Abstract
Synovial sarcoma (SS) is an aggressive soft-tissue malignancy characterised by expression of SS18-SSX fusions, where treatment options are limited. To identify therapeutically actionable genetic dependencies in SS, we performed a series of parallel, high-throughput small interfering RNA (siRNA) screens and compared genetic dependencies in SS tumour cells to those in >130 non-SS tumour cell lines. This identified an unexpected reliance in SS tumour cells upon the DNA damage response kinase, ATR. Clinical ATR inhibitors (ATRi) also elicited a synthetic lethal effect in SS tumour cells and impaired the growth of SS patient-derived xenografts, suggesting this effect might have therapeutic potential. Oncogenic SS18-SSX family fusion genes alter the composition of the BAF chromatin-remodelling complex, causing ejection of wild-type SS18 and the tumour suppressor SMARCB1 from BAF and their eventual degradation. We found that expression of oncogenic SS18-SSX fusion proteins caused profound ATRi sensitivity and a reduction in SS18 and SMARCB1 protein levels but a SSX18-SSX1 Δ71-78 fusion with a C-terminal deletion did not, thus establishing a causative link between oncogenic SS18-SSX fusion genes and ATRi sensitivity. ATRi sensitivity in SS was characterised by an increase in biomarkers of replication fork stress (increased γH2AX, decreased replication fork speed and increased R-loops), an apoptotic response and was found to be dependent upon Cyclin E expression. Finally, we found that combinations with cisplatin or PARP inhibitors enhanced the anti-tumour cell effect of ATRi, suggesting that either single agent ATRi or combination therapy involving ATRi might be further assessed as candidate approaches for SS treatment.
Keywords: Synovial Sarcoma, ATR inhibitor, targeted therapy, replication fork stress, synthetic lethality
Introduction
Synovial sarcoma (SS) is a rare, yet aggressive and difficult to treat type of soft tissue sarcoma (STS) that has a variable age of onset but predominantly affects young adults. Although for patients with localized disease, wide surgical excision combined with radiotherapy can be curative, recurrent disease is common (1). In the metastatic setting, SS patients are treated with cytotoxic chemotherapies, including the topoisomerase inhibitor doxorubicin and/or the alkylating agent ifosfamide. Recently, the multi-kinase inhibitor pazopanib became the first targeted agent to be approved for the treatment of advanced SS after failure of anthracycline containing chemotherapy (2). Despite these multimodal therapy approaches, the outcome of metastatic SS patients remains poor; those with distant metastasis have a 10-year survival rate of only 8.9% compared to 69% for patients with localized tumours (3). These factors highlight that additional, more specific therapeutic approaches with greater efficacy are required to effectively manage this disease.
The main pathological driver event in SS is known, suggesting that in principle at least, mechanism-based targeted approaches to treating SS could be developed. The majority of SS are characterised by a t(X;18) reciprocal chromosomal translocation, often used as a diagnostic biomarker for the disease (4). These t(X;18) translocations fuse the first 10 exons of the SS18 (synovial sarcoma translocation, chromosome 18) gene to the last three exons of one of the SSX (synovial sarcoma, X breakpoint) family of genes, SSX1, SSX2 or SSX4 (4, 5), encoding either SS18-SSX1, SS18-SSX2, or SS18-SSX4 fusion proteins. SS display few other recurrent mutations (6).
A number of studies have aimed to identify the cellular functions of these oncogenic fusions as well as of their wild-type SS18 and SSX counterparts (7, 8). SS18-SSX oncoproteins contribute to the dysregulation of gene expression through association with SWI/SNF (BAF) and Polycomb chromatin remodelling complexes (9–11). BAF complexes mediate nucleosome remodelling via an ATP-dependent process and in doing so modulate transcription (12, 13), DNA repair and the maintenance of genomic integrity (13, 14). SS18-SSX1 fusion proteins displace wild-type SS18 and an additional BAF component, the tumour suppressor SMARCB1, from BAF complexes (7). The displacement of SMARCB1 from BAF leads to its proteasomal degradation, with reduced levels of BAF-associated SMARCB1 being a characteristic of SS tumour cell lines and tumours (7, 15).
Despite an enhanced understanding SS18-SSX function, therapeutic targeting of these oncogenic proteins has not yet been achieved. One of the more recently used approaches to identifying therapeutic targets in cancer has been to identify and exploit genetic dependencies, such as synthetic lethal and gene addiction effects, that are associated with particular cancer driver gene defects. The potential of such an approach is best exemplified by the use of small molecule PARP inhibitors in BRCA1/2 mutant cancers (16, 17). Since the key driver genotype of SS is well-established, we sought to apply a similar approach to identify synthetic lethal interactions in SS. This identified an unexpected dependency in SS tumour cells upon on the kinase ATR (Ataxia Telangiectasia mutated and Rad3-related), a key mediator of the DNA damage response (DDR) (18) that can be exploited with clinical ATR inhibitors.
Materials and Methods
Cell culture
Yamato-SS and Aska-SS cell lines were kindly provided by Kazuyuki Itoh and Norifumi Naka (Osaka Medical Center for Cancer and Cardiovascular Diseases, Osaka, Japan); Akira Kawai (National Cancer Center Hospital, Tokyo, Japan) provided SYO-1 cells and Cinzia Lanzi (Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, Italy) provided CME-1 cells. HS-SY-II cells were obtained from the RIKEN BioResource Center. HCT116 WT and ARID1A-mutant isogenic cell lines have been described previously (19); all other cell lines were supplied by ATCC. Cells were grown in 5% CO2 at 37°C in media described below, supplemented with 15% (HFFI) or 10% (all other cell lines) fetal calf serum (FCS, Gibco) and 1% Pen-Strep (Sigma). Media for cell lines was as follows: Yamato-SS = Dulbecco’s Modified Eagle Medium (DMEM; Gibco); Aska-SS= DMEM; SYO-1= DMEM; CME-1 = RPMI 1640 Medium (Gibco); HS-SY-II = DMEM; HCT116 = McCoy’s 5A Medium (Gibco); U2OS = DMEM; HFF1 = DMEM. All cell lines were obtained between 2010 and 2016 and their identities were confirmed by in-house STR typing and, where appropriate, by PCR/Sanger sequencing confirmation of SS-specific gene fusions. Mycoplasma testing was carried out on each cell line every five passages (all negative). Cells were grown for no longer than 25 passages in total for any experiment.
siRNA screens
Cells lines were reversed transfected with a Dharmacon SMARTpool 384 well plate-arrayed siRNA library designed to target 714 kinases and kinase-related genes, 320 Wnt-pathway associated genes, 80 tumour suppressor genes, and 480 genes recurrently altered in human cancers (Supplementary Table 1) as described in (20). Positive control (siPLK1) and multiple negative controls (siCON1 and siCON2; Dharmacon, catalogue numbers D-001210-01-20 and D-001206-14-20, and AllStar; QIAGEN, catalogue number 1027281) were included on every plate. Transfection reagents were as follows: SYO-1; Aska-SS; Yamato-SS and CME-1 = RNAiMAX (Invitrogen); HS-SY-II = Lipofectamine 2000 (Invitrogen). Screens were performed in triplicate. Cell viability was estimated five days after transfection using CellTiter-Glo assay (Promega). Data processing and quality controls were performed using the cellHTS2 R package as described previously (20, 21). In the case of the VX970 resistance screen (Figure 4), 24h after transfection, VX970 (final concentration 0.75 μM) or vehicle (DMSO) was added to cells. Cells were then exposed to VX970 for four days and viability estimated with CellTiter-Glo assay as above. Drug effect (DE) Z-scores were calculated as described previously (22).
