Abstract
The purpose of this study is to explore Dvl3 variants and their interaction with negative life events on MDD susceptibility in a Chinese Han population. Additionally, we also attempted to identify whether there is an association between Dvl3 variants and pro-inflammatory cytokines. A total of 1102 participants, consisting of 550 patients with MDD and 552 healthy subjects, were recruited for genotyping by TaqMan allelic discrimination assay. Pro-inflammatory cytokine mRNA levels in peripheral blood were measured by QPCR. After the assessment of negative life events by the Life Events Scale, the Dvl3 gene–environment interaction (G × E) and risk factors were evaluated using generalized multifactor dimensionality reduction method (GMDR) and logistic regression analysis, respectively. This study is the first to reveal the interaction between Dvl3 allelic variations and negative life events as well as pro-inflammatory cytokines on MDD susceptibility in a Chinese Han population.
Introduction
Major depressive disorder (MDD), as a severe psychiatric disorder featured by persistent low mood, has a serious effect on physical and psychological health in humans. The World Health Organization has predicted that MDD will be the chief disability-causing illness by 2030, with a high disease burden worldwide1. For evaluation and treatment of this condition, it is important to determine etiological factors and review advances in diagnostic modalities sensitive and specific to MDD. Emerging evidence has indicated that MDD has modest heritability (31–42%)2; therefore, a large number of genome-wide association studies (GWAS) have been conducted to identify candidate genes for this condition. Recently, two relatively large and comprehensive genomic and transcriptomic studies have implicated Dvl3 as a significant risk gene for MDD3,4.
Dvl3 is a gene located on chromosome 3q27. It codes for the disheveled segment polarity protein (Dvl)-3, which takes part in the wingless-related integration site (Wnt) signaling pathway, which is crucial for regulation of hippocampal neurogenesis5. The role of the Wnt signaling pathway in mood disorders was recently reviewed6,7, and Dvl was confirmed to be associated with neural development processes such as dendritic arborization7. Wilkinson et al. found that down-regulation or blockade of Dvl promoted depression-like behavior in depression models8. However, to date, there has been little research on the association between Dvl polymorphism and MDD. There has only been one mega-analysis of GWAS, which revealed a suggestive association between Dvl3 polymorphism (rs1969253) and MDD in individuals of European ancestry (9240 patients with MDD and 9519 control subjects); however, the association did not reach genome-wide significance (P = 4.8 × 10−6)4. Subsequently, Jansen et al. provided further evidence that Dvl3 has an effect on MDD. However, both these studies—which strongly indicated that Dvl3 has an important role in MDD—were conducted among European populations. Therefore, whether there is an association between Dvl3 and MDD among populations of other ethnicities has yet to be confirmed.
Since MDD is a mulitifactorial complex disease, accumulated evidence suggests that both environmental and genetic factors are involved in the etiology of MDD9. Caspi et al. initially provided the evidence regarding the key role of environmental factors in causing depression10; this was followed by several correlated clinical studies which supported the findings11–13. Negative life events, as one of the most well-established environmental risk factors for MDD, refer to loss, divorce, serious illness, interpersonal or family problems, relationships, and social difficulties14. The genetic variation modulates the individual sensitivity to negative environmental influences, making individuals not vulnerable to such events and others vulnerable. Studies on etiology of MDD have illustrated the gene–environment (G × E) interaction of certain risk genes and negative life events was associated with considerably greater risk of developing mood disorders. However, as far as we know, the interaction between Dvl3 polymorphisms and negative life events on MDD susceptibility has not been investigated thus far.
Additionally, over the past several decades, there has been strong and wide support for the cytokine hypothesis, a general hypothesis based on immune–inflammatory system dysfunction. It is one of the more prevalent theories concerning MDD and might provide insights into the pathogenesis of depression and development of biomarkers and, ultimately, more effective depression therapies. The hypothesis15 regards MDD as an environment–neuro–immune disorder. It is believed that physiological or psychological stress could activate the immune system, cause abnormal cytokine production, and thus affect the central nervous system, resulting in emotional changes or disease. Indeed, there is substantial evidence linking MDD to alterations in the inflammatory system, including the presence of elevated levels of Pro-inflammatory cytokines together with other mediators of inflammation16–18. However, to date, little is known about the effect of associations between Dvl variants and immune system responses on MDD.
