Abstract
Residual feed intake (RFI), a measure of feed efficiency, is an important economic and environmental trait in beef production. Selection of low RFI (feed efficient) cattle could maintain levels of production, while decreasing feed costs and methane emissions. However, RFI is a difficult and expensive trait to measure. Identification of single nucleotide polymorphisms (SNPs) associated with RFI may enable rapid, cost effective genomic selection of feed efficient cattle. Genome-wide association studies (GWAS) were conducted in multiple breeds followed by meta-analysis to identify genetic variants associated with RFI and component traits (average daily gain (ADG) and feed intake (FI)) in Irish beef cattle (n = 1492). Expression quantitative trait loci (eQTL) analysis was conducted to identify functional effects of GWAS-identified variants. Twenty-four SNPs were associated (P < 5 × 10−5) with RFI, ADG or FI. The variant rs43555985 exhibited strongest association for RFI (P = 8.28E-06). An eQTL was identified between this variant and GFRA2 (P = 0.0038) where the allele negatively correlated with RFI was associated with increased GFRA2 expression in liver. GFRA2 influences basal metabolic rates, suggesting a mechanism by which genetic variation may contribute to RFI. This study identified SNPs that may be useful both for genomic selection of RFI and for understanding the biology of feed efficiency.
Introduction
Feed can account for more than 75% of variable costs of beef enterprises1. Consequently, selection of cattle that efficiently convert feed to carcass growth would improve farm profits due to reducing expenditure on feed while maintaining protein output2. Moreover, there is pressure on the agricultural industry to reduce methane emissions and improve its environmental footprint, while simultaneously increasing beef output to meet the growing demand for protein worldwide3. Selection for feed efficient cattle could increase beef output while concurrently decreasing methane production, as it has been suggested that low residual feed intake (RFI) (feed efficient) animals emit less methane than their high RFI counterparts4.
RFI is a measure of feed efficiency, defined as the difference between actual and predicted feed intake (FI)5. RFI has been shown to be moderately heritable, with an estimated heritability of 0.332,6, making it an ideal trait for selection as any genetic gain will be maintained and propagated through the cattle herd6. However, calculation of RFI is currently impeded by both the expense and logistics associated with its measurement, involving recording of both FI and body weight gain for each individual animal for a period of 70 days7. Identification of genetic markers for RFI and component traits, such as FI and average daily gain (ADG), and their incorporation into genomic assisted breeding programmes would enable more rapid and cost effective selection of feed efficient cattle8. Indeed, RFI has been incorporated into the Australian dairy industry’s genomic breeding programme9. Unlike the situation that predominates for dairy production systems worldwide, effective identification of selection markers of RFI for beef cattle must take into account a multiplicity of breeds and the mainly crossbred nature of cattle typically utilised within the global beef industry through employing multi-breed populations in order to identify variants of interest10,11. Differences in linkage disequilibrium (LD) between breeds may impact the association of markers and quantitative trait loci across breeds. The use of multiple breeds in a reference population is important to account for this variation in LD between breeds10.
In Ireland, the genomic assisted breeding programme is administered by the Irish Cattle Breeding Federation (ICBF)12. Markers for selection are included on the International Dairy and Beef (IDB) custom genotyping chip, which is based on the Illumina BovineSNP50 genotyping chip providing the IDB chip with genome-wide coverage13,14. As well as single nucleotide polymorphisms (SNPs) that are used for the genomic selection programme, the IDB chip contains SNPs for parentage verification and SNPs included for research purposes only13. This includes a selected subset of SNPs associated with RFI in cattle populations outside of Ireland15–21 which have been added to the IDB chip for research purposes to validate their use as biomarkers of RFI and associated traits in Irish beef cattle (n = 102, Supplementary Table S1).
Genetic markers for RFI can be identified via genome-wide association studies (GWAS). Several GWAS have identified SNPs associated with feed efficiency-related traits in cattle populations, both purebred and crossbred, from North America, South America and Australia16,20–23. Despite considerable interest in identifying markers for RFI, ADG and FI, no published GWAS has been carried out to test for associations between SNPs and these traits in Irish beef cattle. In Ireland, commercial beef cattle are mainly crossbreds, with Charolais (CH), Limousin (LM), Aberdeen Angus (AA), Belgian Blue (BB) and Simmental (SI) breeds predominating genetically12. This breed heterogeneity coupled with the challenges in obtaining sufficient numbers of RFI phenotypes for GWAS are primary reasons for the difficulty in applying GWAS on a large-scale basis for RFI to beef cattle in Ireland and most other beef producing nations.
It is important to identify genetic variants that underlie phenotypic variation. One method to identify SNPs that are implicated in observed variation is by carrying out expression quantitative trait loci (eQTL) studies. eQTL analysis enables investigation of the effect of genotype on gene-expression levels which may in turn affect phenotype24. Previous eQTL analysis carried out in mammary tissue of dairy cattle has identified several eQTLs for milk production traits enabling the identification of genes such as PLAG1 and MGST1 as potentially functional in the development of divergent milk production traits25,26. eQTLs have been identified for temperament in the adrenal cortex of crossbred German cattle27. Despite the ability of eQTL analysis to identify potentially causative genes for complex traits, to the best of the authors’ knowledge no eQTL analysis has been carried out for RFI in beef cattle, or in any other livestock species. Liver and muscle are key tissues of interest with regards to feed efficiency as they are both large, metabolically active tissues accounting for approximately 24% and 25% of basal energy expenditure, respectively28,29. Thus, investigation into the presence of eQTLs in these tissues may aid in unravelling the biology underlying divergence in feed efficiency.
Due to multiple breeds of cattle present in beef production systems, it is important to identify markers for traits that have effects across multiple breeds10. Thus, the objectives of this study were to: (i) perform GWAS for RFI, and its component traits, namely FI and ADG, in different breeds of Irish beef cattle and combine results in order to identify associated SNPs in a mixed breed cohort, (ii) validate a selection of internationally identified markers of RFI present on the IDBv3 chip for utility as selection markers for RFI in Irish cattle and (iii) to investigate the effects of associated variants on gene expression in metabolically important tissues using eQTL analysis, in order to understand the biological mechanisms underlying divergence in RFI and component traits.
Methods
All biological sampling and procedures involving animals within this study were reviewed by the Teagasc Animal Ethics Committee and/or the UCD Animal Research Ethics Committee. All procedures carried out prior to 2013 were licenced by the Irish Department of Health, all procedures carried out since 2013 were licenced by the Irish Health Products Regulatory Authority in accordance with the cruelty to Animals Act 1876 and the European Communities (Amendment of Cruelty to Animals Act 1876) Regulations 2002 and 2005.
Phenotypic data collation
Data were collated for this study from growing bulls (n = 1823), steers (n = 459) and heifers (n = 164), which had previously undergone phenotypic measurement testing in Ireland between 2006 and 2017. The average ages and standard deviations for cattle included in the phenotypic data file were available were available on a group-by-group basis (Supplementary Table S2). Throughout each phenotypic measurement trial, the health of the animals was monitored. Any animal that required treatment was noted and excluded from further analysis. Phenotypes were gathered at the national beef research centre in Teagasc Grange; UCD Lyons Research Farm, University College Dublin and the ICBF national beef performance test station, Tully, Co. Kildare, Ireland. Phenotypes were collected from both purebred and crossbred beef cattle. For crossbred animals to be included in the phenotypic dataset the proportion of genetic material from a single parental breed had to be greater than 50%. Prior to further analysis cattle were grouped by breed. Predominant breeds were LM, CH, SI, BB and AA (n = 737, 499, 413, 191 and 174, respectively), other breeds were represented at smaller numbers. The RFI range for LM, CH, SI, BB and AA was 2.69 to −2.52, 2.70 to −2.48, 2.75 to −2.82, 1.63 to −2.00, and 2.87 to −2.64 respectively.
This resulted in the generation of a phenotypic file consisting of 2,446 cattle. For 429 of these animals, data relating to breed, diet, and methods used to calculate RFI, ADG and FI has been previously described4,28,30–33. Records for remaining cattle were made available by the ICBF from the national beef performance test centre at Tully, Co. Kildare, Ireland.
