Abstract
The fate of carbon sequestered in permafrost is a key concern for future global warming as this large carbon stock is rapidly becoming a net methane source due to widespread thaw. Methane release from permafrost is moderated by methanotrophs, which oxidise 20–60% of this methane before emission to the atmosphere. Despite the importance of methanotrophs to carbon cycling, these microorganisms are under-characterised and have not been studied across a natural permafrost thaw gradient. Here, we examine methanotroph communities from the active layer of a permafrost thaw gradient in Stordalen Mire (Abisko, Sweden) spanning three years, analysing 188 metagenomes and 24 metatranscriptomes paired with in situ biogeochemical data. Methanotroph community composition and activity varied significantly as thaw progressed from intact permafrost palsa, to partially thawed bog and fully thawed fen. Thirteen methanotroph population genomes were recovered, including two novel genomes belonging to the uncultivated upland soil cluster alpha (USCα) group and a novel potentially methanotrophic Hyphomicrobiaceae. Combined analysis of porewater δ13C-CH4 isotopes and methanotroph abundances showed methane oxidation was greatest below the oxic–anoxic interface in the bog. These results detail the direct effect of thaw on autochthonous methanotroph communities, and their consequent changes in population structure, activity and methane moderation potential.
Permafrost, perennially frozen ground ranging in age from two to over 700,000 years old [1], is a significant carbon reservoir, storing 1330–1580 Pg or nearly half the world’s soil carbon [2, 3]. However, rising global temperature is leading to rapid permafrost thaw and release of greenhouse gases (GHGs), resulting in a positive feedback to warming [2]. Current thaw projections under the IPCC’s (Intergovernmental Panel on Climate Change) representative concentration pathway 8.5 predict that between 30 and 99% of near surface (<3.5 m) permafrost will disappear by 2100 [4]. Within this century, 37–174 Pg of carbon is projected to be released to the atmosphere as carbon dioxide (CO2) and the more potent greenhouse gas methane (CH4), due to the microbial degradation of newly thawed soil [2].
Methane flux in thawing permafrost is controlled by microbial methane producers (methanogens) and consumers (methanotrophs), however the environmental factors controlling the distribution of these microorganisms are poorly understood. The next key step towards improving methane emissions models is the incorporation of microbial community data [5–7], which is currently limited by knowledge of the population structure and activity of these communities in the changing natural environment. Aerobic and anaerobic methanotrophs can prevent a large proportion of methane from reaching the atmosphere, with oxidation estimates of 20–60% recorded in permafrost-associated environments [8, 9]. In these systems, aerobic methanotrophs are typically responsible for ameliorating methane release [10], as anaerobic methanotroph abundances are generally below detection levels with very low rates of methane oxidation [11].
Aerobic methanotrophs oxidise methane to methanol using the particulate (pMMO) and/or soluble (sMMO) methane monooxygenase enzymes [12], and are currently restricted to four major lineages, including well-studied members of the Gamma- and Alphaproteobacteria, the intra-aerobic Candidatus Methanomirabilis oxyfera within the NC10 phylum [13], and the verrucomicrobial genera Methylacidiphilum [14, 15] and Methylacidimicrobium [16]. Gammaproteobacterial methanotrophs are organised into 18 characterised genera within the families Methylococcaceae and Methylothermaceae [17–19], whereas alphaproteobacterial methanotrophs are less diverse comprising five genera within the Methylocystaceae and Beijerinckiaceae [18, 20]. The environmental distribution of aerobic methanotrophs is typically examined using the pmoA gene, which encodes subunit A of pMMO. However, Methyloferula and Methylocella within the Beijerinckiaceae lack pMMO, so mmoX, encoding subunit A of the sMMO, must be used as a complementary marker gene [21]. Environmental surveys of pmoA and mmoX sequences have revealed a large diversity of potential methanotrophs outside the cultivated lineages [18]. This novel diversity includes the alphaproteobacterial pmoA group, upland soil cluster alpha (USCα), which is found predominantly in aerobic soils [18], and has recently been identified in permafrost-associated systems [22, 23]. Members of this group have a high affinity for methane and are capable of oxidising methane from the atmosphere [24–26]. Atmospheric methane oxidisers are responsible for the uptake of ~30 Tg of methane (~5% of the total sink) per year, and are the only known biological sink [27–29].
Our understanding of permafrost methanotrophs has focused on intact permafrost where activity is low [10, 22, 30, 31], or artificially thawed incubations [22, 32, 33], with no naturally thawing gradient sites studied to date. Here, the methanotroph communities at Stordalen Mire are examined through metagenomics, metatranscriptomics and paired biogeochemical data, across an environmental thaw gradient, peat depths (surface, mid, deep; spanning the top ~50 cm), and time (September 2010–August 2012). Characterising the presence, genomic potential and activity of methanotrophs across the Stordalen Mire thaw gradient is a significant step towards elucidating the wider role of methanotrophs in the changing Arctic and subarctic environments, and their impact on the global carbon cycle.
Materials and methods
Study site and sample preparation
Sampling of the thaw chronosequence at Stordalen Mire (68° 21′ N, 19° 03′ E, 359 m a.s.l.), porewater and methane flux measurements, and nucleotide extractions and sequencing of DNA were conducted as previously described [34–37]. Metatranscriptome sequencing was performed for select 2010, 2011 and 2012 samples using 240 ng of RNA in the ScriptSeq Complete (Bacteria) low-input library preparation kit (Epicentre). These samples were run on 1/8th of an Illumina NextSeq lane, with initial shallow runs conducted on 1/11th of a HiSeq (Illumina) and MiSeq (Illumina) lane (see Supplementary Information for details).
Genome assembly and binning
All methanotroph population bins were within the set of 1529 bins recovered in ref. [37], except MB1 (see Supplementary Information for details). Population genome MB1 was derived from a co-assembly of 78 palsa samples. Assembly of these samples was conducted using CLC bio’s de novo assembler (QIAGEN, CA), mapping was conducted using BamM v1.3.8-1.50 (Imelfort & Lamberton et al., http://ecogenomics.github.io/BamM), and differential coverage binning was conducted using MetaBAT [38] v3127e20aa4e7 using a 3000 bp contig cut-off. CheckM [39] v1.0.4 determined completeness and contamination of the population genome bins through the identification and quantification of lineage-specific single copy marker genes following the lineage workflow (lineage_wf).
Genome bins were placed in a reference genome tree containing genomes from NCBI (database 2015-11-27) for taxonomic classification and evolutionary analysis using an in-house pipeline, GTDB v0.0.3 (Chaumeil & Parks, https://github.com/Ecogenomics/GTDBNCBI) (Supplementary Information). ARB [40] v6.0.6 was used to visualise the tree, and ITOL [41] was used to refine, with additional cosmetic amendments made in Inkscape (http://inkscape.org). The reported genome relative abundance is the abundance of each lineage within the total community (Supplementary Information).
Methane monooxygenase phylogenetic analysis and data searching
The gene-centric methanotroph analysis using pmoCAB genes and mmoX genes was conducted using Mingle v0.0.10 for tree creation (Parks et al., https://github.com/Ecogenomics/mingle) and GraftM v0.8.1 for data searches[42] (Supplementary Information). The methanotrophy protein sequences and manually annotated protein trees were used as input for the function ‘graftM create’, producing GraftM packages by creating hidden Markov models (HMMs) from an alignment of the sequences. GraftM ‘graft’ used this annotated tree for classification of gene sequences based on translated query read placement. The protein trees were made for phylogenetic analysis using Mingle and incorporated the identified Stordalen proteins, a selection of sequences greater than 160aa that were translated from sequences compiled in Knief [18], along with the contigs for novel groups from Lau et al. [22], Ricke et al. [26] and He et al. [43] when applicable (Supplementary Tables 12–15). Additional protein trees for MxaF/XoxF, RuBisCO (CbbL/RbcL) and NifH were created as above using Mingle, NCBI sequences and the IMG release 4.1 database (Supplementary Tables 16–18), respectively. GraftM gene packages for pmoA and mmoX were run on the raw metagenomes and metatranscriptomes and the results were normalised by total library size and HMM length. GraftM ‘graft’ was run on the NCBI nr database (downloaded in February 2016), and 24,636 SRA files (including amplicon and metagenome files, downloaded March 2017) in order to determine the geographic distribution of the Hyphomicrobiaceae pmoA genes. Where possible, GNU parallel [44] was used to speed data processing.
Metatranscriptomic read processing
Metatranscriptomes were processed using part of the TranscriptM (Frouin et al., https://github.com/elfrouin/transcriptM) pipeline, using Trimmomatic [90] and SortMeRNA [45] for in silico data cleaning (Supplementary Information). Dirseq v0.0.2 (Woodcroft et al., https://github.com/wwood/dirseq) (parameter --ignore-directions) determined the average transcript coverage of the methanotroph genes. Metatranscriptome expression per genome for each sample was calculated using ‘BamM parse -m counts’ (Supplementary Information).
