Abstract
The ethnic drug Melastoma dodecandrum Lour. (MDL) is widely distributed throughout South China, and is the major component of Gong Yan Ping Tablets/Capsules and Zi Di Ning Xue San. Although the pharmacological effects of MDL have been well documented, its chemical profile has not been fully determined. In this study, we have developed a rapid and sensitive UPLC-ESI-Q-Exactive Focus-MS/MS method to characterize the chemical constituents of MDL in the positive and negative ionization modes. A comparison of the chromatographic and spectrometric data obtained using this method with data from databases, the literature and reference standards allowed us to identify or tentatively characterize 109 compounds, including 26 fatty acids, 26 organic acids, 33 flavonoids, six tannins, 10 triterpenoids, two steroids and six other compounds. Notably, 55 of the compounds characterized in this study have never been detected before in this plant. The information obtained in this study therefore enriches our understanding of the chemical composition of MDL and could be used in quality control, pharmacological research and the development of drugs based on MDL. In addition, this study represents the first reported comprehensive analysis of the chemical constituents of MDL.
Keywords: Melastoma dodecandrum Lour., ultra performance liquid chromatography, mass spectrometry, chemical constituents, identification
1. Introduction
Traditional Chinese medicine (TCM) has been used for thousands of years to treat a variety of different diseases. Based on its theoretical therapeutic efficacy and wide range of clinical applications, TCM has received considerable interest from healthcare professionals, as well as those working towards the identification of new therapeutic agents for commercialization. In contrast to the pharmacological characteristics of single agent drugs, multicomponent drugs can exhibit synergistic pharmacological effects, through a “network” approach, where multiple compounds interact with multiple targets, pharmacokinetic or physicochemical synergisms in vivo with interdependent activities to achieve an improved optimal effect [1,2,3]. It is therefore essential to evaluate the chemical composition of each TCM, so that this information can be used to support further studies, such as drug effect, toxicity and metabolism studies.
Melastoma dodecandrum Lour. (MDL) is extensively distributed throughout the southern provinces of China, including Guizhou, Fujian, Zhejiang, Jiangxi and Yunnan. This plant is widely used for its medicinal properties by the Yao, Miao and She people, as well as several other minority groups. Modern pharmacological studies have shown that MDL exhibits several biological effects, including antihypoglycemic, hemostatic, analgesic, anti-inflammatory, blood lipid reducing, antioxidant and liver protection properties [4]. MDL has been used to treat a variety of different ailments, including dysmenorrhea, postpartum abdominal pain, metrorrhagia, leucorrhea, hematochezia, dysentery, carbuncle swollen and boils. To date, only 76 compounds have been isolated from MDL, including organic acids, flavonoids, triterpenoids and steroids [5,6,7,8,9,10,11,12,13]. However, much of the chemical composition of MDL remains unknown, making it difficult to rationalize its bioactivity or evaluate the safety of this material as a therapeutic agent. There is therefore an urgent need to develop an analytical method capable of determining the chemical composition of MDL. With this in mind, the aim of the current study was to establish a rapid and sensitive method for identifying the constituents of MDL. In this study, we used a Q-Exactive Focus MS/MS method to obtain high-resolution mass spectra of the different components. This method was proven to be an advanced, accurate and reliable tool for the comprehensive identification of compounds belonging to a wide range of structural classes [14,15,16,17,18,19,20,21]. Using this method, we tentatively identified a total of 109 compounds, highlighting the efficiency and accuracy of this new technique.
2. Results and Discussion
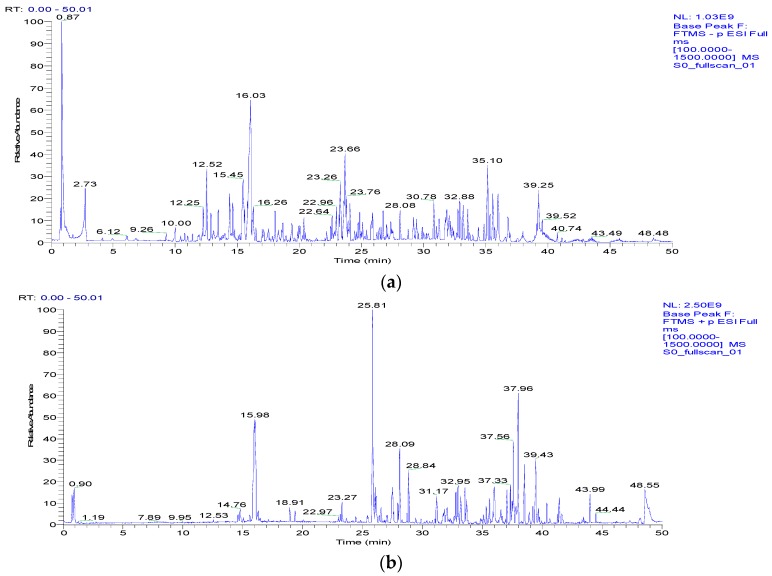
Melastoma dodecandrum Lour. was analyzed in the positive and negative ionization modes using a Q Exactive Focus mass spectrometer, and the base peak chromatogram (BPC) chromatograms for both of these ESI modes are shown in Figure 1. Some of the compounds found in this study were identified based on a comparison of their analytical data (i.e., retention times and high-resolution mass spectra) with those of several reference standards. Thus compounds 10, 34, 35, 46, 68, 79, and 84 were unambiguously identified as gallic acid, luteolin, kaempferide, quercetin, oleanic acid, asiatic acid, and rutin, repectively. Moreover, the fragmentation patterns and pathways of the standards helped further confirm the structures of the derivatives of the reference compounds. Compounds without reference standards were identified by determining the elemental compositions of the precursor and product ions. The molecular formula and rational fragmentation patterns and pathways of these compounds were then identified based on a comparison of these data with chemical databases and the literature as described below in Section 3.5. In this way, we used a UPLC-ESI-Q-Exactive Focus-MS/MS method in combination with available standards, databases and literature data to characterize 109 compounds from MDL. Seven of these compounds were unambiguously identified based on a comparison with the corresponding reference standards. Data for all of these compounds are summarized in Table 1.
Figure 1.
Base peak chromatogram of Melastoma dodecandrum Lour. in negative ion mode (a) and positive ion mode (b) using UPLC-ESI-Q-Exactive Focus-MS/MS.
Table 1.
Tentative identification of the chemical constituents of Melastoma dodecandrum Lour. by UPLC-ESI-Q-Exactive Focus-MS/MS in negative and positive modes.
