedingly poor (4, 5). Hopefully, our findings will stimulate better compliance, increasing the availability of reliable intelligence needed to combat these and other transnational organized crimes.
MATERIALS AND METHODS
Genetic data and species identification
We refer to two different species (sometimes called subspecies) of African elephants: forest (Loxodonta cyclotis) and savannah (Loxodonta africana). These two species are genetically distinct and occupy mostly nonoverlapping regions of Africa (14), although hybridization has been documented in select areas (15).
Elephant reference samples of known geographic origin (mostly dung but also some hair and tissue) were obtained from across Africa and assayed in replicate at 16 dinucleotide microsatellite loci (5). We aimed to collect 30 unique genotypes from each population, collecting samples at a minimum of 1 km apart to increase the likelihood that each sample represented a unique genotype from a separate family group. All populations were separated by at least 1° latitude and/or 1° longitude.
Each reference sample had a minimum of two extracts, and each extract had two or more replicate polymerase chain reactions at each locus. A sample was confirmed homozygous at a locus if one allele was seen in at least three replicates and no other allele was seen in more than one replicate. A sample was assigned heterozygous at a locus if two different alleles were seen in at least two replicates each and no other allele was seen in more than one replicate. If neither of these criteria were met, the sample did not receive a confirmed genotype at that locus. Different reference samples that appear to have the same genotype were identified using CERVUS (16) and collapsed to a single sample with a consensus genotype (5). Those reference samples with 10 or more confirmed loci were included in subsequent analyses.
Reference samples were assigned a species (or, rarely, hybrid status) based on likelihood computations implemented in the program SCAT (Smoothed Continuous Assignment Technique) (14). Reference samples were then grouped into reference locations (populations) based on their known origin, at a resolution of approximately 1° latitude and longitude, with each species having its own collection of populations. (Reference samples identified as hybrids by SCAT were excluded.) A total of 580 unique forest reference samples with 36 distinct population locations and 1264 unique savannah population samples with 55 distinct reference locations comprised the reference data (table S1).
Ivory samples were obtained from 38 ivory seizures made between 2006 and 2016, 37 of which were a half metric ton or larger in total weight (table S2). All ivory samples were assayed in replicate at the same 16 loci used for reference samples, with the same criteria for achieving a confirmed genotype (5). Within each seizure, samples appearing to have the same genotype were identified using CERVUS and collapsed to a single sample with a consensus genotype (5). Those ivory samples with 10 or more confirmed loci were included in subsequent analyses. Species name was assigned to these ivory samples based on likelihood computations implemented in the program SCAT (14).
When referring to seizures, we used the following identifiers: location of seizure, month and year of seizure, and weight of seizure (in metric tons). This identification was unambiguous in all cases except in Hong Kong Oct 2012 1.9t, as there were two seizures fitting that description. However, those two seizures can be distinguished by their known points of export: for one seizure exported from Tanzania and the other from Kenya (Fig. 1B).
To model the probability of allelic dropout in the ivory data (as required for the likelihood calculations in the section “Computation of likelihood ratios”), an additional data set based on serial dilutions of ivory samples was generated. A total of 20 ivory samples (10 forest and 10 savannah elephant samples) were selected for the high quality of their initial assay results, with all 20 samples having 16 confirmed loci. These samples were extracted in duplicate; each extract was serially diluted and assayed in duplicate until complete or almost complete dropout was achieved at all loci across both diluted extracts (thus encompassing all template concentrations likely to be found in the ivory data). This required between 10 and 14 dilutions per sample, with savannah elephant samples; slightly more dilutions were required on average for forest elephant samples. Extract concentration was reduced (multiplicatively) by 0.6 for each of the first four dilutions and by 0.3 for each of the remaining dilutions. A total of 456 replicates from savannah elephant samples and 440 replicates from forest elephant samples were thus generated for each locus. We assumed that this approach approximates allelic dropout from degraded samples because visually degraded samples are avoided in the initial sampling of the seizures and amplification success for genotyped samples is reasonably good (65%).