Biogeochemical analysis
The pmoA and mmoX gene abundances from the metagenome data were averaged across sampling dates into five or 10 cm depth categories and compared to biogeochemical data from the porewater collected on site at the time of sampling, except for the palsa site, which lacked porewater. Measurements included dissolved methane (mM), and methane isotope data (δ13C-CH4), following a previously described procedure [34, 35]. The percent time that each depth category spends below the water table was calculated from manual water table measurements made three to five times per week during the sampling season. Sulphate concentrations were measured at Florida State University (Tallahassee, FL, USA) by ion chromatography on a Dionex ICS-1100 fitted with a 4-mm IonPac AS22 column, with an eluent of 4.5 mM carbonate/1.4 mM bicarbonate and flow rate of 1.2 mL/min (Supplementary Table 5). Methanotroph abundance to water table depth correlations, statistical differences in sulphur concentrations and methanotroph community composition between sites were conducted using ANOVA and paired t-tests in R [46].
Methanotroph population metabolism reconstruction and average amino acid identity
Metabolic reconstructions were facilitated using PROKKA [47] annotations, and submission through the IMG ER genome annotation pipeline [48]. In house pipelines were used to blast the coding regions of each genome to the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology database [49], and key methanotrophy and other metabolic genes were highlighted for incorporation into the metabolic models. BLASTP [50] was used to validate annotation of the gene sequences detected by GraftM. Hydrogenases were found and annotated using HydDB [51]. Metabolic models were created in Inkscape (http://inkscape.org). Average amino acid identity (AAI) facilitated evolutionary distance comparisons with the closest cultured isolates, or between Stordalen genomes. AAI was determined using the CompareM (Parks et al., https://github.com/dparks1134/CompareM) v0.0.17 ‘aai_wf’ on the genome bins, and genomes from IMG. Average nucleotide identity (ANI) was determined using calculate_ani.py (Pritchard, https://github.com/widdowquinn/scripts).
Results and discussion
Identification and distribution of canonical and novel methanotrophs across the thaw gradient
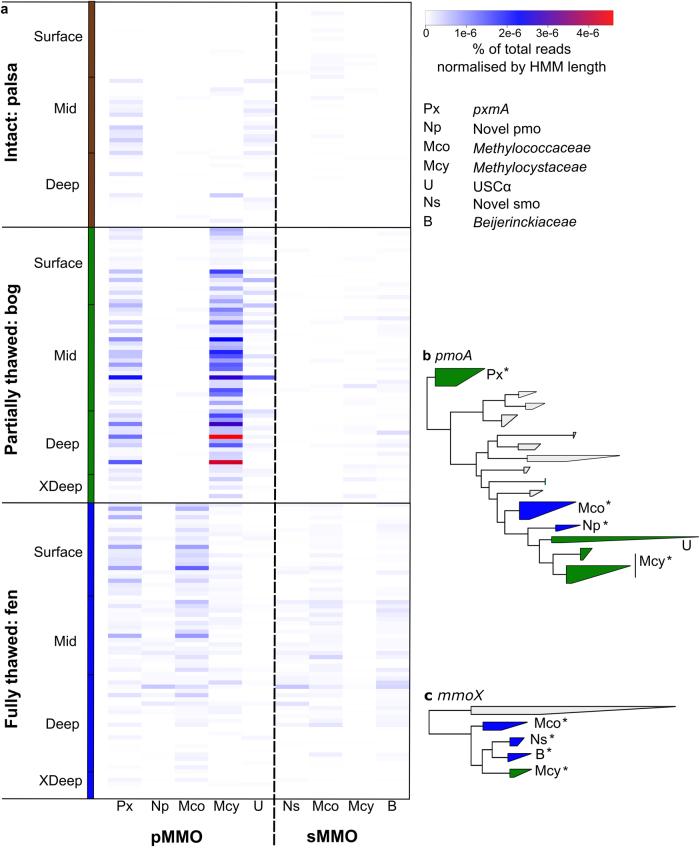
To investigate methanotroph diversity and the succession of the community as thaw progresses from palsa, to bog and fen, 188 raw metagenomes were searched for pMMO (pmoA gene) and sMMO (mmoX gene) reads using GraftM [42]. GraftM classified the reads into taxonomic groups following placement in a curated pmoA or mmoX gene tree (Fig. 1). The bog had the highest abundance of methanotrophs and was dominated by the well-characterised Methylocystaceae. Several members of the Methylocystaceae are suited to soil and bog environments exhibiting low pH and nitrogen, oligotrophy, or variable methane levels where the ability to oxidise atmospheric methane using a high affinity isozyme of the pMMO (pMMO2) is advantageous [52, 53]. In the minerotrophic and pH neutral fen, members of the Methylococcaceae comprised the largest fraction of the low abundance but taxonomically diverse methanotroph community. Methylocystaceae and Methylococcaceae methanotrophs occasionally possess pMMO, with the canonical operon structure of pmoCAB, in addition to a divergent monooxygenase of undetermined function, pXMO, encoded by a reordered pxmABC operon [54]. The pxmA gene was detected in the palsa, fen and bog, indicating the presence of multiple methanotrophic lineages encoding the divergent pXMO. Beijerinckiaceae possessing mmoX appeared in low abundance in both fen and bog (Fig. 1).
Fig. 1.
Methanotroph diversity across the Stordalen Mire thaw gradient. a Heatmap of the relative abundance of methanotrophs as a proportion of the total metagenome based on the particulate methane monooxygenase (pMMO; left) and the soluble methane monooxygenase (sMMO; right). Abundances are indicated by the coloured scale (from white, to blue, to red). The trees used by GraftM to classify the reads are shown for pmoA (b) and mmoX (c). The colour of the clades indicates the environment where these clades are most often found (green = bog, blue = fen). Asterisks indicate significantly different abundances based on one-way ANOVA tests and Bonferroni corrected pairwise t-tests (p value < 0.05; see Supplementary Table 6)
Of the uncharacterised methanotroph clades, USCα had greatest abundance in the bog and was the predominant methanotroph in the palsa. Minimal methane is produced in the oxic palsa environment [35], so methane is likely taken up from the atmosphere. Reads from two additional novel groups, ‘novel pmo’ (pmoA) and ‘novel smo’ (mmoX), were found only in the fen (Fig. 1). The ‘novel pmo’ reads clustered alongside a collection of sequences recently recovered from a Sacramento-San Joaquin Delta wetland metagenome [43]. The ‘novel smo’ group has not previously been recognised.
Interestingly, methanotrophs were often present below the water table in the deep (bog: >5 cm below water table, fen: >10 cm below peat surface) and extra deep (>30 cm below peat surface) metagenomes (Fig. 1), suggesting adaptations to cope with low oxygen conditions. The palsa remains mainly oxic to the depths sampled, whereas the bog includes an oxic surface and then likely becomes primarily anoxic below the water table (maximum depth 20 cm) [55]. Although the fen water table is above the peat surface, sedge roots are known to transport oxygen, which likely supports methanotrophs near live roots [56, 57].
Recovery of methanotroph population genomes
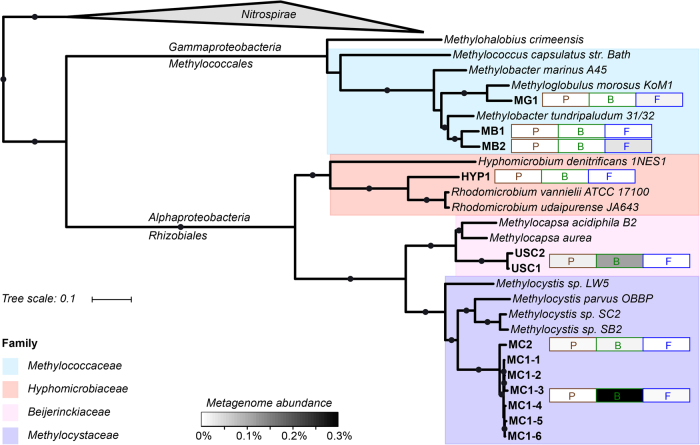
To recover the genomes of canonical and novel methanotrophs, assemblies of the 214 Stordalen Mire metagenomes (188 active layer, and 26 palsa core series) were binned into population genomes using differential coverage binning [37]. This led to the recovery of 1529 medium to high-quality population genomes (>70% completeness, and <10% contamination [37]). Of these genomes, 12 were found to encode pMMO and/or sMMO. An additional genome was recovered from a co-assembly of the palsa samples (n = 78). Phylogenetic analysis of the genomes using a genome tree created from the concatenated alignment of 120 single copy marker genes classified all 13 as proteobacterial. Three of these genomes belonged to the Methylococcaceae (Methylobacter-like MB1-2; Methyloglobulus-like MG1), seven to the Methylocystaceae (Methylocystis-like MC), two to the USCα (USC1 and USC2) and surprisingly one to the Hyphomicrobiaceae (HYP1) (Fig. 2). Six of the Methylocystaceae genomes were recovered in the mid or deep core layers of different bog samples. These genomes had ANI of >99% indicating that they represent the same Methylocystis-like population and will be referred to as MC1 accordingly (Supplementary Fig. 1). One Methylocystis-like genome (MC2) was recovered from a palsa sample, and represented a distinct population at 94% ANI to MC1.