| No. | Tentative Compound | tR (min) | Molecular Formula | Measured m/z | m/z Error in ppm | MS/MS (m/z) | Type of Compounds |
|---|---|---|---|---|---|---|---|
| 1 | Malic acid | 0.99 | C4H6O5 | 133.01422 [M−H]− |
−0.19 | 115.00355 [M − H − H2O]−
71.0138 [C3H3O2]− 89.02435 [M − H − CO2]− |
A |
| 2 | Salicylic acid | 9.34 | C7H6O3 | 137.02438 [M − H]− |
−0.25 | 93.03443 [M – H − CO2]− | B |
| 3 | m-Salicylic acid | 10.00 | C7H6O3 | 137.02437 [M − H]− |
−0.36 | 93.03445 [M – H − CO2]− | B |
| 4 | Citramalic acid | 1.41 | C5H8O5 | 147.02991 [M − H]− |
0.06 | 129.01924 [M – H − H2O]−
101.02427 [M – H − HCOOH]− 85.02937 [C4H5O2]− |
A |
| 5 | Protocatechuic acid | 5.55 | C7H6O4 | 153.01932 [M − H]− |
−0.09 | 109.02937 [M – H − CO2]− | B |
| 6 | Gentisic acid | 9.51 | C7H6O4 | 153.01932 [M − H]− |
−0.09 | 109.02943 [M − H − CO2]−
108.02159 [C6H4O2]− |
B |
| 7 | Pimelic acid | 13.87 | C7H12O4 | 159.0663 [M − H]− |
0.11 | 115.07629 [M – H − CO2]−
97.06577 [C6H9O]− 141.0556 [M − H − H2O]− |
A |
| 8 | Coumaric acid | 13.94 | C9H8O3 | 163.04005 [M − H]− |
−0.08 | 119.05009 [M − H − CO2]− | B |
| 9 # | Vanillic acid | 11.72 | C8H8O4 | 167.03494 [M − H]− |
−0.23 | 152.0114 [M − H − CH3]−
123.045 [M − H − CO2]− 108.02161 [C6H4O2]− |
B |
| 10 *,# | Gallic acid | 2.23 | C7H6O5 | 169.01416 [M − H]− |
−0.51 | 125.02429 [M − H − CO2]−
97.0294 [C5H5O2]− 81.03452 69.03456 [C4H5O]− |
B |
| 11 | Shikimic acid | 1.15 | C7H10O5 | 173.04547 [M − H]− |
−0.44 | 155.03479 [M − H − H2O]−
137.02423 [M – H − 2H2O]− 111.04502 [M − H − H2O − CO2]− 93.03445 [C6H5O]− 73.02941 [C3H5O2]− |
B |
| 12 | 2-Isopropylmalic acid | 11 | C7H12O5 | 175.06120 [M − H]− |
0.03 | 157.05048 [M − H − H2O]−
115.03992 [C5H7O3]− 113.06075 [C6H9O2]− 85.0658 [C5H9O]− |
B |
| 13 | Glucose | 0.89 | C6H12O6 | 179.05605 [M − H]− |
−0.37 | 59.01379 [C2H3O2]−
71.01382 [C3H3O2]− 89.02422 [C3H5O3]− 101.02422 [C4H5O3]− |
F |
| 14 | 2-Hydroxy-3-(2-hydroxyphenyl)propanoic acid | 9.78 | C9H10O4 | 181.05064 [M − H]− |
0.06 | 163.03993 [M − H − H2O]−
135.04512 [M – H − HCOOH]− 119.0501 [M − H − H2O − CO2]− |
B |
| 15 # | Methyl gallate | 10.88 | C8H8O5 | 183.02988 [M − H]− |
−0.11 | 168.00624 [M − H − CH3]−
140.0114 [M − H − CH3 − CO]− 124.01643 [C6H4O3]− |
B |
| 16 | Citric acid | 0.99 | C6H8O7 | 191.01971 [M − H]− |
−0.06 | 111.00864 [M − H − H2O – COOH − OH]−
87.00864 [C3H3O3]− 129.01915 [M − H − H2O − CO2]− 85.02939 |
A |
| 17 # | Ferulic acid | 18.04 | C10H10O4 | 193.05052 [M − H]− |
−0.57 | 178.02702 [M − H − CH3]−
149.06059 [M − H − CO2]− 134.03719 [M − H − CH3 − CO2]− |
B |
| 18 # | Vanillylmandelic acid | 14.47 | C9H10O5 | 197.04573 [M − H]− |
0.93 | 153.05563 [M − H − CH3]−
138.03203 [M − H − CH3-CO2]− 121.0294 [M − H − CH3 − CO2 − OH]− |
B |
| 19 | Sebacic acid | 20.14 | C10H18O4 | 201.11327 [M − H]− |
0.17 | 183.10249 [M − H − H2O]−
139.11273 [M − H − H2O − CO2]− |
A |
| 20 | 1-Oxo-1,2,4-butanetricarboxylic acid | 1.48 | C7H8O7 | 203.01970 [M − H]− |
−0.13 | 141.01923 [M − H − H2O − CO2]−
97.02934 [M − H − H2O − 2CO2]− 69.03453 [C4H5O]− |
A |
| 21 | Undecanedioic acid | 22.12 | C11H20O4 | 215.12874 [M − H]− |
−0.67 | 197.11826 [M − H − H2O]−
153.12842 [M − H − H2O − CO2]− |
A |
| 22 | 2-Hydroxysebacic acid | 16.5 | C10H18O5 | 217.10825 [M − H]− |
0.46 | 199.09734 [M − H − H2O]−
171.10257 [M – H − HCOOH]− 155.10768 [M − H − H2O − CO2]− |
A |
| 23 | Glucoheptonic acid | 0.82 | C7H14O8 | 225.06168 [M − H]− |
0.38 | 179.05602 [C6H11O6]−
161.04546 [C6H9O5]− 87.00864 [C3H3O3]− |
A |
| 24 | Traumatic Acid | 22.97 | C12H20O4 | 227.12894 [M − H]− |
0.24 | 183.13876 [M − H − CO2]−
165.12823 [M − H − H2O − CO2]− |
A |
| 25 | 1-O-galloyl-glycerol | 8.17 | C10H12O7 | 243.05095 [M − H]− |
−0.32 | 169.01408 [M − H − CO2 − 2CH3]−
125.02431 [M – H − 2CO2 − 2CH3]− |
B |
| 26 | Oxododecanedioic acid | 18.35 | C12H20O5 | 243.12381 [M − H]− |
0.05 | 225.1131 [M − H − H2O]−
207.10254 [M − H − H2O]− 181.1234 [C11H17O2]− |
A |
| 27 | Palmitic acid | 39.09 | C16H32O2 | 255.23297 [M − H]− |
0.07 | 237.06160 [M − H − H2O]− | A |
| 28 | Abscisic acid | 19.75 | C15H20O4 | 263.12885 [M − H]− |
−0.12 | 219.13887 [M − H − CO2]−
204.11528 [M − H − CO2 − CH3]− 151.07634 [C9H11O2]− |
B |
| 29 # | Apigenin | 19.79 | C15H10O5 | 269.04559 [M − H]− |
0.17 | 117.03447 [C8H5O]−
151.00354 [C7H3O4]− 107.01373 [C6H3O2]− |
C |
| 30 # | Naringenin | 21.35 | C15H12O5 | 271.06122 [M − H]− |
0.08 | 177.01917 [C9H5O4]−