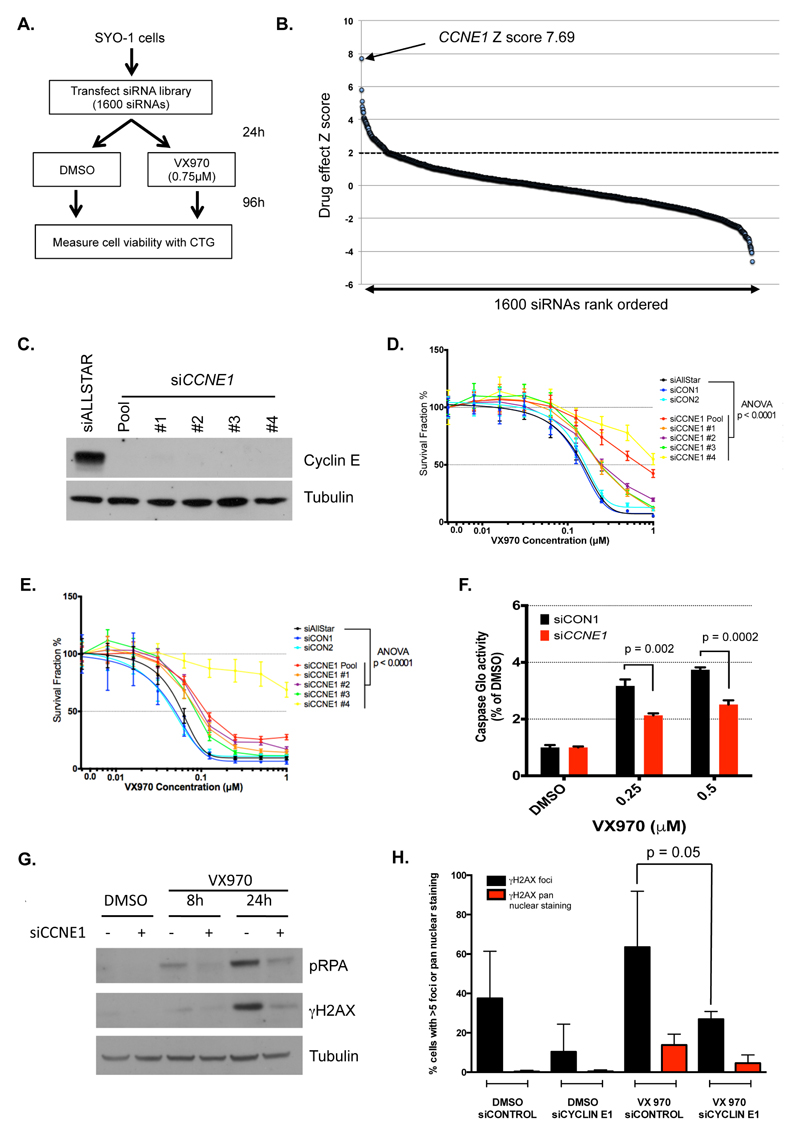
Figure 4. Sensitivity to ATRi in SS tumour cells is Cyclin E dependent.
A. Schematic illustrating VX970 resistance siRNA screen assay design. SYO-1 cells were transfected with the siRNA library and then exposed to either a high concentration of VX970 (0.75 μM) or DMSO. After four days continuous drug exposure, cell viability was assessed. B. Ordered scatter plot illustrating ATRi-resistance-causing effects identified in the screen described in (A). The effect of each siRNA SMARTpool on drug resistance was estimated by the calculation of Drug Effect (DE) Z scores, with DE Z-score > 2 (dashed line) regarded as significant effects. The effect of CCNE1 siRNA SMARTpool is highlighted. C. Western blot illustrating Cyclin E silencing in SYO-1 cells mediated by the CCNE1 SMARTpool (“Pool”) and the four different constituent siRNAs (#1-4). D-E. Dose-response curves showing effect of CCNE1 siRNAs on VX970 sensitivity in SYO-1 (D) and HS-SY-II (E) cells. ANOVA p-values represent 2-way ANOVA for each siRNA compared to non-targeting siAllStar. Error bars represent SD from triplicate experiments. F. Bar chart illustrating effect of siCCNE1 SMARTpool on apoptosis in SYO-1 cells. Apoptosis was measured by Caspase Glo assay after 48 hours of VX970 exposure. Error bars represent standard deviation from triplicate experiments. p values calculated by Student’s t test. G. Western blot showing effect of siCCNE1 SMARTpool on γH2AX and pRPA following 0.5 μM VX970 exposure in SYO-1 cells for 8 or 24 hours. H. Bar chart showing effect of siCCNE1 on accumulation of pan-nuclear γH2AX and foci after 24h exposure to VX970 in SYO-1 cells. Error bars represent standard deviation from triplicate experiments. p value calculated by Student’s t test.
Chemosensitisation screens, dose/response cell survival assays, assessment of apoptosis
Screens were carried out and analysed as in (22). In brief, SYO-1 and HS-SY-II cells were exposed to 0.04 μM VX970 plus a drug library of 79 small molecules (Supplementary Table 2), in a 384-well-plate format. After five days drug exposure, cell viability was estimated with CellTiter-Glo assay (Promega). Screens were carried out in triplicate. For dose/response cell survival assays, cells were plated in 384 well plates at 250-500 cells per well, and appropriate drug concentrations were added 24 hours later. Cell viability was estimated five days later using CellTiter-Glo. For drug combination assays, synergy was determined using MacSynergy II (23). Cell apoptosis was assessed using the Promega Caspase-Glo 3/7 assay as per the manufacturer’s instructions.
Western blotting
Cells were harvested and lysed using NET-N lysis buffer supplemented with protease and phosphatase inhibitors (Roche) and sonicated for five seconds. 10-30 μg of cell lysate was then diluted in NuPage sample reducing agent (Life Technologies) and NuPage SDS sample buffer (Life Technologies). Diluted samples were then boiled for 10 minutes and electrophoresed on a polyacrylamide gel. Electrophoresed proteins were then transferred onto a 0.45 μM nitrocellulose membrane (GE Healthcare), after which membranes were probed with primary antibody overnight at 4°C (primary antibodies list in Supplementary Table 3). Antibody binding was visualised with HRP-conjugated secondary antibody and SuperSignalTM West Pico Chemiluminescent Substrate (Thermo Scientific) and Super-RX X-ray film (Fujifilm).