The management protocol of the ICBF animals is described briefly here. Animals were housed in pens for the duration of their test period which was between 70 and 105 days. A Calan gate system (American Calan, Northwood, NH, USA) was used to record individual animal FI. Bulls were individually offered ad libitum concentrates and 3 kg fresh weight of hay, while steers were offered 8 kg concentrates and 5 kg fresh weight of hay. Hay was offered in order to maintain healthy rumen function and to reflect an Irish commercial high concentrate based dietary regimen. Refused feed was weighed weekly and subtracted from total feed offered in order to calculate total feed consumed. Dry matter intake was then calculated in order to determine FI, which was used for calculation of RFI. Cattle were weighed at the beginning and end of the test period, and every 21 days during the test period. ADG was calculated as the coefficient of linear regression of body weight on time, computed in the software package R34. Mid-test metabolic bodyweight (body weight0.75, MBW) was calculated as body weight0.75 in the middle of the RFI measurement period, which was estimated from the intercept and slope of the regression line after fitting a linear regression through all MBW observations. RFI was calculated for each animal as the difference between actual and predicted FI. Predicted FI for each animal was computed by regressing FI on MBW and ADG. Calculation of predicted FI was calculated for each contemporary group individually.
Genotyping
DNA was isolated for genotyping from one of two tissue types sourced from 429 cattle described previously. Muscle was used when blood was unavailable. Blood samples were obtained by jugular venipuncture at the end of the RFI measurement period, as per Fitzsimons et al.4, and stored at −80 °C prior to use4. Muscle samples were obtained via biopsy of the M. longissimus dorsi following the RFI measurement period, as per Kelly et al.35, and stored at −80 °C before DNA extraction. DNA from blood samples was extracted using the Maxwell 16 Blood DNA kit (Promega, Madison, WI, USA) as per manufacturer’s instructions. DNA was extracted from muscle samples using a phenol-chloroform extraction method. Briefly, 0.1 g of frozen muscle tissue was immersed in 1 mL of Trizol and homogenized using a Precellys 24 homogeniser. 200 µl chloroform was added to the homogenate, which was then centrifuged at room temperature for five minutes at 16,000 g. After centrifugation the aqueous phase was transferred to a new tube. Two volumes of ice cold ethanol were added to the aqueous phase and this mixture was centrifuged at 16,000 g for 15 minutes at 4 °C resulting in the formation of a DNA pellet. The supernatant was removed and the pellet was washed by the addition of 1 ml 70% ethanol and centrifuged at 16,000 g for 5 minutes at 4 °C. Washing was carried out twice. Following washing, any remaining supernatant was removed and the pellet was left to air-dry. The DNA was then re-suspended in 150 µl RNase/Dnase free H2O.
Once DNA was isolated, samples were analysed for quality and quantity using a Nanodrop spectrophotometer. DNA of sufficient quality for genotyping was available for 422 samples. All DNA samples were normalised to a concentration of 50 ng/µl for genotyping analysis. Genotyping was carried out on the IDBv3 chip13 by Weatherby’s Scientific Ltd. (Johnstown, Naas, Co. Kildare, Ireland). The ICBF provided a further genotypes for 1,262 cattle that had been genotyped on the IDBv3 chip by Weatherby’s Scientific.
In addition to the 1,684 animals genotyped directly on the IDBv3 chip, 338 cattle were genotyped on the Illumina Bovine HD genotyping chip. These 338 cattle were imputed to IDBv3 density using Fimpute version 2.236. The reference population used for Fimpute was 50,000 Irish cattle with genotyped parents. Imputation of all 338 cattle was conducted across breed type to reflect the Irish national cattle population.
Once genotyping and imputation were complete the study consisted of 2,022 animals with genotypic data for all IDBv3 markers. This genetic data was uploaded to the SNP Variation Suite (SVS) environment (Golden Helix, Version 7.7.6).
Preparation of files for analysis
Quality control (QC) was carried out on genotypes imported into the SVS environment. SNPs were removed from analysis if they had a call rate of less than 0.80 or a minor allele frequency of less than 0.05. Cattle were removed from analysis if they had a call rate of less than 0.95. Following QC, 2,008 cattle and 44,338 markers remained for analysis. LD pruning was carried out at r2 threshold of 0.5 and 7,841 markers were discarded following pruning37. The remaining 36,496 SNPs that passed all QC measures were acceptable for further analysis (Supplementary Table S3).
The collated phenotypic data were merged with the genotype data, creating a dataset containing 1,822 cattle eligible for analysis. A genomic kinship matrix was computed from the population, which was included as a covariate in the GWAS in order to account for relatedness. From this dataset, cattle from five beef breeds were analysed (n = 1492). The breeds included in the analysis were AA, BB, CH, LM and SI (n = 102, 177, 387, 537 and 289, respectively).
Genome-wide association studies
GWAS were carried out in the SVS environment of Golden Helix using a mixed linear model method, EMMAX38, for each breed individually. GWAS resulted in the generation of summary statistics for each trait of interest, i.e. RFI, ADG and FI, for each breed (AA, BB, CH, LI, and SI).
Meta-Analysis
Following initial breed specific GWAS, meta-analyses were carried out for each trait using the software package METAL39. METAL combines P-value and direction of effect from each GWAS to conduct Z-score method meta-analysis. METAL analysis results in two outputs, the Z-score for each SNP and a P-value for each SNP. A large positive Z-score results in a small P-value providing evidence that the allele positively associated with the trait under test. Conversely, a large negative Z-score results in a small P-value, showing an allele is negatively associated with the trait39. A P-value of less than 5 × 10−5 was used to denote genome-wide significance as per recent GWAS studies22.
Validation of internationally identified RFI SNPs in Irish beef cattle
The inclusion of internationally identified RFI SNPs (n = 102, Supplementary Table S1) in the current study enabled investigation of their role as markers for feed efficiency in an Irish population of beef cattle.
Functional annotation of genes
Functional annotation of candidate genes was carried out to gain insight into the underlying biology of RFI, ADG and FI. Database for Annotation, Visualisation and Integrated Discovery (DAVID, version 6.8)40 was used for functional annotation. From the meta-analysis, a list of candidate genes was generated using Ensembl’s Variant Effect Predictor. The list contained the nearest gene within a 500 kb window to each nominally significant SNP (P < 0.05). DAVID assigned genes to pathways as per the Kyoto Encyclopaedia of Genes and Genomes (KEGG), and determined enrichment of pathways using Fisher’s exact test41. In order to account for multiple testing, a Benjamini-Hochberg correction was applied42. Pathways were deemed to be significant if they obtained a corrected P-value of <0.05. Pathways specifically addressing human diseases and disorders were not included in further analysis of DAVID identified pathways, as these were not relevant to this study.
eQTL analysis
Samples for eQTL analysis were obtained from CH and Holstein-Friesian cattle that had been genotyped as part of the current study and for which RNA-Seq data were available within our group. The RFI range of the CH and Holstein-Friesian cattle were 1.48 to −0.98 and 1.48 to −1.41 respectively. RNA-Seq raw read counts were collated from liver and muscle tissue analyses carried out by our group in published studies29,43 and studies in preparation (Higgins et al., McKenna et al.) related to feed efficiency traits. Forty-two liver samples and 39 muscle samples were brought forward to eQTL analysis. For eQTL identification, liver and muscle samples were analysed separately.
Raw read counts were filtered and genes with more than 10 instances of zero expression were removed from analysis, resulting in 14,588 and 14,309 genes with expression in the liver and muscle, respectively, remaining for eQTL analysis. Filtered raw read counts were normalised using DESeq2′s variance stabilizing transformation (VST) command44. Covariates included in DESeq2 were batch, RFI status (i.e. high or low RFI) and breed. VST normalised counts were brought forward for eQTL analysis.
eQTL analysis was carried out using the R package Matrix eQTL45. Only SNPs that reached genome-wide significance after meta-analysis (n = 24) and their nearest gene were considered for eQTL analysis. If a nearest gene was greater than 100 kilobases away from a SNP, this combination was not included in eQTL analysis. As part of eQTL analysis RFI, breed and sex were included as covariates. SNPs with a MAF of less than 0.1 or with known functions, i.e. missense mutations, were excluded, as were genes that were not expressed in either tissue of interest, i.e. liver or muscle. This resulted in 11 SNPs in the analysis. A Bonferroni correction was applied to account for the 11 SNPs. If an eQTL reached a P-value of 0.0045 or less, it was considered significant after multiple test correction.