Inconsistencies in genotyping of larger alleles at locus FH153
In generating the serial dilution data described above, inconsistencies in genotyping results at locus FH153 were discovered. These inconsistencies affected larger alleles [200 base pairs (bp) or more], which were found almost exclusively in forest elephants. Roughly, the inconsistencies manifested as extract concentration–dependent allele calls, where more highly diluted extracts of a given sample tended to show alleles one or two repeats (2 or 4 bp) shorter than less-diluted extracts of the same sample. This phenomenon did not occur with all forest elephant samples, nor did it occur with all alleles over 200 bp, but it did occur with multiple forest elephant samples from different seizures. These issues were not observed with any savannah elephant samples. (Most samples classified as savannah elephant have alleles of size 185 or smaller at this locus, but a few have alleles as large as 285, probably due to low amounts of forest elephant ancestry in those samples. In the dilution data, the largest allele at this locus in a savannah elephant sample was 173, and in the matching pairs of ivory samples discussed below, the largest allele at this locus in a savannah elephant sample was 177.)
To avoid exposure to this source of genotyping error, the locus FH153 was excluded from consideration for all forest and hybrid elephant samples in all analyses described below. As no evidence of genotyping issues for savannah elephant samples was detected, the locus was retained for analysis of savannah elephant samples. Thus, in the remainder of this document, forest and hybrid elephant samples effectively have 15 loci under consideration, while savannah elephant samples have 16 loci under consideration.
Genotypic matching between seizures
The ivory data were examined for potential genetic matches between samples occurring in different seizures. Also included in this analysis were 10 samples (9 forest and 1 hybrid elephant samples) from a May 2012 poaching event in Garamba National Park, DRC [regarded as its own seizure for matching purposes, although the samples were (bone) tissue obtained from carcasses on the ground].
A total of 5,336,302 genotype pairs from different seizures were analyzed. A pair of samples was termed a matching pair if 10 or more loci were confirmed in both samples and had identical confirmed genotypes, and any discrepancies in confirmed genotype at remaining loci could be explained by allelic dropout (table S3). Loci that were not confirmed in at least one of the two samples under consideration did not explicitly factor in to this matching analysis. Locus FH153 was not considered in forest or hybrid elephant samples.
Computation of likelihood ratios
For each of the matching pairs, we computed forensic genetic likelihood ratios, as described in the remainder of this section. Only one of the paired samples, KKP021-123, was itself a collapsed within-seizure match. For likelihood computations, the two contributing samples KKP021 and KKP123 were each compared to the matched bone sample from the DRC carcasses, rather than pooling their data to make a single comparison. Both samples matched G04, twice confirming the match.
Allele frequencies, population structure corrections, and dropout model parameters depended on the species. This presented a problem of how to handle the hybrid pair. Previous work (15) suggested that the hybrid elephant pair was a backcross to forest elephants, so we therefore treated the hybrid elephant pair as a forest elephant pair when computing likelihood ratios. This approach was not unassailable, but the strength of this particular match should not be in serious doubt: All loci (including FH153 if desired) showed complete agreement for this pair, so any computation taking admixture into account (regardless of the exact amount) would likely arrive at the same conclusion regarding the high strength of the match.
Allele frequencies and population structure
Allele frequencies for each species were estimated by taking the arithmetic average empirical allele frequencies across reference locations for that species with 10 or more samples. This included 17 forest reference locations with 495 total forest elephant samples and 38 savannah reference locations with 1186 total savannah elephant samples. We assume that our 16 loci are unlinked. Thirteen loci are distributed on 11 chromosomes. Two other locus pairs each share a chromosome but are separated by 8 to 10 million bp.
For each species, the population-specific FST estimates (9) were computed with the same 17 forest and 38 savannah elephant reference population locations used in allele frequency estimation. The arithmetic average of the βi across reference locations served as the species-specific estimate of θ used in genotypic probability calculations below (figs. S1 and S2).
Some matching pairs of ivory samples showed an allele that was not present in the reference data used for allele frequency estimation. For these pairs, the allele in question was introduced with frequency θ, and other allele frequencies at that locus were multiplied by 1 − θ before proceeding with the likelihood ratio calculations described below. (In a single case, a matching pair showed two alleles at the same locus that were not in the reference data; each was introduced with frequency θ, and other allele frequencies were multiplied by 1 − 2θ).