Lentiviral SS18-SSX expression
SS18-SSX1, SS18-SSX2 and the SS18-SSX1 Δ71-78 mutant cDNAs were cloned into the pLX301 lentiviral transfer vector (7). For viral infection, cells were plated in 6-well plates and exposed to 0.5 ml lentiviral filtrate. After three days, cells were transferred to a T25 flask containing 1 μg/ml puromycin (Gibco) to remove non-transduced cells. After three days selection in puromycin, cells were harvested and used for further analysis. GFP expression was confirmed in GIPZ-infected cells using an EVOS FL fluorescence microscope (Life Technologies).
FACS analysis
Cells were cultured in 6-well plates and exposed to either 0.5 μM VX970 or DMSO. Cells were pulse-labelled with EdU for 1 hour prior to fixation in ice-cold 100% (v/v) ethanol and stored at -20°C until use. EdU was fluorescently labelled by conjugation to the Alexafluor-488 fluorophore using the Click-IT EdU Flow Cytometry Kit (Life Technologies) according to the manufacturer’s protocol. Total DNA content was assessed by incubation of cells for 15 minutes in 1.43 μM DAPI (Sigma) in PBS. Cell cycle profiles were generated using a BD LSR-II flow cytometer (BD Biosciences) and analysis performed using BD FACSDIVA software V8.01. Flow cytometry was performed according to the manufacturer’s guidelines.
Immunofluorescent imaging
Cells were plated onto poly-L-lysine Cellware 12mm round microscope cover slips (Corning) in 6-well plates and allowed to adhere overnight. After drug exposure, cells were fixed in 4% (v/v) PFA (Sigma) and permeabilised with 0.2% (v/v) Triton x100 (Sigma). Cells were then immunostained with antibodies targeting: γH2AX (Millipore, 05–636); 53BP1 (Novus, NB100-304); RAD51 (Abcam, AB133534); RNA/DNA hybrids (S9.6 ENH001, Kerafast); Nucleolin (Abcam, ab50279); or stained with DAPI. Cells were then fluorescent-labelled with secondary antibodies: goat anti-mouse IgG (ThermoFisher Scientific, Alexa Fluor 555, A-21424); or goat anti-rabbit IgG (ThermoFisher Scientific Alexa Fluor Plus 488, A32731) for 60 min at room temperature before mounting on slides. Slides were imaged at 63 x on a Leica TCS SP2 confocal microscope. Cells were scored positive for γH2AX if greater than five foci could be counted within the DAPI-stained nucleus. Cells with pan-nuclear γH2AX staining were scored separately from those with γH2AX foci. Approximately 120 cells were scored for γH2AX for each condition. For quantification of nuclear S9.6 intensity, 30 individual cells were scored per condition, and ImageJ was used to generate nuclear masks based on DAPI staining. Nuclear S9.6 fluorescence intensity was then determined by subtracting the nucleolin signal and analysing the intensity of the remaining S9.6 signal.
DNA fibre analysis
DNA fibre assays were carried out as described previously (24).
In vivo studies
In vivo efficacy studies were performed using SA13412 patient-derived SS xenografts at Crown Biosciences or using HS-SY-II tumour cells grown subcutaneously in the flank of female NOD SCID mice or Balb/C nude mice at the ICR, London, respectively. When tumours were established (~80 mm3), mice were randomised into treatment and vehicle groups. Animals bearing SA13412 PDXs were treated with either vehicle alone or VX970 (60mg/kg; oral 4 consecutive days/week) (n=10 per group). Animals bearing HS-SY-II tumours were treated with vehicle, VX970 monotherapy (60 mg/kg; oral 4 consecutive days/week), cisplatin monotherapy (3 mg/kg; intraperitoneally once weekly) or the combination (n=6-7 per group). Mice were treated until study end (6-12 weeks) or when the maximum tumour size was reached (defined as tumour size >15 mm in any direction for HS-SY-II experiments; or tumour volume >2000 mm3 for SA13412 PDX experiments). The SA13412 PDX study was carried out in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, USA, and the HS-SY-II experiment in accordance to ARRIVE guidelines, regulations set out in the UK Animals (Scientific Procedures) Act 1986, and in line with a UK Home Office approved project licence held by CJL and approved by the ICR ethics board.
Results
RNA interference profiling of SS cell lines identifies ATR as a candidate genetic dependency
To identify candidate therapeutic targets in SS, we carried out large-scale small interfering RNA (siRNA) screens in five commonly-used SS tumour cell lines; HS-SY-II, Aska-SS, Yamato-SS, CME-1 and SYO-1 (Figure 1A). These cell lines were selected as they harbour either one of the two most common SS-associated fusions (SS18-SSX1 or SS18-SSX2), and were amenable to high-efficiency siRNA transfection in a 384-well plate format (Figure 1B-D). After optimising high-throughput transfection conditions, each cell line was reverse-transfected with a 384-well plate-arrayed siRNA library designed to target 1600 genes (Supplementary Table 1 and Methods), including pharmacologically tractable kinase-coding and kinase-related genes, Wnt-pathway-associated genes given their role in SS (11, 25, 26) and genes recurrently altered in human cancers (27). Cell viability was measured after five days and this data then used to identify those siRNAs that caused significant tumour cell growth inhibition (i.e. genetic dependencies). Robust Z-scores were calculated from three replica screens (Methods and Supplementary Table 4) to estimate the effect of siRNAs on tumour cell inhibition.
