Abstract
Dimethylsulfide (DMS) plays a globally significant role in carbon and sulfur cycling and impacts Earth’s climate because its oxidation products serve as nuclei for cloud formation. While the initial steps of aerobic DMS degradation and the fate of its carbon atoms are reasonably well documented, oxidation of the contained sulfur is largely unexplored. Here, we identified a novel pathway of sulfur compound oxidation in the ubiquitously occurring DMS-degrader Hyphomicrobium denitrificans XT that links the oxidation of the volatile organosulfur compound with that of the inorganic sulfur compound thiosulfate. DMS is first transformed to methanethiol from which sulfide is released and fully oxidized to sulfate. Comparative proteomics indicated thiosulfate as an intermediate of this pathway and pointed at a heterodisulfide reductase (Hdr)-like system acting as a sulfur-oxidizing entity. Indeed, marker exchange mutagenesis of hdr-like genes disrupted the ability of H. denitrificans to metabolize DMS and also prevented formation of sulfate from thiosulfate provided as an additional electron source during chemoorganoheterotrophic growth. Complementation with the hdr-like genes under a constitutive promoter rescued the phenotype on thiosulfate as well as on DMS. The production of sulfate from an organosulfur precursor via the Hdr-like system is previously undocumented and provides a new shunt in the biogeochemical sulfur cycle. Furthermore, our findings fill a long-standing knowledge gap in microbial dissimilatory sulfur metabolism because the Hdr-like pathway is abundant not only in chemoheterotrophs, but also in a wide range of chemo- and photolithoautotrophic sulfur oxidizers acting as key players in global sulfur cycling.
Introduction
In nature, the cycles of inorganic and organic sulfur compounds are intimately interwoven. The most oxidized inorganic form, sulfate (+6), is a ubiquitous electron acceptor used in the absence of energetically more favorable respiratory substrates like oxygen or nitrate. Sulfate respiration results in production of massive amounts of hydrogen sulfide (−2) reaching several hundred million tons annually, mainly in marine sediments and pelagic oxygen minimum zones [1]. Sulfide in turn serves as electron donor for sulfur-oxidizing microorganisms. Sulfur transformations driven by microorganisms also involve inorganic sulfur compounds of intermediate redox states, like thiosulfate (−S–SO3−) or elemental sulfur. In addition, the natural sulfur cycle undergoes a considerable impact by organic sulfur compounds among which the malodorous, volatile dimethylsulfide (CH3–S–CH3, DMS) is especially important.
The main source for the ~300 million tons DMS produced per year is degradation of dimethylsulfoniopropionate, a stress protectant accumulated by some macroalgae and phytoplankton, in the upper mixed layers of the oceans [2–4]. Reduction of dimethylsulfoxide, breakdown of sulfur-containing amino acids and methoxylated aromatic compounds, methylation of sulfide or methanethiol (CH3–SH, MT) as well as anthropogenic emissions from wastewater treatment, animal rendering, kraft pulping, and composting are other sources of DMS not limited to marine environments [4–6]. DMS is credited with a pivotal role in global climate control because its oxidation products initiate cloud formation over the oceans [7]. DMS-degrading microorganisms have vast importance in regulating Earth’s climate, as they prevent the escape of about 90% of DMS to the atmosphere [8]. DMS can serve as a source of carbon and electrons and the sulfur contained can either be assimilated or is excreted as sulfate [9–12], thiosulfate [13], or tetrathionate [14], thereby establishing a tight link to the biogeochemical cycling of inorganic sulfur compounds.
Despite their obvious importance, neither the biochemistry of prokaryotic sulfur oxidation nor the microbial remineralization of DMS is fully understood [15–17]. A particular knowledge gap exists with regard to the intertwinement of both processes. Various DMS degradation pathways have been reported, some of these featuring MT and/or H2S as intermediates [16]. Methanethiol oxidase catalyzes the reaction of MT with oxygen to formaldehyde, hydrogen peroxide, and sulfide [15]. Formaldehyde is either assimilated or oxidized to CO2. The steps involved in sulfide oxidation have either not been elucidated or simply attributed as being the same as in inorganic sulfur compound oxidizers [9, 12, 14, 16, 18].
The major problem with the latter assumption is that the current picture of inorganic sulfur compound oxidation is far from being comprehensive. In a vast number of prokaryotes specialized on lithotrophic growth on reduced sulfur compounds, the periplasmic Sox pathway for thiosulfate oxidation to sulfate [19] is incomplete and lacks the SoxCD component responsible for the oxidation of SoxY-bound sulfane sulfur (R–S−) to the sulfone state (R–SO3−). While a portion of these organisms employs the cytoplasmic Dsr pathway involving dissimilatory sulfite reductase [20], the central sulfur oxidation route is largely unknown for a huge array of other sulfur oxidizers including environmentally and biotechnologically highly relevant organisms like ore-leaching Acidithiobacillus species or thermoacidophilic archaea [17]. Metabolic reconstruction supported by microarray transcript profiling or comparative proteomics predicted the involvement of a heterodisulfide reductase-like HdrC1B1AHypHdrC2B2 complex in these organisms [17, 21–23] (Fig. 1) bearing resemblance to HdrABC from methanogens [24], although no coenzyme M-coenzyme B (CoM-S-S-CoB) heterodisulfide is present in the sulfur oxidizers. Direct experimental evidence for an involvement of an Hdr-like complex in oxidative microbial sulfur metabolism was so far not provided because the majority of the relevant organisms are obligate chemolithoautotrophs that strictly depend on reduced sulfur compounds as electron donors and are not accessible to manipulative genetics.
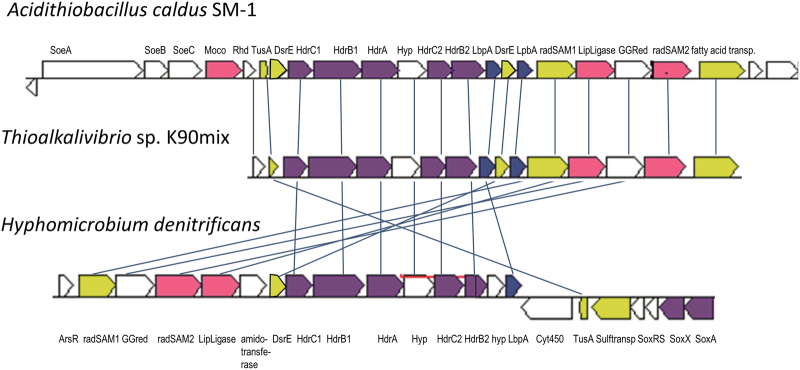
Fig. 1.
Arrangement of hdr-like genes in selected bacteria capable of oxidizing inorganic or volatile organic sulfur compounds. Homologous genes are colored the same between organisms. Kegg/NCBI locus tag identifiers for the regions shown: H. denitrificans, Hden_0682-0703; A. caldus, Atc_2359-2337; Thioalkalivibrio sp. K90mix, TK90_0630-0645. SoeABC, sulfite-oxidizing enzyme [40]; LbpA, lipoate-binding protein; Rhd, rhodanese; GGred, geranylgeranyl reductase-like; radSAM, radical SAM domain-containing protein; LipLigase, lipoate:protein ligase
Here, we addressed the described major knowledge gaps in volatile organosulfur and inorganic sulfur compound degradation. We chose the Alphaproteobacterium Hyphomicrobium denitrificans XT (ATCC 51888) as a model because it contains hdr-like genes (Fig. 1 [17, 25]) and is neither thermo-, acido-, alkali-, nor halophilic. As a typical member of the Hyphomicrobia, this appendaged, budding bacterium is ubiquitous in brackish and fresh waters as well as soils and also found in sewage treatment plants [26]. Most importantly, it is not obligately dependent on sulfur compound oxidation but grows as a restricted facultative methylotroph on substrates like methanol, methylamine (MA), or dimethylamine (DMA). This enabled us to compare the protein equipment of H. denitrificans cells on sulfur-containing and sulfur-free substrates to delineate a probable pathway for DMS oxidation involving thiosulfate as an intermediate and to genetically validate the sulfur-oxidizing function of the Hdr-like proteins in H. denitrificans. The genetic potential for the Hdr pathway of sulfur oxidation is present in a wide spectrum of phylogenetic lineages and may co-exist with DMS/MT-degrading capability.
