Abstract
The cytochrome P450 mono-oxygenase gene, CYP2D6 on chromosome 22q13 (ch22q13), has been inconsistently associated with Parkinson’s disease. Associations with CYP2D6 have either been absent altogether or have involved more than one polymorphism, many of which have the same metabolic effect on gene expression. We examined the association between CYP2D6 polymorphisms and Parkinson’s disease in a case-control study and included 10 polymorphic dinucleotide repeat markers linked to CYP2D6 to determine whether the association was present or due to linkage disequilibrium. There was no association between any polymorphism of CYP2D6 and Parkinson’s disease, but two of 10 dinucleotide repeat markers linked to CYP2D6 were associated with the disease. These results provide evidence to suggest that there may be an unidentified locus for susceptibility to Parkinson’s disease that is in linkage disequilibrium with dinucleotide repeat markers mapping near CYP2D6 on ch22q13.
The CYP2D6 gene on chromosome 22q13 (ch22q13), one of the cytochrome P450 mono-oxygenases, has been associated with Parkinson’s disease (PD) [1], based on metabolic and genetic association studies. Several polymorphisms in the CYP2D6 gene have been associated with PD [2–6], and each of these has a similar effect on the metabolism of debrisoquine hydroxylase (DH) [7–9]. The putative effect of CYP2D6 on PD susceptibility is related to metabolism of an, as yet unidentified, environmental factor. Variant alleles A, B, and D comprise approximately 95% of the individuals with low DH activity and each would be expected to have a similar effect on susceptibility to PD [9]. The most common defective allele, B, is a mutation that would be predicted to disrupt RNA splicing and has been reported to be more frequent among individuals with PD than controls [1–6, 10]. The rarer A allele results in a frameshift in the predicted protein product and was more frequent in patients with PD than controls in one study [3]. The D allele of CYP2D6 involves a deletion that includes the adjacent homologous sequences CYP2D7 and CYP2D8 and was present in Japanese patients more often than in controls [4, 9]. The L allele, recently described [11], has two substituted amino acids that result in increased DH activity. Among Japanese patients with PD, the L allele was more frequent than among controls [6], but the phenotype was biochemically indistinguishable from wild type. The association has not been confirmed and it also differs from the conclusion of another Japanese study [4].
Despite encouraging results, the allelic association of CYP2D6 to PD has been inconsistent [10, 12]. Methodological differences and population admixture may explain weaker associations, but it is also possible that CYP2D6 is in linkage disequilibrium with an unidentified susceptibility locus to PD on ch22q13. In support of that view, we report allelic association between PD and dinucleotide repeat markers linked to CYP2D6 in a population that does not show a significant association with any allele of CYP2D6.
Subjects and Methods
Subjects and Setting
Patients and controls were part of a community-based study of PD, aging, and health in the Washington Heights-Inwood section of New York City [13, 14]. The diagnosis of the idiopathic form of PD was based on published research criteria [15, 16]. Controls were selected randomly from a stratified random sample of 1,800 Medicare recipients in the same community, free of PD, dementia, stroke, or other major neurologic conditions by frequency matching to the distribution of patients by age (<70, 70–80, and >80 years). None of the controls were spouses or relatives of the patients. Patients and controls received interviews and clinical assessments.
Genotyping
Blood DNA was isolated from detergent- and proteinase-digested purified leukocytes or from nuclei produced by hypotonic lysis followed by detergent and proteinase digestion and by organic extraction. Locus-specific amplification of blood DNA was done on patients and controls as previously described [17] with published primers (Genome Database, Welch Laboratory, Johns Hopkins University). Alleles were determined by comparison with reported allele sizes of CEPH reference subject 1347-02. The CYP2D6 B alleles have multiple mutations that are predicted to inactivate the gene product. We assayed for a G-to-A transition at the intron 3/exon 4 junction by BstNI cleavage of a polymerase chain reaction (PCR) amplification product [9]. CYP2D6 A alleles contain a single base deletion and have been detected by generation of a restriction site polymorphism after mismatched PCR primer amplification or by detection of a restriction endonuclease length polymorphism created by introducing a point mutation during PCR amplification. Using this approach, we had rare false positives as determined by direct sequencing of PCR amplification products. To overcome this problem, we detected the single base deletion by determining the size of restriction endonuclease cleaved PCR products on a DNA sequencing gel.