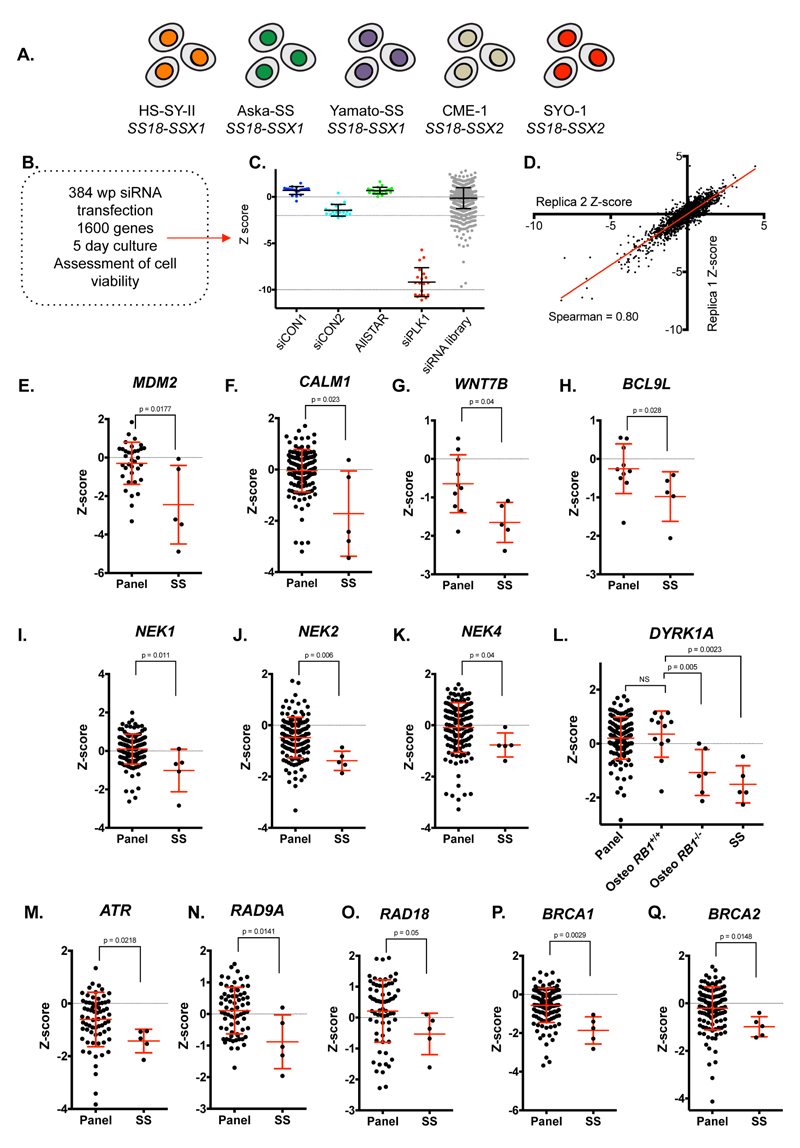
Figure 1. Genetic dependency profiling in synovial sarcoma cells identifies ATR as a candidate genetic dependency.
A. Overview of the five SS cells lines and their characteristic SS18-SSX driver fusions. B. Schematic of siRNA screening procedure and data processing. C. Scatter plot illustrating distribution of siRNA Z-scores for negative (siCON1, siCON2 and AllSTAR) and positive (siPLK1) controls, as well as Z-scores for the library siRNAs in the SYO-1 siRNA screen. Dots represent siRNA Z-scores from individual wells in the library. Each well contained a siRNA SMARTPool designed to target one gene. Error bars represent standard deviation. D. Scatter plot illustrating the correlation of siRNA Z-scores between replica 1 and replica 2 in the SYO-1 screen. E-Q. siRNA Z-score plots illustrating genetic dependencies associated with SS. Error bars represent standard deviation from median effects. p-value represents Mann-Whitney test.
To identify genetic dependencies associated with SS, we compared the siRNA Z-scores from the SS tumour cell line screens to siRNA Z-score profiles of >130 tumour cell lines from a diverse set of cancer histotypes (20). We noted a series of profound SS-specific genetic dependencies, including the proto-oncogene Mouse Double Minute 2 homologue (MDM2), the gene encoding the calcium-binding protein Calmodulin (CALM1) (Figure 1E,F), genes associated with Wnt-signalling (e.g. those encoding the Wnt ligand WNT7B and the β-catenin interacting protein, BCL9L (28) Figure 1G,H) and the druggable Never in Mitosis-A (NIMA) family members (29) NEK1, 2 and 4 (Figure 1I-K). The substantial variation in siRNA Z-scores within the SS tumour cell lines for these genes however suggested that these were not highly penetrant effects (i.e. profound effects in the vast majority of SS tumour cell lines) and might therefore have limited utility as targets in SS. More penetrant genetic dependencies associated with SS included the DREAM complex kinase, DYRK1A (Figure 1L) and the DNA Damage Response (DDR) kinase ATR (Figure 1M, described below). DYRK1A inhibition is synthetic lethal with RB1 (pRb) tumour suppressor defects in osteosarcoma (OS) (20). We found the sensitivity of SS tumour cell lines to DYRK1A siRNA to be of a scale equivalent to that in RB1 null OS tumour cell lines (Figure 1L), suggesting that DYRK1A, which is amenable to small molecule inhibition (20, 30), might be worthy of further assessment as a candidate therapeutic target in SS.
We also found a series of genes involved in the DNA Damage Response network (DDR) (31) to be candidate genetic dependencies in SS, including ATR, the ATR-activating proteins RAD9A and RAD18, and two tumour suppressor genes involved in double strand break repair by homologous recombination (HR), BRCA1 and BRCA2 (Figure 1M-Q), suggesting a reliance upon processes that are associated with the stability and repair of replication forks.
ATR genetic dependency in SS can be elicited with clinical ATR inhibitors
The identification of ATR as a candidate genetic dependency was particularly interesting for a number of reasons: (i) compared to other cancer histologies, we found the five SS tumour cell lines to be amongst the most sensitive tumour cell lines to ATR siRNA and to respond in a relatively consistent fashion (Figure 2A), suggesting a relatively penetrant effect; (ii) relatively little is understood about the sensitivity of SS tumours to small molecule DDR inhibitors, such as ATR inhibitors; and (iii) ATR inhibitors such as VX970 and AZD6738 have recently entered clinical trials for cancer treatment (e.g. clinicaltrials.gov NCT02157792, NCT02223923), suggesting that this genetic dependency could be clinically actionable.
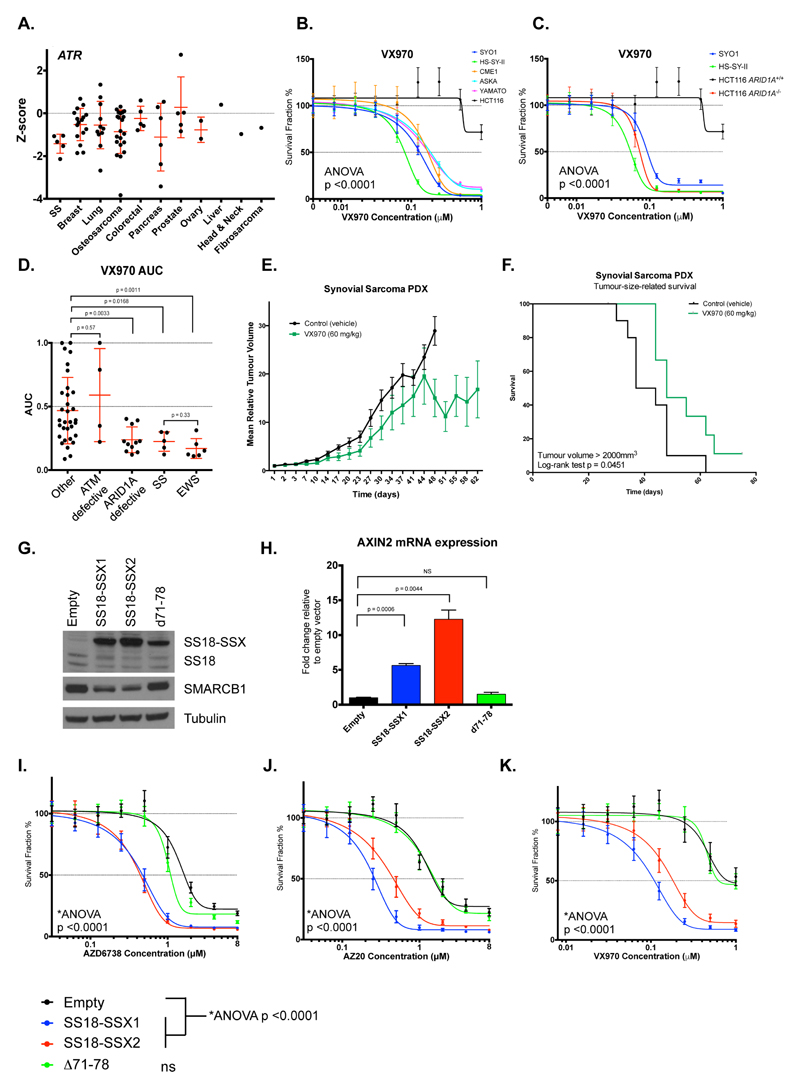
Figure 2. ATRi sensitivity is caused by SS fusion proteins.
