Abstract
We have cloned, purified, and characterized a β-carbonic anhydrase (CA, EC 4.2.1.1), BpsCAβ, from the pathogenic bacterium Burkholderia pseudomallei, responsible for the tropical disease melioidosis. The enzyme showed high catalytic activity for the physiologic CO2 hydration reaction to bicarbonate and protons, with the following kinetic parameters: kcat of 1.6 × 105 s−1 and kcat/KM of 3.4 × 107 M−1 s−1. An inhibition study with a panel of 38 sulfonamides and one sulfamate—including 15 compounds that are used clinically—revealed an interesting structure–activity relationship for the interaction of this enzyme with these inhibitors. Many simple sulfonamides and clinically used agents such as topiramate, sulpiride, celecoxib, valdecoxib, and sulthiame were ineffective BpsCAβ inhibitors (KI > 50 µM). Other drugs, such as ethoxzolamide, dorzolamide, brinzolamide, zonisamide, indisulam, and hydrochlorothiazide were moderately potent micromolar inhibitors. The best inhibition was observed with benzene-1,3-disulfonamides—benzolamide and its analogs acetazolamide and methazolamide—which showed KI in the range of 185–745 nM. The inhibition profile of BpsCAβ is very different from that of the γ-class enzyme from the same pathogen, BpsCAγ. Thus, identifying compounds that would effectively interact with both enzymes is relatively challenging. However, benzolamide was one of the best inhibitors of both of these CAs with KI of 653 and 185 nM, respectively, making it an interesting lead compound for the design of more effective agents, which may be useful tools for understanding the pathogenicity of this bacterium.
Keywords: carbonic anhydrase, metalloenzymes, pathogens, β-class, sulfonamide, Burkholderia pseudomallei
1. Introduction
Our groups started to investigate the biochemical properties, kinetic constants and inhibition profiles of numerous classes of carbonic anhydrases (CAs; EC 4.2.1.1) in different species of pathogenic and non-pathogenic bacteria (see Table 1). CAs are ubiquitous metalloenzymes that catalyze the reversible hydration of carbon dioxide with the production of bicarbonate and protons. The CA superfamily includes seven distinct classes known as the α, β, γ, δ, ζ, η, and θ [1,2,3,4,5,6,7,8,9,10,11,12,13]. The α-, β-, δ-, η-, and perhaps θ-CAs use Zn(II) ions at the active site, the γ-CAs are probably Fe(II) enzymes (but they are also active with bound Zn(II) or Co(II) ions) [14,15,16,17,18,19,20,21], and the ζ-class CAs are cambialistic enzymes, active both with Cd(II) or Zn(II) bound within the active site in order to perform the physiologic reaction catalysis [22,23,24]. The metal ion from the CA active site is coordinated by three His residues in the α-, γ-, δ-, and probably θ-classes, by one His and two Cys residues in β- and ζ-CAs, or by two His and one Gln residues in the η-class, with the fourth ligand being a water molecule/hydroxide ion acting as nucleophile in the catalytic cycle of the enzyme [11,25,26,27,28,29,30]. All CAs identified in animal systems belong to α-class. CAs identified in plants and algae belong to the α-, β-, γ-, δ-, and θ-classes; fungi encode for α- and β-CAs; protozoa encode for α-, β-, or η-CAs. As shown in Table 1, bacteria encode for enzymes belonging to the α-, β-, and γ-CA classes [11,12,27,29,31,32,33]. Moreover, as described in literature, the genome of Gram-negative bacteria belonging to the genera Buchnera and Rickettsia does not encode for any CAs [29]. Thus, the distribution pattern of CAs in bacteria is very fascinating [29]. In the last year, it has been demonstrated in vivo that the inhibition of bacterial CAs influences the pathogenicity and/or the growth of the microorganism [27,34,35,36,37,38,39]. These promising data on live bacteria allow us to propose bacterial CAs inhibition as an approach for obtaining anti-infective agents with a new mechanism of action compared to classical antibiotics. At present, infectious diseases are the second-leading cause of death in the world, and the development of bacterial antibiotic-resistance is an inevitable and widespread phenomenon inherent to most drugs. Several classes of CA inhibitors (CAIs) are known to date, among which the metal complexing anions and sulfonamides and their bioisosteres are the most investigated for the inhibition of the bacterial CAs [12,40,41,42,43,44]. Anions such as the inorganic metal-complexing or more complicated species such as the carboxylates bind to CAs, but generally with less efficiency compared to the sulfonamides [45]. Anions may bind either the tetrahedral geometry of the metal ion or as trigonal–bipyramidal adducts [45]. The antibiotic revolution in medicine was represented by sulfonamides, which were the first antimicrobial drugs (Figure 1) [31,41]. Sulfonamide derivatives that were used clinically, including acetazolamide, methazolamide, ethoxzolamide, dichlorophenamide, dorzolamide, and brinzolamide, bind in a tetrahedral geometry to the Zn(II) ion in deprotonated state, with the nitrogen atom of the sulfonamide moiety coordinated to Zn(II) and an extended network of hydrogen bonds involving amino acid residues Thr199 and Glu106 (numbering system used for the human CA, isoform I) also participating in the anchoring of the inhibitor molecule to the metal ion [31,41,46]. The aromatic/heterocyclic part of the inhibitor interacts with hydrophilic and hydrophobic residues of the cavity. The protein fold in the diverse CA classes (seven CA-classes) is highly different [14,15,16,17,18,19,20,21], mainly due to their different oligomeric state. Successful inhibition studies with sulfonamides should allow the discovery of highly isoform-selective CAIs, which may lead to a new generation of drugs targeting these widespread enzymes.
Table 1.
Bacterial carbonic anhydrases (CAs) investigated by our groups for their biochemical properties, kinetic constants, and inhibition profiles.
| Bacterium | CA | Characteristics | Inhibition | |||
|---|---|---|---|---|---|---|
| Class | Acronym | Pathogenicity | Disease | Sulfonamides | Anions | |
| Vibrio cholerae [8,49,50] | α | VchCAα | + | Cholera | + | + |
| β | VchCAβ | + | + | |||
| γ | VchCAγ | + | + | |||
| Helicobacter pylori [36,51,52] | α | hpαCA | + | Gastritis, gastric | + | + |
| β | hpβCA | ulcers | + | + | ||
| Streptococcus mutans [11,53,54] | β | SmuCA | + | Caries | + | + |
| Haemophilus influenzae | β | HICA | + | Influenza | − | + |
| Neisseria gonorrhoeae | α | NgoCA | + | Gonorrhea | + | + |
| Neisseria sicca | α | NsiCA | + | Septicemia | + | − |
| Porphyromonas gingivalis [11,13,55,56,57] | β | PgiCA β | + | Periodontitis, | + | + |
| γ | PgiCA γ | rheumatoid arthritis | + | + | ||
| Legionella pneumophila [42] | β | LpCA1 | + | Legionellosis | + | + |
| β | LpCA2 | + | + | |||
| Clostridium perfringens [58] | β | CpeCA | + | Food poisoning | − | + |
| Brucella suis [34,59,60] | β | bsCA 1 | + | Brucellosis | + | − |
| β | BsCA II | + | − | |||
| Burkholderia pseudomallei [1,7,61] | β | BpsCAβ | + | Melioidosis | − | + |
| γ | BpsCAγ | + | + | |||
| Salmonella enterica [62,63] | β | stCA 1 | + | Salmonellosis | + | + |
| β | stCA 2 | + | + | |||
| Streptococcus pneumoniae [64] | β | PCA | + | Pneumonia | + | + |
| Mycobacterium tuberculosis [65,66,67] | β | mtCA 1 | + | Tuberculosis | + | − |
| β | mtCA 2 | + | − | |||
| β | mtCA 3 | + | − | |||
| Methanobacterium thermoautotrophicum [68] | β | Cab | − | − | + | + |
| Methanosarcina thermophila [69] | γ | Zn-Cam | − | − | + | + |
| γ | Co-Cam | − | − | + | + | |
| Sulfurihydrogenibium yellowstonense [19,70,71,72] | α | SspCA | − | − | + | + |
| Sulfurihydrogenibium azorense [73,74,75,76] | α | SazCA | − | − | + | + |
| Colwellia psychrerythraea [10,38] | γ | CpsCA | − | − | + | + |
| Pseudoalteromonas haloplanktis [77,78] | γ | PhaCAγ | − | − | + | + |
Figure 1.