Material and methods
Bacterial strains, plasmids primers, and growth conditions
The bacterial strains, plasmids, and primers used in this study are listed in Supplementary Table S1. Escherichia coli BL21 (DE3) ΔiscR [27] was used for recombinant protein production and grown in LB medium. E. coli 10β was used for molecular cloning. H. denitrificans strains were cultivated in minimal media kept at pH 7.2 with 100 mM 3-(N-Morpholino)propanesulfonic acid (MOPS) buffer and containing per liter: 1 g NH4Cl, 0.2 g MgSO4 × 7 H2O, 0.5 g NaH2PO4 × 2 H2O, 1.55 g K2HPO4, and 0.2 ml l−1 trace element solution [28]. Methanol, dimethyl-amine (DMA), methylamine (MA), or dimethylsulfide (DMS) were added as carbon and electron source. Unless otherwise indicated, 200 ml cultures containing 24.4 mM methanol or 50 mM methylamine were shaken in 500-ml Erlenmeyer flasks at 200 rpm and incubated at 30 °C. For growth on DMS, 500-ml serum vials sealed with butyl rubber stoppers containing 100 ml medium were used. Varying concentrations of thiosulfate were supplied in growth experiments. Antibiotics for E. coli and H. denitrificans were used at the following final concentrations: kanamycin 50 µg ml−1, tetracycline 15 µg ml−1, ampicillin 100 µg ml−1, rifampin 50 µg ml−1, streptomycin 200 µg ml−1, and chloramphenicol 25 µg ml−1.
Recombinant DNA techniques
Standard methods for DNA manipulation and cloning were used unless otherwise indicated [29]. Restriction enzymes, T4 ligase, and Q5 DNA polymerase were obtained from New England Biolabs (Ipswich, UK) and used according to the manufacturer’s instructions. Oligonucleotides for cloning were obtained from Eurofins MWG (Ebersberg, Germany). The genotypes of the H. denitrificans mutant strains generated in this study were confirmed by Southern hybridization and PCR. Southern hybridization was performed overnight at 68 °C. DNA probes for Southern hybridization were digoxigenin labeled by PCR as described in ref. [20]. Plasmid DNA from E. coli was purified using the GeneJET Plasmid Miniprep Kit (Thermo Fisher Scientific, Waltham, USA). Chromosomal DNA from H. denitrificans strains was prepared using the First-DNA all-tissue Kit (GEN-IAL GmbH, Troisdorf, Germany).
Transformation of H. denitrificans by electroporation
Electrocompetent cells of H. denitrificans were prepared using a modified protocol that had originally been developed for Hyphomicrobium chloromethanicum [30]. A H. denitrificans culture (400 ml) grown in minimal medium containing 24.4 mM methanol was harvested during early exponential phase at an optical density at 600 nm (OD600) of 0.3 (4000 × g, 10 min, 4 °C). Cells were washed twice with ice-cold water (4000 × g, 10 min, 4 °C), once with ice-cold 10% (v/v) glycerol, and finally resuspended in 800 µl of 10% glycerol. Fifty-microliter aliquots of cells were mixed with 500 ng of plasmid DNA and incubated on ice for 10 min. Electroporation was carried out in 0.1 cm gap cuvettes (Bio-Budget Technologies GmbH, Krefeld, Germany) with a Bio-Rad gene pulser II (Bio-Rad Laboratories) with the following electrical settings: 2.4 kV and 200 Ω at a capacitance of 25 µF. After electroporation, 1 ml of minimal medium containing 24.4 mM methanol was added to the cuvette. Cells were transferred to an Eppendorf tube and incubated at 30 °C for 6 h. Transformants were selected by plating suitable dilutions of electroporated cells onto minimal medium agar containing 24.4 mM MeOH and the appropriate antibiotics. Plates were incubated at 30 °C for up to 14 days. The resulting antibiotic-resistant colonies were screened via PCR.
Construction of H. denitrificans mutant strains
For partial replacement of genes Hden_0691 (hdrA) and Hden_0692 (hyp) by a tetracycline Ω cassette and for markerless deletion of Hden_2748 (tsdA), respectively, splicing by overlap extension (SOE) [31] PCR fragments were constructed using primers Fwd_5′∆hdr, Rev_5′∆hdr, Fwd_3′∆hdr, Rev_3′∆hdr and Fwd_5′∆tsdA, Rev_5′∆tsdA, Fwd_3′∆tsdA and Rev_3′∆tsdA, respectively (see Table S1). The ∆hdr fragment was inserted into plasmid pET-22b (+) via XhoI and NcoI restriction sites resulting in plasmid pET-22b∆hdr. The tetracycline Ω cassette from plasmid pHP45Ω-Tc [32] was inserted into pET-22b∆hdr using HindIII restriction sites. The final construct pET-22b∆hdrTc was electroporated into H. denitrificans Sm200 (Table S1). Transformants were selected on minimal medium plates containing 24.4 mM methanol and the appropriate antibiotics. Double crossover recombinants were Tcr and Smr but Aps because they had lost the vector-encoded ampicillin resistance. The genotype of double crossover recombinants was verified by Southern hybridization experiments. For complementation of the H. denitrificans ∆hdr mutant, plasmid pBBR1p264HdenHdrRif was electroporated in electrocompetent H. denitrificans ∆hdr cells. Transformants were selected on minimal medium plates containing the appropriate antibiotics. The genotypes of recombinants were confirmed by PCR.
For markerless in-frame deletion, the ∆tsdA fragment was inserted into plasmid pk18mobsacB [33] using SphI restriction sites. The tetracycline cassette from pHP45Ω-Tc [32] was inserted into the resulting plasmid pk18mobsacB∆tsdA using SmaI restriction sites. The final construct pk18mobsacB∆tsdATc was electroporated into H. denitrificans Sm200. Transformants were selected on minimal medium plates containing 24.4 mM methanol and the appropriate antibiotics. Single crossover recombinants were Smr, Kmr, and Tcr, verified by PCR screening and plated on minimal medium containing methanol and 10% sucrose. Double crossover recombinants survived in the presence of sucrose due to loss of the vector-encoded levansucrase (SacB).