A. Summary of siRNA Z-scores from the siRNA screen for ATR in cell lines – tumour cell lines are classified by cancer of origin. Error bars represent standard deviation. p-values were calculated using Mann-Whitney test. B. Dose-response curves from five-day survival assays, illustrating the sensitivity of SS tumour cell lines to the clinical ATR inhibitor VX970. ATRi resistant HCT116 cells were used as a negative control. p-value represents 2-way ANOVA compared to HCT116 cells. Error bars represent standard deviation (SD) from triplicate experiments. C. Dose-response curves from five-day survival assays, illustrating the ATRi sensitivity of SS tumour cell lines compared to HCT116 ARID1A+/+ and ARID1A-/- isogenic cell lines. p-value represents 2-way ANOVA compared to HCT116 ARID1A+/+ cells. Error bars represent SD from triplicate experiments. D. Area Under Curve (AUC) values for SS tumour cell lines screened for sensitivity to VX970 in five-day survival assays compared to non-SS tumour cells (“other”), ATM defective, ARID1A defective or Ewing’s sarcoma (EWS) tumour cell lines. p-values represent Mann-Whitney test. Error bars represent standard deviation. E. Tumour inhibition elicited by VX970 treatment in established SS PDX tumours. Mean relative tumour volume plot showing efficacy of VX970 (60mg/kg; oral 4 consecutive days/week) in SA13412 SS PDX models. p-values represent 2-way ANOVA of VX970-treated mean relative tumour volumes versus controls after 48 days treatment. Error bars represent standard error of the mean. F. Kaplan-Meier curves for tumour-size-related survival (defined by a tumour reaching volume of 2000mm3) demonstrating a significant delay in time to reach the tumour size limit in the VX970 treatment group as compared to controls. p-value represents log-rank test. G. Western blot illustrating ectopic expression of SS18-SSX1, SS18-SSX2 and Δ71-78 (d71-78) fusions in HCT116 cells. Ectopic expression of SS18-SSX1 and SS18-SSX2 reduced SMARCB1 protein levels. “Empty” = expression vector without cDNA insert. H. Bar chart illustrating the effect of SS18-SSX1, SS18-SSX2 and Δ71-78 (d71-78) fusions on transcription of the Wnt target gene AXIN2 in HCT116 cells. p-values represent Student’s t-test; error bars represent standard deviation from triplicate experiments. “Empty” = expression vector without cDNA insert. I-K. Dose-response curves illustrating the effect of SS18-SSX1, SS18-SSX2 and Δ71-78 fusion expression on sensitivity of HCT116 cells to AZD6738 (L), AZ20 (M) and VX970 (N) ATR inhibitors. Error bars represent SEM from triplicate experiments. p-values represent two-way ANOVA, ns = not significant (p>0.05 vs. empty). “Empty” = expression vector without cDNA insert.
Having confirmed the sensitivity of SS tumour cell lines to ATR siRNA in post-screen validation experiments (Supplementary Figure 1A-C), we assessed the sensitivity of the five SS tumour cell lines to the clinical ATR inhibitor (ATRi), VX970 (Vertex Pharmaceuticals/Merck KGaA). Compared to previously-validated ATRi resistant HCT116 colorectal cancer cells (32), all five SS cell lines were profoundly sensitive to VX970 (ANOVA p<0.0001), each exhibiting SF50 (concentration required to cause 50% reduction in cell survival) values of ≈ 0.1 µM (Figure 2B). SS tumour cell lines were also more sensitive to ATRi compared to HFF1 fibroblasts and non-tumour epithelial MCF10A cells (Supplementary Figure 1D). Defects in the tumour suppressor gene ARID1A cause ATRi sensitivity, both in vitro and in vivo (32). We found SS tumour cell lines to be as sensitive to VX970 as ARID1A-defective HCT116 cells (HCT116ARID1A–/–) (19) (Figure 2C). Furthermore, when comparing the VX970 sensitivity of SS tumour cell lines to those with other molecular defects associated with ATRi sensitivity, namely ATM gene defects (33), ARID1A mutations (32) and Ewing’s sarcoma (EWS) associated EWS-FLI fusions (34), the SS tumour cell lines showed a similar extent of VX970-sensitivity to EWS tumour cells and ARID1A-defective tumour cells (Figure 1D, Supplementary Figure 1E). This consistent in vitro sensitivity of SS tumour cell lines to ATRi was also observed with other ATR inhibitors, including the clinical ATRi AZD6738 (35) (AstraZeneca) and the toolbox inhibitors AZ20 (35) (AstraZeneca) and VE821 (36) (Vertex Pharmaceuticals) (Supplementary Figure 1F-H), suggesting that these effects were not specific to VX970 and represented a drug class effect. We next assessed whether the clinical ATRi VX970 could inhibit SS tumour growth in vivo. Treatment of mice bearing established patient-derived SS xenografts (PDX SA13412; SS18-SSX1 translocation-positive) with VX970 caused a significant inhibition of tumour growth (ANOVA p<0.0001, Figure 2E) and extended the survival of tumour-bearing mice (log-rank test p=0.0451, Figure 2F).