Fig. 2.
Genome tree of the methanotroph population genomes recovered from Stordalen Mire. Solid circles represent bootstrap support of over 70%. Heatmap bars indicate average relative abundance per environment (P = palsa (brown), B = bog (green), F = fen (blue)) as a percentage of the total metagenome library (Supplementary Information). Raw values and standard deviation are presented in Supplementary Table 8. The highest abundances are observed for MC1 (1.85% in a deep bog sample) and USC1 (1.6% in a mid-depth bog sample)
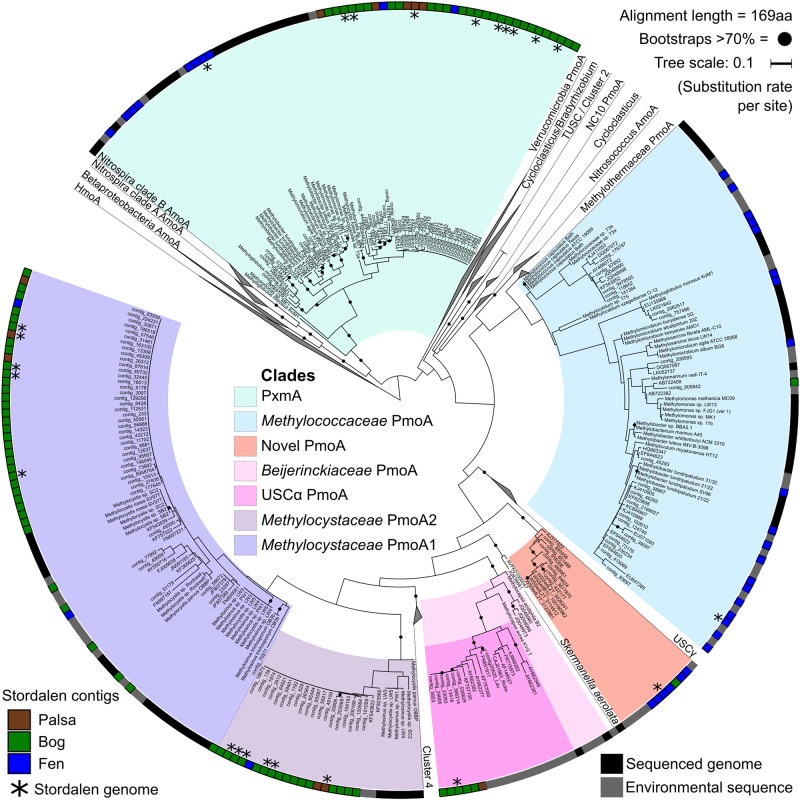
USC1 and USC2 are the first genomic representatives of the USCα lineage [25], as former representation of this group was restricted to partial pmoA sequences and a collection of short (<43 kb) genome fragments [22, 26]. The previous hypothesis that these microorganisms fall within the Beijerinckiaceae was confirmed by placement of USC1 and USC2 in a genome tree [26] (Fig. 2). Average amino acid identity (AAI) of USC1 and USC2 was 72% to their closest isolated taxonomic neighbours, the Methylocapsa spp. [26] (Supplementary Table 1), making it likely these populations belong to a novel species or potentially novel genus within the Beijerinckiaceae [58]. The USCα PmoCAB protein sequences derived from the genomes (USC1 and USC2), and additional sequences from unbinned contigs, clustered with both partial and full length translated USCα pmoA, pmoB and pmoC gene sequences recovered from the active layer of mineral cryosols in the Canadian high Arctic [22] and an acidic forest soil [26] (Fig. 3; Supplementary Fig. 2).
Fig. 3.
Phylogenetic tree of PmoA proteins recovered from Stordalen Mire. This tree is constructed from isolate-derived protein sequences (colour strip = black), Stordalen sequences (colour strip: palsa = brown; bog = green; fen = blue) and translated environmental sequences compiled by Knief [18] (colour strip = grey), with additional sequences from He et al. [43], Ricke et al. [26] and Lau et al. [22] (colour strip = grey). The asterisks indicate sequences that are within population genomes
An additional potential methanotroph genome (HYP1) was recovered from a fen sample, and was distinct from all canonical methanotroph families. Phylogenetic analysis revealed Rhodomicrobium spp. within the Hyphomicrobiaceae as the closest isolated taxonomic neighbours with an AAI of 67.5% (to both R. vannielii and R. udaipurense), indicating likely taxonomic distinction at the genus level (Fig. 2; Supplementary Table 1) [58]. Interestingly, HYP1 encodes novel methanotrophy operons for both pMMO and sMMO. Contamination from poor binning was discounted as these genes were also found in an additional medium-quality Hyphomicrobiaceae genome (HYP2; 60.53% completeness, 5.54% contamination) recovered from a deep fen sample with 80% AAI to HYP1 (Supplementary Figs. 3 and 4). Protein trees showed that HYP1 PmoA and PmoB sequences, and several additional PmoAB sequences from partial genomes and unbinned contigs, clustered closely with the wetland ‘novel pmo’ clade that appears basal to the Methylocystaceae and Beijerinckiaceae (Fig. 3; Supplementary Fig. 2a). Full-length PmoC sequences of this ‘novel pmo’ group have not been recovered until now, and were found to be similarly basal to the Methylocystaceae and Beijerinckiaceae PmoC proteins (Supplementary Fig. 2b). The MmoX from HYP1 and additional unbinned MmoX sequences clustered within the ‘novel smo’ group that formed a monophyletic clade with the Beijerinckiaceae (Supplementary Figs. 5 and 6). Recovery of methanotrophy marker genes in HYP1 suggests that the Hyphomicrobiaceae should be added to the recognised alphaproteobacterial methanotrophic families which were previously limited to the Beijerinckiaceae and Methylocystaceae. To investigate other possible habitats of HYP1, publicly available SRA (Sequence Read Archive) datasets (24,636 sequence files) and the NCBI nr database were searched for pmoA sequences falling within the HYP1 clade. Only one sequence from NCBI nr was identified as belonging to HYP1, and was from a Finnish peat soil (NCBI accession: CAC84777). However, HYP1 was detected (at >1 read hits) in 34 SRA metagenomes from a variety of habitats, including Sacramento-San Joaquin Delta wetland, New York City MTA subway, peat, bog and sand metagenomes (Supplementary Table 2), suggesting that HYP1 is a rare but widely distributed group.
Metabolic characterisation of Stordalen Mire methanotrophs
Metabolic reconstruction of the Stordalen Mire methanotroph genomes revealed diverse functional potential. Methane oxidation pathways, carbon assimilation, dissimilation and other potentially habitat-specific metabolisms were examined in the gammaproteobacterial Methylobacter-like (MB) populations MB1, MB2 and Methyloglobulus-like (MG) population MG1 recovered from the fen samples. MB1 and MB2 encode pMMO (pmoCAB), however MB2 also has sMMO (mmoXYZBCD). MG1 has only pxmA and B that appear consecutively at the end of a contig, indicating that pxmC and the pmoCAB operon are likely in the unrecovered portion (21%) of the genome. MG1 also encodes a hydroxylamine reductase (HAO) allowing for the conversion of hydroxylamine to nitrite, which is likely a toxicity response measure for this population [59]. Methane monooxygenases can co-oxidise ammonia to hydroxylamine, an intermediate toxic to cells unless further processed by an enzyme such as HAO [60]. MB1, MB2 and MG1 all possess the ribulose monophosphate (RuMP) pathway for carbon assimilation, the tetrahydromethanopterin (H4MPT) carbon dissimilation pathway for the oxidation of formaldehyde to formate and nitrogen fixation genes (nifHDK). MB1 also has dissimilatory nitrate reduction, which has been identified in the gammaproteobacterial methanotrophs Methylomicrobium, Methylomonas and Methylobacter tundripaludum [61]. This ability appears to be widespread within the Methylococcaceae, and has been linked to survival in low oxygen conditions where nitrate or nitrite can be used instead of oxygen as terminal electron acceptors in order to direct all available oxygen to methane oxidation [61]. Metabolic reconstruction of MB1-2 and MG and the observed distribution of Methylococcaceae in the metagenomes suggest that these microorganisms can function in the microaerobic environment of the deeper permafrost thaw layers (Fig. 1).