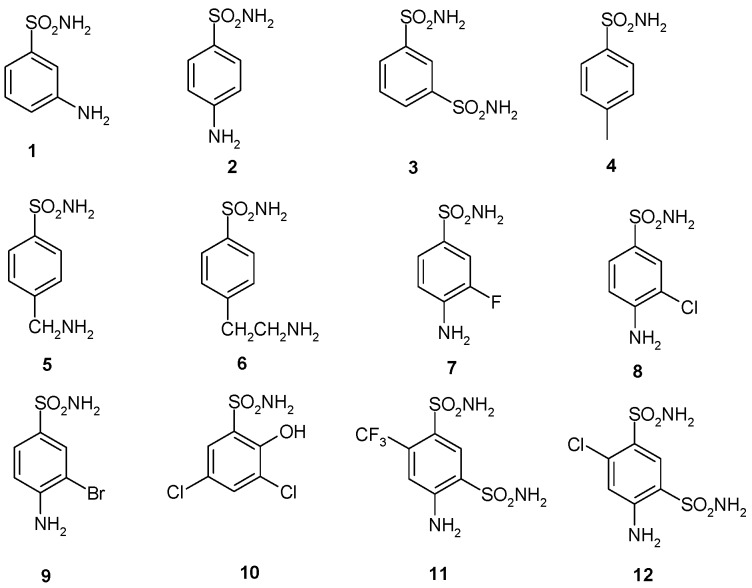
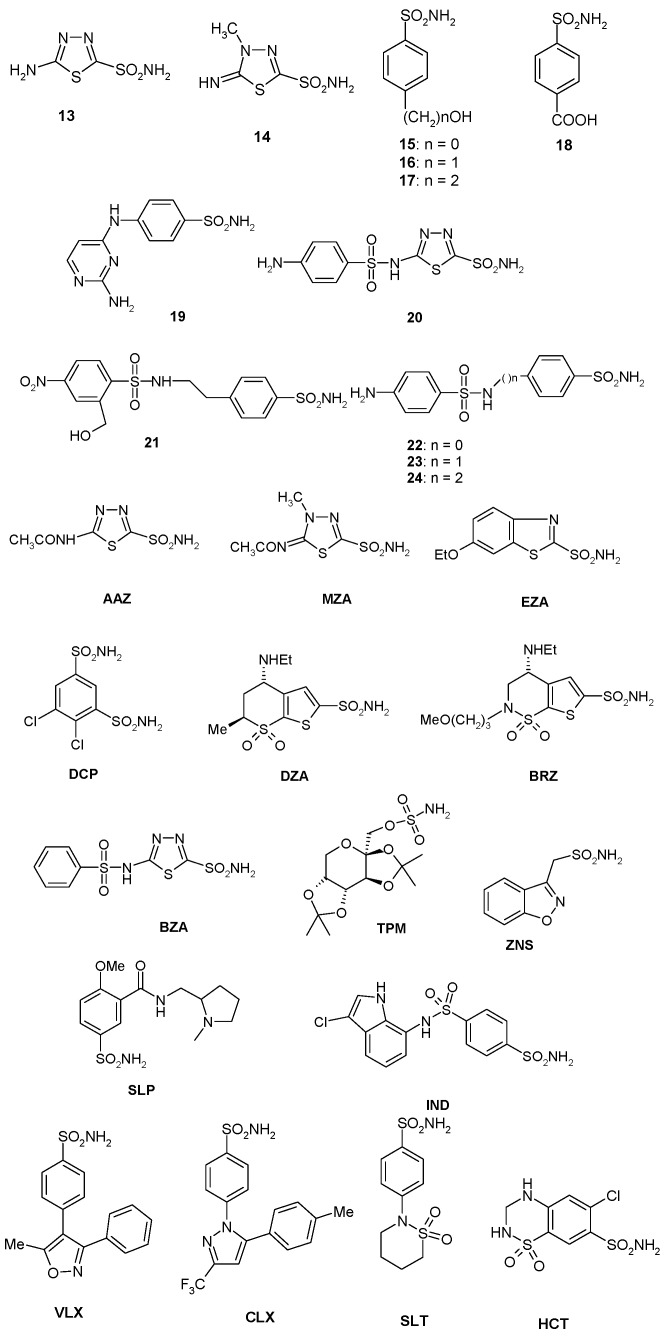
Structure of sulfonamides/sulfamates investigated in the present study. AAZ: acetazolamide; BRZ: brinzolamide; BZA: benzolamide; CLX: celecoxib; DCP: dichlorophenamide; DZA: dorzolamide; EZA: ethoxzolamide; HCT: hydrochlorothiazide; IND: indisulam; MZA: methazolamide; SLP: sulpiride; SLT: sulthiame; TPM: topiramate; VLX: valdecoxib; ZNS: zonisamide.
In this context, we cloned, expressed, and purified the recombinant β-CA (BpsCAβ, Accession number: WP_004550949.1) identified in the genome of Burkholderia pseudomallei, a Gram-negative saprophytic bacteria responsible of melioidosis, which is an endemic disease of tropical and sub-tropical regions [47]. The B. pseudomallei genome encodes for only β- and γ-CAs. In the present manuscript, the inhibition profile of BpsCAβ was investigated using a wide series of sulfonamides/sulfamates. The inhibition profile of BpsCAβ was compared with those obtained for the human α-CAs (hCA I and hCAII) and the other bacterial BpsCAγ previously identified in the genome of Burkholderia pseudomallei [7] and investigated by our groups for its inhibition profiles with anions and sulfonamides. This study may be of interest for designing new types of inhibitors that may have clinical applications, because Burkholderia pseudomallei is fundamentally resistant to penicillin, ampicillin, first-generation and second-generation cephalosporins, macrolides, quinolones, and most aminoglycosides [48].
2. Results and Discussion
2.1. Purification and Protonographic Analysis
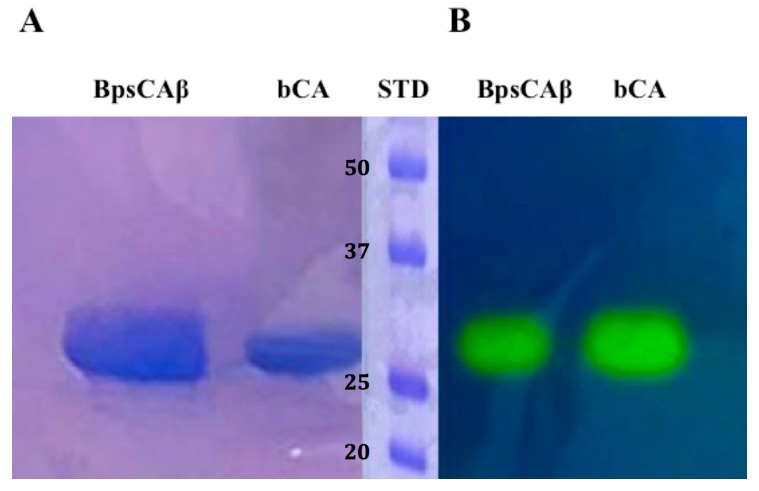
The recombinant BpsCAβ was heterologously expressed as soluble protein in the cytoplasm of the E. coli (DE3) codon plus cells and produced as a fusion protein containing a His-tag tail at its N-terminal amino acid sequence. After sonication and centrifugation, BpsCAβ was purified from the cell extract using the His-select HF Nickel column as the affinity column. The fusion protein showed an apparent molecular weight of about 30 kDa, as indicated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (SDS-PAGE, Figure 2A). The apparent molecular weight of 30 kDa on the SDS-PAGE is due to the SDS concentration, which determines the separation of the subunits of the BpsCAβ. Generally, β-CAs catalyze the hydration of carbon dioxide to bicarbonate and protons when the β-CAs monomers assemble into dimers, tetramers, or octamers. To investigate the hydratase activity of BpsCAβ and bCA (commercial α-CA from bovine erythrocytes, purchased from Sigma) on the SDS-PAGE gel, samples of BpsCAβ or bCA were prepared and loaded on the gel. The protonography technique is based on the monitoring of pH variation in the gel due to the catalyzed conversion of CO2 to bicarbonate and protons. The production of hydrogen ions during the CO2 hydration reaction due to the bCA or BpsCAβ hydratase activity lowers the pH of the solution until the color transition point of the dye (bromothymol blue) is reached (pH 6.8) [79,80,81]. This dye appears blue in its deprotonated form, while its color changes to yellow in the protonated form. The protonated form of bromothymol blue has its peak absorption at 692 nm, thus reflecting yellow light in acidic solutions, and the deprotonated form has its peak absorption at 602 nm, thus reflecting blue light in more basic solutions [79,80,81]. Figure 2 shows the results of the protonographic analysis, and the enzyme activity was detected as yellow bands against the blue background. The protonogram of BpsCAβ (Figure 2B) showed a 30 kDa band of activity. As described in the literature, mammal α-CAs are monomeric, and the protonogram of bCA showed a single band of activity corresponding to a monomer of 30 kDa. However, as mentioned previously, β-CAs are active enzymes for the CO2 hydratase reaction when assembled into dimers, tetramers, or octamers. The yellow bands found corresponding to the inactive monomeric form of BpsCAβ is due to the fact that at the end of the electrophoretic run, the SDS is removed from the gel. This procedure may lead to the rearrangement of BpsCAβ monomers in the gel, and the final result is the reconstitution of the active oligomeric forms of BpsCAβ.