151.00352 [C7H3O4]− 119.05009 [C8H7O]− |
C |
| 31 | Hydroxyhexadecanoic acid | 36.77 | C16H32O3 | 271.22797 [M − H]− |
0.36 | 225.22221 [M – H − HCOOH]− | A |
| 32 | Oleic acid | 39.49 | C18H34O2 | 281.24860 [M − H]− |
−0.01 | 237.06163 [M − H − CO2]− | A |
| 33 | Stearic acid | 41.08 | C18H36O2 | 283.26425 [M − H]− |
−0.01 | 265.14810 [M − H − H2O]−
237.06181 [M – H − HCOOH]− |
A |
| 34 *,# | Kaempferol | 19.95 | C15H10O6 | 285.04047 [M − H]− |
0.02 | 257.04535 [M − H − CO]−
241.05112 [M − H − CO2]− 151.00352 [C7H3O4]− 133.02942 [C8H5O2]− |
C |
| 35 *,# | Luteolin | 21.79 | C15H10O6 | 285.04062 [M − H]− |
0.56 | 239.03464 [M − H − CO − H2O]−
185.06078 [C12H9O2]− 159.04491 [C10H7O2]− 93.03454 [C6H5O]− |
C |
| 36 | Hexadecanedioic acid | 30.38 | C16H30O4 | 285.20709 [M − H]− |
−0.14 | 267.19635 [M − H − H2O]−
223.20653 [M − H − H2O − CO2]− |
A |
| 37 | 3,5-Dihydroxy-hexadecanoic acid | 23.84 | C16H32O4 | 287.22278 [M − H]− |
−0.01 | 269.21246 [M − H − H2O]−
241.21735 [M − H − H2O − CO2]− |
A |
| 38 | Epicatechin | 11.49 | C15H14O6 | 289.07193 [M − H]− |
0.58 | 245.0816 [M − H − CO2]−
203.07123 [C12H11O3]− 137.02423 [C7H5O3]− 109.02934 [C6H5O2]− |
C |
| 39 | Catechin | 13.27 | C15H14O6 | 289.07184 [M − H]− |
0.27 | 245.0089 [M – H − C3H8]−
217.01398 [M − H − C3H8 − CO]− 189.01923 [M − H − C3H8 − 2CO]− 173.0242 [C10H5O3]− 145.0294 [M – H − C3H8 − 2CO − CO2]− |
C |
| 40 | 4,9-Dihydroxy-6,7-dimethoxynaphtho(2,3-d)-1,3-dioxole-5,8-dione | 14.56 | C13H10O8 | 293.03040 [M − H]− |
0.36 | 249.04028 [M − H − CO2]−
225.11308 [C12H17O4]− 162.0321 [M − H − CO2 − CH3 − CO]− |
F |
| 41 | 9-Hode | 32.28 | C18H32O3 | 295.22791 [M − H]− |
0.12 | 277.21716 [M − H − H2O]−
171.1026 [C9H15O3]− |
A |
| 42 | Ricinoleic acid | 32.63 | C18H34O3 | 297.24353 [M − H]− |
0.0 | 183.13885 [C11H19O2]− | A |
| 43 | 2-Glucopyranosyloxybenzoic acid | 9.95 | C13H16O8 | 299.07730 [M − H]− |
0.2 | 137.02425 [M − H − Glc]−
93.03445 [M − H − glc − CO2]− |
B |
| 44 | Hydroxystearic acid | 39.09 | C18H36O3 | 299.25919 [M − H]− |
0.06 | 253.25348 [M − H − HCOOH]−
225.22246 [C15H29O]− |
A |
| 45 # | Ellagic acid | 15.56 | C14H6O8 | 300.99899 [M − H]− |
0 | 283.99619 [M − H − OH]−
245.009 [C12H5O6]− 229.01402 [M − H − CO2 − CO]− 201.01927 [M − H − CO2 − 2CO]− 185.02431 [C11H5O3]− |
D |
| 46 *,# | Quercetin | 19.95 | C15H10O7 | 301.03534 [M − H]− |
−0.12 | 178.99843 [C8H3O5]−
151.00351 [M – H − C6H6 − CO2 − CO]− 107.01373 [M – H − C6H6 − 2CO2 − CO]− |
C |
| 47 | Gallocatechin | 11.12 | C15H14O7 | 305.06683 [M − H]− |
0.52 | 261.0766 [M − H − CO2]−
219.06612 [C12H11O4]− 167.03488 [C8H7O4]− 137.02432 [C7H5O3]− 125.02433 [C6H5O3]− |
C |
| 48 | Eicosanoic acid | 42.84 | C20H40O2 | 311.29562 [M − H]− |
0.22 | 293.17941 [M − H2O]− | A |
| 49 | Glucovanillin | 10.72 | C14H18O8 | 313.09308 [M − H]− |
0.6 | 161.04539 [C6H9O5]−
113.02431 [C5H5O3]− 101.02427 [C4H5O3]− 71.01381 [C3H3O2]− |
F |
| 50 | Octadecanedioic acid | 29.15 | C18H34O4 | 313.23849 [M − H]− |
0.2 | 295.2272 [M − H − H2O]− | A |
| 51 # | 2,3,8-Trihydroxy-7-methoxychromeno[5,4,3-cde]chromene-5,10-dione | 21.54 | C15H8O8 | 315.01489 [M − H]− |
0.80 | 300.99841 [C14H5O8]−
269.10269 161.04578 [C6H9O5]− 71.01388 [C3H3O2]− |
D |
| 52 | Dihydroxystearic acid | 30.79 | C18H36O4 | 315.25403 [M − H]− |
−0.17 | 297.24344 [M − H − H2O]−
201.11363 [M − H − C8H18]− |
A |
| 53 | Digallate | 11.39 | C14H10O9 | 321.02530 [M − H]− |
0.29 | 169.01404 [C7H5O5]−
125.02428 [C6H5O3]− |
B |
| 54 | 2,3-Di-O-methylellagic acid | 20.25 | C16H10O8 | 329.03040 [M − H]− |
0.32 | 314.00681 [M − H − CH3]−
298.98325 [C14H3O8]− 270.98834 [M − H − 2CH3 − CO]− |
D |
| 55 | Woodorien | 9.46 | C14H18O9 | 329.08795 [M − H]− |
0.44 | 167.03482 [M − H − Glc]−
152.01135 [M − H − Glc − CH3]− 121.0294 [M − H – Glc − HCOOH]− 108.02159 [M − H − Glc − CH3 − CO2]− |
B |
| 56 | Galloylglucose | 3.25 | C13H16O10 | 331.06729 [M − H]− |
0.67 | 271.04575 [C11H11O8]−
211.02464 [C9H7O6]− 169.01405 [M − H − Glc]− 125.02432 [C6H5O3]− |
B |
| 57 | Caffeic acid-3-glucoside | 11.08 | C15H18O9 | 341.08798 [M − H]− |
0.52 | 305.06638 [M – H − 2H2O]−
281.06647 [C13H13O7]− 251.05588 [C12H11O6]− 221.04532 [C11H9O5]− 179.03485 [M − H − Glc]− 135.04509 [M − H − Glc − CO2]− |
B |
| 58 | 2-Hydroxy-3,7,8-trimethoxychromeno[5,4,3-cde]chromene-5,10-dione | 23.33 | C17H12O8 | 343.04590 [M − H]− |
−0.13 | 328.0224 [M − H − CH3]−