Using Multimap [18], genetic maps were constructed for the region of interest based on available data. Data were pooled from the latest versions of the CEPH and CHLC databases and from our own typing data for D22S276, D22S279, D22S428, and CYP2D8. Radiation hybrid retention maps were constructed using RHMAP [19]. These maps were in general agreement with previously published maps for the region. To order closely linked markers, we relied on the data compiled for chromosome 22 at the Sanger Center (http://www.sanger.ac.uk/hum22/).
Allele frequencies were determined by counting alleles and calculating sample proportions. Allele frequencies were compared between patients and controls using log likelihood χ2. The objective of this study was to confirm an association between CYP2D6 and PD. Only CYP2D6 and 10 closely linked markers were examined; therefore, an adjustment for multiple variables was not required because the markers are not independent. However, the level of statistical significance was defined as p < 0.01. We also used a multivariate log likelihood model χ2 with a forward stepwise selection process based on the significance of the score statistic. This method removed marker data by testing, based in part on the likelihood ratio statistic, and on maximum partial likelihood estimates. We then used log(1/p) to plot the association over the region.
Results
DNA was available from 121 patients with idiopathic PD, of whom 39 (32.2%) were demented but met criteria for inclusion in the study, and from 138 healthy elderly controls. Patients and controls had a similar age range, but controls were slightly older despite the frequency matching (PD: 71.2 ± 9.9 years; range, 41–91 years; controls: 74.4 ± 6.6 years; range, 59–93 years; p < 0.01). There were more men in the PD group than in the control group (63.7% vs 36.8%; χ2 = 17.6, p < 0.01). Fewer blacks (12%) were in the PD group than whites (60.3%) or Hispanics (27.7%; χ2 = 28.7, p < 0.01). There were no differences in education.
The frequency of the CYP2D6 B allele was similar in patients and controls (PD 18.6% vs controls 17.6%; χ2 = 1.2, p = 0.25). Homozygosity for the B allele was present in 5% of the patients and 3.7% of the controls. A single control and no patients had an A allele, and no other variant alleles of CYP2D6 were identified.
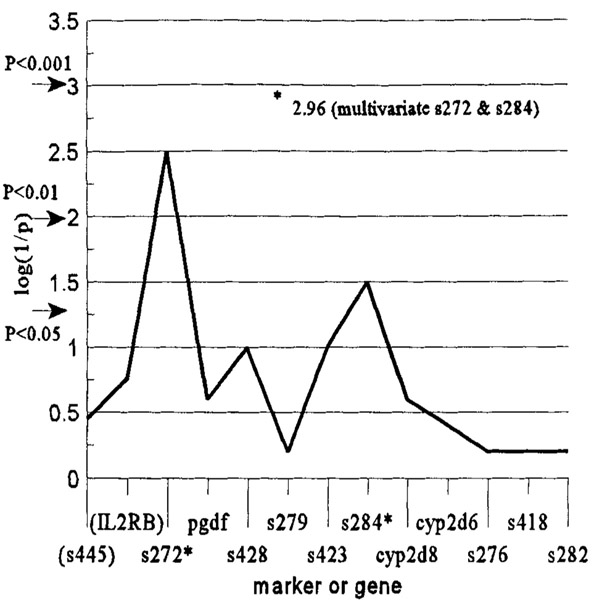
To examine for the possibility of an alternate susceptibility locus near CYP2D6, we compared the allele frequencies in patients and controls in the set of 10 microsatellite dinucleotide repeat markers in the region (Table and Fig). Alleles for adjacent markers D22S272 and D22S284 were significantly different between patients and controls (see Table). The maximum log(1/p) was 2.52, suggesting a possible association in the region. When using the multivariate log likelihood model χ2, only alleles for markers D22S272 and D22S284 were significantly different among patients and controls. The log likelihood χ2 = 13.3, p < 0.001, maximum log(1/p) = 2.96 for the model. This suggests a better approximation of the association in the region spanning both markers D22S272 and D22S284. Markers for these alleles also remained associated after statistical adjustment for ethnic group and age in the multivariate analysis, using the most frequent allele as the indicator. We genotyped two additional dinucleotide repeat markers (D22S445 and D22IL2RB) near D22S272 (see Fig) to better approximate the region of interest, but no allele frequencies for either marker were different in patients and controls.