Dropout model
Allelic dropout probabilities were modeled as locus- and species-specific logistic functions of log average peak height (10). In the serial dilution data, the template proxy is defined for each (extract, dilution factor) pair as the average across loci of the average within-locus peak height, the averages being computed over all replicates of the given extract at the given dilution factor. In the matching ivory data, the definition was equivalent but simpler, as each extract has only one (neat) dilution factor. Again, locus FH153 is included for savannah elephant samples but excluded for forest elephant samples.
Thus, for a given (extract, dilution factor), if we let nl denote the number of replicates for locus l and Hl, the sum of all observed peak heights at locus l, then the template proxy H is
This formulation gives all loci equal weight in determining the template proxy even if the number of replicates varies by locus, which was the case in the matching ivory data but not in the serial dilution data.
To define the training data for the logistic models, we regard the true genotypes of the samples in the dilution data as known and equal to the confirmed genotypes. Therefore, we know which (confirmed) alleles have dropped out in each replicate in the dilution data. We let y denote the binary response variable, which is 1 when a confirmed allele drops out in a given replicate and 0 when it does not drop out, and x the explanatory variable, which is log H when the confirmed genotype at the locus in question is heterozygous and log 2H when it is homozygous. Thus, each replicate at a confirmed homozygous locus contributes one datum point to the training data, and each replicate at a confirmed heterozygous locus contributes two data points to the training data (one for each confirmed allele).
Taking the data for each species separately, for a given locus l, we fit the logistic model
choosing the coefficients β0,l and β1,l by maximum likelihood via the default glm.fit method of the glm function in R (table S5) (17). Dependence on the species is suppressed in the notation.
Having fitted the model parameters, we have the (species-specific) predicted dropout probabilities
for heterozygotes and
for homozygotes. For notational convenience, we define the complementary probabilities
and
We also have the species-specific drop-in probability, determined as the empirical drop-in rate in the dilution data across all replicates
The complementary probability is denoted
There are a couple of technical points to add regarding the template proxy. A large proportion of called alleles had overloaded peak heights, meaning their RFUs (relative fluorescence units) exceeded the threshold that could be accurately measured. This threshold was around 35,000 RFUs. Overloaded peak heights were set equal to 50,000 RFUs when computing template proxy for both dilution data and matching ivory data. A second point pertains to drop-in. In the dilution data, drop-in events were known, and the RFUs of dropped-in alleles were not counted toward the template proxy. In the matching ivory data, all called peaks were counted toward the template proxy, as the true genotypes are not known (all possibilities are allowed). Provided drop-in was rare, this discrepancy should not have been a major factor.
Replicate probabilities
A replicate may have anywhere from zero to three observed alleles. (In the case of three observed alleles, at least one of them is necessarily drop-in, but the observed alleles are still retained in the data if unambiguous calls can be made.)
Let R be a given replicate, regarded as a set containing between zero and three (distinct) alleles, and let H be the template proxy associated to R (that is, associated to the extract of which R is a replicate; there is no need to specify a dilution factor as we have the matching ivory data in mind). In terms of an underlying true genotype g, we have the following replicate probabilities, depending on whether g is homozygous or heterozygous
and
where pc is the frequency of allele c for the species and locus under consideration, and where the indicator functions 1P are defined for any logical proposition P as
The sets R\{a} and R\{a, b} may be empty, and we interpreted the empty product as being 1.
Replicates were regarded as conditionally independent given the underlying genotype and the template proxy, so the above probabilities are multiplied across replicates in the full likelihood computations below.
Genotypic probabilities
We require probabilities for one or two genotypes at a given locus; thus, we require up to four alleles to be considered at once. We let a, b, c, d denote distinct alleles at a given locus, with species-specific frequencies pa, pb, pc, pd computed in the section “Allele frequencies and population structure.” We denote by θ the species-specific FST computed in that same section.
Marginal genotypic probabilities are determined by the Balding-Nichols formulae (18)
and
Joint genotypic probabilities are determined by the Balding-Nichols formula and an assumed relatedness vector k = (k0, k1, k2), where ki is the probability of i shared alleles
Different loci are regarded as unlinked (independent) in the full likelihood computations below.