312.99899 [M – H − 2CH3]− 297.97522 [M − H − 3CH3]− 269.98053 [M − H − 3CH3 − CO]− |
D |
| 59 | Theogallin | 6.68 | C14H16O10 | 343.06720 [M − H]− |
0.38 | 169.01408 [C7H5O5]−
125.02427 [C6H5O3]− |
B |
| 60 | Chlorogenic acid | 9.88 | C16H18O9 | 353.08813 [M − H]− |
0.93 | 191.05602 [C7H11O6]−
179.0349 [C9H7O4]− 135.04512 [C8H7O2]− |
B |
| 61 # | Vitexin | 15.74 | C21H20O10 | 431.09833 [M − H]− |
−0.1 | 341.06631 [C18H13O7]−
311.05603 [C17H11O6]− 283.06088 [C16H11O5]− |
C |
| 62 # | 9,10-Dihydro-10-(4-hydroxyphenyl)-pyrano[2,3-h]epicatechin-8-one | 19.9 | C24H20O8 | 435.10876 [M − H]− |
0.51 | 341.06641 [M − H − Phenol]−
217.01392 [C11H5O5]− 189.01918 [C10H5O4]− 177.01915 [C9H5O4]− |
C |
| 63 | Epicatechin monogallate | 15.92 | C22H18O10 | 441.08286 [M − H]− |
0.31 | 289.07166 [C15H13O6]−
169.0141 [C7H5O5]− 125.02431 [C6H5O3]− |
C |
| 64 | Astragalin | 15.06 | C21H20O11 | 447.09344 [M − H]− |
0.36 | 357.06146 [C18H13O8]−
327.05087 [C17H11O7]− 299.05569 [C16H11O6]− |
C |
| 65 # | Kaempferol-3-glucoside | 16.47 | C21H20O11 | 447.09354 [M − H]− |
0.56 | 285.03983 [M − H − Glc]− | C |
| 66 # | Luteolin-7-glucoside | 17.4 | C21H20O11 | 447.09357 [M − H]− |
0.63 | 284.0325 [M − H − Glc]−
255.02986 [C14H7O5]− 227.03485 [C13H7O4]− |
C |
| 67 # | Ursolic acid | 35.96 | C30H48O3 | 455.35315 [M − H]− |
0.18 | 407.33292 [C29H43O]− | E |
| 68 *,# | Oleanic acid | 36.51 | C30H48O3 | 455.35333 [M − H]− |
0.57 | 407.33292 [C29H43O]− | E |
| 69 # | Quercetin-3-alloside | 16.21 | C21H20O12 | 463.08856 [M − H]− |
0.78 | 300.02731 [M − H − Gal]−
271.02469 [C14H7O6]− 255.02979 [C14H7O5]− |
C |
| 70 | Nigranoic acid | 32.15 | C30H46O4 | 469.33249 [M − H]− |
0.33 | 423.327 [C29H43O2]− | B |
| 71 # | 4-O-(6″-O-p-Coumaroyl-glucopyranosyl)-p-coumaric acid | 18.64 | C24H24O10 | 471.12994 [M − H]− |
0.58 | 307.08206 [C15H15O7]−
163.03993 [C9H7O3]− 119.05008 [C8H7O]− |
B |
| 72 | Corosolic acid | 31.81 | C30H48O4 | 471.34802 [M − H]− |
0.08 | 453.33932 [C30H45O3]− | E |
| 73 # | 7-Hydroxy-3,8-dimethoxy-5,10-dioxo-5,10-dihydro-chromeno[5,4,3-cde]chromen-2-yl 6-deoxymannopyranoside | 18.83 | C22H20O12 | 475.08853 [M − H]− |
0.7 | 460.06451 [M − H − CH3]−
328.02231 [M – H − Rha]− 312.99887 [M − H − Rha − CH3]− 269.98038 [C13H2O7]− |
D |
| 74 | Isorhamnetin-7-O-glucopyranoside | 17.62 | C22H22O12 | 477.10416 [M − H]− |
0.64 | 314.04306 [M − H − Glc]−
271.02448 [M − H − Glc − CH3 − CO]− 243.02959 |
C |
| 75 | Isomyricitrin | 14.78 | C21H20O13 | 479.08301 [M − H]− |
−0.22 | 316.02216 [M − H − Glc]−
271.02454 [C14H7O6]− |
C |
| 76 | 1,6-Bis-O-galloyl-glucose | 11.52 | C20H20O14 | 483.07819 [M − H]− |
0.33 | 465.10410 [M − H − H2O]−
439.08768 [M − H − CO2]− |
B |
| 77 | Quillaic acid | 26.36 | C30H46O5 | 485.32706 [M − H]− |
−0.39 | 407.29538 [M − H − CH3 − H2O]−
241.10255 |
E |
| 78 # | 2-O-(E)-caffeoyl-1-O-p-(E)-coumaroylglucopyrannose | 12.86 | C24H24O11 | 487.12473 [M − H]− |
0.29 | 323.07706 [C15H15O8]−
161.02429 [C9H5O3]− 119.05011 [C8H7O]− |
B |
| 79 *,# | Asiatic acid | 26.85 | C30H48O5 | 487.34271 [M − H]− |
−0.38 | 469.33224 [M − H − H2O]− | E |
| 80 | Oenin | 18.16 | C23H24O12 | 491.11981 [M − H]− |
0.64 | 313.03531 [M − H − Glc − CH]−3
299.01956 [C15H7O7]− 285.04028 [M − H − Glc − CH3 − CO]− |
C |
| 81 | Medicagenic acid | 24.38 | C30H46O6 | 501.32227 [M − H]− |
0.21 | 455.31689 [M − H − HCOOH]− | E |
| 82 | 2″-O-Galloylisovitexin | 17.72 | C28H24O14 | 583.10974 [M − H]− |
0.71 | 431.09842 [C21H19O10]−
341.06635 [C18H13O7]− 311.056 [C17H11O6]− 283.061 [C16H11O5]− |
C |
| 83 | Nicotiflorin | 17.02 | C27H30O15 | 593.15167 [M − H]− |
0.81 | 285.03989 [C15H9O6]−
255.02983 [C14H7O5]− 227.03491 [C13H7O4]− |
C |
| 84 * | Rutin | 15.9 | C27H30O16 | 609.14630 [M − H]− |
0.32 | 300.02737 [C15H8O7]−
271.02469 [C14H7O6]− 255.02977 [C14H7O5]− |
C |
| 85 | 2′-O-Galloylhyperin | 15.56 | C28H24O16 | 615.09943 [M − H]− |
0.44 | 463.08804 [C21H19O12]−
300.02734 [C15H8O7]− 271.02454 [C14H7O6]− 255.02972 [C14H7O5]− |
C |
| 86 | 3-O-trans-p-Coumaroylmaslinic acid | 34.83 | C39H54O6 | 617.38501 [M − H]− |
0.4 | 145.02936 [C9H5O2]− | E |
| 87 | Delphinidin-3-caffeoylglucoside | 18.25 | C30H26O15 | 625.12048 [M − H]− |
0.94 | 463.08832 [C21H19O12]−
300.02731 [C15H8O7]− 271.02472 [C14H7O6]− 255.02985 [C14H7O5]− |
C |
| 88 | 3-O-trans-p-Coumaroyltormentic acid | 31.21 | C39H54O7 | 633.37982 [M − H]− |
0.23 | 163.04001 [C9H7O3]−
145.02939 [C9H5O2]− |
E |