Characterization of phenotypes and quantification of sulfur species
For growth experiments on 0.6 mM DMS, cultures were inoculated to a start OD600 of 0.01 with precultures in late-exponential growth phase cultured on 24.4 mM methanol. DMS was quantified using gas chromatography (GC). An aliquot of 50 µl samples was taken from the headspace and injected into a GC (PerkinElmer Clarus® 480, Rascon FFAP column 25 m × 0.25 micron) equipped with a flame ionization detector. Measurements were conducted at a column temperature of 200 °C, an injector temperature of 150 °C, and a detector temperature of 250 °C. N2 was used as carrier gas. DMS concentrations were calculated by regression analysis based on a seven-point calibration with standard DMS solutions in minimal medium. For growth experiments on thiosulfate, media with 24.4 mM methanol and 2.5 mM thiosulfate were inoculated to a start OD600 of 0.005 with precultures in late-exponential growth phase cultured on the same medium. Thiosulfate, tetrathionate, sulfite, and sulfate were determined by colorimetric, turbidometric, and HPLC methods as described previously [34, 35].
Immunoblot analysis
H. denitrificans cells were resuspended in 1× PBS l (4 mM KH2PO4, 10 mM Na2HPO4, 115 mM NaCl) containing 0.2 mg ml−1 lysozyme and a few grains of DNase and disrupted by sonication (Bandelin Sonopuls UW 2070). After removal of insoluble cell material by centrifugation (16,100 × g, 15 min, 4 °C) protein content was determined with a BCA kit (Thermo Fisher Scientific) and cell extracts were separated by SDS-PAGE. Western analysis was performed using the Transblot SD semi-dry transfer apparatus (Bio-Rad Laboratories) and nitrocellulose membranes (Amersham Protran 0.45 NC, GE Healthcare). HdrA antigens were detected with antisera raised in rabbits (Eurogentec) against recombinant H. denitrificans HdrA purified from E. coli (see Supplementary information for recombinant protein production). Antisera were used at 1:2000 dilution. Binding of α-HdrA was detected with the SignalFireTM ECL reagent system (Cell Signaling Technology).
Proteomic profiling using tandem mass tags
For global proteomic profiling, H. denitrificans were grown in 300 ml minimal medium under two different regimes. In the first case, cultures contained 8 mM dimethylamine and were harvested during exponential growth phase at OD600 of 0.35. In the second case, cultures were pre-grown on 4 mM dimethylamine to an OD600 of 0.2 and then a total of 2.25 mM DMS was added in 0.25–0.5 mM portions. Cultures were harvested in the exponential growth phase before the last added DMS portion was completely used up. Harvesting proceeded at 10,000 × g for 12 min at 4 °C. Cells washed with 1× PBS were resuspended in lysis buffer (1× PBS containing 5% (wt/v) sodium deoxycholate (SDC) and protease inhibitor (cOmplete, Roche Diagnostics)) and disrupted by sonication. Cell debris was removed by centrifugation (16,100 × g, 30 min, 4 °C). The supernatant was adjusted to a protein concentration of 2 mg ml−1 with lysis buffer and stored at −20 °C. Cleared lysates were subjected to in-solution preparation of reduced, alkylated peptides as outlined in Supplementary Information. Peptides were labeled with amine-reactive Thermo Scientific Isobaric Mass Tagging (TMTsixplex) reagents. Experimental details concerning peptide separation, mass spectrometry, data processing, and statistical analysis are given in Supplementary Information. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [36] partner repository with the data set identifier PXD009030.
Identification of hdr-like gene clusters in bacterial genomes
Typical clusters of hdr-like genes were identified in microbial genomes based on BLASTP searches against assembled genomes at NCBI. HdrC1 and HdrB2 amino acid sequences from Acidithiobacillus caldus were used as queries. All hits used in further analysis had an e value of 1e−70 or lower. All positive hits were manually checked for co-occurrence of the other genes in the correct order of appearance (hdrC1B1AhyphdrC2B2). Furthermore, the occurrence of the set of hdr-like genes was checked in the Uncultivated Bacteria and Archaea (UBA) data set [37] using TBLASTN and the amino sequences of the A. caldus Hdr-like proteins as queries with a cutoff in e value of 1e−50. Only those sequences were listed as positive that contained the complete hdr-like gene set on the same contig. The UBA data set, which is the largest currently available set of metagenome-assembled genomes, was also searched with the amino sequence of MtoX from Hyphomicrobium sp. VS [15] in order to identify metagenomes in which mtoX and hdr-like genes co-occur.
Results
Comparative proteomic analysis allows metabolic reconstruction of DMS oxidation in H. denitrificans
Some Hyphomicrobium isolates oxidize thiosulfate and sulfide as additional sources of electrons during chemoorganoheterotrophic growth [12] and several have been reported to grow on DMS as a sole carbon source [9, 11, 12, 15]. Contrary to earlier reports [12], we found that this also holds true for strain XT. All sulfur contained in DMS is released as sulfate (Supplementary Fig. S1).
Detailed analysis of the genome sequence and proteomic comparison of H. denitrificans cells grown on DMS vs. DMA served as the basis for metabolic reconstruction of the DMS oxidation pathway (Fig. 2 and Supplementary Tables S2–S4). The proteome underwent substantial remodeling upon growth on the volatile organic sulfur compound. Thirty-one of 54 proteins that were statistically significantly more abundant during growth on the organosulfur compound are encoded within a range of 86 genes (operon structures Hden_0669–0759), localizing DMS degradation capacity to a defined genetic island. The initial step of DMS oxidation is catalyzed by DMS monooxygenase [9]. In the absence of genes encoding the characterized FMNH2-dependent oxygenase DmoA from Hyphomicrobium sulfonivorans and its partner DmoB, a NADH-dependent FMN oxidoreductase [38], we tentatively assigned Hden_0730 annotated as FMN-dependent-azoreductase for this step. Methanethiol oxidase [15] is localized in the periplasm and releases hydrogen sulfide. Essential components of the thiosulfate-oxidizing Sox multienzyme complex, SoxYZ, SoxXA, and SoxB, were all much more abundant on DMS than on DMA, indicating thiosulfate as an intermediate en route to sulfate. A putative COG2392 membrane protein (also known as YeeE/YedE), Hden_0699, is encoded in the same gene cluster and downstream of soxXA (Fig. 1). Serratia proteins belonging to this group mediate transport of sulfur-containing compounds [39]. Hden_0699 could therefore be involved in transport of sulfur into the cytoplasm, where it is bound by the sulfane–sulfur carrier protein TusA, encoded by the neighboring gene. TusA is a central hub for sulfur in the cytoplasm, not only delivering to the sulfur oxidation pathways in dissimilatory sulfur oxidizers [21], but also distributing to different biosynthetic pathways as has been shown for E. coli [41].
Fig. 2.
Metabolic reconstruction of DMS and thiosulfate metabolism in H. denitrificans XT based on genome sequence analysis, comparative proteomics, and reverse genetics. Fold changes (log2) in protein abundancies are given next to proteins detected as significant in the proteomic study. Prosthetic groups are indicated: orange boxes, [4Fe–4S]; red diamond, non-cubane [4Fe–4S], yellow oval, FAD. DMS, MO DMS monooxygenase; MTO, methanethiol oxidase; TsdA, thiosulfate dehydrogenase
Prominent abundance changes were furthermore observed for the cytoplasmic proteins encoded in the cluster of hdr-like genes (Figs. 1 and 2), indicating that a Hdr-like complex indeed acts as a sulfur-oxidizing unit. Just as the proteins from lithotrophic sulfur oxidizers, the H. denitrificans HdrA-like protein appears to bind one flavin cofactor and only one instead of the six [4Fe–4S] centers in the protein from methanogens (Supplementary Fig. S2). HdrB2 is predicted to contain the two recently characterized non-cubane [4Fe–4S] centers of the heterodisulfide-reducing active site [24] while not all cluster-binding residues are conserved in HdrB1 (Supplementary Fig. S3). The HdrC-like proteins bind two [4Fe–4S] clusters (Supplementary Fig. S4) and may mediate electron transfer between HdrA and the two HdrB-like proteins. Proteins HdrAB1B2C1C2 have previously been co-purified from the hyperthermopilic bacterium Aquifex aeolicus but a functional role was not assigned [42]. The function of the hypothetical protein encoded between genes hdrA and hdrC2 in all hdr-like gene loci from sulfur oxidizers is currently enigmatic. Related proteins are not encoded outside of hdr-like genes clusters, thus there is no relationship to any functionally characterized proteins.