Therefore, we conducted a case-control study to ascertain whether Dvl3 polymorphisms and negative life events as well as their interactions were associated with MDD among northern Chinese Han population. Moreover, considering that the etiology of depression is associated with stress, we also analyzed inflammatory cytokine production in peripheral blood in patients with MDD and discussed the potential connection between inflammatory cytokines and MDD.
Results
Demographic characteristics of the participants
Basic characteristics of participants were displayed in Table 1. No significant difference was found in age, gender or marital status. The mean ages of the patients and control subjects were 44.53 ± 13.53 and 43.19 ± 9.08 years, respectively.
Table 1.
Demographic characteristics of participants.
| Variables | MDD (n = 550) | Control (n = 552) | χ2/t | P value |
|---|---|---|---|---|
| Age, years | 44.53 ± 13.53 | 43.19 ± 9.08 | −1.938 | 0.053 |
| Sex | 3.553 | 0.059 | ||
| Male | 165 (30.00%) | 195 (35.30%) | ||
| Female | 385 (70.00%) | 357 (64.70%) | ||
| Marital status | 5.272 | 0.261 | ||
| Single | 79 (14.40%) | 65 (11.80%) | ||
| Married | 436 (79.30%) | 459 (83.20%) | ||
| Divorced | 31 (5.60%) | 21 (3.80%) | ||
| Widowed | 4 (0.70%) | 7 (1.30%) | ||
| Negative LES score | 9.66 ± 18.37 | 2.68 ± 6.78 | −8.358 | 3.45E-16 |
Data are presented as mean ± standard deviation or number (percentage).
Single-marker association with MDD
The genotypic distributions of all selected Dvl3 polymorphisms conformed to the Hardy–Weinberg equilibrium (P > 0.05). The genotypic distribution pattern and allelic frequencies of the two SNPs among MDD and controls were shown in Table 2. After Bonferroni correction, significant differences in genotypic between the patients and controls were confirmed in rs1709642 (χ2 = 19.464; P = 5.95E-05), and the association between rs1969253 and MDD was not significant. While there was a significant allelic association between rs1709642 (χ2 = 11.320; P = 0.001; OR = 1.333; 95% CI = 1.127–1.576) and MDD, the corresponding association between rs1969253 and MDD was not significant (P = 0.055).
Table 2.
Genotype distribution patterns and allelic frequencies of Dvl polymorphisms among patients with MDD and control subjects.
| SNP | Sample | Genotype (%) | P | Allele (%) | P | Odds ratio (95% CI) | |||
|---|---|---|---|---|---|---|---|---|---|
| rs1969253 | CC | CA | AA | C | A | ||||
| MDD | 123 (22.4) | 302 (54.9) | 125 (22.7) | 0.056 | 548 (49.8) | 552 (50.2) | 0.055 | 1.177 | |
| Control | 162 (29.3) | 271 (49.1) | 119 (21.6) | 595 (53.9) | 509 (46.1) | (0.996–1.392) | |||
| rs1709642 | CC | CT | TT | C | T | ||||
| MDD | 107 (19.5) | 313 (56.9) | 130 (23.6) | 1.19E-04 | 527 (47.9) | 573 (52.1) | 0.002 | 1.333 | |
| Control | 171 (31.0) | 266 (48.2) | 115 (20.8) | 608 (55.1) | 496 (44.9) | (1.127–1.576) | |||
Bold significant p-values were corrected by Bonferroni correction.
Gene–environment interaction analysis on MDD
The gene–environment interaction effects in MDD susceptibility were detected using a GMDR model with age, smoking and alcohol consumption as covariates. Results of the GMDR analysis are summarized in Table 3. The prediction accuracy p-value was determined using the permutation test with 1000 replications. Significant two-locus and three-locus interaction models were observed (p < 0.05). The interaction between the two SNPs relative to negative life events showed a CV consistency of 10/10 and a testing accuracy of 65.74%. As results suggested that there was a significant interaction between Dvl3 polymorphisms and negative life events on MDD susceptibility among the Chinese Han population, this was considered to be the best multi-locus model.
Table 3.
The best gene–environment interaction models obtained by GMDR.
| Locus No. | Best models | PE (%) | CV | P value |
|---|---|---|---|---|
| 2 | rs1969253, negative life events | 35.26 | 10/10 | <0.001 |
| 3 | rs1969253, rs1709642, negative life events |
34.26 | 10/10 | <0.001 |
PE, prediction error; CV, cross validation.