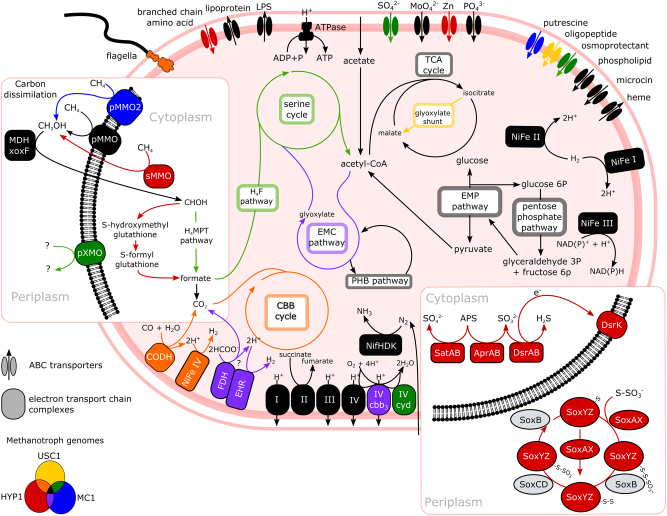
The alphaproteobacterial Stordalen methanotrophs had larger genomes than their gammaproteobacterial counterparts, and had an expanded metabolic diversity indicative of a facultative, rather than obligate, methanotrophic lifestyle (Supplementary Table 4). Metabolic analyses were conducted on the most complete representatives of the alphaproteobacterial lineages, USC1, HYP1 and MC1 (Fig. 4). Similar to USC1 and HYP1, MC1 encodes pMMO genes (pmoCAB) but further possesses a pMMO2 (pmoCAB2) variant that is associated with high affinity methane oxidation [52]. MC1 also encodes the pXMO (pxmABC) homologue, as does USC1. This discovery adds Beijerinckiaceae to the list of pXMO-encoding methanotroph families, and explains the high relative abundance of pxmA genes observed in the palsa environment (Fig. 1).
Fig. 4.
Metabolic reconstruction of the alphaproteobacterial population genomes MC1, HYP1 and USC1. Colours indicate the genome or combination of genomes (Venn diagram) in which the cycle or enzymes are found. Abbreviations: H4F tetrahydrofolate pathway, H4MPT tetrahydromethanopterin pathway, TCA tricarboxylic acid cycle, EMC ethylmalonyl-CoA pathway, EMP Embden–Meyerhof–Parnas pathway (glycolysis), CBB Calvin–Benson–Bassham cycle, PHB polyhydroxybutyrate pathway, LPS lipopolysaccharide, CODH carbon monoxide dehydrogenase, NiFe nickel iron hydrogenase, nitrogenase (NifHDK), pMMO particulate methane monooxygenase, pMMO2 particulate methane monooxygenase isozyme II, sMMO soluble methane monooxygenase, pXMO homologue of particulate methane monooxgyenase, CH3OH methanol, I complex I NADH dehydrogenase, II complex II succinate dehydrogenase, III complex III cytochrome bc1, IV cytochrome c oxidase, IV cbb3 complex IV cytochrome cbb3 oxidase, cyd complex IV cytochrome bd oxidase, FDH formate dehydrogenase, EHR energy converting hydrogenase related (part of formate hydrogenlyase complex), SO42− sulphate, MoO42− molybdate, Zn zinc, PO43− phosphate, CHOH formaldehyde, sulphate adenylyltransferase (SatAB), APS adenosine 5'-phosphosulphate, SO32− sulphite, adenylylsulphate reductase (AprAB), dissimilatory sulphite reductase (DsrAB), H2S hydrogen sulphide, dissimilatory sulphite reductase electron transport complex (DsrK representing the DsrKMJOP complex), SoxYZ/SoxAX/SoxB/SoxCD = sulphur oxidising proteins. See Supplementary Fig. 7 for detailed gene presence/absence and Supplementary Table 3 for the list of additional abbreviations
Genes for the calcium-dependent methanol dehydrogenase (MDH) mxaF-encoded large subunit of the mxaFI-MDH complex, which catalyses the second step of methane oxidation, could not be identified in any of the Stordalen Mire methanotroph genomes. Instead, at least one copy of xoxF, a homologue of mxaF [62], was identified in each genome except MG1. This gene differentiates into five clades (xoxF1-5)[62, 63] and encodes the lanthanide-dependent xoxF–MDH complex known to oxidise methanol in several methanotrophs [62, 64]. The H4MPT carbon dissimilation and tetrahydrofolate (H4F) carbon assimilation pathway, which facilitates entry into the serine cycle [63], are present in USC1 and MC1. Alternatively, HYP1 appears to have a thiol-dependent (glutathione (GSH)-linked) pathway for formaldehyde oxidation to formate using glutathione synthase (gfa), S-(hydroxymethyl)glutathione dehydrogenase (frmA) and S-formylglutathione hydrolase (frmB) genes [63, 65]. This pathway is present as a detoxification and energy generation mechanism in the methylotroph Paracoccus denitrificans [63, 66]. Similar to P. denitrificans and several other alphaproteobacterial methylotrophs [62, 67], in HYP1 these genes are located directly adjacent to a PQQ-dependent alcohol dehydrogenase identified as the xoxF5 type (Supplementary Fig. 8; Supplementary Fig. 9) [62]. This suggests that formaldehyde is produced by the xoxF-MDH of HYP1 and then oxidised to formate using the thiol-dependent pathway.
The ethylmalonyl-CoA (EMC) pathway is used to regenerate glyoxylate from acetyl-CoA for use in the serine cycle [68], and was present in MC1 and HYP1 but not USC1 (Fig. 4). USC1, similar to the Beijerinckiaceae methanotrophs [20], possesses isocitrate lyase (icl), which forms part of the glyoxylate shunt with malate synthase and allows the formation of glyoxylate from isocitrate [68]. The glyoxylate cycle enables the assimilation of acetate, and acetyl-CoA can be produced from acetate using acetate kinase (ack) and phosphotransacetylase (pta) or acetyl-CoA synthetase (acs), which are present in USC1. These genes, in conjunction with demonstrated acetate uptake in isotope studies [69], implicate USCα populations as likely facultative methanotrophs. Additionally, MC1 and HYP1 possess genes involved in transforming acetate to acetyl-CoA (ack, pta, acs), suggesting these populations are also facultative. A facultative methanotrophic lifestyle likely allows USCα and MC1 to survive on acetate, which can diffuse freely across the cell membrane in the acidic conditions of the bog, under methane limited conditions [70].
Although USC1, HYP1 and MC1 contain a near complete EMP pathway, the lack of glucose transporters suggests these populations do not use glucose as a carbon and energy source. Transporters are missing in known methanotrophs and growth of methanotrophs on glucose has not been recorded [20], indicating that this pathway is anabolic in the Stordalen Mire genomes. Poly-β-hydroxybutyrate (PHB) is a storage compound commonly used as a carbon and energy source by methanotrophs under nutrient limiting conditions [71]. Genes for the PHB storage pathway (phaABCZ, bdh, acsA) are present in all three genomes.
HYP1 may have additional and unusual means of energy generation. A complete dissimilatory sulphate reduction pathway was identified, including the canonical markers of dissimilatory sulphite reductase subunits A and B (dsrAB) (Fig. 4). These reductases have high AAI to oxidative dsr genes of the purple non-sulphur photolithotrophic bacteria Rhodomicrobium vannielii (83% WP_013418283) and R. udaipurense (86% KAI93440) [72, 73]. Like Rhodomicrobium spp., HYP1 possesses an incomplete sulphur oxidation system (SOX; soxAXYZD), however sulphur oxidation could still be possible as Rhodomicrobium spp. can use sulphide and thiosulphate [74]. The potential for sulphur oxidation in a methanotroph was recently described in a Methylococcaceae genome from a deep-sea hydrothermal plume [75]. Biogeochemical measurements from the site show that the fen, particularly the fen surface, has significantly higher sulphate concentrations than the bog (Supplementary Table 5; fen surface average = 10.44 µM, average fen = 6.25 µM, bog = 1.83 µM; p value < 10−4), potentially indicating increased sulphur oxidation activity. In the fen, it may be possible for HYP1 to oxidise sulphate that has been reduced during anaerobiosis in the deeper layers [76].
In addition to sulphur, Rhodomicrobium spp. can use hydrogen as an electron donor [74]. HYP1, USC1 and MC1 encode numerous hydrogenases, indicating potential hydrogen use (Fig. 4). Hydrogen uptake and production has been recorded in several verrucomicrobial, alpha- and gammaproteobacterial methanotrophs, and has been linked to nitrogen fixation, which produces the required hydrogen as a by-product [77, 78]. Both HYP1 and USC1 also encode coxLMS for carbon monoxide (CO) oxidation. The ability of these microorganisms to perform CO oxidation could allow CO to be used as both an energy and carbon source [79]. Stable-isotope probing or successful culturing would be required to confirm this metabolism. However, atmospheric CO levels, or CO produced by the photochemical degradation of matter, are sufficient for carboxydotrophs in many soil environments [79]. Several hydrogenotrophic [80], methanotrophic [81] and carbon monoxide oxidising [79] bacteria are known to use these respective pathways to generate energy for carbon fixation using the Calvin–Benson–Bassham (CBB) cycle.