Figure 2.
Comparison between SDS-PAGE and protonography of recombinant B. pseudomallei β-CA (BpsCAβ). (A) SDS-PAGE of bCA and BpsCAβ. The gel was stained with Coomassie blue; (B) Protonogram of α-CA from bovine erythrocytes (bCA) and BpsCAβ. The yellow bands on the blue background correspond to the bCA and BpsCAβ activity, which determine the drop of pH from 8.2 to the transition point of the dye (pH 6.8). Yellow bands were obtained with an incubation time of 15 s. STD: molecular markers starting from the top had the following molecular weights: 50 kDa, 37 kDa, 25 kDA, and 20 kDa (see bold numbers on the figure).
2.2. Kinetic Constants
In Table 2 are shown the rate constants (kcat, KM, and kcat/KM) of the BpsCAβ identified in the genome of Burkholderia pseudomallei and the inhibition constant (KI) using the inhibitor acetazolamide. These constants were compared with the kinetic parameters of the α-CA from the Homo sapiens (isoforms hCA I and hCA II) and with those obtained from BpsCAγ that were reported previously [7]. The catalytic activity values of these enzymes were determined using the “stopped-flow” technique. BpsCAβ showed a kcat of 1.6 × 105 s−1 and a kcat/KM of 3.4 × 107 M−1·s−1. It was slightly less active than the human isoform hCAI (kcat = 2.0 × 105 s−1). Interestingly, the γ-CA from B. pseudomallei showed a kcat = 5.3 × 105 s−1, which is 3.3 times higher than the BpsCAβ (kcat = 1.6 × 105 s−1). This is also evident for the γ-CA from V. cholerae, which was 2.21 times more active than VchCAβ (kcat = 7.39 × 105 s−1). In addition, acetazolamide was found to be a less effective inhibitor for BpsCAβ (KI = X nM) and BpsCAγ (KI = Y nM) [48,49,50] than for hCA II, VchCAα, and hpβCA.
Table 2.
Kinetic parameters for the CO2 hydration reaction catalysed by the human cytosolic isozymes hCA I and II (α-class CAs) at 20 °C and pH 7.5 in 10 mM HEPES buffer and 20 mM Na2SO4, β- and γ-CAs from B. pseudomallei measured at 20 °C, pH 8.3 in 20 mM TRIS buffer and 20 mM NaClO4 [82], α-, β-, and γ-CAs from Vibrio cholerae [8,49,50], and β-CA from Helicobacter pylori [36,51,52]. Acetazolamide inhibition data are also shown.
| Enzyme | Activity Level | Class | kcat (s−1) | kcat/Km (M−1·s−1) | KI (Acetazolamide) (nM) |
|---|---|---|---|---|---|
| hCA I | moderate | α | 2.0 × 105 | 5.0 × 107 | 250 |
| hCA II | very high | α | 1.4 × 106 | 1.5 × 108 | 12 |
| BpsCAβ | moderate | β | 1.6 × 105 | 3.4 × 107 | 745 |
| BpsCAγ | moderate | γ | 5.3 × 105 | 2.5 × 107 | 149 |
| VchCAα | high | α | 8.23 × 105 | 7.0 × 107 | 6.8 |
| VchCAβ | moderate | β | 3.34 × 105 | 4.1 × 107 | 451 |
| VchCAγ | high | γ | 7.39 × 105 | 6.4 × 107 | 473 |
| hpβCA | high | β | 7.1 × 105 | 4.8 × 107 | 40 |
2.3. Sulfonamides and Sulfamates Inhibition Profiles
A panel of simple aromatic and heteroaromatic sulfonamides of types 1–24 and clinically used derivatives such as AAZ–HCT (Figure 1) was used to investigate the sulfonamide/sulfamate inhibition profile of BpsCAβ. Acetazolamide (AAZ), methazolamide (MZA), ethoxzolamide (EZA), and dichlorophenamide (DCP) are the classical systemically acting CAIs [51,52,53,54,55,56,57,58,59]. Dorzolamide (DZA) and brinzolamide (BRZ) are topically-acting anti-glaucoma agents; benzolamide (BZA) is an orphan drug belonging to this class of pharmacological agents; whereas topiramate (TPM), zonisamide (ZNS), and sulthiame (SLT) are widely used antiepileptic drugs [60,61,62,63,64,65]. Sulpiride (SLP), indisulam (IND), valdecoxib (VLX), celecoxib (CLX), and hydrochlorothiazide (HCT) were shown by these groups to belong to this class of pharmacological agents, acting as efficient inhibitors against many enzymes from mammals, plants, and microorganisms such as bacteria, protozoa, and fungi [66,67,68,69,70,71,72,73,74,75,76,77,78,79]. Sulfonamides 1–24 and the clinically used agents investigated in this study were either commercially available or were prepared as reported earlier by our group [21,25,42].
Inhibition data with the 39 investigated sulfonamides/sulfamates against the two enzymes from B. pseudomallei BpsCAβ and BpsCAγ, as well as the human main isoforms hCA I and II (as possible off targets) are shown in Table 3. The following structure–activity relationship (SAR) can be drawn from the data of this table:
Table 3.
Sulfonamides/sulfamates inhibition constants (KI, nM) for the human α-CAs (isoforms hCA I and II) and the β-/γ-CAs identified in the genome of B. pseudomallei, assayed by a CO2 hydrase stopped flow method [7,50].
| Inhibitor | KI, nM * | |||
|---|---|---|---|---|
| hCA I | hCA II | BpsCAγ | BpsCAβ | |
| 1 | 45,400 | 295 | 574 | >50,000 |
| 2 | 25,000 | 240 | 1720 | 3895 |
| 3 | 28,000 | 300 | 1550 | 3170 |
| 4 | 78,500 | 320 | >50,000 | >50,000 |
| 5 | 25,000 | 170 | >50,000 | 3900 |
| 6 | 21,000 | 160 | >50,000 | >50,000 |
| 7 | 8300 | 60 | >50,000 | >50,000 |
| 8 | 9800 | 110 | 12,500 | >50,000 |
| 9 | 6500 | 40 | >50,000 | >50,000 |
| 10 | 6000 | 70 | >50,000 | 4100 |
| 11 | 5800 | 63 | 14,000 | 425 |
| 12 | 8400 | 75 | 23,500 | 307 |
| 13 | 8600 | 60 | 18,400 | 3065 |
| 14 | 9300 | 19 | 1810 | 2500 |
| 15 | 6 | 2 | 9650 | 4345 |
| 16 | 164 | 46 | 10,800 | 2215 |
| 17 | 185 | 50 | 1825 | 417 |
| 18 | 109 | 33 | 1500 | >50000 |
| 19 | 95 | 30 | 1838 | >50000 |
| 20 | 690 | 12 | 1810 | 266 |
| 21 | 55 | 80 | 1335 | 1650 |
| 22 | 21,000 | 125 | 1805 | 5200 |
| 23 | 23,000 | 133 | 1700 | 1500 |
| 24 | 24,000 | 125 | 24,500 | >50,000 |
| AAZ | 250 | 12 | 149 | 745 |
| MZA | 50 | 14 | 1595 | 186 |
| EZA | 25 | 8 | 1865 | 3850 |
| DCP | 1200 | 38 | >50,000 | 529 |
| DZA | 50,000 | 9 | 2260 | 3670 |
| BRZ | 45,000 | 3 | 1270 | 4270 |
| BZA | 15 | 9 | 653 | 185 |
| TPM | 250 | 10 | 3010 | >50,000 |
| ZNS | 56 | 35 | >50,000 | 4060 |
| SLP | 1200 | 40 | 5600 | >50,000 |
| IND | 31 | 15 | 1800 | 4375 |
| VLX | >50,000 | 43 | >50,000 | >50,000 |
| CLX | 50,000 | 21 | >50,000 | >50,000 |
| SLT | 374 | 9 | 8900 | >50,000 |
| HCT | 328 | 290 | >50,000 | 3490 |
* Mean from three different assays. Errors in the range of ±10% of the reported values (data not shown).