Likelihood computations
Let S1 and S2 be two samples of the same species for which we wish to compute a likelihood ratio. To write out the full likelihood computation, we will need to introduce some notation. We denote by L the set of loci that are confirmed in both S1 and S2, and for l ∈ L, we denote by Gl the set of all possible genotypes at locus l (determined by the species-specific allele frequencies). We let i be index samples, e be index extracts of a given sample, and r be index replicates of a given extract at a given locus. By Rielr, we mean replicate r of locus l in extract e of sample i, and by Hie, we mean the template proxy for extract e of sample i. We denote by Ri all replicates for sample i; thus, the observed data are R1, R2.
In terms of the now-defined replicate and genotypic probabilities, we can compute the probability of the observed data R1, R2 under the prosecution and defense hypotheses, that is, the likelihood of those hypotheses given the observed data. Under the prosecution hypothesis Hp, both samples come from the same individual, and hence there is a single underlying genotype. We thus have the likelihood
We regard the defense hypothesis as depending on a relatedness vector k = (k0, k1, k2), the assumed relatedness between the two distinct individuals contributing the two samples under the defense hypotheses. We then have the likelihood
Thus, the likelihood ratio (depending on k) is
Values for LR(k) are recorded in Table 1 for each k in {unrelated, parent-offspring, full sib, half sib}.
Regression
We regressed the observed number of matching pairs of ivory samples between each pair of seizures on explanatory variables capturing the temporal and spatial proximity of the pair of seizures. Seizures that shared a (known) point of export, occurred close together in time, and/or had similar distributions of assigned origin (5) were hypothesized to have a larger expected number of observed matches.
We denote by Y the response variable, which is the observed number of matching pairs of ivory samples (with unrelated likelihood ratio in excess of 107) between a given pair of seizures. Three explanatory variables were included in the regressions. The variable P is binary, taking value 1 if the pair of seizures has a common known point of export and 0 otherwise. The variable T is the time difference, in months, between the seizures. The variable O is a measure of overlap between the distributions of assigned origin of the two seizures, defined as follows.
Seizures were assigned location of origin (5) using the continuous assignment method (14) with Voronoi tessellation–based postprocessing (19). Savannah and forest elephant samples were assigned separately, and no hybrids were assigned. For each species, this produced an approximation to the posterior distribution of the location of origin of each sample of that species in the seizure, represented as weights on a 67 × 67 grid of equally sized cells covering the relevant portion (forest or savannah elephant range) of the continent of Africa. The simple average of these sample-specific distributions over all samples in a seizure can be interpreted as the spatial distribution of the entire seizure, defined on the disjoint union of the two species-specific grids. This allowed a measure of spatial overlap between two seizures to be computed as one minus the total variation distance between their respective spatial distributions. Explicitly, if f1 and f2 denote the pmfs (Poisson probability mass functions) of the spatial distributions of two seizures, as defined above, then the value of the overlap variable for this pair of seizures is
where s ranges over all forest and savannah grid squares.
Overall, only 12 of 703 pairs of seizures had any observed matches, making observed matching a rare event for these regressions. The data also exhibited quasi-complete separation on the port variable: All pairs of seizures with observed matches had a common known point of export, P = 1. Both the small number of cases (12) and the separation issue cause ordinary MLEs (maximum likelihood estimates) to be unsatisfactory (in fact, they fail to even exist due to the separation). We therefore fit the models with the Firth correction (20), implemented in JMP (21).
Estimating the actual number matching between-seizure pairs
We used the hypergeometric distribution to estimate the actual number Mij of matching pairs between seizures i and j. In general, the hypergeometric distribution gives the probability of m successes in n draws, made without replacement from a finite population of size N in which there are M successes.
Seizures i and j have ni and nj genotyped tusks, and mij pairs of tusks are seen to match. A genotyped tusk in one seizure can match at most one genotyped tusk in the other, so the observed number of matches must satisfy 0 ≤ mij ≤ n, where n = min(ni,nj).
Seizures i and j have Ni and Nj total tusks. A tusk in one seizure can match at most one tusk in the other, so the maximum number of matches is N = min(Ni,Nj). The actual number of matching pairs, Mij, must also be at least the observed number, so mij ≤ Mij ≤ N.