| 89 | 1,3,6-Tri-O-galloylglucose | 14.4 | C27H24O18 | 635.08948 [M − H]− |
0.77 | 465.06714 [C20H17O13]−
211.02463 [C9H7O6]− 169.01404 [C7H5O5]− 125.02427 [C6H5O3]− |
B |
| 90 | (3,5,9)-3-Hydroxy-27-{[(2E)-3-(4-hydroxy-3-methoxyphenyl)-2-propenoyl]oxy}olean-12-en-28-oic acid | 35.86 | C40H56O7 | 647.39587 [M − H]− |
0.84 | 632.37152 [C37H52O7]−
453.33835 [C30H45O3]− 175.03993 [C10H7O3]− 133.02946 [C8H5O2]− |
E |
| 91 | 3-O-trans-Feruloyleuscaphic acid | 32.02 | C40H56O8 | 663.39069 [M − H]− |
0.67 | 648.36658 [C39H52O8]−
175.03989 [C10H7O3]− 160.01643 [C9H4O3]− 132.02156 [C8H4O2]− |
E |
| 92 # | 4′-Hydroxyacetophenone | 12.32 | C8H8O2 | 137.05969 [M + H]+ |
−0.01 | 122.03635 [M + H − CH3]+
109.06508 [M + H − CO]+ |
F |
| 93 | Dihydroxyacetophenone | 13.7 | C8H8O3 | 153.05461 [M + H]+ |
−0.06 | 125.05981 [M + H − CO]+
111.04433 [C6H7O2]+ |
F |
| 94 | N-Lauryldiethanolamine | 37.04 | C16H35O2N | 256.26334 [M + H]+ |
−0.6 | 144.1382 [C8H18ON]+
116.1072 [C6H14ON]+ |
F |
| 95 | Licanic acid | 26.99 | C18H28O3 | 293.21085 [M + H]+ |
−0.94 | 275.20044 [M + H − H2O]+
257.19003 [M + H − 2H2O]+ |
A |
| 96 | Kamlolenic acid | 33.51 | C18H30O3 | 295.22678 [M + H]+ |
0.06 | 277.21609 [M + H − H2O]+
259.20532 [M + H − 2H2O]+ 231.21051 [M + H − 3H2O]+ |
A |
| 97 | Diosmetin | 21.99 | C16H12O6 | 301.0705 [M + H]+ |
−0.56 | 286.04681 [M + H − CO3]+
258.05197 [M + H − CH3 − CO]+ |
C |
| 98 # | Sitosterol | 39.21 | C29H50O | 397.38269 [M + H − H2O]+ |
−0.47 | 243.21089 [C18H27]+
175.14799 [C13H19]+ 147.11684 [C11H15]+ |
G |
| 99 # | Stigmasterol | 38.08 | C29H48O | 395.36725 [M + H − H2O]+ |
0.05 | 241.1945 [C18H25]+
199.14772 [C15H19]+ 173.13248 [C13H17]+ |
G |
| 100 | Apigenin-7-O-glucoside | 17.83 | C21H20O10 | 433.1127 [M + H]+ |
−0.51 | 271.05975 [C15H11O5]+
153.01814 [C7H5O4]+ |
C |
| 101 # | Quercetin-3-arabinoside | 16.62 | C20H18O11 | 435.09235 [M + H]+ |
0.36 | 303.04959 [C15H11O7]+
153.01819 [C7H5O4]+ |
C |
| 102 | Luteolin-7-galactoside | 12.52 | C21H20O11 | 449.10745 [M + H]+ |
−0.86 | 287.05453 [M − H − Glc]+
269.044 [M + H − Glc]+ |
C |
| 103 | Pelargonidin-3-O-(6-caffeoyl-glucoside) | 19.91 | C30H26O13 | 595.14459 [M + H]+ |
−0.04 | 433.11255 [M − H − C6H6 − 3CO]+
313.07028 [C17H13O6]+ 163.03888 [C9H7O3]+ |
C |
| 104 | Tiliroside | 20.42 | C30H26O13 | 595.14435 [M + H]+ |
−0.45 | 287.0546 [C15H11O6]+
147.04393 [C9H7O2]+ 119.04929 [C8H7O]+ |
C |
| 105 | Kaempferol-3-(6″-galloylgalactoside) | 16.8 | C28H24O15 | 601.11914 [M + H]+ |
0.57 | 287.05457 [C15H11O6]+
153.01813 [C7H5O4]+ 137.0233 [C7H5O3]+ |
C |
| 106 | Quercetin-3-O-(6″-O-p-coumaroyl)-glucopyranoside | 19.41 | C30H26O14 | 611.13922 [M + H]+ |
−0.51 | 147.04388 [C9H7O2]+
303.04953 [C15H11O7]+ |
C |
| 107 | 2′-O-Galloylhyperin | 15.51 | C28H24O16 | 617.11322 [M + H]+ |
−0.89 | 153.01813 [C7H5O4]+
303.0495 [C15H11O7]+ |
C |
| 108 | Quercetin-3-(6″-caffeoylgalactoside) | 18.29 | C30H26O15 | 627.13409 [M + H]+ |
−0.56 | 163.03873 [C9H7O3]+
303.04941 [C15H11O7]+ |
C |
| 109 # | Casuarinin | 12.61 | C41H28O26 | 937.09332 [M + H]+ |
−0.89 | 153.01817 [C7H5O4]+
171.04417 [C11H7O2]+ 277.03397 [C13H9O7]+ 345.02377 [C16H9O9]+ |
D |
* These compound were unambiguously identified by the use of authentic reference compounds. # These compound were isolated from Melastoma dodecandrum Lour. according to the literature [5,6,7,8,9,10,11,12,13]. Glc, glucopyranosyl, Rha, rhamnopyranosyl. A, fatty acid; B, organic acid; C, flavonoid; D, tannin; E, pentacyclic triterpene; F, others; G, steroid.
2.1. Fragmentation Pattern of Main Compounds
2.1.1. Flavonoids
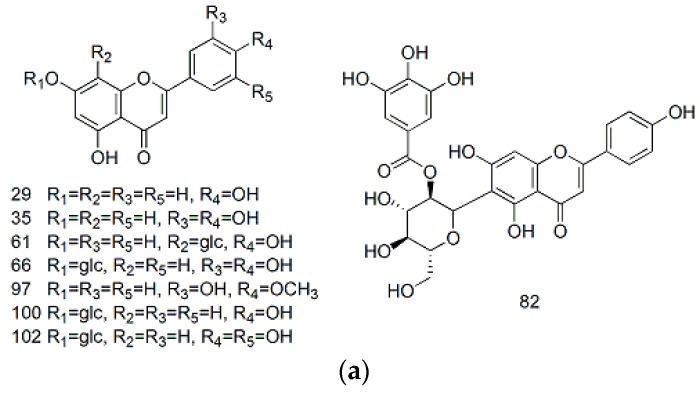
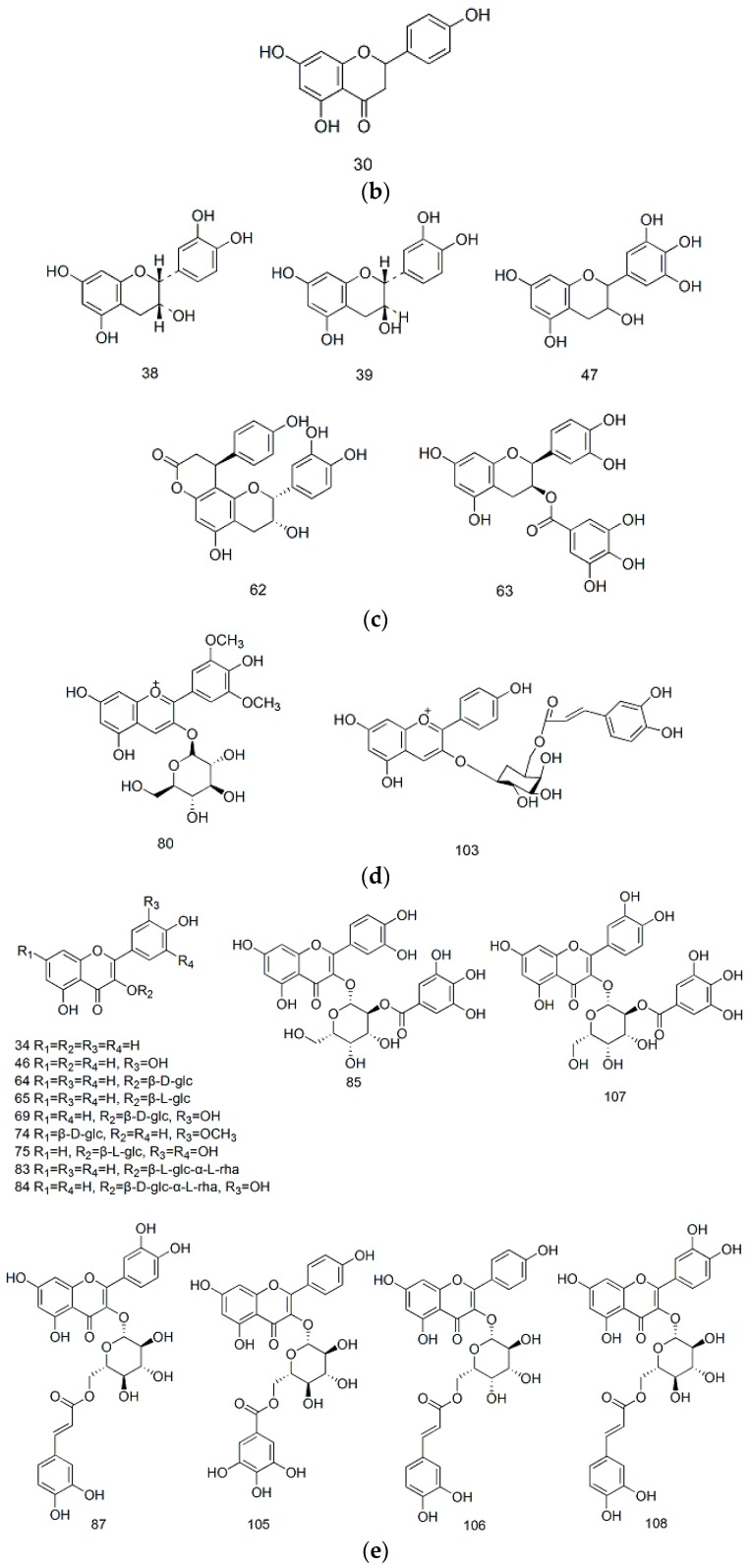
Flavonoids are 2-phenylchromone systems that consist of two benzene rings (A and B) connected by a pyran ring (ring C), which is fused to the A ring. Flavonoids can be classified into several subclasses, including flavones, flavonols, flavanones, flavanonols, anthocyanidins, chalcones, isoflavonoids and flavan-3-ols, depending on the nature of the substituents attached to the different rings. Based on the results of accurate molecular mass measurements and the MS2 fragmentation pathways of the different materials [16,22,23], we characterized a total of 33 different flavonoids (aglycones) in MDL, including eight flavones, one flavanone, five flavan-3-ols, two anthocyanidins and 17 flavonols. The structures of the 33 flavonoids are shown below (Figure 2).