SS18-SSX1 or SS18-SSX2 fusion proteins induce ATRi sensitivity and reduce SMARCB1 and SS18 protein levels
To establish a causative link between the expression of oncogenic SS18-SSX fusions and ATRi sensitivity, we ectopically expressed SS18-SSX1 or SS18-SSX2 cDNAs in cells and assessed ATRi sensitivity. Many cell lines, including HFF1 fibroblasts, were unable to maintain long-term survival in the face of either SS18-SSX1 or SS18-SSX2 cDNA expression, precluding the use of these models in drug sensitivity assays where prolonged cell culture is required. However, we found that ATRi resistant HCT116 cells were able to tolerate lentiviral expression of SS18-SSX1 or SS18-SSX2 cDNA (Supplementary Figure 2A-C). Furthermore, expression of SS18-SSX1 or SS18-SSX2 in HCT116 cells recapitulated three features previously associated with SS fusion gene expression, namely: (i) a modest reduction in endogenous SS18 expression (7); (ii) a reduction in levels of the SWI/SNF component SMARCB1 (7); and (iii) upregulation of the canonical Wnt-pathway target gene AXIN2 (Figure 2G-H), effects replicated in short-term U2OS (osteosarcoma) and HFF1 (fibroblast) cell cultures (Supplementary Figure 2C-E). Amino acid residues at the C-terminus of the SS18-SSX1 fusion protein are critical for the displacement of SMARCB1 from SWI/SNF complexes (7). In comparison to the expression of full-length SS18-SSX1 or SS18-SSX2 fusion proteins, expression of an SS18-SSX1 variant with the final eight residues of SSX1 deleted (Δ71-78) did not cause a reduction in SMARCB1 levels, nor an increase in AXIN2 mRNA (Figure 2G-H). Having established that SS18-SSX fusion expression could recapitulate some of the molecular features associated with SS fusion gene expression, we assessed the effects of these fusions upon ATRi sensitivity. We found both SS18-SSX1 and SS18-SSX2 cDNAs caused a significant (ANOVA p<0.0001) enhancement in sensitivity to a series of distinct ATR inhibitors, whereas the expression of the SS18-SSX1 Δ71-78 mutant isoform did not (Figure 2I-K), establishing a causative link between the expression of oncogenic fusion genes and ATRi sensitivity. Furthermore, siRNA mediated gene silencing of SMARCB1 in HCT116 cells also caused VX970 sensitivity (ANOVA, p<0.001, Supplementary Figure 2F,G), suggesting that the SWI/SNF defect caused by SS fusion genes could, in principle, be responsible for ATR inhibitor sensitivity.
As defects in DNA damage response pathways, such as ATM and ATR activation, have been associated with increased ATRi sensitivity (33, 37), we assessed whether defects in ATM/ATR activation following DNA damage in SS tumour cell lines could explain the ATRi sensitivity. However, we did not detect a profound dysfunction in the ability of SS tumour cells to elicit ATR signalling in response to either cisplatin or hydroxyurea, or ATM signalling in response to ionising radiation (IR) (Supplementary Figure 3A-C). Furthermore, we found that SS tumour cells generated nuclear RAD51 foci in response to ATRi, hydroxyurea or IR (Supplementary Figure 3D), suggesting that a defect in RAD51-mediated DNA repair processes might not explain ATRi sensitivity. ATRi sensitivity has also been associated with a reduction in chromatin bound topoisomerase levels (32). Again, we did not observe a clear defect in chromatin-TOP2A levels that could explain the profound ATRi sensitivity (Supplementary Figure 3E-J).
ATRi causes apoptosis and replication fork stress in SS tumour cells
In addition to ATR, our siRNA screens suggested that SS tumour cells were also reliant upon additional proteins involved in the replication fork stress response, including RAD9A and RAD18 (Figure 1N-O). As well as finding that SS tumour cells exposed to ATRi exhibited biomarkers of an apoptotic response (caspase 3/7 and PARP1 cleavage (Figure 3A-B)), we noted that ATRi induced pan-nuclear phosphorylation of histone H2AX (γH2AX), which is a biomarker of replication fork stress (Figure 3B-E). To assess the replication fork stress phenotype in more detail, we used DNA fibre analysis (24) and found that VX970 exposure caused a significant reduction in replication fork speed in SYO-1 SS tumour cells (p<0.0001, paired t-test, Figure 3F, Supplementary Figure 4A-B). In addition, ectopic expression of SS18-SSX1 fusion protein in HCT116 cells caused a modest but significant decrease in replication fork speed (p<0.0001, paired t-test), an effect enhanced by exposure to VX970 (Figure 3G, Supplementary Figure 4C). This suggested that the SS18-SSX1 fusion protein might cause an increase in replication fork stress, which is in turn enhanced by ATR inhibition. One cause of replication fork stress is steric interference between DNA replication and transcriptional machineries, often associated with R-loops, DNA-RNA hybrid structures that activate ATR-mediated DNA damage responses (38). We reasoned that an enhanced transcriptional programme caused by SS fusion expression might cause R-loops. R-loop frequency can be estimated by the immunohistochemical detection of DNA-RNA hybrid structures (38). We found that the exposure of SYO-1 cells to VX970 enhanced the R-loop IHC signal (p<0.001, Student’s t-test, Figure 3H). The expression of the SS18-SSX1 fusion in HCT116 cells also caused a modest but significant increase in R-loop IHC signal (p<0.001, Student’s t-test), which was enhanced by the addition of VX970 (p<0.001, Student’s t-test, Figure 3I).
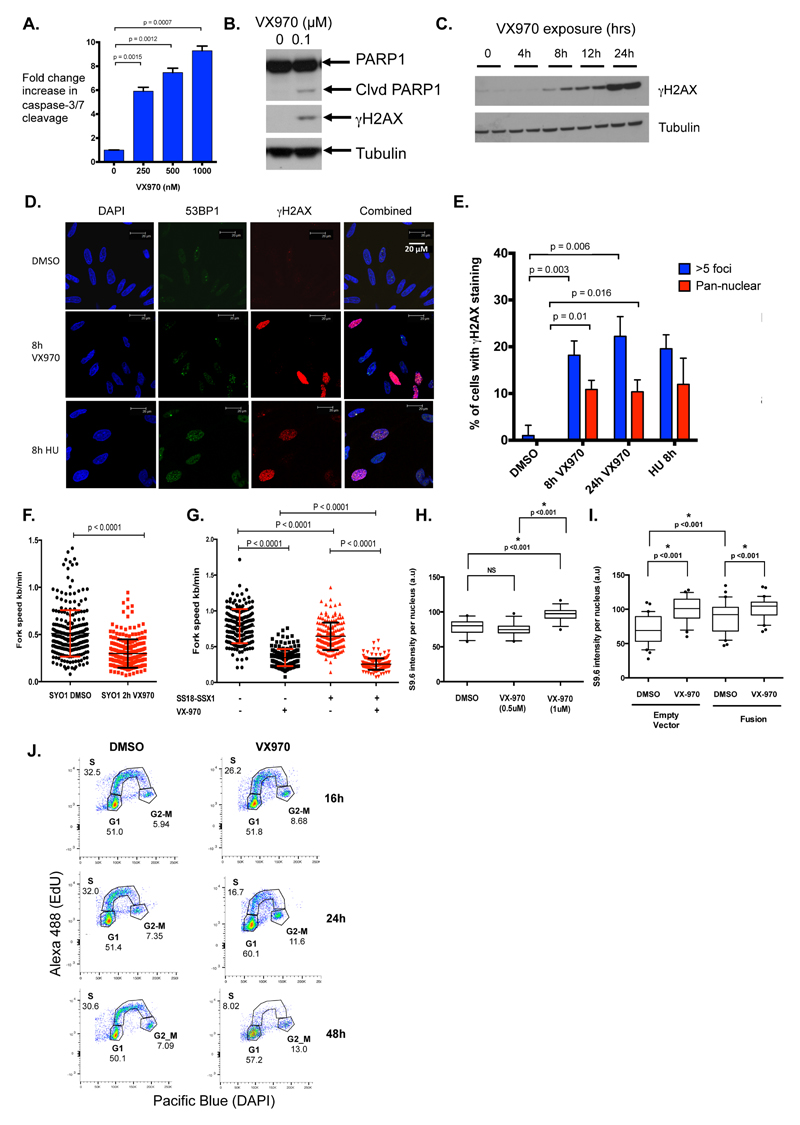
Figure 3. ATR inhibition causes apoptosis and replication fork stress in SS tumour cells.