Most likely, the Hdr-like proteins form sulfite which would then be exported to the periplasm, probably by Hden_0720, a TauE-like sulfite transporter [43]. Dedicated sulfite-oxidizing enzymes like the cytoplasmically oriented sulfite-oxidizing enzyme SoeABC [40] or periplasmic Sor-related sulfite dehydrogenases [44] are not encoded in H. denitrificans XT, but once in the periplasm, hydrogensulfite could be removed by spontaneous chemical reaction with hydrogen sulfide [45] released in the MT oxidase reaction, thus regenerating thiosulfate as a substrate for the Sox system. Another possibility is oxidation of sulfite by the combined action of SoxXA, SoxYZ, and SoxB as has been described in vitro for the Paracoccus pantotrophus proteins [46].
Thiosulfate oxidation in H. denitrificans wild-type and mutant strains
To gain deeper insights into the suggested connection between metabolism of inorganic and organic sulfur compounds in H. denitrificans, we first assessed thiosulfate consumption by the wild-type (wt) strain. In contrast to older reports [12], it was well able to degrade thiosulfate. Besides the Sox proteins, H. denitrificans encodes a second thiosulfate-metabolizing enzyme, the tetrathionate-forming diheme cytochrome c thiosulfate dehydrogenase, TsdA [47]. H. denitrificans wt cultures growing chemoorganoheterotrophically in a medium buffered to pH 7.2 and containing 5 mM thiosulfate started thiosulfate consumption in stationary phase and produced stoichiometric amounts of tetrathionate (Fig. 3a). Basic published strategies for introduction of plasmids into H. denitrificans [48, 49] served as the starting point for reverse genetics enabling targeted gene knockouts and in-frame deletion of the tsdA gene. The ΔtsdA strain did not degrade thiosulfate under the described conditions and thiosulfate dehydrogenase was thus identified as the sole tetrathionate-forming enzyme in the organism (Fig. 3b). Tetrathionate is a dead-end product and not metabolized any further. When 0.5, 1, or 2.5 mM tetrathionate was added to methanol or methylamine-containing medium, concentrations stayed essentially the same even until late stationary phase (not shown).
Fig. 3.

a Stoichiometric formation of tetrathionate from thiosulfate by H. denitrificans XT on 50 mM methylamine and 5 mM thiosulfate. b H. denitrificans ∆tsdA is unable to form tetrathionate from thiosulfate. Medium contained 24.4 mM methanol and 5 mM thiosulfate. Thiosulfate (○), tetrathionate (Δ), and OD600 (□) are given. Inocula had been grown on thiosulfate-free methanol-containing medium. All data are the average of two independent biological replicates. Errors bars indicate SD
The picture completely changed when cultures were grown in the presence of only 2.5 mM thiosulfate. Now the wild-type and the ΔtsdA strain did no longer form tetrathionate but both quantitatively oxidized thiosulfate to sulfate (Fig. 4). Metabolic reconstruction (Fig. 2) predicted the Hdr-like proteins to be involved in the latter process. This concept was supported by the presence of the HdrA-like protein in methylamine/thiosulfate, but not in MA-grown cells (Fig. 5). Proof was provided by our finding that sulfate formation absolutely required the presence of a functional Hdr-like complex and was no longer possible in H. denitrificans strain Δhdr (Fig. 6a) in which part of the hdrA-like gene and the subsequent gene for a hypothetical protein were replaced by a tetracycline Ω cartridge preventing transcription and translation of the affected genes and all genes located downstream in the same transcriptional unit [32]. It should be noted that the strain lacking a functional Hdr-like system still degraded thiosulfate albeit at a much lower rate than the wild type. Tetrathionate was formed in stoichiometric amounts in this case (Fig. 6a). When the hdr-like gene cluster from dsrE to lbpA (Fig. 1) was reintroduced in trans on broad host range plasmid pBBR1p264 [50] such that it was expressed under a strong constitutive promoter from Gluconobacter oxydans, thiosulfate oxidation and sulfate formation capacities were completely restored (Fig. 6a).
Fig. 4.

Growth of H. denitrificans XT (boxes, □, ■) and strain ΔtsdA (circles, ○, ●) on 24.4 mM methanol and 2.5 mM thiosulfate. Precultures were grown on methanol-containing medium with 2.5 mM thiosulfate. In the middle panel, open circles and boxes refer to sulfate, closed circles and boxes give tetrathionate concentrations. All data are the average of two independent biological replicates. Errors bars indicate SD
Fig. 5.

Western blot analysis with antiserum against HdrA (37.6 kDa) performed with crude extracts of H. denitrificans wild type grown on methylamine plus thiosulfate (MA + S2O32−, 14.4 μg protein), methylamine (MA, 17.8 μg protein), and dimethylsulfide (DMS, 22.5 μg protein). Lane 4 was loaded with extract (17.5 μg protein) of H. denitrificans expressing plasmid-encoded genes dsrEhdrClB1AhyphdrC2B2hyplbpA from a constitutive promoter. These cells were grown on methylamine. M, marker proteins. The antiserum was raised against HdrA produced recombinantly in E. coli
Fig. 6.
Growth on and consumption of (a) thiosulfate in methanol-containing medium or (b) DMS by H. denitrificans wt (□), Δhdr (○), and the complemented Δhdr+ strain (Δ). Consumption of thiosulfate and DMS, respectively, is shown in the upper panels. Stoichiometric production of sulfate from thiosulfate for the wt and the Δhdr+ strain is indicated by filled symbols in the upper left panel. Tetrathionate concentrations are shown in the middle left panel. Neither thiosulfate and tetrathionate nor sulfite were detected as intermediates during growth of the wild type on DMS, even with very sensitive HPLC methods. All data are the average of two independent biological replicates. Errors bars indicate SD
DMS oxidation in Hyphomicrobiumdenitrificans XT
Finally, H. denitrificans wild type, the ΔtsdA and Δhdr deletion strains, and the ∆hdr+-complemented strain were compared on DMS as the sole carbon and energy source. The ΔtsdA strain did not show any differences to the wild type excluding tetrathionate as an intermediate (Supplementary Fig. S4). The Δhdr strain, however, proved completely unable to metabolize DMS and the complemented strain carrying the hdr-like genes in trans regained the ability to grow on DMS and to form sulfate (Fig. 6b), clearly identifying the proteins encoded by H. dentirificans hdr-like genes as essential for downstream processing of the sulfur contained in DMS.