(i) Some sulfonamides, including 1, 4, 6–9, 18, 19, 24, SLP, VLX, CLX, SLT, and the sulfamate TPM did not inhibit BpsCAβ significantly up to 50 µM concentration of inhibitor within the assay system (Table 3). They include a variety of chemotypes, such as simple benzenesulfonamides with compact meta- or para-substituents (1, 4, 6, 18), halogen sulfanilamides (7–9), the compounds with elongated molecules 19 and 24, as well as the clinically used compounds with a rather bulky scaffold such as the two coxibs, sulpiride and the sugar sulfamate TPM. It is thus difficult to draw a precise SAR for this limited number of inhibitors with such diverse structural features in their molecules.
(ii) A large number of the investigated sulfonamides were moderately potent inhibitors of BpsCAβ, with inhibition constants in the micromolar range. They include: 2, 3, 5, 10, 13–16, 21–23, EZA, DZA, BRZ, ZNS, IND, and HCT (Table 3), and their KI range between 1500 and 5200 nM. Several interesting SAR data can be observed: the shift of the amino moiety from meta (as in 1) to para (as in 2) led to a net increase in the inhibitory action, as sulfanilamide 2 is moderately potent whereas metanilamide 1 was devoid of activity. The same activity was shown by 5, which has an extra methylene moiety compared to sulfanilamide, but the increase of the linker to two methylene groups (as in 6) leads to the complete loss of activity. It is also interesting to note that the introduction of halogens in the sulfanilamide 2 scaffold (as in 7–9) led to a loss of activity against BpsCAβ, although the halogenated sulfonamides were more effective hCA I and II inhibitors compared to sulfanilamide itself (Table 3). For the phenol/alcohols 15–17 on the other hand, the increase of the linker between the OH moiety and the aromatic ring from 0 to 2 led to an increase of the BpsCAβ inhibitory action, with the best inhibitor being the hydroxyethyl derivative 17 (KI of 417 nM). However, for the similar subseries 22–24, the SAR is more complex, with 23 acting as the best inhibitor and the shorter or the longer molecule congeners (22 and 24) showing a decrease (or a loss) of activity (Table 3). The bulky clinically used derivatives EZA, DZA, BRZ, ZNS, IND, and HCT also belong to this category of CAIs with medium potency inhibitory action against BpsCAβ
(iii) The most efficient BpsCAβ inhibitors detected among the investigated sulfonamides were 11, 12, and DCP (belonging to the benzene-1,3-disulfonamide class of compounds), 17 (already mentioned above), 20, and BZA (benzolamide-type CAIs possessing an elongated molecule and two sulfonamide functionalities), AAZ, and MZA. These sulfonamides showed KI in the range of 185–745 nM (Table 3). The following SAR is obvious: the deacetylated derivatives of AAZ (13) and MZA (14) were much less effective BpsCAβ inhibitors compared to the clinically used drugs. This is a general phenomenon seen for many other CAs investigated earlier [65,66,67,68,69,70,71,72,73,74,75,76,77,78,79]. The best BpsCAβ inhibitors were MZA and BZA, with an almost identical inhibition constant (185–186 nM). It is interesting to note that MZA—which differs only by a CH2 moiety from AAZ—was four times stronger as an inhibitor than acetazolamide. Considering benzolamide (BZA), the introduction of a 4-amino moiety as in aminobenzolamide (20), led to a small decrease of the inhibitory activity from 185 nM to 266 nM (Table 3). Such compounds belong to a class of disulfonamide CAIs, which are highly effective against many CAs [80]. As mentioned above, the same efficient inhibitory action was observed for the benzene-1,3-disulfonamides DCP, 11, and 12, which incorporate two primary sulfonamide moieties in their scaffolds. Overall, the main conclusion is that several important CAI scaffolds lead to effective BpsCAβ inhibition, but compounds with an inhibition constant <100 nM were not identified in this study.
(iv) The inhibition profile of BpsCAβ differs substantially from that of the γ-class enzyme from the same pathogen (BpsCAγ) that was recently investigated by this group [7]. A relatively limited number of sulfonamides that have been investigated act as effective CAIs against both the β- and γ-CAs from this bacterium (Table 3). Among them are 20, AAZ, MZA, and BZA, with the latter compound having the most promising profile (KI of 653 and 185 nM, respectively). Thus, BZA might be considered an important lead compound for obtaining more effective CAIs targeting Burkholderia pseudomallei.
3. Materials and Methods
3.1. Gene Identification and Cloning
The identification of the gene encoding B. pseudomallei β-CA (BpsCAβ) was performed as described by Del Prete et al. [61] Briefly, the β-CA gene of B. pseudomallei with the accession number WP_004189176.1 was identified running the ‘‘BLAST’’ program, using the nucleotide sequences of bacterial β-CAs as query sequence. The GeneArt Company (Invitrogen)—specializing in gene synthesis—designed the synthetic BpsCAβ gene (BpsCAβ-DNA) encoding for the BpsCAβ (protein of 256 amino acid residues) containing four base-pair sequences (CACC) necessary for directional cloning at the 5′ end of the PfCAdom gene. The recovered PfCAdom gene and the linearized expression vector (pET-100/D-TOPO) were ligated by T4 DNA ligase to form the expression vector pET-100/BpsCAβ.
3.2. Expression and Purification of the Recombinant BpsCAβ
BL21-CodonPlus(DE3)-RIPL competent cells (Agilent) were transformed with pET-100/BpsCAβ, grown at 37 °C, and induced with 1 mM isopropil-β-d-1-tiogalattopiranoside (IPTG). After 30 min, ZnSO4 (0.5 mM) was added to the culture medium and cells were grown for an additional 3 h. Subsequently, cells were harvested and re-suspended in the following buffer: 50 mM Tris/HCl, pH 8.0, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), and 1 mM benzamidine. Cells were then disrupted by sonication at 4 °C. After centrifugation at 12,000× g for 45 min, the supernatant was incubated with His Select HF nickel affinity gel resin (Sigma) equilibrated in lysis buffer for 30 min. Following centrifugation at 2000× g, the resin was washed in buffer (50 mM Tris/HCl, pH 8.3, 500 mM KCl, 20 mM imidazole). The protein was eluted with the wash buffer containing 300 mM imidazole. The collected fractions were dialyzed against 50 mM Tris/HCl, pH 8.3. At this stage of purification, the protein was at least 95% pure and the obtained recovery was of about 20 mg of the recombinant protein.
3.3. Protonography
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed as described by Laemmli [83]. Wells of 12% SDS-gel were loaded with bCA or BpsCAβ mixed with Laemmli loading buffer without 2-mercaptoethanol and without boiling the samples in order to avoid protein denaturation. The gel was run at 180 V until the dye front ran off the gel. Following the electrophoresis, the gel was removed from glass plates and soaked in 2.5% Triton X-100 for 1 h on a shaker and then twice in 100 mM Tris, pH 8.2 containing 10% isopropanol for 10 min. Subsequently, the gel was incubated in 0.1% bromothymol blue in 100 mM Tris, pH 8.2 for 30 min and then immersed in CO2-saturated ddH2O to visualize the hydratase activity of the enzyme. The assay was performed at room temperature and the CO2-saturated solution was prepared by bubbling CO2 into 200 mL distilled water for approximately 3 h. The local decrease in pH due to the presence of CA activity was evidenced by the formation of yellow bands due to the change of the indicator color from blue (alkaline pH) to yellow (acidic pH) [79,80,81].