A. Bar chart illustrating a dose-dependent increase in Caspase Glo activity in SYO-1 cells exposed to VX970 for 48h. Caspase Glo luminescence was normalized to cell viability as determined by CellTiter-Glo, and calculated as fold change compared to DMSO-exposed cells. p-values represent student’s t-test. Error bars represent standard deviation. B. Western blot illustrating accumulation of cleaved PARP1 and γH2AX upon VX970 exposure in SYO-1 SS cells. C. Western blot illustrating accumulation of γH2AX after exposure of HS-SY-II cells to 500 nM VX970. Two independently-derived lysates were generated at each time point as shown. D. Representative confocal microscopy images illustrating elevated 53BP1 (green) and γH2AX (red) foci/pan nuclear staining in SYO-1 cells following exposure to 500 nM VX970 (8 hours) or 2 mM hydroxyurea (HU; positive control), compared to DMSO-exposed controls. E. Bar chart showing percentage of SYO-1 cells with elevated γH2AX levels following exposure to 500 nM VX970 (h = hours of exposure). 100 images were captured and scored for >5 γH2AX foci or pan-nuclear γH2AX staining. HU exposure was used as a positive control. All p-values were calculated using student’s t-tests. Error bars represent standard deviation from triplicate experiments. F. Replication fork rates as measured from DNA fibre monitoring in SYO-1 cells exposed to DMSO or 500 nM VX970 for 2 hours. At least 100 tracks were measured for each condition. Average fork speed SYO-1 exposed to DMSO = 0.51 kb/min, average fork speed SYO-1 exposed to VX970 = 0.3 kb/min. Error bars represent standard deviation, p-value represents paired t-test. G. Replication fork rates as measured from DNA fibre monitoring in HCT116 cells expressing ectopic SS18-SSX1 cDNA exposed to DMSO or 500 nM VX970 for 2 hours. At least 100 tracks were measured for each condition. Error bars represent standard deviation, p-values represent paired t-test. H. Box and whisker plot of R-loop (S9.6 nuclear staining) intensity in SYO-1 cells exposed to VX970 for 4 hours. Nuclear S9.6 staining intensity levels were corrected for S9.6 nucleolar intensity. 30 individual cells were scored for each condition. Box represents 10-90% of data. p-values represent student’s t-test. I. Box and whisker plot of R-loop (S9.6 nuclear staining) intensity in HCT116 cells expressing ectopic SS18-SSX1 cDNA (“fusion”) exposed to DMSO or 500 nM VX970 for 4 hours. p-values represent student’s t-test. “Empty vector” = expression vector without cDNA insert. J. Cell cycle profiles for EdU incorporation in SYO-1 cells exposed to 0.5 μM VX970. DAPI was used to estimate DNA content. Numbers indicate fraction of cells present in the different cell cycle phases after 16, 24 and 48 hours VX970 exposure.
As previous studies have demonstrated that ATRi prevents normal replication of DNA under oncogene-induced replication fork stress (39), we performed FACS cell-cycle analysis to determine whether a similar effect operated in SS cells. SYO-1 cells were pulse-labelled with EdU, which is incorporated into DNA during active DNA synthesis. Cells exposed to VX970 exhibited a profound reduction in the fraction of cells in active S-phase (from 30% to 8% after 48 hours VX970 exposure (Figure 3J, Supplementary Figure 4D)), suggesting that ATRi exposure in SS tumour cells impaired DNA replication, consistent with our previous observations.
ATRi sensitivity in SS tumour cells is Cyclin E dependent
To gain more insight into the processes involved in ATRi sensitivity, we took a relatively unbiased approach using an siRNA “resistance” screen to identify genes that could reverse the ATRi sensitivity phenotype in a SS tumour cell line. SYO-1 cells were reverse-transfected in a 384-well plate format with the siRNA library described earlier, and 24h later exposed to either a relatively high concentration of VX970 (0.75 μM) or vehicle (DMSO) for four continuous days (Figure 4A). Results of three highly correlated replica screens were combined in the final analysis. By comparing siRNA effects in VX970-exposed vs. DMSO-exposed cells we identified those siRNAs that caused VX970 resistance (see Methods), quantifying resistance-causing effects as Drug Effect (DE) Z-scores (Supplementary Table 5, Figure 4B). The most profound ATRi resistance-causing effect identified in this screen was caused by siRNA designed to target the Cyclin E encoding gene CCNE1 (Figure 4B, DE Z-score 7.7). Multiple independent CCNE1 siRNAs derived from the SMARTpool reduced Cyclin E protein expression (Figure 4C), and caused a statistically significant reduction in ATRi sensitivity in subsequent dose-response survival assays in both SYO-1 and HS-SY-II SS cells (Figure 4D-E, ANOVA p<0.0001). We found that the CCNE1 siRNA SMARTpool used in the siRNA screen also reduced the extent of VX970-induced apoptosis in SYO-1 cells (Figure 4F). As Cyclin E has been implicated in enhancing replication fork stress (40, 41), we next tested the ability of CCNE1 siRNA to reverse the γH2AX response to ATRi exposure. We found that CCNE1 siRNA reduced both γH2AX and RPA phosphorylation in response to ATRi exposure (Figure 4G), an observation also reproduced by immunofluorescent detection of γH2AX (Figure 4H). However, Cyclin E protein expression itself was not regulated by ectopic expression of SS18-SSX1, SS18-SSX2 or SS18-SSX1 Δ71-78 fusions (Supplementary Figure 4E). Several likely explanations might account for the role of Cyclin E in these phenotypes; the presence of a SS fusion gene might create a genomic context (e.g. a chromatin remodelling event) that allows Cyclin E to mediate replication fork stress. Alternatively, the reduction in Cyclin E expression in SS tumour cells might independently reduce or delay entry into S phase, thus minimising the effects of an ATR inhibitor that would otherwise mediate its cytotoxic effects by targeting cells undergoing DNA replication. Indeed, we found that CCNE1 siRNA reduced the fraction of SS tumour cells in S phase (Supplementary Figure 4F,G), although such an observation might not completely discount the possibility that Cyclin E in S-phase might also play a role in ATRi sensitivity in SS cells