Figure 2.
Structure of flavones (a); flavanones (b); flavan-3-ols (c); anthocyanidins (d); and flavonols (e).
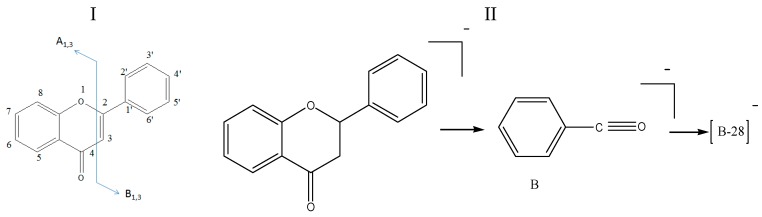
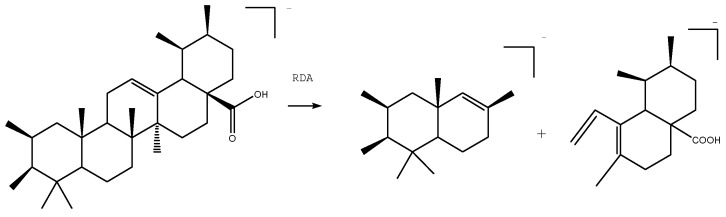
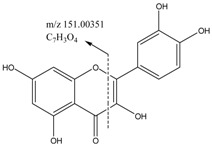
Two different fragmentation patterns and pathways were observed for the flavonoids (I and II, shown in Figure 3). The retro-Diels–Alder (RDA) fragmentation (I) would result in the formation of A 1, 3 and B 1, 3 as the main fragment ions of the flavonoid moiety (aglycones) because of the X 1, 3 cleavage of the C ring. The substituent groups on the parent compounds were determined based on the compositions of the A 1, 3 and B 1, 3 fragments. The fragmentation of the parent compound according to Pattern I resulted in high-intensity fragment ions, whereas the fragmentation according to pattern II result in low-intensity fragment ions. Furthermore, the main fragmentation patterns of flavonoids (glycosides) typically consist of fragments associated with deglycosylation, demethylation and decarboxylation yielding [M − H − Glc]−, [M − H − CH3]− and [M − H − CO2]− ions, respectively. The structures of the fragments resulting from the RDA fragmentation are shown in Table 2.
Figure 3.
Fragmentation pattern I and II.
Table 2.
RDA fragmentation pathways of compound 29, 30, 46, and 63.
| Compound No. | Mass Spectra of MS/MS | RDA Fragmentation Pathway |
|---|---|---|
| 29 |  |
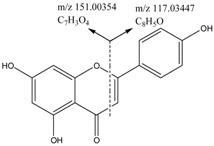 |
| 30 | 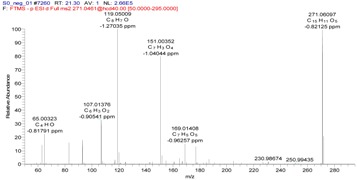 |
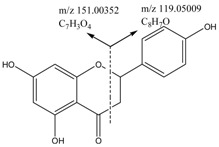 |
| 46 | 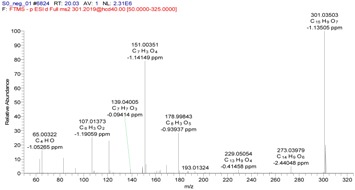 |
 |
| 63 | 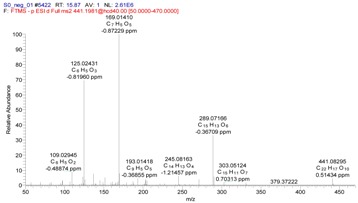 |
 |
2.1.2. Pentacyclic Triterpenes
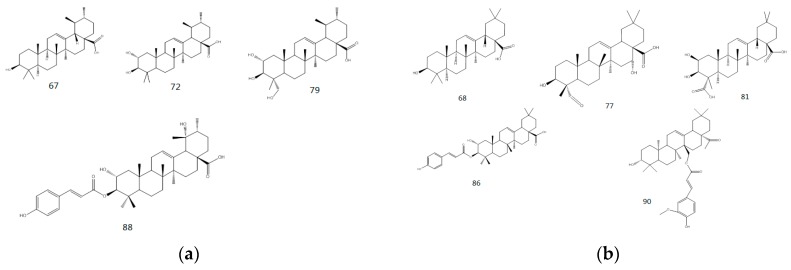
Pentacyclic triterpenes are wildly distributed in Nature and consist of five rings, which are typically referred to as the A, B, C, D and E rings. Pentacyclic triterpenes can be divided into several different categories, depending on the nature of their E ring. In this study, we characterized two different types of pentacyclic triterpenes, including ursane- and oleanane-type pentacyclic triterpenes. The endo-double bond in pentacyclic triterpenes can readily undergo a RDA reaction during MS analysis (shown in Figure 4). Dehydration and decarboxylation are also observed as common fragmentation pathways in these systems during MS analysis [21,22,23]. Depending on the different substituents attached to their endo-double bonds and their accurate mass measurements, we were able to fully characterize all of the pentacyclic triterpenes found in MDL. As shown in Figure 5, we characterized a total of nine pentacyclic triterpenes using high-resolution MS2 mass spectrometry, including four ursane-type and five oleanane-type compounds pentacyclic triterpenes.
Figure 4.
RDA fragmentation pathway of pentacyclic triterpene with endo-double bond.
Figure 5.
Structure of ursane-type (67, 72, 79, 88) and oleanane-type (68, 77, 81, 86, 90) pentacyclic triterpene. This figure is fuzzy, please replace it with sharper figure.
2.1.3. Tannins
The main structural features of tannins include their gallic acid ester moieties (or polymer) and glucose core (or other polyols). The MS2 spectra of the tannins revealed fragment ions with m/z values of 1091, 939, 769, 617, 599, 447 and 277. The m/z differences between these fragment ions were 152 and 170 atomic mass units (amu), which indicated that the fragments ions were produced by the successive removal of O-galloylhyperin and gallic acid anions. The fragment ion observed with an m/z value of 169 was characteristic of the fragment ions derived from O-galloylhyperin, whereas the fragment ion observed with an m/z value of 125 was attributed to the loss of CO2 from gallic acid [17]. A total of six other tannins were characterized in this way using high-resolution MS2 mass spectrometry. The structure of tannins characterized were shown in Figure 6.
Figure 6.
Structure of tannins.