Furthermore, we assessed the causative factors selected by GMDR using logistic regression analysis which incorporated age, smoking and alcohol consumption as covariates. Results were summarized in Table 4. Individuals with allele A−(CC) (OR = 1.927; 95% CI = 1.072–3.464) or A+(AC, AA) (OR = 2.518; 95% CI = 1.266–5.008) of rs1969253 and demonstrating a high negative life events score were in higher risk of developing MDD compared to the others. While subjects carrying the T+ allele (CT, TT) of rs1709642 with a high negative life events score (OR = 3.025; 95% CI = 1.471–6.223) were more susceptible to MDD relative to the rest of the study population.
Table 4.
Interaction between Dvl3 polymorphisms and negative life events.
| Variables | MDD | Control | OR(95% CI) | P value |
|---|---|---|---|---|
| A − rs1969253 | ||||
| and LN | 91 | 137 | 1 | |
| A− and HN | 32 | 25 | 1.927 (1.072–3.464) | 0.028 |
| A+ and LN | 254 | 342 | 1.118 (0.819–1.526) | 0.481 |
| A+ and HN | 173 | 48 | 2.518 (1.266–5.008) | 0.008 |
| rs1709642 | ||||
| T− and LN | 84 | 146 | 1 | |
| T− and HN | 23 | 25 | 1.599 (0.854–2.992) | 0.142 |
| T+ and LN | 261 | 333 | 1.362 (0.996–1.864) | 0.053 |
| T+ and HN | 182 | 48 | 3.025 (1.471–6.223) | 0.003 |
LN, low negative life events; HN, high negative life events.
Pro-inflammatory cytokines in peripheral blood
First, upon intergroup comparison of mRNA expression levels in peripheral blood, patients with MDD were found to exhibit significantly higher mRNA levels of Pro-inflammatory cytokines IL-1β, TNF-α, and IL-6than the control subjects (Fig. 1). Second, in the MDD group, no difference was observed in mRNA expression levels of IL-1β, TNF-α, orIL-6 between patients with low and high negative life events (Fig. 2). Finally, upon comparing Pro-inflammatory cytokine mRNA levels among patients with MDD with different Dvl3 polymorphisms, individuals carrying alleles A+(AC, AA) of rs1969253 and T+(CT, TT) of rs1709642 were found to exhibit significantly higher TNF-α mRNA levels than the remaining patients (Fig. 3).
Figure 1.
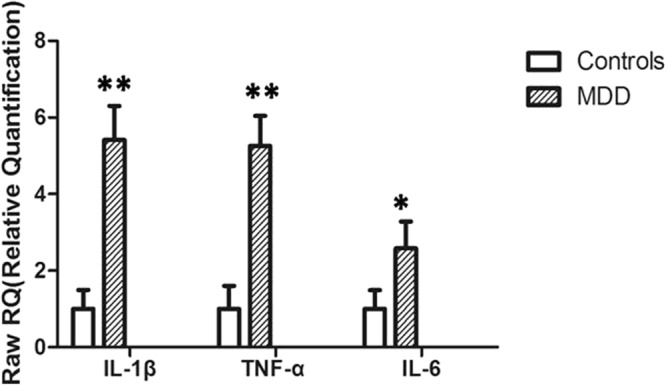
mRNA levels of pro-inflammatory cytokines (mean ± standard deviation) in peripheral blood in patients with MDD and control subjects (n = 100, each). **P < 0.01; *P < 0.05.
Figure 2.
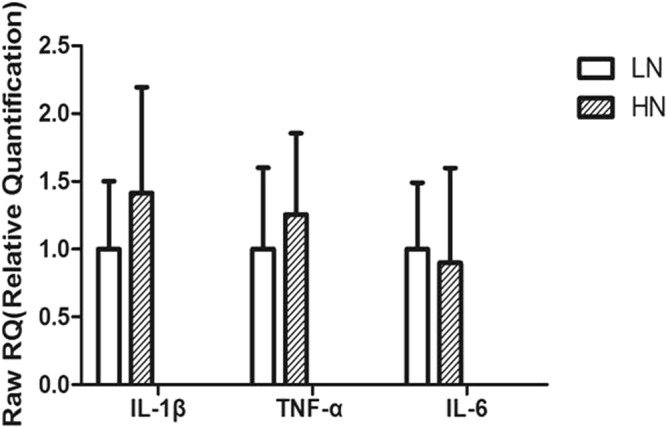
mRNA levels of pro-inflammatory cytokines (mean ± standard deviation) among patients with MDD with LN and HN (n = 100, each). LN, low negative life events; HN, high negative life events (n = 100).