Results
GWAS and meta-analysis for RFI
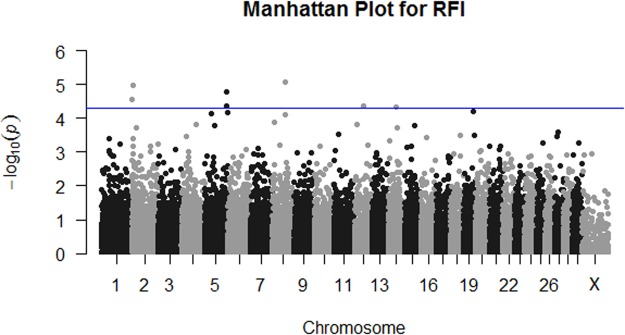
GWAS results generated for RFI by meta-analysis are plotted in Fig. 1. Seven SNPs achieved genome-wide significance for RFI (Table 1). The SNP most associated with RFI was rs43555985, located at chromosome 8 position 69,658,202, a non-coding region 53.4 kb upstream from GFRA2. Two variants were within the start-stop coordinates of genes; intronic variant rs110418027 in SMC1B and 3′ untranslated region (UTR) variant rs43691372 in DIS3. The remaining four SNPs are located in the non-coding region of the Bovine genome and their distance to nearest gene is specified in Table 1. The per breed GWAS results for each of these RFI associated variants are illustrated in Table 2 and Supplementary Table S4. Functional annotation of genes containing or near to nominally significant SNPs for RFI using DAVID did not identify any enriched pathways that survived Benjamini-Hochberg correction42.
Figure 1.
Manhattan plot represents meta-analysis results for RFI, which combined GWAS carried out for five cohorts of Irish beef cattle. The blue line indicates P-value < 5 × 10−5.
Table 1.
SNPs which reached significance (P < 5 × 10−5) in a multi-breed population of beef cattle after meta-analysis of GWAS results for each respective trait.
| SNP I.D. | Trait of interest | Chr_mb | Zscore | P-value | Nearby gene | SNP location relative to gene |
|---|---|---|---|---|---|---|
| rs43555985 | RFI | 8_69 | −4.458 | 8.28E-06 | GFRA2 | 53.4 kb upstream |
| rs41638273 | RFI | 2_6 | 4.4 | 1.08E-05 | SLC40A1 | 15.7 kb upstream |
| rs109695205 | RFI | 5_113 | 4.313 | 1.61E-05 | NFAM1 | 26.5 kb upstream |
| rs110161277 | RFI | 2_2 | 4.192 | 2.76E-05 | PLEKHB2 | 143.8 kb downstream |
| rs110418027 | RFI | 5_116 | −4.089 | 4.34E-05 | SMC1B | Intron variant |
| rs43691372 | RFI | 12_47 | 4.082 | 4.47E-05 | DIS3 | 3′ UTR variant |
| rs42820242 | RFI | 14_44 | 4.081 | 4.48E-05 | IL7 | 104.2 kb downstream |
| rs386023985 | ADG | 19_48 | −6.593 | 4.32E-11 | ERN1 | 7.9 kb upstream |
| rs135897656 | ADG | 3_119 | 6.195 | 5.83E-10 | CSF2RA | Intron variant |
| rs136457441 | ADG | 19_28 | 5.936 | 2.93E-09 | RPL26 | Missense variant |
| rs110660154 | ADG | 1_19 | 5.314 | 1.08E-07 | SPATA16 | 265 kb downstream |
| rs110780286 | ADG | 18_15 | 4.492 | 7.06E-06 | ITFG1 | Intron variant |
| rs382426807 | ADG | 19_43 | 4.473 | 7.70E-06 | STAT5A | Synonymous variant |
| rs41595251 | ADG | 9_91 | −4.375 | 1.22E-05 | OPRM1 | 269 kb upstream |
| rs110590483 | ADG | 11_39 | −4.243 | 2.21E-05 | CCDC85A | 509 kb downstream |
| rs109252082 | ADG | 19_53 | 4.124 | 3.72E-05 | TBC1D16 | Intron variant |
| rs41592667 | ADG | 9_35 | 4.12 | 3.78E-05 | FRK | 88 kb downstream |
| rs41630180 | ADG | 17_1 | −4.097 | 4.18E-05 | TLL1 | Intron variant |
| rs41614223 | ADG | 9_27 | −4.086 | 4.39E-05 | NKAIN2 | 8.8 kb downstream |
| rs137576435 | ADG | 19_12 | −4.079 | 4.52E-05 | BCAS3 | Intron variant |
| rs136789347 | ADG | 23_52 | −4.069 | 4.72E-05 | OR5M10 | 13.8 kb upstream |
| IDBV32000008978 | FI | 20_67 | 4.355 | 1.33E-05 | ADAMTS16 | Synonymous variant |
| rs55617218 | FI | 19_14 | −4.205 | 2.61E-05 | HNF1B | Intron variant |
| rs109691080 | FI | 1_6 | 4.084 | 4.43E-05 | MAP3K7CL | 58.9 kb upstream |
SNP: Single nucleotide polymorphism; RFI: Residual feed intake; ADG: Average daily gain; FI: Feed intake; Chr_mb: Chromosome_megabase.
Table 2.
Individual breed GWAS results for all genetic variants that reached genome-wide significance following meta-analysis.
| Trait | SNP ID | Meta-analysis P-value | Direction of Effect | AA P-value | BB P-value | CH P-value | LM P-value | SI P-value |
|---|---|---|---|---|---|---|---|---|
| RFI | rs43555985 | 8.28E-06 | −−−−− | 2.14E-01 | 1.18E-01 | 5.30E-01 | 1.24E-03 | 2.39E-03 |
| rs41638273 | 1.08E-05 | +++++ | 1.65E-01 | 3.16E-03 | 5.79E-03 | 9.03E-02 | 1.74E-01 | |
| rs109695205 | 1.61E-05 | +++++ | 3.02E-02 | 4.12E-02 | 1.09E-01 | 4.04E-02 | 2.35E-02 | |
| rs110161277 | 2.76E-05 | +++++ | 1.68E-01 | 5.24E-03 | 1.41E-02 | 1.50E-01 | 8.58E-02 | |
| rs110418027 | 4.34E-05 | −−−−− | 7.77E-02 | 8.29E-02 | 4.74E-02 | 3.92E-02 | 7.50E-02 | |
| rs43691372 | 4.47E-05 | +++++ | 1.74E-01 | 5.36E-02 | 6.36E-01 | 8.67E-03 | 4.65E-03 | |
| rs42820242 | 4.48E-05 | +++++ | 1.49E-01 | 2.74E-01 | 7.48E-02 | 1.06E-02 | 4.41E-02 | |
| ADG | rs386023985 | 4.32E-11 | −−−−− | 3.43E-01 | 3.82E-01 | 4.71E-04 | 1.16E-07 | 1.30E-03 |
| rs135897656 | 5.83E-10 | +++++ | 1.54E-01 | 1.70E-01 | 3.64E-07 | 1.19E-06 | 3.01E-01 | |
| rs136457441 | 2.93E-09 | +−+++ | 4.58E-01 | −9.64E-01 | 4.55E-06 | 1.88E-05 | 1.03E-02 | |
| rs110660154 | 1.08E-07 | +++++ | 3.69E-02 | −3.56E-01 | 5.20E-05 | 9.81E-02 | 5.63E-03 | |
| rs110780286 | 7.06E-06 | +++++ | 2.13E-01 | −9.79E-03 | 3.94E-01 | 9.86E-02 | 4.21E-04 | |
| rs382426807 | 7.70E-06 | +−+−+ | 5.84E-01 | −7.69E-01 | 1.50E-07 | 5.65E-06 | 6.85E-01 | |
| rs41595251 | 1.22E-05 | −−−−−− | 1.83E-01 | −9.94E-01 | 1.17E-01 | 4.61E-05 | 1.70E-02 | |
| rs110590483 | 2.21E-05 | −−−−− | 3.60E-01 | 1.69E-01 | 5.64E-03 | 5.12E-01 | 2.27E-03 | |
| rs109252082 | 3.72E-05 | +++++ | 8.27E-02 | 5.49E-01 | 1.26E-03 | 5.09E-01 | 1.07E-02 | |
| rs41592667 | 3.78E-05 | +++++ | 4.92E-01 | −6.02E-01 | 7.18E-03 | 8.14E-02 | 6.75E-03 | |
| rs41630180 | 4.18E-05 | −−−−− | 8.08E-01 | −1.87E-03 | 1.25E-01 | 6.19E-04 | 2.61E-01 | |
| rs41614223 | 4.39E-05 | −−−−− | 2.94E−01 | −7.84E-03 | 9.24E-04 | 6.27E-02 | 5.16E-01 | |
| rs137576435 | 4.52E-05 | −−−−−− | 3.95E-01 | 4.11E-01 | 2.03E-02 | 7.64E-01 | 1.66E-04 | |
| rs136789347 | 4.72E-05 | −−−−−− | 1.99E-01 | 6.66E-01 | 1.35E-01 | 1.77E-04 | 5.03E-02 | |
| FI | IDBV32000008978 | 1.33E-05 | ++++− | 6.00E−03 | 1.69E-03 | 7.68E-03 | 3.35E-02 | 8.59E-01 |
| rs55617218 | 2.61E-05 | +−−−− | 4.22E-01 | 1.75E-01 | 2.83E-01 | 1.92E-04 | 8.14E-03 | |
| rs109691080 | 4.43E-05 | −++++ | 6.89E-01 | 4.45E-01 | 1.41E-04 | 1.88E-01 | 6.53E-03 |
SNP: single nucleotide polymorphism; RFI: residual feed intake; ADG: average daily gain; Direction of effect: direction of effect of the Illumina A allele; FI: feed intake; AA: Aberdeen Angus; BB: Belgian Blue; CH: Charolais; LM: Limousin; SI: Simmental.