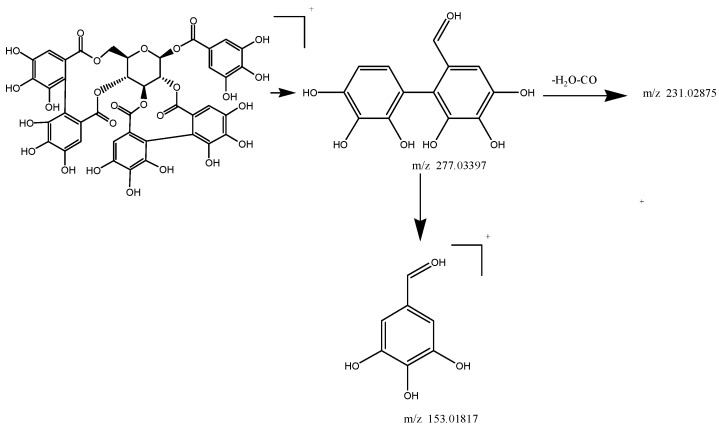
Taking compound 109 as a representative example, the analysis of this compound in the positive ionization mode (tR = 12.61 min) gave an [M + H]+ ion with an m/z value of 937.09332, indicating a molecular formula of C41H28O26 (Δ = −0.89 ppm) (Table 2). The MS2 spectrum of compound 109 contained five fragment ions with m/z values of 345.02377, 277.03397, 231.02875, 171.04417 and 153.01817. The fragment pathway for this compound is shown in Figure 7. Based on these results, compound 109 was characterized as casuarinin.
Figure 7.
Fragmentation pathway of compound 109.
2.1.4. Organic Acids
Compound 4 (tR = 1.41 min) was analyzed in the negative ionization mode and gave an [M − H]− ion with an m/z value of 147.02991, which indicated a molecular formula of C5H8O5 (Δ = 0.06 ppm) (Table 1). The MS2 spectrum of compound 4 gave three fragment ions with m/z values of 129.01924 [M − H − H2O]−, 101.02427 [M – H − HCOOH]− and 85.02937 [M − H − H2O − CO2]−. This fragmentation process was used to confirm the identities of the other organic acids, resulting in the characterization of 26 compounds as organic acids.
3. Materials and Methods
3.1. Chemicals and Reagents
Acetonitrile and methanol (HPLC grade) were purchased from Fisher Scientific (Waltham, MA, USA). Distilled water was purchased from Watson’s Food & Beverage Co., Ltd. (Guangzhou, China). Formic acid (MS grade) was purchased from Fisher Scientific (Waltham, MA, USA). Reference standards of oleanolic acid (94.9%, batch No. 110709-201206), kaempferol (93.2%, batch No. 110861-201209), luteolin (100%, batch No. 111520-200504) and quercetin (97.4%, batch No. 100081-200907) were purchased from the National Institutes for Food and Drug Control (Beijing, China). A reference standard of asiatic acid (99%, batch No. 20150901) was purchased from CRM/RM center of China (Beijing, China). Reference standards of rutin (98%, batch No. 153-18-4) and gallic acid (98.5%, batch No. 149-91-7) were purchased from Chengdu-PUSH Bio-Technology Co., Ltd. (Chengdou, Sichuan, China).
3.2. Plant Materials and Sample Preparation
The whole plants of MDL were collected from Yunnan Province in China and authenticated by Professor Yaojun Yang (Beijing University of Chinese Medicine, Beijing, China). Voucher specimens (DN001) of the plant were deposited at the authors’ laboratory. The samples were dried and powdered, before being sieved through a 40-mesh sieve. A sample of the powder (approximately 2.0 g) was suspended in 25 mL of methanol, and the resulting mixture was subjected to ultrasonic treatment for 30 min before being cooled to room temperature. Methanol was added to compensate for the lost weight and the resulting mixture was filtered through a 0.22-μm PTFE syringe filter. The filtrate was collected and subjected to centrifugation (13,000 rpm, 10 min). The supernatant was then transferred to an autosampler vial for analysis by UPLC-ESI-Q-Exactive Focus-MS/MS.
3.3. UPLC-ESI-Q-Exactive Focus-MS/MS Analysis
UPLC analysis was performed on a Thermo Scientific Ultimate 3000 system (Sunnyvale, CA, USA) equipped with a binary solvent delivery manager and a sample manger. Chromatographic separations were performed on a Thermo Scientific Hypersil GOLD C18 column (100 mm × 2.1 mm, 1.9 μm). The column temperature was maintained at 40 °C. A mobile phase consisting of 0.1% formic acid in water (A) and acetonitrile (B) was used to elute the column according to the optimized gradient program, as follows: 98% A from 0 to 5 min; 98%–80% A from 5 to 15 min; 80%–40% A from 15 to 30 min; 40%–2% A from 30 to 40 min; 2% A from 40 to 47 min; 2%–98% A from 47 to 47.1 min; 98% A from 47.1 to 50 min. The flow rate was set at 0.3 mL/min. An injection volume of 5 μL was used for the reference compounds and then analytical samples. For MS detection, the operating parameters were as follows: spray voltage, +3500 V/–3200 V; atomization temp, 350 °C; sheath gas pressure, 35 arb; aux gas pressure, 10 arb; capillary temperature, 320 °C; S-lens RF, 60 V; resolution, MS full scan 70,000 FWHM, MS/MS full scan 15,000 FWHM; scan range, m/z 100–1500 for MS; m/z 30–1500 for MS/MS; scanning mode, fullscan-ddms2.
3.4. Optimization of Analytical Conditions
To obtain better chromatographic separation and mass spectrometric detection, we evaluated three different mobile phase systems, including aqueous methanol, aqueous acetonitrile and aqueous acetonitrile-formic acid solutions. The aqueous acetonitrile solution resulted in the best separation of the major components of MDL. Furthermore, the addition of 0.1% formic acid to this mobile phase resulted in a considerable improvement in the symmetry properties of most of the chromatographic peaks. We also varied the flow rate (0.25, 0.3, and 0.35 mL/min), column temperature (30, 35, and 40 °C) and injection volume (2, 3, and 5 μL) during method development. The results of these optimization experiments established the following conditions for the chromatographic separation of the different components of MDL: mobile phase, aqueous acetonitrile containing 0.1% formic; flow rate, 0.3 mL/min; column temperature, 40 °C; and injection volume, 5 μL.
3.5. Structure Analysis Procedure
In the positive and negative scan mode, based on the high-accuracy precursor ions and product ions obtained from Q-Exactive Focus-MS/MS, the elemental compositions were calculated when the maximum tolerance of mass error for all the precursor ions and product ions was set at 1.5 ppm, which can satisfy the requirements for positive identification. Based on the elemental compositions of the precursors, the most rational molecular formula was sought in different chemical databases such as the Spectral Database for Organic Compounds SDBS (http://sdbs.db.aist.go.jp), m/z cloud (https://www.mzcloud.org) and ChemSpider (http://www.chemspider.com). Meanwhile by searching literature sources, such as PubMed of the U.S. National Library Medicine and the National Institutes of Health, Scifinder Scholar of the American Chemical Society, Science Direct of Elsevier and Chinese National Knowledge Infrastructure (CNKI) of Tsinghua University, all components reported in the literatures on MDL and plants of the same family were summarized in a Microsoft Office Excel table to establish an in-house library [5,6,7,8,9,10,11,12,13] for searching the most rational molecular formula. When several matching compounds with the same formula were found, the fragmentation patterns and pathways of the compounds were analyzed and then validated by Mass Frontier 7.0 (Thermo Scientific) for positive identification.
4. Conclusions
A new UPLC-ESI-Q-Exactive Focus-MS/MS method was developed to analyze the chemical constituents of MDL based on their mass spectral fragmentation patterns. This new method resulted in the characterization of 109 compounds. The results of this study therefore provide an important reference to improve our understanding of the composition of MDL. We found that flavonoids are the main components of MDL, especially the flavonols, which possess a wide range of interesting pharmacological activities, such as anticancer, antibacterial, and antiviral activities. In terms of their structural characteristics, the triterpenoids found in MDL were ursane- and oleanane-type systems. Several tannins and steroids were also found in MDL. In addition to the fatty acids found in MDL, we found 55 other compounds that have never been reported in MDL besides fatty acids. Further studies pertaining to the chemical constituents in Melastoma dodecandrum Lour. are currently underway in our laboratory. Moreover, The study shows that, with the application of the UPLC-ESI-Q-Exactive Focus-MS/MS to characterizing the constituents of MDL, this method offers a rapid, sensitive and high throughput methodology for the identification of constituents of TCM prescriptions and herbal medicines.