3.4. Kinetic and Inhibition Assay
An Applied Photophysics stopped-flow instrument has been used for assaying the CA-catalyzed CO2 hydration activity. [82] Phenol red (at a concentration of 0.2 mM) was used as indicator, working at the absorbance maximum of 557 nm, with 20 mM TRIS (pH 8.3) as buffer, and 20 mM NaClO4 (for maintaining constant the ionic strength), following the initial rates of the CA-catalyzed CO2 hydration reaction for a period of 10–100 s. The CO2 concentrations ranged from 1.7 to 17 mM for the determination of the kinetic parameters (by Lineweaver–Burk plots) and inhibition constants. For each inhibitor, at least six traces of the initial 5%–10% of the reaction were used to determine the initial velocity. The uncatalyzed rates were determined in the same manner and subtracted from the total observed rates. Stock solutions of inhibitor (10–100 mM) were prepared in distilled-deionized water, and dilutions up to 0.01 mM were done thereafter with the assay buffer. Inhibitor and enzyme solutions were preincubated together for 15 min at room temperature prior to assay in order to allow for the formation of the E-I (Enzyme-Inhibitor) complex or for the eventual active site-mediated hydrolysis of the inhibitor. The inhibition constants were obtained by non-linear least-squares methods using PRISM 3 and the Cheng–Prusoff equation (as reported earlier), and represent the mean from at least three different determinations. All CA isoforms were recombinant ones obtained in-house. All salts/small molecules were of the highest purity available, from Sigma-Aldrich (Milan, Italy).
4. Conclusions
We have cloned, purified, and characterized a β-CA from the pathogenic bacterium Burkholderia pseudomallei, responsible for the tropical disease melioidosis, named here BpsCAβ. The enzyme showed a relatively high catalytic activity for the physiologic CO2 hydration reaction to bicarbonate and protons, with the following kinetic parameters: kcat of 1.6 × 105 s−1 and kcat/KM of 3.4 × 107 M−1·s−1. An inhibition study with a panel of 38 sulfonamides and one sulfamate—including many agents that are used clinically—revealed an interesting structure–activity relationship for the interaction of this enzyme with its inhibitors. Many simple sulfonamides and clinically used agents such as topiramate, sulpiride, celecoxib, valdecoxib, and sulthiame were ineffective BpsCAβ inhibitors (KI > 50 µM). Other drugs, such as ethoxzolamide, dorzolamide, brinzolamide, zonisamide, indisulam, and hydrochlorothiazide were moderately potent micromolar inhibitors. The most promising inhibition was observed using benzene-1,3-disulfonamides, benzolamide and its analogs, acetazolamide, and methazolamide, which showed KI in the range of 185–745 nM. The inhibition profile of BpsCAβ differs substantially from that of the γ-class enzyme from the same pathogen (BpsCAγ), making it rather difficult to propose compounds that would effectively interact with both enzymes. However, benzolamide was one of the most promising inhibitors of both these CAs, with KI of 653 and 185 nM, respectively. Thus, benzolamide is an interesting lead compound for the design of more effective such agents, which may be useful tools for understanding the pathogenicity of this bacterium.
Acknowledgments
We acknowledge the Australian Research Council for funding (DP160102681).
Author Contributions
Daniela Vullo performed the enzyme kinetic and all the inhibition assays; Sonia Del Prete, Vincenzo Carginale and Pietro Di Fonzo performed the cloning, expression, purification and protonography of the recombinant bacterial enzyme; W. Alexander Donald edited and revised the English of the manuscript; Clemente Capasso and Claudiu T. Supuran wrote, edited and supervised the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the recombinant protein (BpsCAβ) and compounds are available from the authors.
References
- 1.Vullo D., Del Prete S., Osman S.M., AlOthman Z., Capasso C., Donald W.A., Supuran C.T. Burkholderia pseudomallei γ-carbonic anhydrase is strongly activated by amino acids and amines. Bioorg. Med. Chem. Lett. 2017;27:77–80. doi: 10.1016/j.bmcl.2016.11.027. [DOI] [PubMed] [Google Scholar]
- 2.Vullo D., Del Prete S., De Luca V., Carginale V., Ferraroni M., Dedeoglu N., Osman S.M., AlOthman Z., Capasso C., Supuran C.T. Anion inhibition studies of the β-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Bioorg. Med. Chem. Lett. 2016;26:1406–1410. doi: 10.1016/j.bmcl.2016.01.072. [DOI] [PubMed] [Google Scholar]
- 3.Vullo D., Del Prete S., Capasso C., Supuran C.T. Carbonic anhydrase activators: Activation of the β-carbonic anhydrase from Malassezia globosa with amines and amino acids. Bioorg. Med. Chem. Lett. 2016;26:1381–1385. doi: 10.1016/j.bmcl.2016.01.078. [DOI] [PubMed] [Google Scholar]
- 4.Supuran C.T., Capasso C. New light on bacterial carbonic anhydrases phylogeny based on the analysis of signal peptide sequences. J. Enzyme Inhib. Med. Chem. 2016;31:1254–1260. doi: 10.1080/14756366.2016.1201479. [DOI] [PubMed] [Google Scholar]
- 5.Ozensoy Guler O., Capasso C., Supuran C.T. A magnificent enzyme superfamily: Carbonic anhydrases, their purification and characterization. J. Enzyme Inhib. Med. Chem. 2016;31:689–694. doi: 10.3109/14756366.2015.1059333. [DOI] [PubMed] [Google Scholar]
- 6.Melis C., Meleddu R., Angeli A., Distinto S., Bianco G., Capasso C., Cottiglia F., Angius R., Supuran C.T., Maccioni E. Isatin: A privileged scaffold for the design of carbonic anhydrase inhibitors. J. Enzyme Inhib. Med. Chem. 2017;32:68–73. doi: 10.1080/14756366.2016.1235042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Del Prete S., Vullo D., Di Fonzo P., Osman S.M., AlOthman Z., Donald W.A., Supuran C.T., Capasso C. Sulfonamide inhibition profile of the γ-carbonic anhydrase identified in the genome of the pathogenic bacterium Burkholderia pseudomallei the etiological agent responsible of melioidosis. Bioorg. Med. Chem. Lett. 2017;27:490–495. doi: 10.1016/j.bmcl.2016.12.035. [DOI] [PubMed] [Google Scholar]
- 8.Del Prete S., Vullo D., De Luca V., Carginale V., di Fonzo P., Osman S.M., AlOthman Z., Supuran C.T., Capasso C. Anion inhibition profiles of α-, β- and γ-carbonic anhydrases from the pathogenic bacterium Vibrio cholerae. Bioorg. Med. Chem. 2016;24:3413–3417. doi: 10.1016/j.bmc.2016.05.029. [DOI] [PubMed] [Google Scholar]
- 9.Del Prete S., De Luca V., Vullo D., Osman S.M., AlOthman Z., Carginale V., Supuran C.T., Capasso C. A new procedure for the cloning, expression and purification of the β-carbonic anhydrase from the pathogenic yeast Malassezia globosa, an anti-dandruff drug target. J. Enzyme Inhib. Med. Chem. 2016;31:1156–1161. doi: 10.3109/14756366.2015.1102137. [DOI] [PubMed] [Google Scholar]
- 10.De Luca V., Vullo D., Del Prete S., Carginale V., Osman S.M., AlOthman Z., Supuran C.T., Capasso C. Cloning, characterization and anion inhibition studies of a γ-carbonic anhydrase from the antarctic bacterium Colwellia psychrerythraea. Bioorg. Med. Chem. 2016;24:835–840. doi: 10.1016/j.bmc.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Capasso C., Supuran C.T. An overview of the carbonic anhydrases from two pathogens of the oral cavity: Streptococcus mutans and Porphyromonas gingivalis. Curr. Top. Med. Chem. 2016;16:2359–2368. doi: 10.2174/1568026616666160413135522. [DOI] [PubMed] [Google Scholar]