We want to estimate Mij on the basis of mij, and the maximum likelihood estimate is the value that maximizes the probability of mij. The probability is given by the hypergeometric distribution
where (n − mij) ≤ (N − Mij). The estimated number of matches will satisfy mij ≤ ≤ N and ≤ (N − n + mij).
Estimates for Mij and the statistics used for those estimates are shown in table S5.
Supplementary Material
Acknowledgments
We thank H. J. Kim for help with the figure and manuscript preparation; R. Booth and C. Mailand for help with the DNA analyses and logistics; R. Waples and B. Clark for the comments on the manuscript; and C. Morris, G. Peters, and K. Miles for the information pertaining to some of the seizures in this paper. H. J. Kim assisted in manuscript preparation. Funding: This work was supported by the Paul G. Allen Family Foundation, the Bureau of International Narcotics and Law Enforcement Affairs of the U.S. State Department, the U.N. Office on Drugs and Crime, the World Bank, the Woodtiger Fund, the Wildcat Foundation, the INTERPOL, the Bosack Kruger Charitable Foundation, Paul and Yaffa Maritz, the U.S. Fish and Wildlife Service African Elephant Conservation Act, and the National Institute of Justice (grant no. 2011-DN-13X-K541). Disclaimer: The opinions, findings, conclusions, and recommendations expressed in this publication are those of the authors and do not necessarily reflect those of the agencies or donors that funded this work. The following governments agreed to provide and share results from their ivory seizures: Malaysia, Thailand, the Philippines, Singapore, Cameroon, Taiwan, Hong Kong, Kenya, Uganda, Sri Lanka, Malawi, and Togo. Author contributions: S.K.W. and S.T. conceived the study. Samples were acquired by S.K.W., S.T., Y.H., M.Y.O., and F.A.T.S. Genotyping was conducted by A.T., M.W., Y.H., and S.T. Statistical design and analyses were conducted by S.K.W., B.S.W., and J.B. Manuscript preparation was done by S.K.W. Manuscript edits were done by S.K.W., B.S.W., and J.B. Competing interests: The authors declare that they have no competing interest. Data and materials availability: All data needed to evaluate the conclusion in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. However, ivory samples are subject to restricted access (see www.whitehouse.gov/the-press-office/2014/02/11/fact-sheet-national-strategycombating-wildlife-trafficking-commercial-b for the U.S. regulatory conditions currently governing trade in ivory, which may also apply to availability of samples).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/9/eaat0625/DC1
Table S1A. Western forest elephant reference locations.
Table S1B. Central forest elephant reference locations.
Table S1C. Southern savannah elephant reference locations.
Table S1D. Eastern savannah elephant reference locations.
Table S1E. Northern savannah elephant reference locations.
Table S2A. Seizures 2002–2012.
Table S2B. Seizures 2013–2016.
Table S3A. Matching pairs of savannah elephant samples.
Table S3B. Matching pairs of forest and hybrid elephant samples.
Table S4. Dropout model parameters.
Table S5. Estimated total number of matching samples (Mij) for each seizure pair.
Fig. S1. Population-specific FST estimates for forest elephants.
Fig. S2. Population-specific FST estimates for savannah elephants.
REFERENCES AND NOTES
- 1.C. May, Transnational Crime and the Developing World (2017); http://www.gfintegrity.org/report/transnational-crime-and-the-developing-world.
- 2.UNODC, World Wildlife Crime Report: Trafficking in protected species (2016).
- 3.Underwood F. M., Burn R. W., Milliken T., Dissecting the illegal ivory trade: An analysis of ivory seizures. PLOS ONE 8, e76539 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CITES SC69 Doc.51.1. Report on the conservation status of African and Asian elephants, trade in elephant specimens, the African Elephant Action Plan, and MIKE and ETIS (2017); https://cites.org/sites/default/files/eng/com/sc/69/E-SC69-51-01.pdf.