Figure 3.
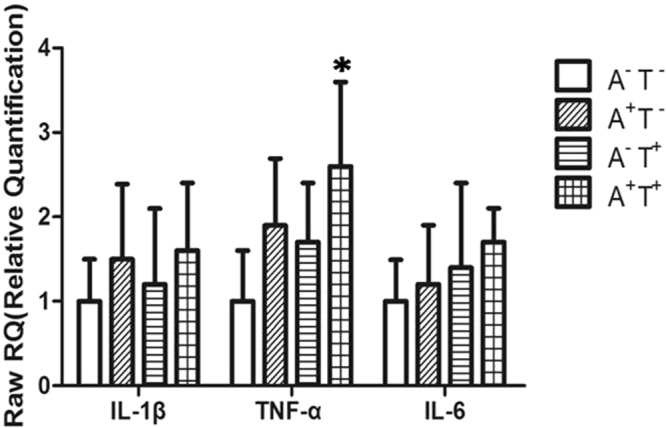
mRNA levels of pro-inflammatory cytokines (mean ± standard deviation) among patients with major depressive disorder with different Dvl3 polymorphisms. A−, allele CC of rs1969253 (n = 25); A+, allele AC, AA of rs1969253 (n = 25); T-, allele CC of rs1709642 (n = 25); and T+, allele CT, TT of rs1709642 (n = 25). *P < 0.05.
Discussion
In 2013 a PGC GWAS for MDD firstly threw a light on Dvl3 and caught our attention, but the following studies by PGC GWAS did not verify the significance of Dvl3. Whereas Jansen et al. provided further evidence19 that Dvl3 has an effect on MDD. In the present study, we attempted to investigate the role of Dvl3 in susceptibility to MDD among the Chinese Han population. We found significant differences in genotypic and allelic frequencies of Dvl3 polymorphisms between patients with MDD and control subjects and also confirmed one possible susceptibility loci—rs1709642—for onset of MDD. After Bonferroni correction, the current study found no association of Dvl3 rs1969253 with MDD at the single-locus level. According to our results, individuals with genotypes T+ (CT, TT) of rs1709642 when exposed to high negative life events were more likely to suffer from MDD than the remaining subjects. Similarly, in a recent gene expression study (MDD, 882; remitted MDD, 635; and control, 331), Jansen et al. observed that Dvl3 expression was up-regulated among patients with current MDD. Another larger gene expression study reported that Dvl3 polymorphism (rs1969253) was associated with MDD among individuals of European ancestry, although the association failed to reach genome-wide significance. We suspected that the inconsistent result may be arisen due to sample differences: the number of MDD attacks, the proportion of melancholic MDD and different stages of MDD, for example. In addition, it is still unclear to which differences in sample immune response20,21, neuroplasticity, or other factors contribute to the failure to replicate genetic effects of Dvl3 gene in the CONVERGE sample. Some scholars22 proposed lack of consideration of such environmental effects may be one prominent reason for the failure of replication in candidate gene studies and GWAS into the genetic background of depression.
Studies targeted depression as a product of multiple networks, where the activity of the pathways may be influenced by both genetic disposition and environmental exposure, mostly by individual-specific environmental factors23,24. Response to stress is regarded as one of the most important hypotheses for the etiology of MDD25. Existing literature confirms that negative life events are closely related to the incidence and severity of MDD26. However, thus far, no information on G × E interaction of Dvl3 in the risk of developing MDD has been found. Our results revealed significant interaction effects between Dvl3 polymorphisms and negative life events with regard to the risk of developing MDD. We observed two best models: a two-factor model indicating interactions between rs1969253 and negative life events and a three-factor model indicating interactions betweenrs1969253–rs1709642 and negative life events. This result helps explain why our study just like the previous study4 on Dvl3 polymorphism rs1969253 and MDD failed to achieve genome-wide significance —environmental factors such as negative life events should be considered jointly with genetic polymorphisms. The present result regarding rs1709642 is an important discovery which remains to be further verified. For all we know, this study is the first to survey the impact of interactions between allelic variations in Dvl3 and negative life events on the susceptibility of MDD.