- 12.Supuran C.T., Capasso C. The η-class carbonic anhydrases as drug targets for antimalarial agents. Expert Opin. Ther. Targets. 2015;19:551–563. doi: 10.1517/14728222.2014.991312. [DOI] [PubMed] [Google Scholar]
- 13.Del Prete S., Vullo D., De Luca V., AlOthman Z., Osman S.M., Supuran C.T., Capasso C. Biochemical characterization of recombinant β-carbonic anhydrase (PgiCAb) identified in the genome of the oral pathogenic bacterium Porphyromonas gingivalis. J. Enzyme Inhib. Med. Chem. 2015;30:366–370. doi: 10.3109/14756366.2014.931383. [DOI] [PubMed] [Google Scholar]
- 14.Pinard M.A., Lotlikar S.R., Boone C.D., Vullo D., Supuran C.T., Patrauchan M.A., McKenna R. Structure and inhibition studies of a type II β-carbonic anhydrase psCA3 from Pseudomonas aeruginosa. Bioorg. Med. Chem. 2015;23:4831–4838. doi: 10.1016/j.bmc.2015.05.029. [DOI] [PubMed] [Google Scholar]
- 15.Ferraroni M., Del Prete S., Vullo D., Capasso C., Supuran C.T. Crystal structure and kinetic studies of a tetrameric type II β-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Acta Crystallogr. D Biol. Crystallogr. 2015;71:2449–2456. doi: 10.1107/S1399004715018635. [DOI] [PubMed] [Google Scholar]
- 16.De Simone G., Monti S.M., Alterio V., Buonanno M., De Luca V., Rossi M., Carginale V., Supuran C.T., Capasso C., Di Fiore A. Crystal structure of the most catalytically effective carbonic anhydrase enzyme known, sazca from the thermophilic bacterium Sulfurihydrogenibium azorense. Bioorg. Med. Chem. Lett. 2015;25:2002–2006. doi: 10.1016/j.bmcl.2015.02.068. [DOI] [PubMed] [Google Scholar]
- 17.Zolnowska B., Slawinski J., Pogorzelska A., Chojnacki J., Vullo D., Supuran C.T. Carbonic anhydrase inhibitors. Synthesis, and molecular structure of novel series N-substituted N′-(2-arylmethylthio-4-chloro-5-methylbenzenesulfonyl)guanidines and their inhibition of human cytosolic isozymes I and II and the transmembrane tumor-associated isozymes IX and XII. Eur. J. Med. Chem. 2014;71:135–147. doi: 10.1016/j.ejmech.2013.10.081. [DOI] [PubMed] [Google Scholar]
- 18.De Luca L., Ferro S., Damiano F.M., Supuran C.T., Vullo D., Chimirri A., Gitto R. Structure-based screening for the discovery of new carbonic anhydrase VII inhibitors. Eur. J. Med. Chem. 2014;71:105–111. doi: 10.1016/j.ejmech.2013.10.071. [DOI] [PubMed] [Google Scholar]
- 19.Di Fiore A., Capasso C., De Luca V., Monti S.M., Carginale V., Supuran C.T., Scozzafava A., Pedone C., Rossi M., De Simone G. X-ray structure of the first “extremo-α-carbonic anhydrase”, a dimeric enzyme from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Acta Crystallogr. D Biol. Crystallogr. 2013;69:1150–1159. doi: 10.1107/S0907444913007208. [DOI] [PubMed] [Google Scholar]
- 20.Supuran C.T. Structure-based drug discovery of carbonic anhydrase inhibitors. J. Enzyme Inhib. Med. Chem. 2012;27:759–772. doi: 10.3109/14756366.2012.672983. [DOI] [PubMed] [Google Scholar]
- 21.Supuran C.T. Carbonic anhydrases—An overview. Curr. Pharm. Des. 2008;14:603–614. doi: 10.2174/138161208783877884. [DOI] [PubMed] [Google Scholar]
- 22.Supuran C.T. Advances in structure-based drug discovery of carbonic anhydrase inhibitors. Expert Opin. Drug Discov. 2017;12:61–88. doi: 10.1080/17460441.2017.1253677. [DOI] [PubMed] [Google Scholar]
- 23.Bhatt A., Mahon B.P., Cruzeiro V.W., Cornelio B., Laronze-Cochard M., Ceruso M., Sapi J., Rance G.A., Khlobystov A.N., Fontana A., et al. Structure-activity relationships of benzenesulfonamide-based inhibitors towards carbonic anhydrase isoform specificity. Chembiochem. 2017;18:213–222. doi: 10.1002/cbic.201600513. [DOI] [PubMed] [Google Scholar]
- 24.Alterio V., Langella E., Viparelli F., Vullo D., Ascione G., Dathan N.A., Morel F.M., Supuran C.T., De Simone G., Monti S.M. Structural and inhibition insights into carbonic anhydrase CDCA1 from the marine diatom Thalassiosira weissflogii. Biochimie. 2012;94:1232–1241. doi: 10.1016/j.biochi.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Supuran C.T. Structure and function of carbonic anhydrases. Biochem. J. 2016;473:2023–2032. doi: 10.1042/BCJ20160115. [DOI] [PubMed] [Google Scholar]
- 26.Buzas G.M., Supuran C.T. The history and rationale of using carbonic anhydrase inhibitors in the treatment of peptic ulcers. In memoriam ioan puscas (1932–2015) J. Enzyme Inhib. Med. Chem. 2016;31:527–533. doi: 10.3109/14756366.2015.1051042. [DOI] [PubMed] [Google Scholar]
- 27.Capasso C., Supuran C.T. An overview of the selectivity and efficiency of the bacterial carbonic anhydrase inhibitors. Curr. Med. Chem. 2015;22:2130–2139. doi: 10.2174/0929867321666141012174921. [DOI] [PubMed] [Google Scholar]
- 28.Carta F., Supuran C.T., Scozzafava A. Sulfonamides and their isosters as carbonic anhydrase inhibitors. Future Med. Chem. 2014;6:1149–1165. doi: 10.4155/fmc.14.68. [DOI] [PubMed] [Google Scholar]
- 29.Capasso C., Supuran C.T. An overview of the α-, β- and γ-carbonic anhydrases from bacteria: Can bacterial carbonic anhydrases shed new light on evolution of bacteria? J. Enzyme Inhib. Med. Chem. 2015;30:325–332. doi: 10.3109/14756366.2014.910202. [DOI] [PubMed] [Google Scholar]
- 30.Supuran C.T. Carbonic anhydrases: Novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Discov. 2008;7:168–181. doi: 10.1038/nrd2467. [DOI] [PubMed] [Google Scholar]
- 31.Capasso C., Supuran C.T. Bacterial, fungal and protozoan carbonic anhydrases as drug targets. Expert Opin. Ther. Targets. 2015;19:1689–1704. doi: 10.1517/14728222.2015.1067685. [DOI] [PubMed] [Google Scholar]
- 32.Capasso C., Supuran C.T. Sulfa and trimethoprim-like drugs—Antimetabolites acting as carbonic anhydrase, dihydropteroate synthase and dihydrofolate reductase inhibitors. J. Enzyme Inhib. Med. Chem. 2014;29:379–387. doi: 10.3109/14756366.2013.787422. [DOI] [PubMed] [Google Scholar]
- 33.Capasso C., Supuran C.T. Anti-infective carbonic anhydrase inhibitors: A patent and literature review. Expert Opin. Ther. Pat. 2013;23:693–704. doi: 10.1517/13543776.2013.778245. [DOI] [PubMed] [Google Scholar]
- 34.Joseph P., Ouahrani-Bettache S., Montero J.L., Nishimori I., Minakuchi T., Vullo D., Scozzafava A., Winum J.Y., Kohler S., Supuran C.T. A new β-carbonic anhydrase from Brucella suis, its cloning, characterization, and inhibition with sulfonamides and sulfamates, leading to impaired pathogen growth. Bioorg. Med. Chem. 2011;19:1172–1178. doi: 10.1016/j.bmc.2010.12.048. [DOI] [PubMed] [Google Scholar]
- 35.Modak J.K., Liu Y.C., Machuca M.A., Supuran C.T., Roujeinikova A. Structural basis for the inhibition of Helicobacter pylori α-carbonic anhydrase by sulfonamides. PLoS ONE. 2015;10:e0127149. doi: 10.1371/journal.pone.0127149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishimori I., Onishi S., Takeuchi H., Supuran C.T. The α and β classes carbonic anhydrases from Helicobacter pylori as novel drug targets. Curr. Pharm. Des. 2008;14:622–630. doi: 10.2174/138161208783877875. [DOI] [PubMed] [Google Scholar]