GWAS and meta-analysis for ADG
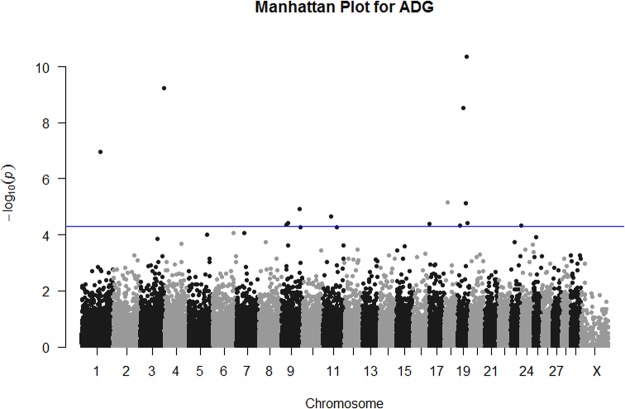
GWAS results for ADG are illustrated in Fig. 2. A total of 14 SNPs reached genome-wide significance for ADG. The most significantly associated SNP was rs386023985 which is located at chromosome 19 position 48,916,589, 7.9 kb upstream from the ERN1 gene. One missense variant, rs136457441 in RPL26, was associated with ADG. One associated variant was synonymous, rs382426807 in STAT5A. Five intronic variants were associated with ADG in the genes: CSFRA2, ITFG1, TBC1D16, TLL1 and BCAS3. The remaining 5 associated SNPs were located upstream or downstream of genes as indicated in Table 1. Individual breed GWAS results for the SNPs associated with ADG following meta-analysis are outlined in Table 2 and Supplementary Table S4.
Figure 2.
Manhattan plot of meta-analysis results for ADG. Meta-analysis was carried out on GWAS results generated for five breeds of Irish beef cattle. The blue line indicates P-value < 5 × 10−5.
Functional annotation of genes nearest to nominally significant SNPs for ADG identified 7 pathways that were significantly enriched following Benjamini-Hochberg correction42 (Table 3). The thyroid hormone signalling pathway was the most enriched pathway (corrected P = 0.01).
Table 3.
Significant KEGG pathways identified for each trait in a multi-breed population of beef cattle following meta-analysis of GWAS results.
| Trait | Biological Process | B-H P-value | Number of genes |
|---|---|---|---|
| ADG | Thyroid hormone signalling pathway | 0.010 | 20 |
| ADG | cGMP-PKG signalling pathway | 0.011 | 25 |
| ADG | Vascular smooth muscle contraction | 0.015 | 18 |
| ADG | Retrograde endocannabinoid signalling pathway | 0.013 | 18 |
| ADG | Focal adhesion | 0.027 | 27 |
| ADG | cAMP signalling pathway | 0.026 | 26 |
| ADG | Adherens junction | 0.029 | 13 |
| FI | Axon guidance | 0.001 | 23 |
| FI | Thyroid hormone signalling pathway | 0.015 | 19 |
B-H P-value: Benjamini-Hochberg corrected P-value; ADG: average daily gain; FI: feed intake. Pathways were designated as significant if they reached Benjamini-Hochberg corrected P < 0.05.
GWAS and meta-analysis for FI
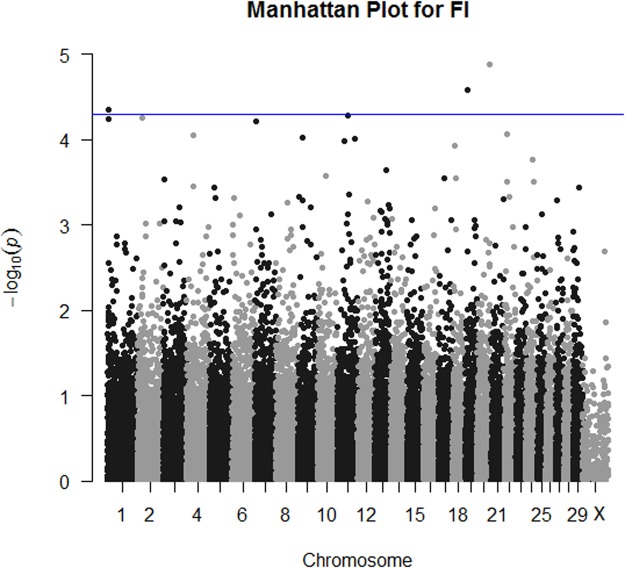
GWAS results for FI are plotted in Fig. 3. Three SNPs reached genome-wide significance for FI (Table 1). Individual breed GWAS results for these variants are presented in Table 2 and Supplementary Table S2. The SNP most associated with FI was IDBV32000008978 located at chromosome 20 position 67,944,737. This is a synonymous variant in ADAMTS16. The other two were an intronic variant in HNF1B and a variant located 58.9 kb upstream of MAP3K7CL. Functional annotation of the FI SNP results identified two pathways that were significantly enriched after correction; axon guidance and the thyroid hormone signalling pathway (Table 3).
Figure 3.
Manhattan plot of FI meta-analysis of GWAS results for Irish beef cattle. The blue line indicates P-value < 5 × 10−5.
Validation of internationally identified SNPs in an Irish cattle population
Of the 102 internationally identified RFI SNPs included on the custom IDBv3 genotyping chip, 71 passed all QC measures and were included in the GWAS for RFI in the current study. Two of these SNPs, rs29014641 and rs109500421, were nominally significant in our study but did not survive multiple test correction for this subset of SNPs. This subset of SNPs was not exhaustive for RFI and did not include all variants within quantitative trait loci (QTLs) as identified by Nkrumah, et al.18, for example. However, a post-hoc search for genotyped SNPs within those regions which found that no genetic variants within these QTLs reached genome-wide significance following meta-analysis.
eQTL analysis of SNPs identified as significant from meta-analysis
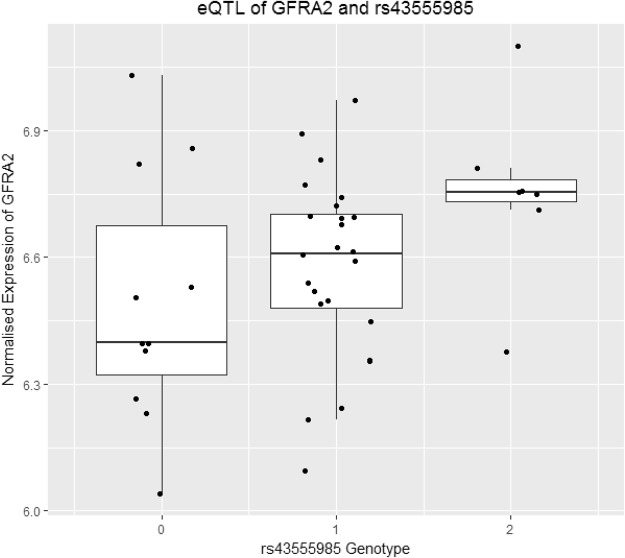
Table 4 contains results of eQTL analysis. One cis-eQTL was detected in liver, between rs43555985, the top associated SNP from the RFI GWAS, and GFRA2 (P = 0.0038; survives multiple test correction). eQTL analysis indicated that the minor allele of rs4355985 is associated with increased expression of GFRA2 (Fig. 4). The same minor allele is associated with lower RFI in the GWAS. The effect of GFRA2 expression on RFI is presented in Supplementary Fig. S5 on a per genotype basis.