As shown in Fig. 2, crucial steps of DMS/MT and thiosulfate oxidation occur in different cellular compartments and it is thus not immediately obvious why a block in downstream thiosulfate oxidation would completely prevent degradation of DMS. Here, we need to take into account that sulfide would accumulate in the course of the MT oxidase-catalyzed reaction in the absence of a functional cytoplasmic sulfur oxidation system. MT oxidase is subject to strong feedback inhibition by sulfide [15, 51], precluding any substantial MT and thus also DMS oxidation unless sulfide is efficiently removed. In addition to the Hdr-like pathway, H. denitrificans contains the equipment for sulfide detoxification in the cytoplasm via sulfide:quinone oxidoreductase (SqrB, Hden_0718), a rhodanese-type sulfur transferase (Hden_0719, Rhd) and persulfide dioxygenase (Pdo, Hden_740) [52] (Fig. 7). Although SqrB and Pdo are significantly more abundant on DMS than in its absence (Table S2), their presence in the Δhdr strain is obviously neither sufficient to sustain growth on this substrate nor are they involved in any significant production of sulfate.
Fig. 7.

Cytoplasmic sulfur oxidation pathways. For clarity, reactions are not given with exact stoichiometries. In cases A and B, the electrons released by formation of hydrogensulfite can be fed into respiratory or photosynthetic electron transport chains. In both cases, the TusA protein is proposed as a central sulfur carrier in the cytoplasm and collects sulfur stemming from the oxidation of organic and/or reduced inorganic sulfur compounds [21]. Initial steps take place outside of the cytoplasm. A Oxidation of (protein-bound) persulfide sulfur by the Hdr-like system discovered in this work. B Oxidation of protein-bound persulfide sulfur by the Dsr system involving dissimilatory sulfite reductase. The immediate sulfur substrate carrier for this enzyme is the protein DsrC [17]. C Sulfide detoxification in the cytoplasm via sulfide:quinone oxidoreductase (SQR), rhodanese (RHD), and sulfur/persulfide dioxygenase (PDO). Electrons resulting from the SQR-catalyzed reaction can be fed into the quinone pool. Sulfite is formed by direct reaction with molecular oxygen preventing energy conservation [52]. GSH (GSSH), reduced glutathione (persulfide). D Some archaeal and bacterial sulfur oxidizers contain sulfur oxygenase reductases (SOR). These enzymes usually catalyze a dioxygen-dependent disproportionation of elemental sulfur/and or poylsulfides to hydrogensulfite, thiosulfate, and hydrogen sulfide. Here, SOR is depicted without reductase activity as has been described for Thioalkalivibrio paradoxus [53]
Diversity of the Hdr-like system
To investigate the diversity of microorganisms in the environment that contain the novel sulfur-oxidizing heterodisulfide reductase-like system identified in H. denitrificans by reverse genetics, we checked all genome and metagenome sequences available through the National Center for Biotechnology Information. Stringent criteria were applied and only those organisms were listed that contain all six core hdr-like genes in the same order of appearance (Table S5). The gene set was identified in Archaea as well as in the domain Bacteria. The greatest number of organisms and metagenomes containing hdr-like genes is found among the Proteobacteria. With very few exceptions, all characterized organisms containing hdr-like genes are either established chemo- and photolithotrophs oxidizing reduced inorganic sulfur compounds or they have been reported as oxidizing mineral sulfides (FeS, FeS2, and others) or organic sulfur compounds (DMS). In some instances, the ability of the respective organism to oxidize sulfur compounds has not been tested or reported. Notably, all of these organisms stem from sulfur-dominated habitats like solfataras or ocean sediments and are therefore likely sulfur compound oxidizers. Our database search revealed the occurrence of hdr-like genes in several assembled metagenomes assigned as unclassified Sulfolobales, Proteobacteria, Alpha-, Beta-, or Gammaproteobacteria. The respective metagenomics studies focused on subseafloor or subsurface aquifers, sediments and rocks/rock porewater as well as mine wastewater again emphasizing that the activity of organisms containing the Hdr-like system for sulfur oxidation may considerably impact the transformation of sulfur compounds at these sites.
A previous database survey focusing on the linkage between genes encoding sulfurtransferases and genes encoding putative cytoplasmic sulfur oxidation systems revealed an almost mutually exclusive occurrence of hdr-like genes and those for the Dsr system of sulfur oxidation [17]. Two organisms (Thioalkalivibrio sulfidophilus and Thioalkalivibrio nitratireducens) contained both genes sets. As evident from Table S5, the same holds true for two metagenomes assigned to the Betaproteobacteria and one gammaproteobacterial metagenome assigned to the Chromatiales. In total, only a 5.5% fraction of all analyzed sulfur oxidizer genomes contains dsr as well as hdr-like genes sets. This implies that using either one of the pathways is the rule rather than the exception.
The genetic capacity for sulfur oxidation in the cytoplasm via the Hdr-like and/or the Dsr system (Fig. 7a, b) can (but does not have to) be combined with the genetic equipment for sulfide detoxification involving Sqr, Rhd, and Pdo [52] (Fig. 7c) and/or cytoplasmically located sulfur oxygenase reductase (SOR), an enzyme that catalyzes oxygen-dependent disproportionation of elemental sulfur and/or polysulfides [53] (Fig. 7d). As already noted above, H. denitrificans encodes Hdr-like proteins as well as Sqr, Rhd, and Pdo. Acidithiobacillus albertensis DSM 14366T may serve as an example containing hdr-like genes (BLW97_RS11375-RS11335), the pathway involving Pdo (BLW97_RS13940) and SOR (BLW97_RS13410). T. nitratireducens DSM 14787T is equipped with all four different genes sets (hdr-like: TVNIR_3249-3244, dsr: TVNIR-0860-0847, pdo: TVNIR_2193, sor: TVNIR_2721 and TVNIR_1174).
Linkage of aerobic DMS and MT oxidation and Hdr-like system
When DMS is used as a carbon and electron source under aerobic conditions, the first step is either catalyzed by a DMS monooxygenase or by an oxygen-independent methyltransferase [15, 16, 54]. Both reactions result in the formation of methanethiol. The presence of the recently identified methanethiol oxidase, MTO [15], can therefore be taken as an ideal indicator for all organisms pursuing the described pathways and also covers those microorganisms that might exclusively degrade methanethiol. We checked all organisms listed by Eyice and coworkers as containing MTO [15] for the co-occurrence of the hdr-like gene cluster and thereby detected an intertwinement of aerobic DMS degradation with Hdr-based sulfate formation not only for Hyphomicrobium species, but also for a number of Thioalkalivibrio species (Table 1). This finding expands the linked occurrence of aerobic DMS/MT degradation and sulfate formation by an Hdr-like system from ubiquitous environments colonized by mesophiles like Hyphomicrobium to habitats dominated by extreme conditions. Our database survey further revealed a set of other chemo- and phototrophs, including magnetotactic bacteria, Thiobacillus, and Thiothrix species as well as purple sulfur bacteria, that combine MT oxidase and the Dsr pathway of sulfur oxidation (Table 1) verifying that the final steps of sulfate formation in organosulfur compounds oxidizers are indeed catalyzed by the same modules as in bacteria restricted to oxidation of inorganic sulfur compounds.
Table 1.