Consistent with previous findings, Dvl3 has emerged as a promising genetic risk factor for MDD in the present study. It has been reported that Dvl3 transcript levels are down-regulated in the nucleus accumbens and frontal region8,27 in individuals with MDD and up-regulated in leukocytes in individuals reporting social isolation28. There is evidence that Dvl influences MDD both through intermediate Dvl3 expression in the brain and the Wnt or NF-κB (nuclear factor kappa-light -chain-enhancer of activated B cells) pathway29,30, which is closely related with neuro-immune interactions. There are now multiple lines of evidence indicating that pro- and/or anti-inflammatory cytokine levels are altered in the serum or cerebrospinal fluid31,32 in patients with depression. The statement that IL-1β, IL-6, and TNF-α levels are increased in the serum and/or plasma in patients with depression33–36 is widely accepted to date. For this reason, in the present study, we attempted to identify whether there is an association between Dvl3 variants and Pro-inflammatory cytokines. Another remarkable finding of the present study is the association between Dvl3 variants and inflammatory response. Genetic polymorphism of Dvl3 (rs1969253 and rs1709642) was associated with increased TNF-α levels. This effect was independent of a history of negative life events. Although the patho-physiological role of TNF-α in MDD is not clear, we could exclude the possibility that Dvl3 mutations influence TNF-α level through the NF-κB pathway. There has been substantial research on the interconnectedness of NF-κB and TNF, including some studies involving knockout mouse models37. Although Dvl3 is not likely to be a major susceptibility factor for MDD, its clinical value might not be negligible. Analysis of Dvl3 polymorphisms might help identify groups of patients with MDD who are likely to respond to anti-inflammatory agents.
The present results should be considered on the basis of several limitations. First, our selected SNPs were located in the same block of Dvl and might not be representative of genetic information in other regions of the gene. Second, the results of life events assessment using LES, to some extent, might be influenced by subjective interpretation. Third, this study merely included patients and control subjects of Chinese Han origin from Northern China, and the sample size was relatively small. It is, therefore, difficult to generalize these results to other populations.
Materials and Methods
Participants
A total of 1102 participants from a single hospital, which consisted of 550 first-episode depressive patients with MDD and 552 healthy control subjects, were recruited for the study between February 2014 and December 2016. All participants were of Chinese Han origin and were living in the same geographical area in the north of China. Basic characteristics of participants were displayed in Table 1. Cases were diagnosed with MDD by at least two psychiatrists according to the Fourth Edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV)34,36–38. The patients all had first-episode depressive disorder and not a single patient received any antidepressant treatment within 4 weeks preceding assessment. Only patients with a minimum HAMD score of 21 were included in this study.
Subjects with a history of brain organic mental disorders, a family history of genetic disease, mental retardation or dementia, immunodeficiency disease, or those who recently received blood transfusion treatment were excluded from the study. Healthy subjects with a family history of mental disorders were also excluded from participating. Subjects who failed to provide sufficient information or continue the study prematurely were also excluded. All procedures of our research were conducted according to the guidelines established by the National Institutes of Health, and every effort was made to minimize suffering. This study was approved by Harbin Medical University Research Ethics Committee, Harbin, China. Written informed consent forms were obtained from all participants in our study.
Outcome measures
Evaluation of negative life events
The Life Events Scale (LES) developed by Yang and Zhang was used to evaluate negative life events, which includes 48 items classified into 3 domains: family life (28 items), work (13 items), and social and other aspects (7 items). The validity of LES has previously been confirmed in a Chinese population39. The negative events mainly refer to serious illness, relationship breakdown, housing difficulties, unemployment, social difficulties, and financial crises. The LES was filled by interviewers according to their own experience and the scores for positive, negative and total life events were calculated. The scale assesses four aspects of the event: time of occurrence (absent = 1; over a year ago = 2; within the past year = 3; or chronic = 4), character (good = 1 or bad = 2), influence on mood (absent = 1; mild = 2;moderate = 3; severe = 4; or extreme = 5), and duration of influence (≤3 months = 1; 3–6 months = 2; 6–12 months = 3; or >12 months = 4). The 75% percentile (a score of 6) was considered as the cutoff value for high and low level categories.