Acknowledgments
This research was financially supported by Beijing Key Laboratory for Quality Evaluation of Traditional Chinese Medicine (No. BZ0386).
Author Contributions
Jinfeng Wang and Ruichao Lin designed the research, performed the experimental work, analyzed data and wrote the manuscript. Ziyao Jia, Yutong Wang and Xi Liu made substantial contributions to acquisition of data and interpretation of data. Zhihao Zhang gave suggestions for writing. Linghua Wang prepared the samples and drew the chemical structures. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compound 10, 34, 35, 46, 68, 79, and 84 are available from the authors.
References
- 1.Zhang Z., Wang X., Wang J., Jia Z., Liu Y., Xie X., Wang C., Jia W. Metabonomics Approach to Assessing the Metabolism Variation and Endoexogenous Metabolic Interaction of Ginsenosides in Cold Stress Rats. J. Proteome Res. 2016;15:1842–1852. doi: 10.1021/acs.jproteome.6b00015. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Z.H., Vaziri N.D., Wei F., Cheng X.L., Bai X., Zhao Y.Y. An integrated lipidomics and metabolomics reveal nephroprotective effect and biochemical mechanism of Rheum officinale in chronic renal failure. Sci. Rep. 2016;6:22151. doi: 10.1038/srep22151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y., Zhang Z., Li S., Ye X., Li X., He K. Synergy effects of herb extracts: Pharmacokinetics and pharmacodynamic basis. Fitoterapia. 2014;92:133–147. doi: 10.1016/j.fitote.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Yu Z.C., Lin X.X., Su J.Q., Lin Q.J., Chen Z.D. Advance in Melastoma dodecandrum Lour. Researches. Med. Plant. 2011;2:63–67. [Google Scholar]
- 5.Zhang R.Z. Master’s Thesis. Fudan University; Shanghai, China: 2013. Studies on the Chemical Constituents of Melastoma dodecandrum L. and Achillea alpine. [Google Scholar]
- 6.Cheng M. Master’s Thesis. Jinan University; Guangzhou, Guangdong, China: 2015. Studies on the Chemical Constituents of Melastoma dodecandrum L. [Google Scholar]
- 7.Lin S., Li Y.C., Guo Y.Y., Guo S.M., Que H.Q., Qi Y.P. Chemical constituents of Melastoma dodecandrum L. (II) Chin. Tradit. Herb. Drugs. 2009;40:1192–1195. [Google Scholar]
- 8.Tang M., Liao B.Z., Lin S., Deng S.S. Chemical constituents of Melastoma dodecandrum L. Chin. Tradit. Herb. Drugs. 2008;39:1149–1151. [Google Scholar]
- 9.Zeng R.X., Zhang Q.H., Guan Y.M., Chen L.H., Wen S.J., Lu X.P. Analysis of monosaccharide composition in polysaccharides extract from Melastoma dodecandrum L. by precolumn derivatization HPLC. Chin. J. Exp. Tradit. Med. Formulae. 2015;21:73–76. [Google Scholar]
- 10.Cao D., Ma Z.Q., Jiang Y., Zhao C.J., Lin R.C. Chemical Composition of Melastoma dodecandrum Lour. Inf. Tradit. Chin. Med. 2016;33:11–14. [Google Scholar]
- 11.Yang D., Ma Q.Y., Liu Y.Q., Ding Z.T., Zhou J., Zhao Y.X. Chemical Constituents from Melastoma dodecandrum Lour. Nat. Prod. Res. Dev. 2010;22:940–944. [Google Scholar]
- 12.Zhang C., Fang Y.X. Studies of the Chemical Constituents of Chinese Herb Melastoma dodecandrum Lour. China J. Chin. Mater. Media. 2003;28:49–51. [PubMed] [Google Scholar]
- 13.Zhang C. Master’s Thesis. Guangdong University of Technology; Guangzhou, China: 2003. Studies on Chemical Constituents and Pharmacological Activities of Chinese Herbal Melastoma dodecandrum Lour. [Google Scholar]
- 14.Kouloura E., Skaltsounis A.L., Michel S., Halabalaki M. Ion tree-based structure elucidation of acetophenone dimers (AtA) from Acronychia pedunculata and their identification in extracts by liquid chromatography electrospray ionization LTQ-Orbitrap mass spectrometry. J. Mass Spectrom. 2015;50:495–512. doi: 10.1002/jms.3556. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J.Y., Wang F., Zhang H., Lu J.Q., Qiao Y.J. Rapid identification of polymethoxylated flavonoids in traditional Chinese medicines with a practical strategy of stepwise mass defect filtering coupled to diagnostic product ions analysis based on a hybrid LTQ-Orbitrap mass spectrometer. Phytochem. Anal. 2014;25:405–414. doi: 10.1002/pca.2508. [DOI] [PubMed] [Google Scholar]
- 16.Ru L., Wei S., Xue Q., Jia L., Hong L., Min Y. Chemical profiling of Scutellaria barbata by ultra high performance liquid chromatography coupled with hybrid quadrupole orbitrap mass spectrometry. J. Chin. Pharm. Sci. 2015;24:635–646. [Google Scholar]
- 17.Dong H.J., Chen X.H., Zeng R. Rapid analysis on chemical constituents in roots of Rheum pumilum by UPLC coupled with hybrid quadrupole-orbit trap MS. Chin. Tradit. Herb. Drugs. 2016;47:2428–2435. [Google Scholar]
- 18.Wang S.S., Xu H.Y., Ma Y.N., Wang X.G., Shi Y., Huang B., Tang S.H., Zhang Y., Li D.F., Liang R.X., et al. Characterization and rapid identification of chemical constituents of NaoXinTong capsules by UHPLC-linear ion trap/Orbitrap mass spectrometry. J. Pharm. Biomed. Anal. 2015;111:104–118. doi: 10.1016/j.jpba.2015.01.020. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y.Y., Cheng X.L., Zhang Y., Chao X., Zhao Y., Lin R.C., Sun W.J. A fast and sensitive HPLC-MS/MS analysis and preliminary pharmacokinetic characterization of ergone in rats. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2010;878:29–33. doi: 10.1016/j.jchromb.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Y.Y., Qin X.Y., Cheng X.L., Liu X.Y., Lin R.C., Zhang Y., Li X.Y., Sun X.L., Sun W.J. Rapid resolution liquid chromatography-mass spectrometry and high-performance liquid chromatography-fluorescence detection for metabolism and pharmacokinetic studies of ergosta-4,6,8(14),22-tetraen-3-one. Anal. Chim. Acta. 2010;675:199–206. doi: 10.1016/j.aca.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 21.Guo L.X., Li R., Liu K., Yang J., Li H.J., Li S.L., Liu J.Q., Liu L.F., Xin G.Z. Structural characterization and discrimination of Chinese medicinal materials with multiple botanical origins based on metabolite profiling and chemometrics analysis: Clematidis Radix et Rhizoma as a case study. J. Chromatogr. A. 2015;1425:129–140. doi: 10.1016/j.chroma.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 22.Mao Q., Bai M., Xu J.D., Kong M., Zhu L.Y., Zhu H., Wang Q., Li S.L. Discrimination of leaves of Panax ginseng and P. quinquefolius by ultrahigh performance liquid chromatography quadrupole/time-of-flight mass spectrometry based metabolomics approach. J. Pharm. Biomed. Anal. 2014;97:129–140. doi: 10.1016/j.jpba.2014.04.032. [DOI] [PubMed] [Google Scholar]
- 23.Duan L., Guo L., Liu K., Liu E.H., Li P. Characterization and classification of seven citrus herbs by liquid chromatography-quadrupole time-of-flight mass spectrometry and genetic algorithm optimized support vector machines. J. Chromatogr. A. 2014;1339:118–127. doi: 10.1016/j.chroma.2014.02.091. [DOI] [PubMed] [Google Scholar]