Occurrence of mto, dsr, and hdr genes in genome-sequenced mto-containing prokaryotes
| Organism/group | MTO | Dsr complexa | Hdr complexb |
|---|---|---|---|
| α-Proteobacteria | |||
| Hyphomicrobiaceae | |||
| Hyphomicrobium denitrificans ATCC 51888T | Hden_0743 | No | Hden_0689-0694 |
| Magnetospira sp. QH-2 | MGMAQ_0496 | MGMAQ_2110-2122 | No |
| β-Proteobacteria | |||
| Burkholderiaceae | |||
| Burkholderiales bacterium JOSHI_001 | BurJ1DRAFT_0065 | BurJ1DRAFT_3336-3324 | No |
| Thiobacillaceae | |||
| Thiobacillus denitrificans ATCC 23644T | B059DRAFT_02518 | B059DRAFT_02130-02118 | No |
| Thiobacillus thioparus DSM 505T | B058_RS0108880 | B058_RS0104165- 4100 | No |
| γ-Proteobacteria | |||
| Chromatiaceae | |||
| Chromtiaceae bacterium 2141T.STBD.0c.01a | B1781_RS11525, B1781_RS11000 | B1781_RS05315-375 | No |
| Thiocapsa roseopersicina 1711 (DSM 217T) | SAMN05421783_102223 | SAMN05421783_10260-246 | No |
| Thiorhodococcus drewsii AZ1 (DSM 15006T) | ThidrDRAFT_0542 (wrongly stated in ref. [15]: ThidrDRAFT_3297 | ThiDRAFT_2036-2022 | No |
| Ectothiorhodospiraceae | |||
| Thioalkalivibrio sp. AL5 | F574_RS0114010 | No | F574_RS0107460-7435 |
| Thioalkalivibrio sp. ALJ2 | F468_RS0112785 | No | F468_RS0104125-4150 |
| Thioalkalivibrio sp. ALJ4 | C936_RS0113555 | No | C936_RS0107740-7765 |
| Thioalkalivibrio sp. ALJ5 | C937_RS0112700 | No | C937_RS0107450-7425 |
| Thioalkalivibrio sp. ALJ9 | G317_RS0100035 | No | G317_RS0106450-6475 |
| Thioalkalivibrio sp. ARh4 | F465_RS0114060 | No | F465_RS0107820-7795 |
| Thioalkalivibrio versutus AL2 (DSM 13738T) | C164DRAFT_2790 | No | C164DRAFT_0453-0448 C164DRAFT_3541-3546 |
| Thiotrichaceae | |||
| Thiothrix disciformis DSM 14473T | A3IEDRAFT_00868 | A3IEDRAFT_03610--03597 | No |
aThe dsr gene cluster comprises dsrABCEFHMKLJOPN
bThe cluster consists of hdrC1B1A-hyp-hdrC2B2
Discussion
The findings presented here close several major knowledge gaps in prokaryotic oxidation of organic and inorganic sulfur compounds. Our study revealed a linkage of organosulfur compound and inorganic sulfur compound degradation present not only in H. denitrificans, but also in a range of haloalkaliphiles (Table 1). Our findings add a new aspect to the current understanding of organosulfur compound oxidation because an intertwinement of organic and inorganic sulfur compound oxidation via Hdr-like proteins is so far unprecedented and provides a novel shunt in the biogeochemical sulfur cycle. H. denitrificans occurs in a wide range of natural and industrial habitats and although its specific contribution to global DMS removal has not been determined, the possibility of sulfate formation from DMS via the combination of initial monooxygenase-catalyzed steps and the Hdr-like pathway by single organisms should no longer be underestimated.
Overall, DMS oxidation and concomitant assimilation of carbon into cell material in these organisms can be summarized as:
For H. denitrificans, thiosulfate produced in the periplasm was firmly established as an intermediate of this process. Just as externally provided thiosulfate, it is initially degraded in the periplasm by the well-established Sox system. This multienzyme complex releases sulfate and SoxY-bound sulfane sulfur (Fig. 2). The cytoplasmic Hdr-like proteins proved to be indispensable for further processing of sulfane sulfur, a finding that we consider a major breakthrough because our experiments provide first direct evidence for the function of Hdr-like proteins in any sulfur oxidizer.
The reaction mechanism of as well as the fate of the electrons released by the Hdr-like system is currently unresolved. The similarity with components of archaeal heterodisulfide reductase points at (protein-bound) persulfides (RSS−) and/or disulfides (RSSR) as possible intermediates in the reaction cycle. The most probable end product is sulfite, the same product as formed by the much better studied Dsr system [20] (Fig. 7). Database analyses showed that genes encoding the Hdr-like and Dsr systems occur almost mutually exclusively in chemo- and phototrophic sulfur oxidizers (ref. [17] and Table 1 and S5)
In H. denitrificans, production of Hdr-like proteins appears dependent on the presence of oxidizable substrates as shown by comparative proteome and immunoblot analyses (Figs. 2 and 5). Induction of the hyphomicrobial hdr-like genes is probably mediated by the ArsR-type regulator encoded by the first gene in the operon (Fig. 1). ArsR/SmtB family of metal (toxic ion) responsive repressors are typically autoregulated and part of the operon that contains genes involved in metabolism of the ion itself [39].
Our results disclose proteins encoded in hdr-like gene loci as essential components of a novel cytoplasmic sulfur oxidation pathway and show that the pathway is present in a wide range of mesophilic and extremophilic bacteria and archaea. Homologous genes were not only found in cultivated prokaryotes previously recognized for their potential to oxidize reduced sulfur compounds, but also in a number of organisms for which this capacity had not been reported. In addition, metagenomes from several sulfur-dominated sites contained the typical arrangement of hdr-like genes. So far, identification of bacteria and archaea associated with oxidative dissimilatory sulfur metabolism and the ability to oxidize sulfane sulfur to sulfite in the environment has almost exclusively focused on testing for the presence of sulfite reductase (DsrAB) genes [55–59]. Our work strongly indicates that general conclusions on the biogeochemical sulfur cycle based on sequence data should integrate the new Hdr-like pathway (Fig. 7). Given its occurrence in a wide range of extremely thermo-, alkali-, and/or acidophilic prokaryotes, this applies especially to habitats characterized by extreme conditions.
Prokaryotes harboring the equipment for inorganic sulfur compound oxidation as well as volatile organosulfur compound degradation via the Hdr-like pathway impact biogeochemical processes in terrestrial and marine sediments, subsurface ecosystems, aquifers, wastewater treatment plants as well as other industrial habitats. Our findings will certainly better inform future microbial trait-based ecosystem models relevant for the prediction of sulfur-based biogeochemical processes and the natural sulfur cycle.
Data deposition
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (http://www.ebi.ac.uk/pride) with the data set identifier PXD009030.
Electronic supplementary material
Acknowledgements
We thank Renate Zigann for excellent technical assistance. Vera Waffenschmidt, Mara Kotthoff, Luise Göbbels, and Julia Finkensieper supported phenotypic analyses of H. denitrificans strains during internships in CD’s laboratory. Hendrik Schäfer provided valuable hints regarding handling and growth of Hyphomicrobium strains. The rifampicin resistance cartridge was kindly provided by Eric Kofoid, Microbiology/CBS, UC Davis. LC-MS measurements and data processing were performed at the Core Facility Mass Spectrometry, Institute of Biochemistry and Molecular Biology, University of Bonn. This work was funded by the Deutsche Forschungsgemeinschaft (Grant Da 351/8-1).
Author contributions:
TK and CD designed the research; TK performed the research, TK and CD analyzed data and wrote the paper.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0209-7) contains supplementary material, which is available to authorized users.