Table.
Association Between CYP2D6 Alleles and Dinucleotide Repeat Marker Alleles on Chromosome 22q13 and Parkinson’s Disease
| Marker or Genea |
Relative Location (cM) |
Log Likelihood Ratio χ2 |
p |
|---|---|---|---|
| s272 | −2.0 | 8.84 | 0.003 |
| PDGF-β | −1.7 | 1.61 | 0.21 |
| s428 | −1.3 | 2.65 | 0.11 |
| s279 | −0.9 | 0.25 | 0.62 |
| s423 | −0.6 | 2.51 | 0.11 |
| s284 | −0.3 | 4.69 | 0.03 |
| CYP2D8 | 0 | 0.68 | 0.11 |
| CYP2D6 B | 0 | 1.28 | 0.25 |
| s276 | +0.4 | 0.31 | 0.58 |
| s418 | +1.4 | 0.35 | 0.55 |
| s282 | +2.3 | 0.26 | 0.61 |
Location refers to the distance from the CYP2D6 locus. In the multivariate analysis, only D22S272 and D22S284 remained significantly different between patients and controls.
PDGF-β = platelet-derived growth factor-β.
Fig.
Association between markers on chromosome 22q13 and Parkinson’s disease. Statistical significance values for p < 0.05, p < 0.01, and p < 0.001 are illustrated by arrows. The p values correspond to the log likelihood ratio χ2. Initially, D22(S445) and D22(IL2RB) were not included, but these two markers were added later to more fully examine the model. Also note that the asterisk indicates the log(1/p) for the multivariate model that is described in Results. The p value was <0.0011.
Discussion
Because the reports of associations between different polymorphisms of CYP2D6 and PD have been inconsistent, it is unlikely that this gene affects PD susceptibility. We also found no evidence for an association between CYP2D6 and PD, as have other recent investigators [20]. However, we provide an alternative explanation by suggesting that there may be an unidentified locus for susceptibility to PD that is in linkage disequilibrium with CYP2D6. The frequency of alleles for two nearby dinucleotide repeat markers was different in patients compared with controls from the same community. Investigations of familial PD have been limited, but linkage studies of familial PD have generally excluded CYP2D6 as a susceptibility locus. However, one family segregation study that excluded CYP2D6 as the disease gene did report a weak association with PD with onset after age 60 [21]. Although it is possible that linkage might be detected in a larger study of familial PD with markers near D22S272 or D22S284, it remains uncertain whether the genetic etiology of familial PD differs from that of the sporadic form.
The use of dinucleotide markers to detect linkage disequilibrium in case–control studies is problematic. Linkage is only one of the reasons for detecting allelic association. Case–control designs are particularly sensitive to sample admixture and clustered sampling [22]. Although stratification by ethnic group and adjustment in multivariate analyses may reduce the effects of admixture to some extent, they cannot be eliminated. Using a multivariate model to investigate the association indicated that the region spanning both markers provided strongest evidence for an association [p = 0.0011, log(1/p) = 2.96]. The statistical significance of this finding alone would be considered modest, because we used dinucleotide markers in unrelated individuals. There is no definitive level of statistical significance that can assure that any association is valid in case–control studies. Thomson [23] has suggested that for genetic studies of complex traits, which usually include related individuals, association should be judged by the degree of statistical significance. For example, a single study with a p value of < 0.001 or several independent studies with p values of 0.05 to 0.01 should be viewed as suggestive for the presence of a genetic locus. No such guidelines exist for case–control studies. However, we consider our findings, combined with earlier reports [2–6] identifying CYP2D6 as the genetic locus, suggestive of an unidentified genetic locus for PD in the region of ch22q13.