DNA extraction and genotyping methods
Genomic DNA was isolated from collected EDTA-anticoagulated blood samples using the AxyPrepTM Blood Genomic DNA Miniprep Kit (Axygen, Union City, CA, USA). Using the Haploview program40, we selected two tag single-nucleotide polymorphisms (SNPs; rs1969253 and rs1709642) from the whole Dvl gene. Primers for polymerase chain reaction (PCR) amplification were designed with Primer 5.0 software (Premier, Canada), and final specific primers were checked using NCBI-BLAST (http://www.ncbi.nlm.nih.gov/). The ABI PRISM 7900 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) and ABI 3730 DNA sequencer (Applied Biosystems) were used for genotyping and purifying.
Quantitative PCR (qPCR)
Messenger RNA (mRNA) expression levels of interleukin (IL)-1β, tumor necrosis factor (TNF)-α, andIL-6 were determined by qPCR assay using SYBR Green PCR Master mix reagent (TaKaRa, Otsu, Shiga, Japan). Briefly, RNA was extracted using a blood/liquid sample total RNA rapid extraction kit (BioTeke Corporation, Beijing, China). First-strand cDNA synthesis was performed with 1 μg of total RNA usingthe PrimeScriptTM RT reagent kit (Takara, Dalian, China) following the manufacturer’s instructions. The quality and quantity of total RNA and genomic DNA were evaluated by 1.5% agarose gel electrophoresis and measured with a UVS-99 densitometer (ACTGene, Piscataway, NJ, USA). Relative expression levels were normalized against β-actin and analyzed by the 2-ΔΔCt method [ΔΔCt = (Ct Target - Ct Reference) sample - (Ct Target- Ct Reference) control].
Statistical analysis
Pearson’s chi-square test was used to perform Hardy–Weinberg equilibrium tests, and pairwise linkage disequilibrium. Using SPSS version 20.0 (International Business Machines, Armonk, NY, USA), the Student’s t-test was used to compare the means of continuous variables between cases and controls, and the differences in the distribution of categorical variables were evaluated using the chi-square test. Bonferroni correction was applied to the p-value to correct for multiple testing and a corrected p-value values ≤ 0.05 (two-tailed) was considered as being statistically significant.
Gene–environment interactions were analyzed using generalized multifactor dimensionality reduction (GMDR) software (version 1.0.0) which was designed by the Computational Genetics Laboratory, Dartmouth Medical School, Lebanon, NH, USA41. The best gene-environment interaction model based on the values arising from cross-validation (CV) consistency and accuracy testing were selected. A permutation test with 1000 replications was used to measure empirical p-values thereby substantiating the significance of the model. In GMDR analysis, a p-value was corrected for multiple testing by permutation test and a corrected p-value < 0.05 (two-tailed) was considered to be statistically significant
For validating the results of GMDR, odds ratios (OR; with 95% confidence intervals) of risk factors were computed by logistic regression analysis using SPSS version 20.0. To narrow down the number of possible combinations, only dominant models were subjected to further analysis. The Bonferroni method was used to correct P values for multiple comparisons.
Conclusion
For all we know, this is the first report that there was an effect modification between Dvl3 variation and negative life events on MDD susceptibility in a Chinese Han population. This study is also the first to reveal the underlying genetic architecture of Dvl3, including genetic loci frequencies, effect sizes and models of action. The candidate SNPs could regulate Dvl3 expression, and even modulate the effects of negative life events; therefore, their effects converged to a signifcant modulation on the development of MDD. These factors are important determinants for the success in identifying genetic associations for disease(s) with complex traits. Although their specific mechanism still needs further study, the present results confirm Dvl3 as a susceptibility factor for MDD as well as a modifier of inflammatory response.
Acknowledgements
We thank the patients and families for their support and participation. We are also grateful to all the subjects who participated in this study. This research was supported by the National Natural Science Foundation of China (81773536) to Prof. Yanjie Yang.
Author Contributions
Y.J.Y., D.P.C. and X.H.Q. designed the study, D.H., J.R.Y., J.S.M. and J.Z. participated in the acquisition of data, which were analyzed by J.S.M., X.Y.Z. and B.B., J.Z. wrote the article, Y.J.Y., X.H.Q. and X.Z.Z. critically reviewed it. All authors assisted in the revision process, and gave approval for the final version of the article to be published.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jian Zhang, Jiarun Yang and Dong Han contributed equally.
Contributor Information
Yanjie Yang, Email: yanjie1965@163.com.