Table 4.
Results from eQTL analysis of genome-wide significant SNPs in liver and muscle.
| SNP | Nearest Gene | Trait | Liver P-value | Muscle P-value |
|---|---|---|---|---|
| rs43555985 | GFRA2 | RFI | 0.0038* | 0.25 |
| rs109695205 | NFAM1 | RFI | 0.95 | 0.55 |
| rs110418027 | SMC1B | RFI | 0.15 | Not expressed |
| rs43691372 | DIS3 | RFI | 0.96 | 0.16 |
| rs386023985 | ERN1 | ADG | 0.99 | 0.23 |
| rs110780286 | ITFG1 | ADG | 0.83 | 0.23 |
| rs382426807 | STAT5A | ADG | 0.94 | 0.72 |
| rs41592667 | FRK | ADG | 0.22 | 0.19 |
| rs41630180 | TLL1 | ADG | Not expressed | 0.33 |
| rs137576435 | BCAS3 | ADG | 0.25 | 0.93 |
| rs109691080 | MAP3K7CL | FI | Not expressed | 0.80 |
SNP: Single nucleotide polymorphism; ADG: average daily gain; FI: feed intake; RFI: residual feed intake, *eQTLs were designated as significant if they reached P < 0.0045.
Figure 4.
Boxplot representing the relationship between rs43555985 genotypes and normalised liver gene-expression of GFRA2. Presence of the minor allele of rs43555985 is correlated with increased expression of GFRA2. 0: GG; 1: GA; 2: AA.
Discussion
Despite the economic and environmental benefits of RFI, the trait or indeed any measure of feed efficiency, is not widely adopted within breeding programmes for beef cattle due to the difficulty and expense associated with measuring feed intake10. The identification of robust genetic markers of RFI applicable to several breeds, as well as crossbred cattle, would enable the traits inclusion in genomic breeding programmes. This study sought to identify SNPs associated with RFI that could be applicable to Irish beef production enterprises as well as uncovering novel markers of potential use to international beef producers. To unravel the underlying biology causing phenotypic variation in feed efficiency related traits, we carried out eQTL analysis of GWAS-identified variants to study their effect on local gene expression.
rs43555985 was associated with RFI and is an eQTL of the GFRA2 gene in liver tissue. The minor allele of this SNP was associated lower RFI within each of the individual breed GWAS and following meta-analysis. The minor allele of rs43555985 was also associated with increased expression of GFRA2 following eQTL analysis. GFRA2 is a cell-surface receptor that facilitates binding of a member of the glial cell-derived neurotrophic factor family. GFRA2 knock-out mice are unable to digest food correctly, have impaired salivary secretion and gut motility and exhibit a slower growth rate than wild-type mice while having an increased basal metabolic rate46. If increased GFRA2 expression is associated with improved feed efficiency, the mechanism may involve lowering metabolic rates. Increased metabolic rate leads to increased energy requirements to carry out biological processes and to maintain physiological homeostasis, resulting in less consumed energy being used for growth47. It has been illustrated previously that high-RFI lambs have a higher basal metabolic rate than their low-RFI (more desirable) counterparts and low-RFI heifers exhibited lower metabolic rates than their high-RFI (inefficient) counterparts48,49. Further investigation and validation of rs43555985 prior to inclusion in genomic breeding programmes is required. Furthermore, rs43555985 is also located 79.9 kb upstream from XPO7. Post-hoc eQTL analysis for this gene illustrated that there is no statistically significant relationship between rs43555985 genotype and XPO7 expression.
The second most statistically significant SNP for RFI, rs41638273, maps to a region of chromosome 2 that contains the SLC40A1 gene. This region is also the site of a QTL for RFI which contains the myostatin gene50. Specific mutations in the myostatin gene have been linked with increased muscle growth traits51. Improved feed efficiency was associated with double muscled Angus steers by Cafe et al.52 when compared to lesser muscled counterparts52. DIS3, a gene nearby to a variant associated with RFI in the current study, encodes a protein involved in RNA metabolism53 and has been linked with feed conversion efficiency in pigs54.
The minor allele of rs386023985 was negatively associated with ADG following GWAS meta-analysis in the current study. Similarly, this variant was negatively associated with ADG within each individual breed GWAS conducted, this SNP also reached genome-wide significance within the LM individual breed GWAS. rs386023985 has not previously been associated with ADG or other growth traits in cattle. The gene nearest to rs386023985 is ERN1 (IRE1), a sensor of metabolic stress, is involved in the unfolded protein response55.
Copy number variation in RPL26, a ribosomal protein gene, has been linked to RFI divergence in Holstein cows56. A variant identified in this study, rs136457441, is a missense variant in RPL26 causing an isoleucine to threonine change at amino acid position 67 in the RPL26 protein. This variant, associated with ADG following meta-analysis in the current study and reached genome-wide significance for ADG within the CH individual breed GWAS, is located in exon 3 of RPL26. rs136457441 has been designated as tolerated by the Sorting Tolerant from Intolerant (SIFT) algorithm, which predicts whether amino acid substitutions effect protein function57. Further investigation into the functional effect of this mutation is required to elucidate its biological role in ADG.
An exonic variant associated with ADG is rs382426807, a synonymous variant in STAT5A. This gene encodes a transcription factor that can be activated as part of the somatotropic axis, which is the pathway involved in the secretion of growth hormone and skeletal muscle growth58. STAT5A has been associated with increased live weight gain in Polish Black-and-White bulls59 and increased expression of the growth hormone receptor, an upstream activator of STAT5A, has been previously demonstrated in efficient beef heifers by Kelly et al.60.
Five variants associated with ADG were located in introns of the following genes: TLL1, CSF2RA, ITFG1, TBC1D16 and BCAS3. TLL1 encodes a member of the tolloid family metalloproteases that have been previously implicated in the cleavage and development of myostatin in humans61. Myostatin in its normal state negatively regulates muscle growth. The production of aberrant myostatin protein isoforms results in the development of the double muscle phenotype51.
CSF2RA encodes for a granulocyte/macrophage colony stimulating factor62. ITFG1, the gene within which rs110780286 is located, is involved in T-cell differentiation and may induce the production of anti-inflammatory cytokines63. It has been previously illustrated that immune genes and immune pathways are associated with variation in feed efficiency and ADG in cattle64,65. Several groups have suggested that the immune system plays a key role in weight gain and feed efficiency in cattle. For example, Reynolds et al.64 found that steers with higher ADG have lower immunity related gene expression64 and it has been demonstrated that cattle with poor feed efficiency had increased activation of their immune system65. It is possible that cattle with poor feed efficiency and low ADG are experiencing chronic inflammation which results in poor feed efficiency which has been suggested previously by Alexandre et al.65 following analysis of beef cattle divergent in RFI and by Mani et al.66 upon investigation of inflammation in RFI divergent pigs65,66.
rs41595251 is associated with ADG and is a variant located upstream from OPRM1, the µ-opioid receptor gene, on chromosome 9. OPRM1 has been associated with increased food intake in humans67. ADG-associated SNP rs136789347 is nearby to OR5M10 which encodes for an olfactory receptor in humans68. Olfactory receptors have been suggested as one method by which the endocannabinoid system stimulates the feeding drive in mice69. rs41614223 is located downstream from the transcriptional start site of NKAIN2, which produces a Sodium-Potassium ATPase involved in action potential generation in neurons70. Each of these genes, OPRM1, OR5M10 and NKAIN2 have a neurological function. There is evidence from bovine studies71,72 that there is significant neurological control of food consumption. The association of these neurological genes with ADG in this cohort of beef cattle may further indicate that feeding behaviour in cattle may also be subject to some degree of neurological control73. Further investigation is required to investigate the role neurological systems play in modulating the development of divergent RFI and related traits in cattle.
rs41592667 is upstream of FRK which encodes for tyrosine-protein kinase FRK. Gene sets enriched for cell cycle-related genes, similar to FRK, have previously been shown to be associated with feed intake and feed efficiency in beef cattle74. TBC1D16, a gene which encodes for a GTPase and contains the intronic variant rs109252082, has been associated with growth rate in pigs75. Despite these SNPs not being associated with feed efficiency or component traits prior to the current study, they are near to, or within, genes that have been associated with feed efficiency related traits previously.