- 37.Nishimori I., Vullo D., Minakuchi T., Morimoto K., Onishi S., Scozzafava A., Supuran C.T. Carbonic anhydrase inhibitors: Cloning and sulfonamide inhibition studies of a carboxyterminal truncated α-carbonic anhydrase from Helicobacter pylori. Bioorg. Med. Chem. Lett. 2006;16:2182–2188. doi: 10.1016/j.bmcl.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 38.Vullo D., De Luca V., Del Prete S., Carginale V., Scozzafava A., Osman S.M., AlOthman Z., Capasso C., Supuran C.T. Sulfonamide inhibition studies of the γ-carbonic anhydrase from the antarctic bacterium Colwellia psychrerythraea. Bioorg. Med. Chem. Lett. 2016;26:1253–1259. doi: 10.1016/j.bmcl.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 39.Del Prete S., Isik S., Vullo D., De Luca V., Carginale V., Scozzafava A., Supuran C.T., Capasso C. DNA cloning, characterization, and inhibition studies of an α-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. J. Med. Chem. 2012;55:10742–10748. doi: 10.1021/jm301611m. [DOI] [PubMed] [Google Scholar]
- 40.Supuran C.T. Carbonic anhydrases: From biomedical applications of the inhibitors and activators to biotechnological use for CO2 capture. J. Enzyme Inhib. Med. Chem. 2013;28:229–230. doi: 10.3109/14756366.2013.761876. [DOI] [PubMed] [Google Scholar]
- 41.Capasso C., Supuran C. Inhibition of bacterial carbonic anhydrases as a novel approach to escape drug resistance. Curr. Top. Med. Chem. 2017;17:1237–1248. doi: 10.2174/1568026617666170104101058. [DOI] [PubMed] [Google Scholar]
- 42.Supuran C.T. Legionella pneumophila carbonic anhydrases: Underexplored antibacterial drug targets. Pathogens. 2016;5:44. doi: 10.3390/pathogens5020044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Supuran C.T. How many carbonic anhydrase inhibition mechanisms exist? J. Enzyme Inhib. Med. Chem. 2016;31:345–360. doi: 10.3109/14756366.2015.1122001. [DOI] [PubMed] [Google Scholar]
- 44.Bejaoui M., Pantazi E., De Luca V., Panisello A., Folch-Puy E., Hotter G., Capasso C., Supuran C.T., Rosello-Catafau J. Correction: Carbonic anhydrase protects fatty liver grafts against ischemic reperfusion damage. PLoS ONE. 2015;10:e0139411. doi: 10.1371/journal.pone.0139411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Simone G., Supuran C.T. (In)organic anions as carbonic anhydrase inhibitors. J. Inorg. Biochem. 2012;111:117–129. doi: 10.1016/j.jinorgbio.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 46.Supuran C.T. Bacterial carbonic anhydrases as drug targets: Toward novel antibiotics? Front Pharmacol. 2011;2:34. doi: 10.3389/fphar.2011.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stephens D.P., Thomas J.H., Ward L.M., Currie B.J. Melioidosis causing critical illness: A review of 24 years of experience from the royal darwin hospital ICU. Crit. Care Med. 2016;44:1500–1505. doi: 10.1097/CCM.0000000000001668. [DOI] [PubMed] [Google Scholar]
- 48.Gilad J., Schwartz D., Amsalem Y. Clinical features and laboratory diagnosis of infection with the potential bioterrorism agents burkholderia mallei and Burkholderia pseudomallei. Int. J. Biomed. Sci. 2007;3:144–152. [PMC free article] [PubMed] [Google Scholar]
- 49.Del Prete S., Vullo D., De Luca V., Carginale V., Osman S.M., AlOthman Z., Supuran C.T., Capasso C. Comparison of the sulfonamide inhibition profiles of the α-, β- and γ-carbonic anhydrases from the pathogenic bacterium Vibrio cholerae. Bioorg. Med. Chem. Lett. 2016;26:1941–1946. doi: 10.1016/j.bmcl.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 50.Del Prete S., Vullo D., De Luca V., Carginale V., Ferraroni M., Osman S.M., AlOthman Z., Supuran C.T., Capasso C. Sulfonamide inhibition studies of the β-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Bioorg. Med. Chem. 2016;24:1115–1120. doi: 10.1016/j.bmc.2016.01.037. [DOI] [PubMed] [Google Scholar]
- 51.Modak J.K., Liu Y.C., Supuran C.T., Roujeinikova A. Structure-activity relationship for sulfonamide inhibition of Helicobacter pylori α-carbonic anhydrase. J. Med. Chem. 2016;59:11098–11109. doi: 10.1021/acs.jmedchem.6b01333. [DOI] [PubMed] [Google Scholar]
- 52.Morishita S., Nishimori I., Minakuchi T., Onishi S., Takeuchi H., Sugiura T., Vullo D., Scozzafava A., Supuran C.T. Cloning, polymorphism, and inhibition of β-carbonic anhydrase of Helicobacter pylori. J. Gastroenterol. 2008;43:849–857. doi: 10.1007/s00535-008-2240-3. [DOI] [PubMed] [Google Scholar]
- 53.Dedeoglu N., DeLuca V., Isik S., Yildirim H., Kockar F., Capasso C., Supuran C.T. Sulfonamide inhibition study of the β-class carbonic anhydrase from the caries producing pathogen Streptococcus mutans. Bioorg. Med. Chem. Lett. 2015;25:2291–2297. doi: 10.1016/j.bmcl.2015.04.037. [DOI] [PubMed] [Google Scholar]
- 54.Dedeoglu N., De Luca V., Isik S., Yildirim H., Kockar F., Capasso C., Supuran C.T. Cloning, characterization and anion inhibition study of a β-class carbonic anhydrase from the caries producing pathogen Streptococcus mutans. Bioorg. Med. Chem. 2015;23:2995–3001. doi: 10.1016/j.bmc.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 55.Alafeefy A.M., Ceruso M., Al-Tamimi A.M., Del Prete S., Supuran C.T., Capasso C. Inhibition studies of quinazoline-sulfonamide derivatives against the γ-CA (PgiCA) from the pathogenic bacterium, Porphyromonas gingivalis. J. Enzyme Inhib. Med. Chem. 2015;30:592–596. doi: 10.3109/14756366.2014.957202. [DOI] [PubMed] [Google Scholar]
- 56.Vullo D., Del Prete S., Osman S.M., De Luca V., Scozzafava A., Alothman Z., Supuran C.T., Capasso C. Sulfonamide inhibition studies of the γ-carbonic anhydrase from the oral pathogen Porphyromonas gingivalis. Bioorg. Med. Chem. Lett. 2014;24:240–244. doi: 10.1016/j.bmcl.2013.11.030. [DOI] [PubMed] [Google Scholar]
- 57.Del Prete S., Vullo D., De Luca V., Carginale V., Scozzafava A., Supuran C.T., Capasso C. A highly catalytically active γ-carbonic anhydrase from the pathogenic anaerobe Porphyromonas gingivalis and its inhibition profile with anions and small molecules. Bioorg. Med. Chem. Lett. 2013;23:4067–4071. doi: 10.1016/j.bmcl.2013.05.063. [DOI] [PubMed] [Google Scholar]
- 58.Vullo D., Sai Kumar R.S., Scozzafava A., Capasso C., Ferry J.G., Supuran C.T. Anion inhibition studies of a β-carbonic anhydrase from Clostridium perfringens. Bioorg. Med. Chem. Lett. 2013;23:6706–6710. doi: 10.1016/j.bmcl.2013.10.037. [DOI] [PubMed] [Google Scholar]
- 59.Maresca A., Scozzafava A., Kohler S., Winum J.Y., Supuran C.T. Inhibition of β-carbonic anhydrases from the bacterial pathogen Brucella suis with inorganic anions. J. Inorg. Biochem. 2012;110:36–39. doi: 10.1016/j.jinorgbio.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 60.Vullo D., Nishimori I., Scozzafava A., Kohler S., Winum J.Y., Supuran C.T. Inhibition studies of a β-carbonic anhydrase from Brucella suis with a series of water soluble glycosyl sulfanilamides. Bioorg. Med. Chem. Lett. 2010;20:2178–2182. doi: 10.1016/j.bmcl.2010.02.042. [DOI] [PubMed] [Google Scholar]