Depin Cao, Email: caodp211@163.com.
Xiaohui Qiu, Email: qiuxiaohui@foxmail.com.
References
- 1.Organization., W. H. Depression Fact Sheet. WHO, http://www.who.int/mediacentre/factsheets /fs369/en/ (April, 2016).
- 2.Kessler RC, Merikangas KR, Wang PS. Prevalence, comorbidity, and service utilization for mood disorders in the United States at the beginning of the twenty-first century. Annual review of clinical psychology. 2007;3:137–158. doi: 10.1146/annurev.clinpsy.3.022806.091444. [DOI] [PubMed] [Google Scholar]
- 3.Jansen R, et al. Gene expression in major depressive disorder. Molecular psychiatry. 2016;21:339–347. doi: 10.1038/mp.2015.57. [DOI] [PubMed] [Google Scholar]
- 4.Major Depressive Disorder Working Group of the Psychiatric, G. C. et al. A mega-analysis of genome-wide association studies for major depressive disorder. Molecular psychiatry18, 497–511, 10.1038/mp.2012.21 (2013). [DOI] [PMC free article] [PubMed]
- 5.Hussaini SM, et al. Wnt signaling in neuropsychiatric disorders: ties with adult hippocampal neurogenesis and behavior. Neuroscience and biobehavioral reviews. 2014;47:369–383. doi: 10.1016/j.neubiorev.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pilar-Cuellar F, et al. Neural plasticity and proliferation in the generation of antidepressant effects: hippocampal implication. Neural plasticity. 2013;2013:537265. doi: 10.1155/2013/537265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sani G, et al. The wnt pathway in mood disorders. Current neuropharmacology. 2012;10:239–253. doi: 10.2174/157015912803217279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilkinson MB, et al. A novel role of the WNT-dishevelled-GSK3beta signaling cascade in the mouse nucleus accumbens in a social defeat model of depression. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2011;31:9084–9092. doi: 10.1523/jneurosci.0039-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopizzo N, et al. Gene-environment interaction in major depression: focus on experience-dependent biological systems. Frontiers in psychiatry. 2015;6:68. doi: 10.3389/fpsyt.2015.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caspi A, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 11.Uher R. The role of genetic variation in the causation of mental illness: an evolution-informed framework. Molecular psychiatry. 2009;14:1072–1082. doi: 10.1038/mp.2009.85. [DOI] [PubMed] [Google Scholar]
- 12.Walther DJ, et al. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science. 2003;299:76. doi: 10.1126/science.1078197. [DOI] [PubMed] [Google Scholar]
- 13.Facchinetti F, Fava M, Fioroni L, Genazzani AD, Genazzani AR. Stressful life events and affective disorders inhibit pulsatile LH secretion in hypothalamic amenorrhea. Psychoneuroendocrinology. 1993;18:397–404. doi: 10.1016/0306-4530(93)90014-C. [DOI] [PubMed] [Google Scholar]
- 14.Paykel E. S. Life events and affective disorders. Acta Psychiatrica Scandinavica. 2003;108(s418):61–66. doi: 10.1034/j.1600-0447.108.s418.13.x. [DOI] [PubMed] [Google Scholar]
- 15.Maes M, Bosmans E, Meltzer HY, Scharpé S, Suy E. Interleukin-1 beta: a putative mediator of HPA axis hyperactivity in major depression? American Journal of Psychiatry. 1993;150:1189. doi: 10.1176/ajp.150.8.1189. [DOI] [PubMed] [Google Scholar]
- 16.Maes M, et al. The inflammatory & neurodegenerative (I&ND) hypothesis of depression: leads for future research and new drug developments in depression. Metabolic brain disease. 2009;24:27–53. doi: 10.1007/s11011-008-9118-1. [DOI] [PubMed] [Google Scholar]
- 17.Miller AH, Maletic V, Raison CL. Inflammation and its discontents: the role of cytokines in the pathophysiology of major depression. Biological psychiatry. 2009;65:732–741. doi: 10.1016/j.biopsych.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uher R, et al. An inflammatory biomarker as a differential predictor of outcome of depression treatment with escitalopram and nortriptyline. The American journal of psychiatry. 2014;171:1278–1286. doi: 10.1176/appi.ajp.2014.14010094. [DOI] [PubMed] [Google Scholar]
- 19.Jansen R, et al. Gene expression in major depressive disorder. Molecular psychiatry. 2016;21:444. doi: 10.1038/mp.2015.94. [DOI] [PubMed] [Google Scholar]