HNF1B, nearby to a variant associated with FI, has previously been identified as differentially expressed in Holstein cattle divergent for RFI76, and this gene is a target of miR-802, which has been identified as upregulated in high RFI cattle77. The silencing of HNF1B in mice leads to impaired insulin sensitivity78. However, previous work by our group has shown that RFI divergent beef cattle have similar levels of insulin sensitivity and it is unlikely that insulin sensitivity plays a role in RFI divergence28. Further work is required to understand the contribution of HNF1B to the development of divergence in FI. In this study the variant IDBV332000008978 was associated with FI. This variant is a synonymous variant within the ADAMTS16 gene which is a member of ADAMTS protease family and has previously been identified as associated with FCR in pigs79.
Following functional gene set enrichment analysis using DAVID, the thyroid hormone signalling pathway was found to be most enriched for ADG. Thyroid hormones play a key role in the regulation of basal metabolism in mammals80, although, it has been demonstrated previously that the levels of thyroid hormones are not related to RFI status in heifers81. However, in a study of dairy cattle is was reported that low levels of thyroid hormones are associated with lower RFI82. Further investigation into the role of the thyroid hormone signalling pathway is warranted to further elucidate the role this biological mechanism plays in the divergence of RFI in cattle. The retrograde endocannabinoid signalling pathway was also found to reach significance level in the list of nominal significant genes for ADG. It has been demonstrated that the endocannabinoid system plays a role in inducing food intake and modulating energy expenditure and feed intake in mice83,84. It is possible that alterations in genes in the retrograde endocannabinoid pathway may also stimulate or inhibit feeding behaviours in cattle which may impact on feed efficiency. It has been observed previously that low RFI cattle have fewer daily feeding events and have a lower eating rate than high RFI cattle85. Focal adhesion was another pathway found to be enriched for nominally significant ADG associated SNPs. Focal adhesion is a pathway involved in cell motility, proliferation and survival. This pathway is dependent upon focal adhesion kinase86. PTK2, the gene encoding for focal adhesion kinase has been previously noted as downregulated in high-RFI animals from a population of dairy cattle87.
Conclusion
In this study we illustrate genome-wide associations between SNPs and RFI and its component traits in beef cattle. In total, we identified 24 SNPs as reaching statistical significance for RFI, ADG and FI in a multi-breed cohort of beef cattle. Several of the SNPs identified in this study are located nearby or within genes related to immune function, muscle growth and development, and neurological pathways. The identification of a novel eQTL for RFI at GFRA2 also represents an insight into the biology of feed efficiency.
Due to the small sample size of our individual breed GWAS, which we used meta-analysis to overcome, all identified SNPs and the eQTL must be validated, both in larger Irish and international populations before incorporation into genomic assisted beef cattle breeding programmes. Furthermore, validation is required in larger reference populations to account for the LD and genetic heterogeneity which exists between breeds of cattle.
An additional method which may have been employed to increase sample size could have been single step GWAS (ssGWAS)88–90. ssGWAS incorporates genotypes, phenotypes and pedigree information to calculate genomic estimated breeding values for animals with or without genotypes88.
It is important to ensure that the SNPs influence these traits and have no negative impact on other economically important production traits. SNPs with a validated desirable effect can be included in Irish and international genomic assisted breeding programmes to facilitate the rapid and cost effective selection of more feed efficient beef cattle.
Electronic supplementary material
Acknowledgements
This work was funded by the IdentiFEED project (13/S/519), supported by the Irish Department of Agriculture, Food and the Marine (DAFM) under the National Development Plan 2007–2013. The authors declare no conflict of interest.
Author Contributions
Conceived and designed experiments: S.W., D.M., M.McG., D.K. Collated data: M.H., C.F. Performed analyses: M.H. Contributed to data/analyses: S.W., D.M., C.F., D.K., M.McG., M.McC., S.C., C.M. Results interpretation and preparation of the paper: M.H., S.W., D.M. All authors aided with manuscript edits. All authors agree to be accountable for all aspects of the work.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sinéad M. Waters and Derek W. Morris jointly supervised this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-32374-6.
References
- 1.Finneran E, et al. Simulation Modelling of the Cost of Producing and Utilising Feeds for Ruminants on Irish Farms. Journal of Farm Management. 2010;14:95–116. [Google Scholar]
- 2.Berry DP, Crowley JJ. Cell Biology Symposium: genetics of feed efficiency in dairy and beef cattle. Journal of Animal Science. 2013;91:1594–1613. doi: 10.2527/jas.2012-5862. [DOI] [PubMed] [Google Scholar]
- 3.Ripple WJ, et al. Ruminants, climate change and climate policy. Nature Climate Change. 2014;4:2–5. doi: 10.1038/nclimate2081. [DOI] [Google Scholar]
- 4.Fitzsimons C, Kenny D, Deighton M, Fahey A, McGee M. Methane emissions, body composition, and rumen fermentation traits of beef heifers differing in residual feed intake. Journal of Animal Science. 2013;91:5789–5800. doi: 10.2527/jas.2013-6956. [DOI] [PubMed] [Google Scholar]
- 5.Koch RM, Swiger LA, Chambers D, Gregory KE. Efficiency of feed use in beef cattle. Journal of Animal Science. 1963;22:486–494. doi: 10.2527/jas1963.222486x. [DOI] [Google Scholar]
- 6. Kenny, D. A., Fitzsimons, C., Waters, S. M. & McGee, M. Improving feed efficiency of beef cattle; current state of the art and future challenges. Animal In Press (2018). [DOI] [PubMed]
- 7.Nielsen MK, et al. Review: Life-cycle, total-industry genetic improvement of feed efficiency in beef cattle: Blueprint for the Beef Improvement Federation The Professional Animal. Scientist. 2013;29:559–565. [Google Scholar]
- 8.Cole J, VanRaden P. Possibilities in an age of genomics: The future of selection indices. Journal of Dairy Science. 2017;101:3686–3701. doi: 10.3168/jds.2017-13335. [DOI] [PubMed] [Google Scholar]
- 9.Pryce JE, et al. Hot topic: Definition and implementation of a breeding value for feed efficiency in dairy cows. Journal of Dairy Science. 2015;98:7340–7350. doi: 10.3168/jds.2015-9621. [DOI] [PubMed] [Google Scholar]
- 10.Hayes BJ, Lewin HA, Goddard ME. The future of livestock breeding: genomic selection for efficiency, reduced emissions intensity, and adaptation. Trends in Genetics. 2013;29:206–214. doi: 10.1016/j.tig.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 11.Rolf MM, et al. Genomics in the United States beef industry. Livestock Science. 2014;166:84–93. doi: 10.1016/j.livsci.2014.06.005. [DOI] [Google Scholar]
- 12.Wickham B, et al. Industrial perspective: capturing the benefits of genomics to Irish cattle breeding. Animal Production Science. 2012;52:172–179. doi: 10.1071/AN11166. [DOI] [Google Scholar]
- 13.Mullen, M. P. et al. Development of a custom SNP chip for dairy and beef cattle breeding, parentage and research. Interbull Bulletin (2013).
- 14.Inc., I. BovineSNP50 Genotyping BeadChip, https://www.illumina.com/Documents/products/datasheets/datasheet_bovine_snp5O.pdf (2016).