Before the identification of a causal polymorphism, allelic association should be interpreted cautiously. Failure to replicate an association in a different population does not preclude an association between a polymorphism and a disease susceptibility locus, but it does greatly reduce the likelihood that the polymorphism has any causal relationship with disease susceptibility. The precise location of a disease gene, relative to an associated marker allele, is also difficult to predict based on measures of disequilibrium, particularly when there may be more than one founder haplotype.
Several candidate genes have been localized to the region near D22S272 and D22S284. The implicated region contains more than 1 million base pairs of DNA and would be expected to contain more than 50 genes, most of which have not been identified. Genes that have been identified within several centimorgans of CYP2D6 include somatostatin receptor 3, interleukin-2 receptor b, platelet-derived growth factor-β polypeptide, adenylosuccinate lyase, N-acetylgalactosaminidase, thyroid autoantigen-ku, aconitase hydroxylase, diaphorase-cytochrome-b5 reductase, and the peripheral benzodiazepine receptor [24]. The collective findings suggest that a continued effort toward localization and positional cloning of the gene responsible for PD susceptibility on ch22q13 is warranted.
Acknowledgments
This study was supported by Federal grants from the National Institutes of Health, NS32527(R.M.), AG07232 (R.M.), and ES06831 (J.G.), and by grants from the Parkinson’s Disease Foundation and the John and Evelyn Kossak Foundation, Inc.
We are grateful for helpful comments and suggestions, and we thank Brenda Alfaro, BA, and Helen Mejia, BA, for their contribution to the interviews and the collection and organization of data.
References
- 1.Barbeau A, Cloutier T, Roy M, et al. Ecogenetics of Parkinson’s disease: 4-hydroxylation of debrisoquine. Lancet 1985;2:1213–1216 [DOI] [PubMed] [Google Scholar]
- 2.Armstrong M, Daly AK, Cholerton S, et al. Mutant debrisoquine hydroxylation genes in Parkinson’s disease. Lancet 1992; 339:1017–1018 [DOI] [PubMed] [Google Scholar]
- 3.Smith CA, Gough AC, Leigh PN, et al. Debrisoquine hydroxylase gene polymorphism and susceptibility to Parkinson’s disease. Lancet 1992;339:1375–1377 [DOI] [PubMed] [Google Scholar]
- 4.Kondo I, Kanazawa I. Association of Xba I allele (Xba I 44kb) of the human cytochrome P-450dbl(CYP2D6) gene in Japanese patients with idiopathic Parkinson’s disease In: Nagatsu T, Narabayashi H, Yoshida M, eds. Parkinson’s disease: from clinical aspects to molecular basis. New York: Springer-Verlag, 1991:111–117 [Google Scholar]
- 5.Kurth MC, Kurth JH. Variant cytochrome P450 CYP2D6 allelic frequencies in Parkinson’s disease. Am J Med Genet 1993; 48:166–168 [DOI] [PubMed] [Google Scholar]
- 6.Tsuneoka Y, Matsuo Y, Iwahashi K, et al. A novel cytochrome P-450IID6 mutant gene associated with Parkinson’s disease. J Biochem 1993;114:263–266 [DOI] [PubMed] [Google Scholar]
- 7.Daily AK, Armstrong M, Monkman SC, et al. Genetic and metabolic criteria for the assignment of debrisoquine 4-hydroxylation (cytochrome P4502D6) phenotypes. Pharmacogenetics 1991;1:33–41 [DOI] [PubMed] [Google Scholar]