Ribulose bisphosphate carboxylase (RuBisCO) is the key enzyme and genetic marker for the CBB cycle. HYP1 encodes RuBisCO form IV, a RuBisCO-like protein of unknown function [82], and form II (cbbM). Form II is known to operate under high CO2 and low O2 concentrations [82, 83], which would suit fixation in the deeper fen layers (Supplementary Fig. 10). Of the other genomes, MC1 possesses only form IV, whereas USC1, similar to the characterised methanotrophic Beijerinckiaceae [84], encodes the catalytic form I (both cbbL and S; Supplementary Fig. 11). HYP1, MC1 and USC1 are also predicted to be capable of nitrogen fixation, as phylogenetic analysis of the NifH proteins suggests they belong to the functional Type 1D clade (Supplementary Fig. 12). Given the wide distribution of nitrogen fixation genes in the Stordalen Mire methanotrophs, it appears that this metabolism may be a selective advantage for methanotrophs in the nitrogen limited thaw environment.
Metatranscriptomic analysis of in situ activity
Gene presence in metagenomes reveals the metabolic potential of the community but does not indicate which genes are active at the time of sampling. In order to investigate the activity of methanotrophs in the system, expression of key genes for methane oxidation (pmoA and mmoX) were examined in 24 metatranscriptomes spanning different sites and depths. Despite high abundance in the bog metagenomes, low relative transcript expression was observed for the Methylocystaceae (Fig. 5). Low activity of Methylocystaceae has been recorded in other environments [85], and is likely a consequence of harsher environmental conditions in the low pH and ombrotrophic bog that would favour stress-tolerance over productivity [23, 86]. In the fen, the diversity of methanotrophs was not reflected in the metatranscriptomes, which were dominated by Methylococcaceae even in the deeper layers (Fig. 5). This unexpected prevalence of Methylococcaceae activity at depth has been observed in Arctic soils, which further supports hypotheses that these microorganisms are active under microaerophilic conditions [61, 87]. Methylococcaceae pmoA transcripts in the fen also comprised a greater proportion of the total metatranscriptome reads than in the bog or palsa, indicating that methanotrophs make up more of the active community in the fully thawed site. This suggests that Methylococcaceae are oxidising a substantial amount of CH4 and/or responding to increased CH4 availability in the fen, which has the highest CH4 flux of all three sites [35].
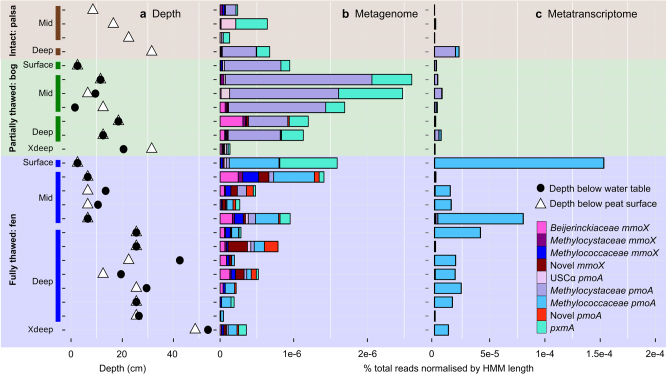
Fig. 5.
Methanotroph abundance and activity in the 24 samples with paired metagenomes and metatranscriptomes from Stordalen Mire. For spatial orientation, distance from the water table and peat surface is shown in a. The metagenome abundances are indicated in b and the transcript expression in c. Methanotroph pmoA and mmoX read abundances are presented as a percentage of total reads normalised by HMM length for both metagenomes and metatranscriptomes
Consistent with previous findings from isolates [68] and similar environments [23, 43, 85], very few pxmA or mmoX transcripts were detected in the Stordalen Mire metatranscriptomes. The ‘novel pmo’ and ‘novel smo’ of the HYP1 group had minimal or no transcript expression (12 ‘novel pmo’ reads detected in one surface fen sample, and 25 ‘novel smo’ reads in one mid fen sample). USCα had very low pmoA expression in the palsa core mid depth sample only (112 reads detected), which fits with low expression observed in samples of an Arctic cryosol environment [22]. Transcript expression of the second step of methanotrophy, methanol oxidation to formaldehyde, was high for xoxF, with virtually no representation in the metatranscriptome reads of mxaF (Supplementary Fig. 13). This strongly suggests that the xoxF form of MDH is the most important complex for methanol oxidation at Stordalen Mire, which supports recent findings that this form is likely dominant in lanthanide non-limiting environments [64]. Further, research into methylotroph and methanotroph co-cultures has revealed that high mxaF expression over xoxF can be linked to cross-feeding and syntrophy between these microorganisms [88]. The dominance of xoxF–MDH expression at Stordalen suggests that lanthanide is non-limiting in this environment, and potentially that methanol is not being secreted for use by syntrophic partners.
Mapping metatranscriptomes to the population genomes enabled transcription analysis of entire metabolic pathways for specific lineages, and revealed the functionality of methanotrophs beyond the expression of marker genes for primary methane oxidation. The highest relative transcript expression was 0.27% for the MC1 population genome in a bog mid-depth sample, and 0.24% for MC2 in a palsa deep (calculated as proportion of total transcript reads in the metatranscriptome; Supplementary Table 7). MC1 and MC2 had high transcript expression for a variety of genes in the bog, and some expression in the fen and one palsa sample, confirming active carbon assimilation (Supplementary Fig. 14). Activity through transcript expression was also observed for MB1 and MB2 in the fen, which had high transcript coverage of pmoCAB genes and downstream methane processing genes (Supplementary Fig. 14). xoxF was consistently expressed in MC1-2 and MB1-2, indicating use of the xoxF–MDH for methanol oxidation in these populations. Surprisingly, MC1-2 and MB1-2 appeared to be actively fixing nitrogen through high expression of nifH, reaffirming the importance of this function in environments with low nitrogen availability [23]. The novel metabolic inferences could not be confirmed for MG1, HYP1 and USC1 due to limited transcript expression across all metatranscriptome samples. It is likely that these populations were inactive, or below detection, at the time of sampling.
Relationship between methanotrophs and biogeochemistry
Previous work at the site revealed that average CH4 flux increases as thaw progresses, from minimal emission in the palsa, to ~1.46 mg CH4 m2/h and ~8.75 mg CH4 m2/h in the bog and fen, respectively [35]. Here, methanotroph metagenome abundances, determined using the pmoA and mmoX genes, were analysed alongside depth resolved biogeochemical data from the sites to evaluate relationships between community and methane oxidation. At the fen and bog sites the concentration and δ13C signature of dissolved CH4 was analysed in porewater samples collected in parallel with the peat samples. The δ13C signature of CH4 is influenced by the combined effect of the methanogenic pathway (acetoclastic versus hydrogenotrophic) that produced the CH4, and the activity of the methanotrophs [35, 89]). The highest abundance of methanotrophs occurred in the region of the peat profiles where there was maximum dissolved CH4, but still periodic oxygen availability either due to varying water tables in the bog [57] or root transport in the fen [56] (bog = 15–20 cm, fen = 10–15 cm; Fig. 6).
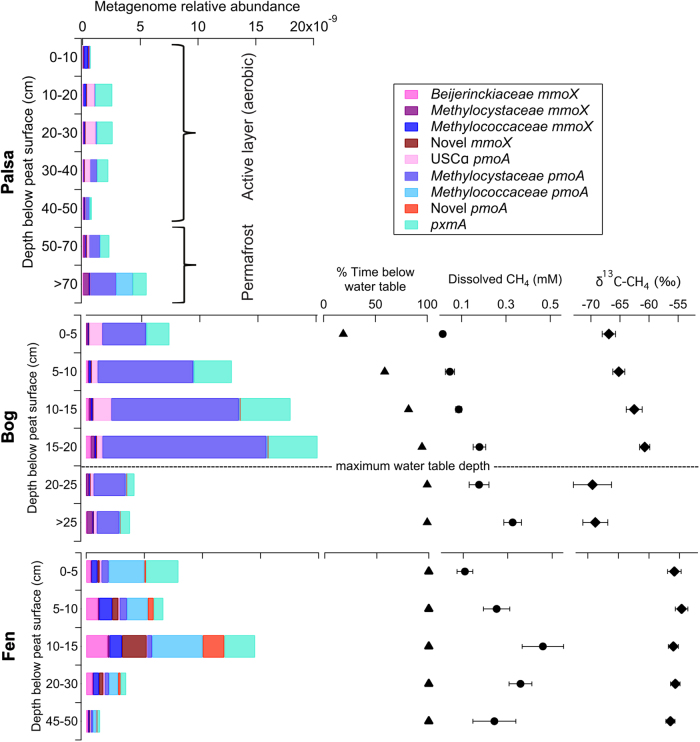
Fig. 6.