- 61.Del Prete S., Vullo D., Di Fonzo P., Osman S.M., AlOthman Z., Supuran C.T., Capasso C. Anion inhibition profiles of the γ-carbonic anhydrase from the pathogenic bacterium Burkholderia pseudomallei responsible of melioidosis and highly drug resistant to common antibiotics. Bioorg. Med. Chem. 2017;25:575–580. doi: 10.1016/j.bmc.2016.11.021. [DOI] [PubMed] [Google Scholar]
- 62.Vullo D., Nishimori I., Minakuchi T., Scozzafava A., Supuran C.T. Inhibition studies with anions and small molecules of two novel β-carbonic anhydrases from the bacterial pathogen Salmonella enterica serovar typhimurium. Bioorg. Med. Chem. Lett. 2011;21:3591–3595. doi: 10.1016/j.bmcl.2011.04.105. [DOI] [PubMed] [Google Scholar]
- 63.Nishimori I., Minakuchi T., Vullo D., Scozzafava A., Supuran C.T. Inhibition studies of the β-carbonic anhydrases from the bacterial pathogen Salmonella enterica serovar typhimurium with sulfonamides and sulfamates. Bioorg. Med. Chem. 2011;19:5023–5030. doi: 10.1016/j.bmc.2011.06.038. [DOI] [PubMed] [Google Scholar]
- 64.Burghout P., Vullo D., Scozzafava A., Hermans P.W., Supuran C.T. Inhibition of the β-carbonic anhydrase from Streptococcus pneumoniae by inorganic anions and small molecules: Toward innovative drug design of antiinfectives? Bioorg. Med. Chem. 2011;19:243–248. doi: 10.1016/j.bmc.2010.11.031. [DOI] [PubMed] [Google Scholar]
- 65.Ceruso M., Vullo D., Scozzafava A., Supuran C.T. Sulfonamides incorporating fluorine and 1,3,5-triazine moieties are effective inhibitors of three β-class carbonic anhydrases from Mycobacterium tuberculosis. J. Enzyme Inhib. Med. Chem. 2014;29:686–689. doi: 10.3109/14756366.2013.842233. [DOI] [PubMed] [Google Scholar]
- 66.Nishimori I., Minakuchi T., Maresca A., Carta F., Scozzafava A., Supuran C.T. The β-carbonic anhydrases from Mycobacterium tuberculosis as drug targets. Curr. Pharm. Des. 2010;16:3300–3309. doi: 10.2174/138161210793429814. [DOI] [PubMed] [Google Scholar]
- 67.Carta F., Maresca A., Covarrubias A.S., Mowbray S.L., Jones T.A., Supuran C.T. Carbonic anhydrase inhibitors. Characterization and inhibition studies of the most active β-carbonic anhydrase from Mycobacterium tuberculosis, rv3588c. Bioorg. Med. Chem. Lett. 2009;19:6649–6654. doi: 10.1016/j.bmcl.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 68.Innocenti A., Zimmerman S., Ferry J.G., Scozzafava A., Supuran C.T. Carbonic anhydrase inhibitors. Inhibition of the β-class enzyme from the methanoarchaeon Methanobacterium thermoautotrophicum (Cab) with anions. Bioorg. Med. Chem. Lett. 2004;14:4563–4567. doi: 10.1016/j.bmcl.2004.06.073. [DOI] [PubMed] [Google Scholar]
- 69.Innocenti A., Zimmerman S., Ferry J.G., Scozzafava A., Supuran C.T. Carbonic anhydrase inhibitors. Inhibition of the zinc and cobalt γ-class enzyme from the archaeon Methanosarcina thermophila with anions. Bioorg. Med. Chem. Lett. 2004;14:3327–3331. doi: 10.1016/j.bmcl.2004.03.101. [DOI] [PubMed] [Google Scholar]
- 70.Vullo D., De Luca V., Scozzafava A., Carginale V., Rossi M., Supuran C.T., Capasso C. The first activation study of a bacterial carbonic anhydrase (CA). The thermostable α-CA from Sulfurihydrogenibium yellowstonense YO3AOP1 is highly activated by amino acids and amines. Bioorg. Med. Chem. Lett. 2012;22:6324–6327. doi: 10.1016/j.bmcl.2012.08.088. [DOI] [PubMed] [Google Scholar]
- 71.Vullo D., Luca V.D., Scozzafava A., Carginale V., Rossi M., Supuran C.T., Capasso C. The α-carbonic anhydrase from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1 is highly susceptible to inhibition by sulfonamides. Bioorg. Med. Chem. 2013;21:1534–1538. doi: 10.1016/j.bmc.2012.07.024. [DOI] [PubMed] [Google Scholar]
- 72.De Luca V., Vullo D., Scozzafava A., Carginale V., Rossi M., Supuran C.T., Capasso C. Anion inhibition studies of an α-carbonic anhydrase from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Bioorg. Med. Chem. Lett. 2012;22:5630–5634. doi: 10.1016/j.bmcl.2012.06.106. [DOI] [PubMed] [Google Scholar]
- 73.Alafeefy A.M., Abdel-Aziz H.A., Vullo D., Al-Tamimi A.M., Al-Jaber N.A., Capasso C., Supuran C.T. Inhibition of carbonic anhydrases from the extremophilic bacteria Sulfurihydrogenibium yellostonense (Sspca) and S. azorense (Sazca) with a new series of sulfonamides incorporating aroylhydrazone-, [1,2,4]triazolo[3,4-b][1,3,4]thiadiazinyl- or 2-(cyanophenylmethylene)-1,3,4-thiadiazol-3(2H)-yl moieties. Bioorg. Med. Chem. 2014;22:141–147. doi: 10.1016/j.bmc.2013.11.042. [DOI] [PubMed] [Google Scholar]
- 74.Vullo D., De Luca V., Scozzafava A., Carginale V., Rossi M., Supuran C.T., Capasso C. The extremo-α-carbonic anhydrase from the thermophilic bacterium Sulfurihydrogenibium azorense is highly inhibited by sulfonamides. Bioorg. Med. Chem. 2013;21:4521–4525. doi: 10.1016/j.bmc.2013.05.042. [DOI] [PubMed] [Google Scholar]
- 75.Akdemir A., Vullo D., De Luca V., Scozzafava A., Carginale V., Rossi M., Supuran C.T., Capasso C. The extremo-α-carbonic anhydrase (CA) from Sulfurihydrogenibium azorense, the fastest ca known, is highly activated by amino acids and amines. Bioorg. Med. Chem. Lett. 2013;23:1087–1090. doi: 10.1016/j.bmcl.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 76.Vullo D., De Luca V., Scozzafava A., Carginale V., Rossi M., Supuran C.T., Capasso C. Anion inhibition studies of the fastest carbonic anhydrase (CA) known, the extremo-ca from the bacterium Sulfurihydrogenibium azorense. Bioorg. Med. Chem. Lett. 2012;22:7142–7145. doi: 10.1016/j.bmcl.2012.09.065. [DOI] [PubMed] [Google Scholar]
- 77.Vullo D., De Luca V., Del Prete S., Carginale V., Scozzafava A., Capasso C., Supuran C.T. Sulfonamide inhibition studies of the γ-carbonic anhydrase from the antarctic bacterium pseudoalteromonas haloplanktis. Bioorg. Med. Chem. Lett. 2015;25:3550–3555. doi: 10.1016/j.bmcl.2015.06.079. [DOI] [PubMed] [Google Scholar]
- 78.De Luca V., Vullo D., Del Prete S., Carginale V., Scozzafava A., Osman S.M., AlOthman Z., Supuran C.T., Capasso C. Cloning, characterization and anion inhibition studies of a new γ-carbonic anhydrase from the antarctic bacterium Pseudoalteromonas haloplanktis. Bioorg. Med. Chem. 2015;23:4405–4409. doi: 10.1016/j.bmc.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 79.Del Prete S., De Luca V., Supuran C.T., Capasso C. Protonography, a technique applicable for the analysis of η-carbonic anhydrase activity. J. Enzyme Inhib. Med. Chem. 2015:1–5. doi: 10.3109/14756366.2014.990963. [DOI] [PubMed] [Google Scholar]
- 80.Del Prete S., De Luca V., Iandolo E., Supuran C.T., Capasso C. Protonography, a powerful tool for analyzing the activity and the oligomeric state of the γ-carbonic anhydrase identified in the genome of Porphyromonas gingivalis. Bioorg. Med. Chem. 2015;23:3747–3750. doi: 10.1016/j.bmc.2015.03.080. [DOI] [PubMed] [Google Scholar]
- 81.De Luca V., Del Prete S., Supuran C.T., Capasso C. Protonography, a new technique for the analysis of carbonic anhydrase activity. J. Enzyme Inhib. Med. Chem. 2015;30:277–282. doi: 10.3109/14756366.2014.917085. [DOI] [PubMed] [Google Scholar]
- 82.Khalifah R.G. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J. Biol. Chem. 1971;246:2561–2573. [PubMed] [Google Scholar]
- 83.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]