- 8.Gough AC, Miles JS, Spurr NK, et al. Identification of the primary defect at the cytochrome P450 CYP2D locus. Nature 1990;347:773–776 [DOI] [PubMed] [Google Scholar]
- 9.Heim M, Meyer UA. Genotyping of poor metabolizers of debrisoquine by allele-specific PCR amplification. Lancet 1990;336:529–532 [DOI] [PubMed] [Google Scholar]
- 10.Agundez JAG, Jimenez-Jimenez FJ, Luengo A, et al. Association between the oxidative polymorphism and early onset Parkinson’s disease. Clin Pharmacol Ther 1995;57:291–298 [DOI] [PubMed] [Google Scholar]
- 11.Johansson I, Lundqvist E, Bertilsson L, et al. Inherited amplification of an active gene in the cytochrome P450 CYP2D locus as a cause of ultra-rapid metabolism of debrisoquine. Proc Natl Acad Sci USA 1993;90:11825–11829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandy MS, Armstrong M, Tanner CM, et al. CYP2D6 allelic frequencies in young-onset Parkinson’s disease. Neurology 1996;47:225–230 [DOI] [PubMed] [Google Scholar]
- 13.Logroscino G, Marder K, Cote L, et al. Dietary lipids and antioxidants in Parkinson’s disease: a population-based, case–control study. Ann Neurol 1996;39:89–94 [DOI] [PubMed] [Google Scholar]
- 14.Mayeux R, Marder K, Cote LJ, et al. The frequency of idiopathic Parkinson’s disease by age, ethnic group, and sex in northern Manhattan, 1988–1993. Am J Epidemiol 1995; 142: 820–827 [DOI] [PubMed] [Google Scholar]
- 15.Hughes AJ, Daniel SE, Kilford L, Lees AJ. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: a clinicopathological study of 100 cases. J Neurol Neurosurg Psychiatry 1992;55:181–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes AJ, Ben-Shlomo Y, Kilford L, Lees AJ. What features improve the accuracy of clinical diagnosis in Parkinson’s disease: a clinicopathologic study. Neurology 1992;42:1142–1146 [DOI] [PubMed] [Google Scholar]
- 17.Lasser DM, Wilhelmsen KC, Nygaard TG, Tantravahi U. Characterization of microsatellite polymorphisms DXS691 and DXS692: genetic mapping to Xq26.2-Xq27 and Xq25-Xq26.2. Genomics 1993;16:785–786 [DOI] [PubMed] [Google Scholar]
- 18.Matise T, Perlin M, Chakravarti A. Automated construction of genetic linkage maps using an expert system (Multimap): a human genome linkage map. Nature Genet 1994;6:384–390 [DOI] [PubMed] [Google Scholar]
- 19.Boehnke M, Lange K, Cox DR. Statistical methods for multipoint radiation hybrid mapping. Am J Hum Genet 1991;49:1174–1188 [PMC free article] [PubMed] [Google Scholar]
- 20.Diederich N, Hilger C, Goetz CG, et al. Genetic variability of the CYP 2D6 gene is not a risk factor for sporadic Parkinson’s disease. Ann Neurol 1996;40:463–465 [DOI] [PubMed] [Google Scholar]
- 21.Platé-Bordeneuve V, Davis MB, Maraganore DM, et al. Debrisoquine hydroxylase gene polymorphism in familial Parkinson’s disease. J Neurol Neurosurg Psychiatry 1994;57:911–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ott J Analysis of human genetic linkage. Baltimore: Johns Hopkins University Press, 1991:241–242 [Google Scholar]
- 23.Thompson G Identifying complex disease genes: progress and paradigms. Nature Genet 1994;8:108–110 [DOI] [PubMed] [Google Scholar]
- 24.Kim UJ, Shizuya H, Kang HL, et al. A bacterial artificial chromosome-based framework contig map of human chromosome 22q. Proc Natl Acad Sci USA 1996;93:6297–6301 [DOI] [PMC free article] [PubMed] [Google Scholar]