Depth profiles of palsa, bog and fen methanotroph community abundances (as pmoA and mmoX gene reads normalised by total library and HMM length) and relationship to water table, dissolved CH4 concentration and δ13C signature (no porewater present at the palsa site). Metagenome and porewater chemistry data from 2011 and 2012 was averaged across all dates by depth category; error bars represent ± 1 standard error. This shows a link between methanotroph abundances and dissolved CH4 concentration across the thaw gradient. At the bog site, δ13C-CH4 patterns track the depth distribution of the methanotroph community, with the heaviest (most oxidised) CH4 occurring just above the maximum water table depth where methanogen populations are highest and the lightest (least oxidised) CH4 occurring in permanently inundated peat where methanotroph abundances are low
Since hydrogenotrophic methanogens were dominant in the bog samples [35], most of the variation in δ13C-CH4 at this site was likely due to variation in methane oxidation (not variation in production pathway), with less negative δ13C-CH4 values indicating the preferential use of lighter 12CH4 by methanotrophs, and consequently greater CH4 oxidation. While methanotroph transcript expression in the bog revealed no depth associated trends (Fig. 5), comparison of methanotroph abundances and δ13C-CH4 suggest that methanotrophy increased with depth across the region of peat that is periodically above the water table and decreased in the permanently inundated peat (Fig. 6). The heaviest (most oxidised) CH4 and the highest methanotroph abundances occurred in peat that is inundated >90% of the time (15–20 cm) (Fig. 6). This was surprising, as CH4 oxidation was expected to be greatest nearer the theoretical optimal oxygen and CH4 conditions of the oxic–anoxic boundary at the average water table depth (6 cm) [85]. Instead, it appears that CH4 concentration, which increases with depth, is the key driver of methanotroph community patterns and that a highly variable water table in the bog allows infrequent oxic events that provide sufficient oxygen to support specialised methanotroph populations.
In the fen, methanotroph abundances correlated with distance from the water table (Supplementary Fig. 15), however low variability in δ13C-CH4 indicated no clear isotopic evidence for activity of methanotrophs even though the highest methanotroph abundances were found between 10 and 15 cm, and Methylococcaceae were active (Figs. 5 and 6). This disconnection of abundances, activity and δ13C-CH4 may be related to the capacity of plant roots to provide substrates for methanogenesis within the peat, a conduit for oxygen into the peat and CH4 out of the peat, which could consequently bypass oxidation and disrupt clear methane oxidation gradients [57].
Conclusion
The methanotrophs found within the Stordalen Mire thaw gradient include canonical and novel populations that encode diverse metabolisms. The low CH4 environment of the palsa is populated by the likely facultative and high affinity atmospheric methane-oxidisers USCα. The Methylocystaceae (MC1 and MC2) in the bog can cope with fluctuating CH4 conditions due to the pMMO isozymes (pMMO and pMMO2) and capability for acetate uptake. In the high CH4 fen, the obligate Methylococcaceae (MB1 and MB2) are dominant and active, while the metabolically diverse HYP1 are in low abundance and appear relatively inactive. It is evident that an evolutionarily complex, diverse and shifting methanotroph community is at the forefront of climate change as permafrost thaws. The low abundance of USC1 and HYP1 precluded their enrichment and visualisation, however it is hoped that the metabolic inferences determined from these genomes will guide future efforts to target and eventually isolate these elusive microorganisms, and experimentally confirm the extent of their ecological impact.
Data availability
Data used in this manuscript are submitted under NCBI BioProject accession number PRJNA386568.
Electronic supplementary material
Acknowledgements
We thank the IsoGenie consortium field teams for sampling efforts in 2010–2012, particularly Tyler Logan, and the Abisko Scientific Research Station for site support. We thank members of the wider IsoGenie consortium for feedback on early stages of the manuscript, particularly Matthew Sullivan and Ruth Varner. We thank Serene Low, Margaret Butler and Brian Kemish for sequencing, coordination and computing support. This work was funded by the US Department of Energy Office of Biological and Environmental research, grants DE-SC0004632, DE-SC0010580 and DESC0016440. CMS is supported by the Australian Government Research Training Program (RTP) Scholarship, and GWT is supported by University of Queensland Vice-Chancellor’s Research Focused Fellowship.
Author contributions
GWT, JPC, SF, SRS, PMC and VIR designed the study. VIR coordinated sampling, and SBH, JPC, SF and GWT collected samples. CMS, BJW, JAB, PNE and GWT designed and/or performed bioinformatics analyses. CKM, SBH, JPC, PMC and SRS designed and/or performed porewater/isotope measurements. CMS, CKM, BJW, VIR, SRS, PMC, JPC, SBH and GWT integrated bioinformatics and biogeochemical data. CMS and GWT wrote the paper with contributions from CKM. All co-authors edited, reviewed and approved the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0065-5) contains supplementary material, which is available to authorized users.
References
- 1.Froese DG, Westgate JA, Reyes AV, Enkin RJ, Preece SJ. Ancient permafrost and a future, warmer Arctic. Science. 2008;321:1648. doi: 10.1126/science.1157525. [DOI] [PubMed] [Google Scholar]
- 2.Schuur EAG, Mcguire AD, Schadel C, Grosse G, Harden JW, Hayes DJ, et al. Climate change and the permafrost carbon feedback. Nature. 2015;520:171–9. doi: 10.1038/nature14338. [DOI] [PubMed] [Google Scholar]
- 3.Tarnocai C, Riley WJ, Canadell JG, Schuur EAG, Kuhry P, Mazhitova G, Zimov S. Soil organic carbon pools in the northern circumpolar permafrost region. Glob Biogeochem Cycles. 2009;23:GB2023. doi: 10.1029/2008GB003327. [DOI] [Google Scholar]
- 4.Koven CD, Riley WJ, Stern A. Analysis of permafrost thermal dynamics and response to climate change in the CMIP5 earth system models. J Clim. 2013;26:1877–1900. doi: 10.1175/JCLI-D-12-00228.1. [DOI] [Google Scholar]
- 5.Nazaries L, Murrell JC, Millard P, Baggs L, Singh BK. Methane, microbes and models: fundamental understanding of the soil methane cycle for future predictions. Environ Microbiol. 2013;15:2395–417. doi: 10.1111/1462-2920.12149. [DOI] [PubMed] [Google Scholar]
- 6.Oh Y, Stackhouse B, Lau MCY, Xu X, Trugman AT, Moch J, et al. A scalable model for methane consumption in arctic mineral soils. Geophys Res Lett. 2016;43:5143–50. doi: 10.1002/2016GL069049. [DOI] [Google Scholar]
- 7.Xu X, Yuan F, Hanson PJ, Wullschleger SD, Thornton PE, Riley WJ, et al. Reviews and syntheses: Four decades of modeling methane cycling in terrestrial ecosystems. Biogeosciences. 2016;13:3735–55. doi: 10.5194/bg-13-3735-2016. [DOI] [Google Scholar]
- 8.Moosavi SC, Crill PM. CH4 oxidation by tundra wetlands as measured by a selective inhibitor technique. J Geophys Res: Atmos. 1998;103:29093–106. doi: 10.1029/97JD03519. [DOI] [Google Scholar]
- 9.Popp TJ, Chanton JP, Whiting GJ, Grant N. Evaluation of methane oxidation in the rhizosphere of a Carex dominated fen in northcentral Alberta, Canada. Biogeochemistry. 2000;51:259–81. doi: 10.1023/A:1006452609284. [DOI] [Google Scholar]
- 10.Liebner S, Rublack K, Stuehrmann T, Wagner D. Diversity of aerobic methanotrophic bacteria in a permafrost active layer soil of the Lena Delta, Siberia. Microb Ecol. 2009;57:25–35. doi: 10.1007/s00248-008-9411-x. [DOI] [PubMed] [Google Scholar]