- 20.Guo CC, Hyett MP, Nguyen VT, Parker GB, Breakspear MJ. Distinct neurobiological signatures of brain connectivity in depression subtypes during natural viewing of emotionally salient films. Psychological medicine. 2016;46:1535–1545. doi: 10.1017/S0033291716000179. [DOI] [PubMed] [Google Scholar]
- 21.Roca M, et al. Cognitive function after clinical remission in patients with melancholic and non-melancholic depression: a 6 month follow-up study. Journal of affective disorders. 2015;171:85–92. doi: 10.1016/j.jad.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 22.Gonda X, et al. Significance of risk polymorphisms for depression depends on stress exposure. Scientific reports. 2018;8:3946. doi: 10.1038/s41598-018-22221-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levinson DF. The genetics of depression: a review. Biological psychiatry. 2006;60:84–92. doi: 10.1016/j.biopsych.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 24.Kendler KS, Baker JH. Genetic influences on measures of the environment: a systematic review. Psychological medicine. 2007;37:615–626. doi: 10.1017/s0033291706009524. [DOI] [PubMed] [Google Scholar]
- 25.Palazidou E. The neurobiology of depression. British medical bulletin. 2012;101:127–145. doi: 10.1093/bmb/lds004. [DOI] [PubMed] [Google Scholar]
- 26.Liu Z, et al. Negative life events and corticotropin-releasing-hormone receptor1 gene in recurrent major depressive disorder. Scientific reports. 2013;3:1548. doi: 10.1038/srep01548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pirooznia M, et al. Metamoodics: meta-analysis and bioinformatics resource for mood disorders. Molecular psychiatry. 2014;19:748–749. doi: 10.1038/mp.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cole SW, et al. Social regulation of gene expression in human leukocytes. Genome biology. 2007;8:R189. doi: 10.1186/gb-2007-8-9-r189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng N, Ye Y, Wang W, Li L. Dishevelled interacts with p65 and acts as a repressor of NF-kappaB-mediated transcription. Cell research. 2010;20:1117–1127. doi: 10.1038/cr.2010.108. [DOI] [PubMed] [Google Scholar]
- 30.Hoeflich KP, et al. Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature. 2000;406:86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- 31.Thomas AJ, et al. Increase in interleukin-1beta in late-life depression. The American journal of psychiatry. 2005;162:175–177. doi: 10.1176/appi.ajp.162.1.175. [DOI] [PubMed] [Google Scholar]
- 32.Carpenter LL, Heninger GR, Malison RT, Tyrka AR, Price LH. Cerebrospinal fluid interleukin (IL)-6 in unipolar major depression. Journal of affective disorders. 2004;79:285–289. doi: 10.1016/S0165-0327(02)00460-3. [DOI] [PubMed] [Google Scholar]
- 33.Dowlati Y, et al. A meta-analysis of cytokines in major depression. Biological psychiatry. 2010;67:446–457. doi: 10.1016/j.biopsych.2009.09.033. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Ho RC, Mak A. Interleukin (IL)-6, tumour necrosis factor alpha (TNF-alpha) and soluble interleukin-2 receptors (sIL-2R) are elevated in patients with major depressive disorder: a meta-analysis and meta-regression. Journal of affective disorders. 2012;139:230–239. doi: 10.1016/j.jad.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Valkanova V, Ebmeier KP, Allan CL. CRP, IL-6 and depression: a systematic review and meta-analysis of longitudinal studies. Journal of affective disorders. 2013;150:736–744. doi: 10.1016/j.jad.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Cattaneo A, et al. Inflammation and neuronal plasticity: a link between childhood trauma and depression pathogenesis. Frontiers in cellular neuroscience. 2015;9:40. doi: 10.3389/fncel.2015.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayden MS, Ghosh S. Regulation of NF-kappaB by TNF family cytokines. Seminars in immunology. 2014;26:253–266. doi: 10.1016/j.smim.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 4th ed. Text Revision, Washinton (2000).
- 39.Yang DS, Yl Z. Life event scale(LES). Rating scales for mental health. Chin. Ment. Health. 1999;12:101–106. [Google Scholar]
- 40.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics (Oxford, England) 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 41.Ritchie MD, et al. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. American journal of human genetics. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]