- 15.Rolf MM, et al. Genome-wide association analysis for feed efficiency in Angus cattle. Animal Genetics. 2012;43:367–374. doi: 10.1111/j.1365-2052.2011.02273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abo-Ismail MK, et al. Single nucleotide polymorphisms for feed efficiency and performance in crossbred beef cattle. BMC Genetics. 2014;15:14. doi: 10.1186/1471-2156-15-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherman EL, Nkrumah JD, Murdoch BM, Moore SS. Identification of polymorphisms influencing feed intake and efficiency in beef cattle. Animal Genetics. 2008;39:225–231. doi: 10.1111/j.1365-2052.2008.01704.x. [DOI] [PubMed] [Google Scholar]
- 18.Nkrumah JD, et al. Primary genome scan to identify putative quantitative trait loci for feedlot growth rate, feed intake, and feed efficiency of beef cattle. Journal of Animal Science. 2007;85:3170–3181. doi: 10.2527/jas.2007-0234. [DOI] [PubMed] [Google Scholar]
- 19.Serão NVL, et al. Single nucleotide polymorphisms and haplotypes associated with feed efficiency in beef cattle. BMC Genetics. 2013;14:94–94. doi: 10.1186/1471-2156-14-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barendse W, et al. A validated whole-genome association study of efficient food conversion in cattle. Genetics. 2007;176:1893–1905. doi: 10.1534/genetics.107.072637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexandre PA, et al. Bovine NR1I3 gene polymorphisms and its association with feed efficiency traits in Nellore cattle. Meta Gene. 2014;2:206–217. doi: 10.1016/j.mgene.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seabury CM, et al. Genome-wide association study for feed efficiency and growth traits in U.S. beef cattle. BMC Genomics. 2017;18:386. doi: 10.1186/s12864-017-3754-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber K, et al. Accuracy of genomic breeding values in multibreed beef cattle populations derived from deregressed breeding values and phenotypes. Journal of Animal Science. 2012;90:4177–4190. doi: 10.2527/jas.2011-4586. [DOI] [PubMed] [Google Scholar]
- 24.Nica A. C., Dermitzakis E. T. Expression quantitative trait loci: present and future. Philosophical Transactions of the Royal Society B: Biological Sciences. 2013;368(1620):20120362–20120362. doi: 10.1098/rstb.2012.0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fink, T. et al. Functional confirmation of PLAG1 as the candidate causative gene underlying major pleiotropic effects on body weight and milk characteristics. Scientific Reports (2017). [DOI] [PMC free article] [PubMed]
- 26.Littlejohn MD, et al. Sequence-based Association Analysis Reveals an MGST1 eQTL with Pleiotropic Effects on Bovine Milk Composition. Scientific Reports. 2016;6:25376. doi: 10.1038/srep25376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brand B, et al. Adrenal cortex expression quantitative trait loci in a German Holstein × Charolais cross. BMC Genetics. 2016;17:135. doi: 10.1186/s12863-016-0442-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fitzsimons C, Kenny DA, Waters SM, Earley B, McGee M. Effects of phenotypic residual feed intake on response to a glucose tolerance test and gene expression in the insulin signaling pathway in longissimus dorsi in beef cattle. Journal of Animal Science. 2014;92:4616–4631. doi: 10.2527/jas.2014-7699. [DOI] [PubMed] [Google Scholar]
- 29.Keogh K, Kenny DA, Cormican P, Kelly AK, Waters SM. Effect of dietary restriction and subsequent re-alimentation on the transcriptional profile of hepatic tissue in cattle. BMC Genomics. 2016;17:244. doi: 10.1186/s12864-016-2578-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coyle S., Fitzsimons C., Kenny D. A., Kelly A. K., McGee M. 1482 Repeatability of feed efficiency in steers offered a high-concentrate diet. Journal of Animal Science. 2016;94(suppl_5):719–719. [Google Scholar]
- 31.Clarke A, et al. Intake, growth and carcass traits in male progeny of sires differing in genetic merit for beef production. Animal. 2009;3:791–801. doi: 10.1017/S1751731109004200. [DOI] [PubMed] [Google Scholar]
- 32.Keogh K, Waters S, Kelly A, Kenny D. Feed restriction and subsequent realimentation in Holstein Friesian bulls: I. Effect on animal performance; muscle, fat, and linear body measurements; and slaughter characteristics. Journal of Animal Science. 2015;93:3578–3589. doi: 10.2527/jas.2014-8470. [DOI] [PubMed] [Google Scholar]
- 33.Lawrence P, Kenny D, Earley B, McGee M. Grazed grass herbage intake and performance of beef heifers with predetermined phenotypic residual feed intake classification. Animal. 2012;6:1648–1661. doi: 10.1017/S1751731112000559. [DOI] [PubMed] [Google Scholar]
- 34.Team, R. C. R: A language and environment for statistical computing., http://www.r-project.org/ (2014).
- 35.Kelly AK, et al. mRNA expression of genes regulating oxidative phosphorylation in the muscle of beef cattle divergently ranked on residual feed intake. Physiological Genomics. 2011;43:12–23. doi: 10.1152/physiolgenomics.00213.2009. [DOI] [PubMed] [Google Scholar]
- 36.Sargolzaei M, Chesnais JP, Schenkel FS. A new approach for efficient genotype imputation using information from relatives. BMC Genomics. 2014;15:478. doi: 10.1186/1471-2164-15-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laurie CC, et al. Quality control and quality assurance in genotypic data for genome‐wide association studies. Genetic Epidemiology. 2010;34:591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nature Genetics. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang DW, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biology. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 43.Keogh K, et al. Effect of Dietary Restriction and Subsequent Re-Alimentation on the Transcriptional Profile of Bovine Skeletal Muscle. PLoS One. 2016;11:e0149373. doi: 10.1371/journal.pone.0149373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossi J, et al. Alimentary tract innervation deficits and dysfunction in mice lacking GDNF family receptor α2. Journal of Clinical Investigation. 2003;112:707–716. doi: 10.1172/JCI200317995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herd RM, Arthur PF. Physiological basis for residual feed intake. Journal of Animal Science. 2009;87:E64–71. doi: 10.2527/jas.2008-1345. [DOI] [PubMed] [Google Scholar]
- 48.Zhang X, et al. Association of residual feed intake with growth and slaughtering performance, blood metabolism, and body composition in growing lambs. Scientific Reports. 2017;7:12681. doi: 10.1038/s41598-017-13042-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gonano CV, et al. The relationship between feed efficiency and the circadian profile of blood plasma analytes measured in beef heifers at different physiological stages. Animal. 2014;8:1684–1698. doi: 10.1017/S1751731114001463. [DOI] [PubMed] [Google Scholar]
- 50.Saatchi Mahdi, Schnabel Robert D, Taylor Jeremy F, Garrick Dorian J. Large-effect pleiotropic or closely linked QTL segregate within and across ten US cattle breeds. BMC Genomics. 2014;15(1):442. doi: 10.1186/1471-2164-15-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grobet L, et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nature Genetics. 1997;17:71–74. doi: 10.1038/ng0997-71. [DOI] [PubMed] [Google Scholar]
- 52.Cafe L, McKiernan W, Robinson D. Selection for increased muscling improved feed efficiency and carcass characteristics of Angus steers. Animal Production Science. 2014;54:1412–1416. [Google Scholar]
- 53.Dziembowski A, Lorentzen E, Conti E, Séraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nature Structural and Molecular Biology. 2007;14:15. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 54.Horodyska J, Hamill RM, Varley PF, Reyer H, Wimmers K. Genome-wide association analysis and functional annotation of positional candidate genes for feed conversion efficiency and growth rate in pigs. PLoS One. 2017;12:e0173482. doi: 10.1371/journal.pone.0173482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shan B, et al. The metabolic ER stress sensor IRE1 [alpha] suppresses alternative activation of macrophages and impairs energy expenditure in obesity. Nature Immunology. 2017;18:519–529. doi: 10.1038/ni.3709. [DOI] [PubMed] [Google Scholar]