- 11.Blazewicz SJ, Petersen DG, Waldrop MP, Firestone MK. Anaerobic oxidation of methane in tropical and boreal soils: ecological significance in terrestrial methane cycling. J Geophys Res: Biogeosci. 2012;117:G02033. doi: 10.1029/2011JG001864. [DOI] [Google Scholar]
- 12.Hanson RS, Hanson TE. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–71. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–8. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 14.Dunfield PF, Yuryev A, Senin P, Smirnova AV, Stott MB, Hou S, et al. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature. 2007;450:879–82. doi: 10.1038/nature06411. [DOI] [PubMed] [Google Scholar]
- 15.Pol A, Heijmans K, Harhangi HR, Tedesco D, Jetten MS, Op den Camp HJ. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature. 2007;450:874–8. doi: 10.1038/nature06222. [DOI] [PubMed] [Google Scholar]
- 16.van Teeseling MC, Pol A, Harhangi HR, van der Zwart S, Jetten MS, Op den Camp HJ, et al. Expanding the verrucomicrobial methanotrophic world: description of three novel species of methylacidimicrobium gen. nov. Appl Environ Microbiol. 2014;80:6782–91. doi: 10.1128/AEM.01838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heyer J, Berger U, Hardt M, Dunfield PF. Methylohalobius crimeensis gen. nov., sp. nov., a moderately halophilic, methanotrophic bacterium isolated from hypersaline lakes of Crimea. Int J Syst Evol Microbiol. 2005;55:1817–26. doi: 10.1099/ijs.0.63213-0. [DOI] [PubMed] [Google Scholar]
- 18.Knief C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front Microbiol. 2015;6:1346. doi: 10.3389/fmicb.2015.01346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skennerton CT, Ward LM, Michel A, Metcalfe K, Valiente C, Mullin S, et al. Genomic reconstruction of an uncultured hydrothermal vent gammaproteobacterial methanotroph (family Methylothermaceae) indicates multiple adaptations to oxygen limitation. Front Microbiol. 2015;6:1425. doi: 10.3389/fmicb.2015.01425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamas I, Smirnova AV, He Z, Dunfield PF. The (d)evolution of methanotrophy in the Beijerinckiaceaea - a comparative genomics analysis. ISME J. 2014;8:369–82. doi: 10.1038/ismej.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vorobev AV, Baani M, Doronina NV, Brady AL, Liesack W, Dunfield PF, et al. Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase. Int J Syst Evol Microbiol. 2011;61:2456–63. doi: 10.1099/ijs.0.028118-0. [DOI] [PubMed] [Google Scholar]
- 22.Lau MCY, Stackhouse BT, Layton AC, Chauhan A, Vishnivetskaya TA, Chourey K, et al. An active atmospheric methane sink in high Arctic mineral cryosols. ISME J. 2015;9:1880–91. doi: 10.1038/ismej.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liebner S, Svenning MM. Environmental transcription of mmoX by methane-oxidizing proteobacteria in a subarctic palsa peatland. Appl Environ Microbiol. 2013;79:701–6. doi: 10.1128/AEM.02292-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henckel T, Jackel U, Schnell S, Conrad R. Molecular analyses of novel methanotrophic communities in forest soil that oxidize atmospheric methane. Appl Environ Microbiol. 2000;66:1801–8. doi: 10.1128/AEM.66.5.1801-1808.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes AJ, Roslev P, Mcdonald IR, Iversen N, Henriksen K, Murrell JC. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl Environ Microbiol. 1999;65:3312–8. doi: 10.1128/aem.65.8.3312-3318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ricke P, Kube M, Nakagawa S, Erkel C, Reinhardt R, Liesack W. First genome data from uncultured upland soil cluster alpha methanotrophs provide further evidence for a close phylogenetic relationship to Methylocapsa acidiphila B2 and for high-affinity methanotrophy involving particulate methane monooxygenase. Appl Environ Microbiol. 2005;71:7472–82. doi: 10.1128/AEM.71.11.7472-7482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crill PM. Seasonal patterns of methane uptake and carbon dioxide release by a temperate woodland soil. Glob Biogeochem Cycles. 1991;5:319–34. doi: 10.1029/91GB02466. [DOI] [Google Scholar]
- 28.Kirschke S, Bousquet P, Ciais P, Saunois M, Canadell JG, Dlugokencky EJ, et al. Three decades of global methane sources and sinks. Nat Geosci. 2013;6:813. doi: 10.1038/ngeo1955. [DOI] [Google Scholar]
- 29.Kolb S. The quest for atmospheric methane oxidizers in forest soils. Environ Microbiol Rep. 2009;1:336–46. doi: 10.1111/j.1758-2229.2009.00047.x. [DOI] [PubMed] [Google Scholar]
- 30.Barbier BA, Dziduch I, Liebner S, Ganzert L, Lantuit H, Pollard W, et al. Methane-cycling communities in a permafrost-affected soil on Herschel Island, Western Canadian Arctic: active layer profiling of mcrA and pmoA genes. FEMS Microbiol Ecol. 2012;82:287–302. doi: 10.1111/j.1574-6941.2012.01332.x. [DOI] [PubMed] [Google Scholar]
- 31.Tveit A, Schwacke R, Svenning MM, Urich T. Organic carbon transformations in high-Arctic peat soils: key functions and microorganisms. ISME J. 2013;7:299–311. doi: 10.1038/ismej.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stackhouse BT, Vishnivetskaya TA, Layton A, Chauhan A, Pfiffner S, Mykytczuk NC, et al. Effects of simulated spring thaw of permafrost from mineral cryosol on CO2 emissions and atmospheric CH4 uptake. J Geophys Res: Biogeosci. 2015;120:1764–84. doi: 10.1002/2015JG003004. [DOI] [Google Scholar]
- 33.Tveit AT, Urich T, Frenzel P, Svenning MM. Metabolic and trophic interactions modulate methane production by Arctic peat microbiota in response to warming. Proc Natl Acad Sci USA. 2015;112:E2507–E2516. doi: 10.1073/pnas.1420797112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hodgkins SB, Tfaily MM, Mccalley CK, Logan TA, Crill PM, Saleska SR, et al. Changes in peat chemistry associated with permafrost thaw increase greenhouse gas production. Proc Natl Acad Sci USA. 2014;111:5819–24. doi: 10.1073/pnas.1314641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mccalley CK, Woodcroft BJ, Hodgkins SB, Wehr RA, Kim EH, Mondav R, et al. Methane dynamics regulated by microbial community response to permafrost thaw. Nature. 2014;514:478–81. doi: 10.1038/nature13798. [DOI] [PubMed] [Google Scholar]
- 36.Mondav R, Woodcroft BJ, Kim EH, Mccalley CK, Hodgkins SB, Crill PM, et al. Discovery of a novel methanogen prevalent in thawing permafrost. Nat Commun. 2014;5:3212. doi: 10.1038/ncomms4212. [DOI] [PubMed] [Google Scholar]
- 37.Woodcroft BJ, Singleton CM, Boyd JA, Evans PN, Emerson JB, Zayed AAF, et al. Genome-centric view of carbon processing in thawing permafrost. Nature. 2018 (in press). [DOI] [PubMed]
- 38.Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. Peer J. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ludwig W, Strunk O, Westram R, Richter L, Meier H, kumar Y, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–71. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letunic I, Bork P. Interactive tree of life (iTOL)v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–5. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyd JA, Woodcroft BJ, Tyson GW. GraftM: a tool for scalable, phylogenetically informed classification of genes within metagenomes. Nucleic Acids Res. 2018;46:e59. doi: 10.1093/nar/gky174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He S, Malfatti SA, Mcfarland JW, Anderson FE, Pati A, Huntemann M, et al. Patterns in wetland microbial community composition and functional gene repertoire associated with methane emissions. mBio. 2015;6:e00066–15. doi: 10.1128/mBio.00066-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tange O. Gnu parallel-the command-line power tool. USENIX Mag. 2011;36:42–47. [Google Scholar]
- 45.Kopylova E, Noe L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–7. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 46.Team, R. C. R language definition. Vienna, Austria: R foundation for statistical computing; 2000.