References
- 1.Bowles MW, Mogollon JM, Kasten S, Zabel M, Hinrichs KU. Global rates of marine sulfate reduction and implications for sub-sea-floor metabolic activities. Science. 2014;344:889–91. doi: 10.1126/science.1249213. [DOI] [PubMed] [Google Scholar]
- 2.Curson AR, Todd JD, Sullivan MJ, Johnston AW. Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nat Rev Microbiol. 2011;9:849–59. doi: 10.1038/nrmicro2653. [DOI] [PubMed] [Google Scholar]
- 3.Kettle AJ, Andreae MO. Flux of dimethylsulfide from the oceans: a comparison of updated datasets and flux models. J Geophys Res. 2000;105:26793–808. doi: 10.1029/2000JD900252. [DOI] [Google Scholar]
- 4.Lomans BP, van der Drift C, Pol A, Op den Camp HJ. Microbial cycling of volatile organic sulfur compounds. Cell Mol Life Sci. 2002;59:575–88. doi: 10.1007/s00018-002-8450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrión O, Pratscher J, Curson ARJ, Williams BT, Rostant WG, Murrell JC, et al. Methanethiol-dependent dimethylsulfide production in soil environments. ISME J. 2017;11:2379–90. doi: 10.1038/ismej.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayes AC, Liss SN, Allen DG. Growth kinetics of Hyphomicrobium and Thiobacillus spp. in mixed cultures degrading dimethyl sulfide and methanol. Appl Environ Microbiol. 2010;76:5423–31. doi: 10.1128/AEM.00076-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charlson R, Lovelock J, Andreae M, Warren S. Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature. 1987;326:655–61. doi: 10.1038/326655a0. [DOI] [Google Scholar]
- 8.Kiene RP, Bates TS. Biological removal of dimethyl sulphide from sea water. Nature. 1990;345:702–5. doi: 10.1038/345702a0. [DOI] [Google Scholar]
- 9.De Bont JAM, van Dijken JP, Harder W. Dimethyl sulphoxide and dimethyl sulphide as a carbon, sulphur and energy source for growth of Hyphomicrobium S. J Gen Microbiol. 1981;127:315–23. [Google Scholar]
- 10.Kanagawa T, Kelly DP. Breakdown of dimethylsulphide by mixed cultures and by Thiobacillus thioparus. FEMS Microbiol Lett. 1986;34:13. doi: 10.1111/j.1574-6968.1986.tb01340.x. [DOI] [Google Scholar]
- 11.Pol A, Op den Camp HJ, Mees SG, Kersten MA, van der Drift C. Isolation of a dimethylsulfide-utilizing Hyphomicrobium species and its application in biofiltration of polluted air. Biodegradation. 1994;5:105–12. doi: 10.1007/BF00700635. [DOI] [PubMed] [Google Scholar]
- 12.Suylen GMH, Stefess GC, Kuenen JG. Chemolithotrophic potential of Hyphomicrobium species capable of growth on methylated sulphur compounds. Arch Microbiol. 1986;146:192–8. doi: 10.1007/BF00402350. [DOI] [Google Scholar]
- 13.de Zwart JMM, Sluis JMR, Kuenen JG. Competition for dimethyl sulfide and hydrogen sulfide by Methylophaga sulfidovorans and Thiobacillus thioparus T5 in continuous cultures. Appl Environ Microbiol. 1997;63:3318–22. doi: 10.1128/aem.63.8.3318-3322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boden R, Kelly DP, Murrell JC, Schäfer H. Oxidation of dimethylsulfide to tetrathionate by Methylophaga thiooxidans sp. nov.: a new link in the sulfur cycle. Environ Microbiol. 2010;12:2688–99. doi: 10.1111/j.1462-2920.2010.02238.x. [DOI] [PubMed] [Google Scholar]
- 15.Eyice O, Myronova N, Pol A, Carrión O, Todd JD, Smith TJ, et al. Bacterial SBP56 identified as a Cu-dependent methanethiol oxidase widely distributed in the biosphere. ISME J. 2018;12:145–60. doi: 10.1038/ismej.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schäfer H, Myronova N, Boden R. Microbial degradation of dimethylsulphide and related C1-sulphur compounds: organisms and pathways controlling fluxes of sulphur in the biosphere. J Exp Bot. 2010;61:315–34. doi: 10.1093/jxb/erp355. [DOI] [PubMed] [Google Scholar]
- 17.Venceslau SS, Stockdreher Y, Dahl C, Pereira IAC. The "bacterial heterodisulfide" DsrC is a key protein in dissimilatory sulfur metabolism. Biochim Biophys Acta. 2014;1837:1148–64. doi: 10.1016/j.bbabio.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Smith NA, Kelly DP. Mechanism of oxidation of dimethyl disulphide by Thiobacillus thioparus strain E6. J Gen Microbiol. 1988;134:3031–9. [Google Scholar]
- 19.Dahl C, Friedrich CG, Kletzin A. Sulfur oxidation in prokaryotes. Encyclopedia of life sciences (ELS). Chichester: John Wiley & Sons, Ltd.; 2008. 10.1002/9780470015902.a9780470021155.
- 20.Dahl C, Engels S, Pott-Sperling AS, Schulte A, Sander J, Lübbe Y, et al. Novel genes of the dsr gene cluster and evidence for close interaction of Dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum. J Bacteriol. 2005;187:1392–404. doi: 10.1128/JB.187.4.1392-1404.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dahl C. Cytoplasmic sulfur trafficking in sulfur-oxidizing prokaryotes. IUBMB Life. 2015;67:268–74. doi: 10.1002/iub.1371. [DOI] [PubMed] [Google Scholar]
- 22.Osorio H, Mangold S, Denis Y, Nancucheo I, Esparza M, Johnson DB, et al. Anaerobic sulfur metabolism coupled to dissimilatory iron reduction in the extremophile Acidithiobacillus ferrooxidans. Appl Environ Microbiol. 2013;79:2172–81. doi: 10.1128/AEM.03057-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quatrini R, Appia-Ayme C, Denis Y, Jedlicki E, Holmes DS, Bonnefoy V. Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics. 2009;10:394. doi: 10.1186/1471-2164-10-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner T, Koch J, Ermler U, Shima S. Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction. Science. 2017;357:699–703. doi: 10.1126/science.aan0425. [DOI] [PubMed] [Google Scholar]
- 25.Ossa DMH, Oliveira RR, Murakami MT, Vincentini R, Costa-Filho AJ, Alexandrino F, et al. Expression, purification and spectroscopic analysis of an HdrC: an iron-sulfur cluster-containing protein from Acidithiobacillus ferrooxidans. Process Biochem. 2011;46:1335–41. doi: 10.1016/j.procbio.2011.03.001. [DOI] [Google Scholar]
- 26.Gliesche CG, Fesefeldt A, Hirsch P. Genus I. Hyphomicrobium Stutzer and Hartleb 1898, 17AL. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, editors. Bergey's manual of systematic bacteriology. 2 ed. New York, NY: Springer; 2005. pp. 476–94. [Google Scholar]
- 27.Akhtar MK, Jones PR. Deletion of iscR stimulates recombinant clostridial Fe-Fe hydrogenase activity and H2-accumulation in Escherichia coli BL21(DE3) Appl Microbiol Biotechnol. 2008;78:853–62. doi: 10.1007/s00253-008-1377-6. [DOI] [PubMed] [Google Scholar]
- 28.Vishniac W, Santer M. The thiobacilli. Bacteriol Rev. 1957;21:195–213. doi: 10.1128/br.21.3.195-213.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ausubel FA, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, et al. Current protocols in molecular biology. New York, NY: John Wiley & Sons; 1997. [Google Scholar]