- 56.Hou Y, et al. Analysis of copy number variations in Holstein cows identify potential mechanisms contributing to differences in residual feed intake. Functional & Integrative Genomics. 2012;12:717–723. doi: 10.1007/s10142-012-0295-y. [DOI] [PubMed] [Google Scholar]
- 57.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 58.Renaville R, Hammadi M, Portetelle D. Role of the somatotropic axis in the mammalian metabolism. Domestic Animal Endocrinology. 2002;23:351–360. doi: 10.1016/S0739-7240(02)00170-4. [DOI] [PubMed] [Google Scholar]
- 59.Oprządek J, Flisikowski K. Polymorphisms at loci of leptin (LEP), Pit1 and STAT5A and their association with growth, feed conversion. Animal Science Papers and Reports. 2003;21:135–145. [Google Scholar]
- 60.Kelly A, et al. Expression of key genes of the somatotropic axis in longissimus dorsi muscle of beef heifers phenotypically divergent for residual feed intake. Journal of Animal Science. 2013;91:159–167. doi: 10.2527/jas.2012-5557. [DOI] [PubMed] [Google Scholar]
- 61.Lee S-J. Genetic Analysis of the Role of Proteolysis in the Activation of Latent Myostatin. PLoS One. 2008;3:e1628. doi: 10.1371/journal.pone.0001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suzuki T, et al. Familial pulmonary alveolar proteinosis caused by mutations in CSF2RA. Journal of Experimental Medicine. 2008;205:2703–2710. doi: 10.1084/jem.20080990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fiscella M, et al. TIP, a T-cell factor identified using high-throughput screening increases survival in a graft-versus-host disease model. Nature Biotechnology. 2003;21:302. doi: 10.1038/nbt797. [DOI] [PubMed] [Google Scholar]
- 64.Reynolds J. G., Foote A. P., Freetly H. C., Oliver W. T., Lindholm-Perry A. K. Relationships between inflammation- and immunity-related transcript abundance in the rumen and jejunum of beef steers with divergent average daily gain. Animal Genetics. 2017;48(4):447–449. doi: 10.1111/age.12546. [DOI] [PubMed] [Google Scholar]
- 65.Alexandre PA, et al. Liver transcriptomic networks reveal main biological processes associated with feed efficiency in beef cattle. BMC Genomics. 2015;16:1073. doi: 10.1186/s12864-015-2292-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mani V, et al. Intestinal integrity, endotoxin transport and detoxification in pigs divergently selected for residual feed intake. Journal of Animal Science. 2013;91:2141–2150. doi: 10.2527/jas.2012-6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davis CA, et al. Dopamine for “wanting” and opioids for “liking”: a comparison of obese adults with and without binge eating. Obesity. 2009;17:1220–1225. doi: 10.1038/oby.2009.52. [DOI] [PubMed] [Google Scholar]
- 68.Tobar H, Moreno P, Vélez P. Highly conserved regions in the 5′ region of human olfactory receptor genes. Genetic and Molecular Research. 2009;8:117–128. doi: 10.4238/vol8-1gmr550. [DOI] [PubMed] [Google Scholar]
- 69.Soria-Gómez E, et al. The endocannabinoid system controls food intake via olfactory processes. Nature Neuroscience. 2014;17:407–415. doi: 10.1038/nn.3647. [DOI] [PubMed] [Google Scholar]
- 70.Gorokhova S, Bibert S, Geering K, Heintz N. A novel family of transmembrane proteins interacting with β subunits of the Na,K-ATPase. Human Molecular Genetics. 2007;16:2394–2410. doi: 10.1093/hmg/ddm167. [DOI] [PubMed] [Google Scholar]
- 71.Perkins S, et al. Residual feed intake studies in Angus-sired cattle reveal a potential role for hypothalamic gene expression in regulating feed efficiency. Journal of Animal Science. 2014;92:549–560. doi: 10.2527/jas.2013-7019. [DOI] [PubMed] [Google Scholar]
- 72.Alam T, Bahar B, Waters SM, McGee M, Sweeney T. Analysis of multiple polymorphisms in the bovine neuropeptide Y5 receptor gene and structural modelling of the encoded protein. Molecular Biology Reports. 2012;39:4411–4421. doi: 10.1007/s11033-011-1229-9. [DOI] [PubMed] [Google Scholar]
- 73.Fitzsimons, C., McGee, M., Waters, S. M. & Kenny, D. A. In Biology of Domestic Animals (eds Scanes, C. G. & Hill, R. A.) Ch. 6, (CRC Press. Boca Raton, 2017).
- 74.Sherman EL, Nkrumah JD, Moore SS. Whole genome single nucleotide polymorphism associations with feed intake and feed efficiency in beef cattle. Journal of Animal Science. 2010;88:16–22. doi: 10.2527/jas.2008-1759. [DOI] [PubMed] [Google Scholar]
- 75.Puig-Oliveras A, et al. A Co-Association Network Analysis of the Genetic Determination of Pig Conformation, Growth and Fatness. PLoS One. 2014;9:e114862. doi: 10.1371/journal.pone.0114862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xi YM, Yang Z, Wu F, Han ZY, Wang GL. Gene expression profiling of hormonal regulation related to the residual feed intake of Holstein cattle. Biochemical and Biophysical Research Communications. 2015;465:19–25. doi: 10.1016/j.bbrc.2015.07.092. [DOI] [PubMed] [Google Scholar]
- 77.Al-Husseini W, et al. Characterization and profiling of liver microRNAs by RNA-sequencing in cattle divergently selected for residual feed intake. Asian-Australasian Journal of Animal Sciences. 2016;29:1371. doi: 10.5713/ajas.15.0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kornfeld J-W, et al. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature. 2013;494:111–115. doi: 10.1038/nature11793. [DOI] [PubMed] [Google Scholar]
- 79.Sahana G, Kadlecová V, Hornshøj H, Nielsen B, Christensen OF. A genome-wide association scan in pig identifies novel regions associated with feed efficiency trait. Journal of Animal Science. 2013;91:1041–1050. doi: 10.2527/jas.2012-5643. [DOI] [PubMed] [Google Scholar]
- 80.Mullur R, Liu Y-Y, Brent GA. Thyroid hormone regulation of metabolism. Physiological Reviews. 2014;94:355–382. doi: 10.1152/physrev.00030.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kelly AK, et al. Repeatability of feed efficiency, carcass ultrasound, feeding behavior, and blood metabolic variables in finishing heifers divergently selected for residual feed intake. Journal of Animal Science. 2010;88:3214–3225. doi: 10.2527/jas.2009-2700. [DOI] [PubMed] [Google Scholar]
- 82.Dechow C.D., Baumrucker C.R., Bruckmaier R.M., Blum J.W. Blood plasma traits associated with genetic merit for feed utilization in Holstein cows. Journal of Dairy Science. 2017;100(10):8232–8238. doi: 10.3168/jds.2016-12502. [DOI] [PubMed] [Google Scholar]
- 83.Jo Y-H, Chen Y-JJ, Chua SC, Jr., Talmage DA, Role LW. Integration of Endocannabinoid and Leptin Signaling in an Appetite-Related Neural Circuit. Neuron. 2005;48:1055–1066. doi: 10.1016/j.neuron.2005.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Matias I, Di Marzo V. Endocannabinoids and the control of energy balance. Trends in Endocrinology &. Metabolism. 2007;18:27–37. doi: 10.1016/j.tem.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 85.Kelly AK, et al. Effect of divergence in residual feed intake on feeding behavior, blood metabolic variables, and body composition traits in growing beef heifers. Journal of Animal Science. 2010;88:109–123. doi: 10.2527/jas.2009-2196. [DOI] [PubMed] [Google Scholar]
- 86.Zachary I. Focal adhesion kinase. The International Journal of Biochemistry & Cell Biology. 1997;29:929–934. doi: 10.1016/S1357-2725(97)00008-3. [DOI] [PubMed] [Google Scholar]
- 87.Salleh MS, et al. RNA-Seq transcriptomics and pathway analyses reveal potential regulatory genes and molecular mechanisms in high- and low-residual feed intake in Nordic dairy cattle. BMC Genomics. 2017;18:258. doi: 10.1186/s12864-017-3622-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang H, et al. Genome-wide association mapping including phenotypes from relatives without genotypes in a single-step (ssGWAS) for 6-week body weight in broiler chickens. Frontiers in Genetics. 2014;5:134. doi: 10.3389/fgene.2014.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang H, Misztal I, Aguilar I, Legarra A, Muir WM. Genome-wide association mapping including phenotypes from relatives without genotypes. Genetic Research (Camb) 2012;94:73–83. doi: 10.1017/S0016672312000274. [DOI] [PubMed] [Google Scholar]
- 90.Lu Y, et al. Genome-wide association analyses based on a multiple-trait approach for modeling feed efficiency. Journal of Dairy Science. 2018;101:3140–3154. doi: 10.3168/jds.2017-13364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.