- 5.Wasser S. K., Brown L., Mailand C., Mondol S., Clark W., Laurie C., Weir B. S., Genetic assignment of large seizures of elephant ivory reveals Africa’s major poaching hotspots. Science 349, 84–87 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wasser S. K., Clark W. J., Drori O., Kisamo E. S., Mailand C., Mutayoba B., Stephens M., Combating the illegal trade in African elephant ivory with DNA forensics. Conserv. Biol. 22, 1065–1071 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Buckleton J. S., Evett I. W., Weir B. S., Setting bounds for the likelihood ratio when multiple hypotheses are postulated. Sci. Justice 38, 23–26 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Curran J. M., Gill P., Bill M. R., Interpretation of repeat measurement DNA evidence allowing for multiple contributors and population substructure. Forensic Sci. Int. 148, 47–53 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Buckleton J., Curran J., Goudet J., Taylor D., Thiery A., Weir B. S., Population-specific FST values for forensic STR markers: A worldwide survey. Forensic Sci. Int. Genet. 23, 91–100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tvedebrink T., Eriksen P. S., Mogensen H. S., Morling N., Estimating the probability of allelic drop-out of STR alleles in forensic genetics. Forensic Sci. Int. Genet. 3, 222–226 (2009). [DOI] [PubMed] [Google Scholar]
- 11.The Economist, Do dope-smugglers also peddle ivory? (2017); https://www.economist.com/news/middle-east-and-africa/21716651-emerging-links-between-two-nasty-trades-do-dope-smugglers-also-peddle-ivory.
- 12.B. Christy, Forensic Technology Helps Case Against West African Ivory Dealer Accused as a Trafficker (2014); https://news.nationalgeographic.com/news/2014/06/140603-ivory-trafficking-elephants-togo-lome-laga-world/.
- 13.Cerling T. E., Barnette J. E., Chesson L. A., Douglas-Hamilton I., Gobush K. S., Uno K. T., Wasser S. K., Xu X., Radiocarbon dating of seized ivory confirms rapid decline in African elephant populations and provides insight into illegal trade. Proc. Natl. Acad. Sci. U.S.A. 113, 13330–13335 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wasser S. K., Shedlock A. M., Comstock K., Ostrander E. A., Mutayoba B., Stephens M., Assigning African elephant DNA to geographic region of origin: Applications to the ivory trade. Proc. Natl. Acad. Sci. U.S.A. 101, 14847–14852 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mondol S., Moltke I., Hart J., Keigwin M., Brown L., Stephens M., Wasser S. K., New evidence for hybrid zones of forest and savanna elephants in Central and West Africa. Mol. Ecol. 24, 6134–6147 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Kalinowski S. T., Taper M. L., Marshall T. C., Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16, 1099–1106 (2007). [DOI] [PubMed] [Google Scholar]
- 17.R Development Core Team, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2017). [Google Scholar]
- 18.Balding D. J., Nichols R. A., DNA profile match probability calculation: How to allow for population stratification, relatedness, database selection and single bands. Forensic Sci. Int. 64, 125–140 (1994). [DOI] [PubMed] [Google Scholar]
- 19.Wasser S., Mailand C., Booth R., Mutayoba B., Kisamo E., Clark B., Stephens M., Using DNA to track the origin of the largest ivory seizure since the 1989 trade ban. Proc. Natl. Acad. Sci. U.S.A. 104, 4228–4233 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Firth D., Bias reduction of maximum likelihood estimates. Biometrika 80, 27–38 (1993). [Google Scholar]
- 21.JMP 13.2.0. SAS Institute Inc (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/9/eaat0625/DC1
Table S1A. Western forest elephant reference locations.
Table S1B. Central forest elephant reference locations.
Table S1C. Southern savannah elephant reference locations.
Table S1D. Eastern savannah elephant reference locations.
Table S1E. Northern savannah elephant reference locations.
Table S2A. Seizures 2002–2012.
Table S2B. Seizures 2013–2016.
Table S3A. Matching pairs of savannah elephant samples.
Table S3B. Matching pairs of forest and hybrid elephant samples.
Table S4. Dropout model parameters.
Table S5. Estimated total number of matching samples (Mij) for each seizure pair.
Fig. S1. Population-specific FST estimates for forest elephants.
Fig. S2. Population-specific FST estimates for savannah elephants.