- 30.Borodina E, McDonald IR, Murrell JC. Chloromethane-dependent expression of the cmu gene cluster of Hyphomicrobium chloromethanicum. Appl Environ Microbiol. 2004;70:4177–86. doi: 10.1128/AEM.70.7.4177-4186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horton RM. PCR mediated recombination and mutagenesis: SOEing together tailor-made genes. Mol Biotechnol. 1995;3:93–99. doi: 10.1007/BF02789105. [DOI] [PubMed] [Google Scholar]
- 32.Fellay R, Frey J, Krisch HM. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vivo insertional mutagenesis of Gram-negative bacteria. Gene. 1987;52:147–54. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 33.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 34.Dahl C. Insertional gene inactivation in a phototrophic sulphur bacterium: APS-reductase-deficient mutants of Chromatium vinosum. Microbiology. 1996;142:3363–72. doi: 10.1099/13500872-142-12-3363. [DOI] [PubMed] [Google Scholar]
- 35.Franz B, Gehrke T, Lichtenberg H, Hormes J, Dahl C, Prange A. Unexpected extracellular and intracellular sulfur species during growth of Allochromatium vinosum with reduced sulfur compounds. Microbiology. 2009;155:2766–74. doi: 10.1099/mic.0.027904-0. [DOI] [PubMed] [Google Scholar]
- 36.Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–56. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parks DH, Rinke C, Chuvochina M, Chaumeil PA, Woodcroft BJ, Evans PN, et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2:1533–42. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- 38.Boden R, Borodina E, Wood AP, Kelly DP, Murrell JC, Schäfer H. Purification and characterization of dimethylsulfide monooxygenase from Hyphomicrobium sulfonivorans. J Bacteriol. 2011;193:1250–8. doi: 10.1128/JB.00977-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gristwood T, McNeil MB, Clulow JS, Salmond GP, Fineran PC. PigS and PigP regulate prodigiosin biosynthesis in Serratia via differential control of divergent operons, which include predicted transporters of sulfur-containing molecules. J Bacteriol. 2011;193:1076–85. doi: 10.1128/JB.00352-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dahl C, Franz B, Hensen D, Kesselheim A, Zigann R. Sulfite oxidation in the purple sulfur bacterium Allochromatium vinosum: identification of SoeABC as a major player and relevance of SoxYZ in the process. Microbiology. 2013;159:2626–38. doi: 10.1099/mic.0.071019-0. [DOI] [PubMed] [Google Scholar]
- 41.Dahl JU, Radon C, Buhning M, Nimtz M, Leichert LI, Denis Y, et al. The sulfur carrier protein TusA has a pleiotropic role in Escherichia coli that also affects molybdenum cofactor biosynthesis. J Biol Chem. 2013;288:5426–42. doi: 10.1074/jbc.M112.431569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boughanemi S, Lyonnet J, Infossi P, Bauzan M, Kosta A, Lignon S, et al. Microbial oxidative sulfur metabolism: biochemical evidence of the membrane-bound heterodisulfide reductase-like complex of the bacterium Aquifex aeolicus. FEMS Microbiol Lett. 2016;363:fnw156. doi: 10.1093/femsle/fnw156. [DOI] [PubMed] [Google Scholar]
- 43.Weinitschke S, Denger K, Cook AM, Smits THM. The DUF81 protein TauE in Cupriavidus necator H16, a sulfite exporter in the metabolism of C2 sulfonates. Microbiology. 2007;153:3055–60. doi: 10.1099/mic.0.2007/009845-0. [DOI] [PubMed] [Google Scholar]
- 44.Kappler U, Enemark JH. Sulfite-oxidizing enzymes. J Biol Inorg Chem. 2014;20:253–64. doi: 10.1007/s00775-014-1197-3. [DOI] [PubMed] [Google Scholar]
- 45.Heunisch GW. Stoichiometry of the reaction of sulfites with hydrogen sulfide ion. Inorg Chem. 1977;16:1411–3. doi: 10.1021/ic50172a033. [DOI] [Google Scholar]
- 46.Friedrich CG, Quentmeier A, Bardischewsky F, Rother D, Kraft R, Kostka S, et al. Novel genes coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17. J Bacteriol. 2000;182:4677–87. doi: 10.1128/JB.182.17.4677-4687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brito JA, Denkmann K, Pereira IAC, Archer M, Dahl C. Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum: Structural and functional insights into thiosulfate oxidation. J Biol Chem. 2015;290:9222–38. doi: 10.1074/jbc.M114.623397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dijkhuizen L, Harder W, de BL, van BA, Clement W, Bron S, et al. Genetic manipulation of the restricted facultative methylotroph Hyphomicrobium X by the R-plasmid-mediated introduction of the Escherichia coli pdh genes. Arch Microbiol. 1984;139:311–8. doi: 10.1007/BF00408372. [DOI] [PubMed] [Google Scholar]
- 49.Gliesche CG. Transformation of methylotrophic bacteria by electroporation. Can J Microbiol. 1997;43:197–201. doi: 10.1139/m97-026. [DOI] [PubMed] [Google Scholar]
- 50.Kallnik V, Meyer M, Deppenmeier U, Schweiger P. Construction of expression vectors for protein production in Gluconobacter oxydans. J Biotechnol. 2010;150:460–5. doi: 10.1016/j.jbiotec.2010.10.069. [DOI] [PubMed] [Google Scholar]
- 51.Suylen GMH, Large PJ, van Dijken JP, Kuenen JG. Methyl mercaptan oxidase, a key enzyme in the metabolism of methylated sulfur compounds by Hyphomicrobium EG. J Gen Microbiol. 1987;133:2989–97. [Google Scholar]
- 52.Xia Y, Lu C, Hou N, Xin Y, Liu J, Liu H, et al. Sulfide production and oxidation by heterotrophic bacteria under aerobic conditions. ISME J. 2017;11:2754–66. doi: 10.1038/ismej.2017.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rühl P, Poll U, Braun J, Klingl A, Kletzin A. A sulfur oxygenase from the haloalkaliphilic bacterium Thioalkalivibrio paradoxus with atypically low reductase activity. J Bacteriol. 2017;199:e00675–16. doi: 10.1128/JB.00675-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Visscher PT, Taylor BF. A new meachnism for the aerobic catabolism of dimethyl sulfide. Appl Environ Microbiol. 1993;59:3784–9. doi: 10.1128/aem.59.11.3784-3789.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun. 2016;7:13219. doi: 10.1038/ncomms13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anantharaman K, Hausmann B, Jungbluth SP, Kantor RS, Lavy A, Warren LA, et al. Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. ISME J. 2018. 10.1038/s41396-41018-40078-41390. [DOI] [PMC free article] [PubMed]
- 57.Hausmann B, Pelikan C, Herbold CW, Kostlbacher S, Albertsen M, Eichorst SA, et al. Peatland Acidobacteria with a dissimilatory sulfur metabolism. ISME J. 2018. 10.1038/s41396-41018-40077-41391. [DOI] [PMC free article] [PubMed]
- 58.Loy A, Duller S, Baranyi C, Muámann M, Ott J, Sharon I, et al. Reverse dissimilatory sulfite reductase as phylogenetic marker for a subgroup of sulfur-oxidizing prokaryotes. Environ Microbiol. 2009;11:289–99. doi: 10.1111/j.1462-2920.2008.01760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Probst AJ, Ladd B, Jarett JK, Geller-McGrath DE, Sieber CMK, Emerson JB, et al. Differential depth distribution of microbial function and putative symbionts through sediment-hosted aquifers in the deep terrestrial subsurface. Nat Microbiol. 2018;3:328–36. doi: 10.1038/s41564-017-0098-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.