- 47.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 48.Markowitz VM, Mavromatis K, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics. 2009;25:2271–8. doi: 10.1093/bioinformatics/btp393. [DOI] [PubMed] [Google Scholar]
- 49.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 51.Søndergaard D, Pedersen CNS, Greening C. HydDB: a web tool for hydrogenase classification and analysis. Sci Rep. 2016;6:34212. doi: 10.1038/srep34212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baani M, Liesack W. Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proc Natl Acad Sci USA. 2008;105:10203–8. doi: 10.1073/pnas.0702643105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belova SE, Baani M, Suzina NE, Bodelier PLE, Liesack W, Dedysh SN. Acetate utilization as a survival strategy of peat-inhabiting Methylocystis spp. Environ Microbiol Rep. 2011;3:36–46. doi: 10.1111/j.1758-2229.2010.00180.x. [DOI] [PubMed] [Google Scholar]
- 54.Tavormina PL, Orphan VJ, Kalyuzhnaya MG, Jetten MSM, Klotz MG. A novel family of functional operons encoding methane/ammonia monooxygenase-related proteins in gammaproteobacterial methanotrophs. Environ Microbiol Rep. 2011;3:91–100. doi: 10.1111/j.1758-2229.2010.00192.x. [DOI] [PubMed] [Google Scholar]
- 55.Gray ND, McCann CM, Christgen B, Ahammad SZ, Roberts JA, Graham DW. Soil geochemistry confines microbial abundances across an arctic landscape; implications for net carbon exchange with the atmosphere. Biogeochemistry. 2014;120:1–11. doi: 10.1007/s10533-014-9997-7. [DOI] [Google Scholar]
- 56.Cheema S, Zeyer J, Henneberger R. Methanotrophic and methanogenic communities in Swiss alpine fens dominated by Carex rostrata and Eriophorum angustifolium. Appl Environ Microbiol. 2015;81:5832–44. doi: 10.1128/AEM.01519-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whalen SC. Biogeochemistry of methane exchange between natural wetlands and the atmosphere. Environ Eng Sci. 2005;22:73–94. doi: 10.1089/ees.2005.22.73. [DOI] [Google Scholar]
- 58.Luo C, Rodriguez-R LM, Konstantinidis KT. MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res. 2014;42:e73. doi: 10.1093/nar/gku169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Campbell MA, Nyerges G, Kozlowski JA, Poret-Peterson AT, Stein LY, Klotz MG. Model of the molecular basis for hydroxylamine oxidation and nitrous oxide production in methanotrophic bacteria. FEMS Microbiol Lett. 2011;322:82–89. doi: 10.1111/j.1574-6968.2011.02340.x. [DOI] [PubMed] [Google Scholar]
- 60.Crossman LC, Moir JWB, Enticknap JJ, Richardson DJ, Spiro S. Heterologous expression of heterotrophic nitrification genes. Microbiology. 1997;143:3775–83. doi: 10.1099/00221287-143-12-3775. [DOI] [PubMed] [Google Scholar]
- 61.Kits KD, Campbell DJ, Remus Rosana A, Stein LY. Diverse electron sources support denitrification under hypoxia in the obligate methanotroph Methylomicrobium album strain BG8. Front Microbiol. 2015;6:1072. doi: 10.3389/fmicb.2015.01072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keltjens JT, Pol A, Reimann J, Op den Camp HJ. PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl Microbiol Biotechnol. 2014;98:6163–83. doi: 10.1007/s00253-014-5766-8. [DOI] [PubMed] [Google Scholar]
- 63.Chistoserdova L. Modularity of methylotrophy, revisited. Environ Microbiol. 2011;13:2603–22. doi: 10.1111/j.1462-2920.2011.02464.x. [DOI] [PubMed] [Google Scholar]
- 64.Chu F, Lidstrom ME. XoxF acts as the predominant methanol dehydrogenase in the type I methanotroph Methylomicrobium buryatense. J Bacteriol. 2016;198:1317–25. doi: 10.1128/JB.00959-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen NH, Djoko KY, Veyrier FJ, Mcewan AG. Formaldehyde stress responses in bacterial pathogens. Front Microbiol. 2016;7:257. doi: 10.3389/fmicb.2016.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ras J, van Ophem PW, Reijnders WN, van Spanning RJ, Duine JA, Stouthamer AH, et al. Isolation, sequencing, and mutagenesis of the gene encoding NAD- and glutathione-dependent formaldehyde dehydrogenase (GD-FALDH) from Paracoccus denitrificans, in which GD-FALDH is essential for methylotrophic growth. J Bacteriol. 1995;177:247–51. doi: 10.1128/jb.177.1.247-251.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dziewit L, Czarnecki J, Prochwicz E, Wibberg D, Schlüter A, Pühler A, et al. Genome-guided insight into the methylotrophy of Paracoccus aminophilus JCM 7686. Front Microbiol. 2015;6:852. doi: 10.3389/fmicb.2015.00852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vorobev A, Jagadevan S, Jain S, Anantharaman K, Dick GJ, Vuilleumier S, et al. Genomic and transcriptomic analyses of the facultative methanotroph Methylocystis sp. strain SB2 grown on methane or ethanol. Appl Environ Microbiol. 2014;80:3044–52. doi: 10.1128/AEM.00218-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pratscher J, Dumont MG, Conrad R. Assimilation of acetate by the putative atmospheric methane oxidizers belonging to the USCα clade. Environ Microbiol. 2011;13:2692–701. doi: 10.1111/j.1462-2920.2011.02537.x. [DOI] [PubMed] [Google Scholar]
- 70.Semrau JD, Dispirito AA, Vuilleumier S. Facultative methanotrophy: false leads, true results, and suggestions for future research. FEMS Microbiol Lett. 2011;323:1–12. doi: 10.1111/j.1574-6968.2011.02315.x. [DOI] [PubMed] [Google Scholar]
- 71.Pieja AJ, Sundstrom ER, Criddle CS. Poly-3-hydroxybutyrate metabolism in the type II methanotroph Methylocystis parvus OBBP. Appl Environ Microbiol. 2011;77:6012–9. doi: 10.1128/AEM.00509-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ghosh W, Dam B. Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse Bacteria and Archaea. FEMS Microbiol Rev. 2009;33:999–1043. doi: 10.1111/j.1574-6976.2009.00187.x. [DOI] [PubMed] [Google Scholar]
- 73.Stockdreher Y, Sturm M, Josten M, Sahl HG, Dobler N, Zigann R, et al. New proteins involved in sulfur trafficking in the cytoplasm of Allochromatium vinosum. J Biol Chem. 2014;289:12390–403. doi: 10.1074/jbc.M113.536425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ramana VV, Raj PS, Tushar L, Sasikala Ch, Ramana ChV. Rhodomicrobium udaipurense sp. nov., a psychrotolerant, phototrophic alphaproteobacterium isolated from a freshwater stream. Int J Syst Evol Microbiol. 2013;63:2684–9. doi: 10.1099/ijs.0.046409-0. [DOI] [PubMed] [Google Scholar]
- 75.Anantharaman K, Breier JA, Dick GJ. Metagenomic resolution of microbial functions in deep-sea hydrothermal plumes across the Eastern Lau Spreading Center. ISME J. 2016;10:225–39. doi: 10.1038/ismej.2015.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pester M, Knorr KH, Friedrich M, Wagner M, Loy A. Sulfate-reducing microorganisms in wetlands – fameless actors in carbon cycling and climate change. Front Microbiol. 2012;3:72. doi: 10.3389/fmicb.2012.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Csáki R, Hanczár T, Bodrossy L, Murrell JC, Kovács KL. Molecular characterization of structural genes coding for a membrane bound hydrogenase in Methylococcus capsulatus (Bath) FEMS Microbiol Lett. 2006;205:203–7. doi: 10.1111/j.1574-6968.2001.tb10948.x. [DOI] [PubMed] [Google Scholar]
- 78.Mohammadi S, Pol A, van Alen TA, Jetten MS, Op den Camp HJ. Methylacidiphilum fumariolicum SolV, a thermoacidophilic ‘Knallgas’ methanotroph with both an oxygen-sensitive and -insensitive hydrogenase. ISME J. 2016;11:945–58. doi: 10.1038/ismej.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.King GM, Weber CF. Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat Rev Microbiol. 2007;5:107–18. doi: 10.1038/nrmicro1595. [DOI] [PubMed] [Google Scholar]
- 80.Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB, et al. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 2016;10:761–77. doi: 10.1038/ismej.2015.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khadem AF, Pol A, Wieczorek A, Mohammadi SS, Francoijs KJ, Stunnenberg HG, et al. Autotrophic methanotrophy in Verrucomicrobia: Methylacidiphilum fumariolicum solv uses the Calvin-Benson-Bassham cycle for carbon dioxide fixation. J Bacteriol. 2011;193:4438–46. doi: 10.1128/JB.00407-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tabita FR, Satagopan S, Hanson TE, Kreel NE, Scott SS. Distinct form I, II, III, and IV RuBisCO proteins from the three kingdoms of life provide clues about RuBisCO evolution and structure/function relationships. J Exp Bot. 2008;59:1515–24. doi: 10.1093/jxb/erm361. [DOI] [PubMed] [Google Scholar]
- 83.Tsai YCC, Lapina MC, Bhushan S, Mueller-Cajar O. Identification and characterization of multiple RuBisCO activases in chemoautotrophic bacteria. Nat Commun. 2015;6:8883. doi: 10.1038/ncomms9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dedysh SN, Naumoff DG, Vorobev AV, Kyrpides N, Woyke T, Shapiro N, et al. Draft genome sequence of Methyloferula stellata AR4, an obligate methanotroph possessing only a soluble methane monooxygenase. Genome Announc. 2015;3:e01555–4. doi: 10.1128/genomeA.01555-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reim A, Luke C, Krause S, Pratscher J, Frenzel P. One millimetre makes the difference: high-resolution analysis of methane-oxidizing bacteria and their specific activity at the oxic-anoxic interface in a flooded paddy soil. ISME J. 2012;6:2128–39. doi: 10.1038/ismej.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ho A, Di Lonardo DP, Bodelier PLE. Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol Ecol. 2017;93:fix006. doi: 10.1093/femsec/fix006. [DOI] [PubMed] [Google Scholar]
- 87.Tveit AT, Urich T, Svenning MM. Metatranscriptomic analysis of Arctic peat soil microbiota. Appl Environ Microbiol. 2014;80:5761–72. doi: 10.1128/AEM.01030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Krause SMB, Johnson T, Samadhi Karunaratne Y, Fu Y, Beck DA, Chistoserdova L, et al. Lanthanide-dependent cross-feeding of methane-derived carbon is linked by microbial community interactions. Proc Natl Acad Sci USA. 2017;114:358–63. doi: 10.1073/pnas.1619871114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Neumann RB, Blazewicz SJ, Conaway CH, Turetsky MR, Waldrop MP. Modeling CH4 and CO2 cycling using porewater stable isotopes in a thermokarst bog in Interior Alaska: results from three conceptual reaction networks. Biogeochemistry. 2016;127:57–87. doi: 10.1007/s10533-015-0168-2. [DOI] [Google Scholar]
- 90.Bolger Anthony M., Lohse Marc, Usadel Bjoern. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used in this manuscript are submitted under NCBI BioProject accession number PRJNA386568.