Identification of candidate drug combinations with ATR inhibitors in SS
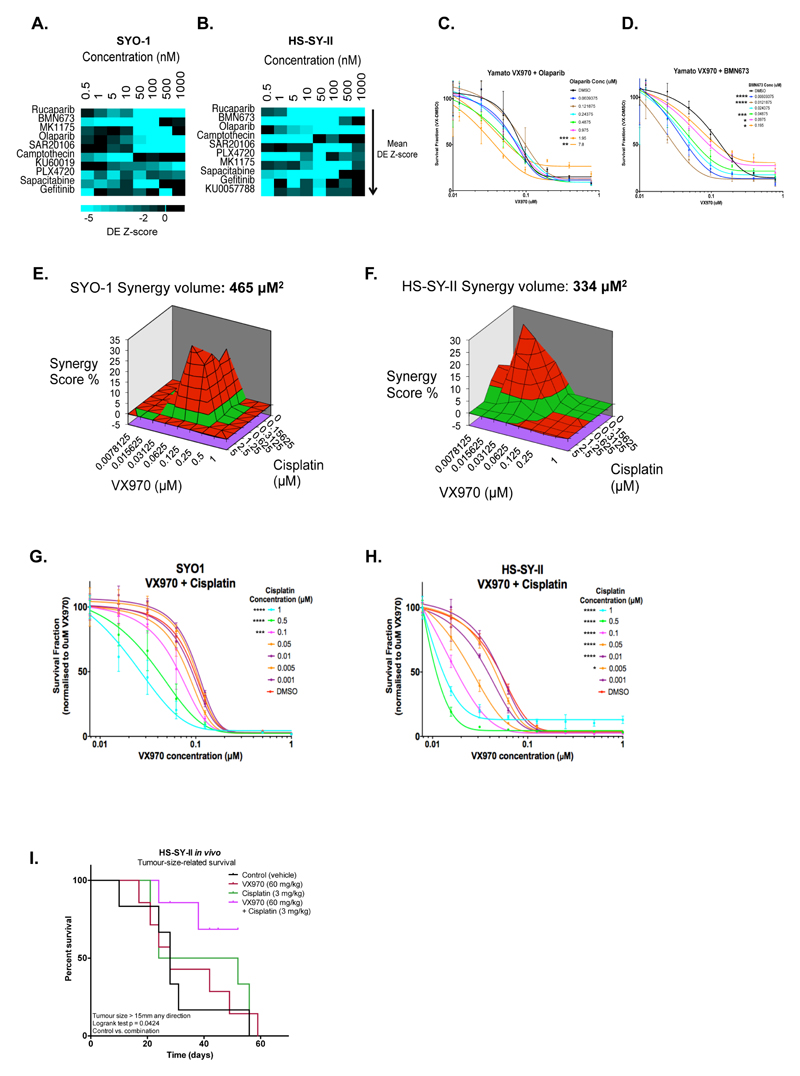
To improve the likelihood of a significant and sustained clinical response to ATR-targeted therapy, we also assessed the effects of combination therapy approaches with ATRi in SS tumour cells. To do this, we used high-throughput chemosensitisation screens in SYO-1 and HS-SY-II cells to identify ATRi combinatorial effects. Here, we used a bespoke drug library encompassing 79 different small molecules, which are either already used in cancer treatment or are in late-stage development (see Methods). To maximize the possibility of identifying drug combination effects, each small molecule in the library was used in eight concentrations (see Methods). From this screen, we identified several drugs that sensitised both SYO-1 and HS-SY-II cells to VX970. These included the three clinical PARP1 inhibitors rucaparib, olaparib and talazoparib (BMN673), the WEE1 kinase inhibitor MK1775, the CHK1 kinase inhibitor SAR20106, the Topoisomerase inhibitor camptothecin and the replication inhibitor sapacitabine (Figure 5A-B), all agents that either cause and/or enhance replication fork stress. In validation experiments in a third SS tumour cell line, Yamato-SS, we found that the addition of either olaparib or talazoparib enhanced the effects of VX970 (Figure 5C-D). Synergistic effects of combined ATR and PARP inhibition have been reported in other tumour types as well (42–44).
Figure 5. Identification of candidate drug combinations with ATR inhibitors in SS.
A-B. Heat maps summarising the results from high-throughput chemosensitisation screens using VX970 in SYO-1 (A) and HS-SY-II (B) cells. Drug sensitisation effects are shown as Drug Effect (DE) Z-scores for each concentration of the top 10 chemosensitising molecules out of 79 library small molecules used in combination with VX970. Concentration (nM) denotes the concentration of the library small molecules used in combination with VX970. Library small molecules are rank ordered top to bottom according to average VX970 DE Z-scores; library small molecules causing VX970 chemosensitisation are ranked at the top of the heat map. C-D. Dose-response curves in Yamato-SS cells exposed to escalating concentrations of VX970 combined with the PARP inhibitors olaparib (C) or talazoparib (BMN673) (D) for five days. Error bars represent standard deviation. p-values were calculated with a 2-way ANOVA compared to cells exposed to VX970 alone. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. E-F. MacSynergy plots for SYO-1 (E) and HS-SY-II (F) SS cells exposed to escalating concentrations of VX970 combined with cisplatin. 3D synergy plots represent synergy volumes in µM2; volumes > 120 µM2 were considered as synergistic (details in Methods) and are shown in bold. G-H. Dose-response curves for SYO-1 (G) and HS-SY-II (H) cells exposed to escalating concentrations of VX970 combined with cisplatin. Error bars represent standard deviation. p-values were calculated with a 2-way ANOVA compared to cells exposed to VX970 alone. *p < 0.05; ***p < 0.001; ****p < 0.0001. I. Kaplan-Meier survival curves for tumour-size-related survival (defined by a tumour reaching the tumour size limit of 15 mm in any one direction) demonstrating a significant delay in time to reach the tumour size limit in the combined treatment group as compared to controls. p-value calculated by log-rank test.
We also assessed known synergistic combinations with ATRi, as well as assessing the effects of combining ATRi with standard-of-care agents used in SS. For example, in non-SS tumour cells, the combination of VX970 with platinum salts has been reported as synergistic (33); we found that the combination of VX970 and cisplatin generated a synergistic effect on cell inhibition in both SYO-1 and HS-SY-II cells, generating synergy volumes of 465 μM2 and 334 μM2, respectively (Figure 5E-H). This combinatorial effect also elicited a survival benefit in mice bearing HS-SY-II SS tumour xenografts (p=0.0424, log-rank test, Figure 5I). Finally, we also assessed combinations of VX970 when used with cytotoxic agents used in the treatment of SS (doxorubicin and the alkylating chemotherapeutic cyclophosphamide) as well as the targeted drug used in SS treatment pazopanib (2). We observed largely non-synergistic effects in each of these cases, although in HS-SY-II cells, the combination of VX970 plus the active derivate of cyclophosphamide (4-HC) caused a mild synergistic effect (Supplementary Figure 5).
Discussion
In this study, high-throughput siRNA screening of SS cell lines identified a selective and novel dependency upon the DDR-related kinase ATR. Validation studies confirmed a synthetic lethal interaction between SS18-SSX fusion genes and ATR and established that ATRi might be used to exploit this effect.
The sensitivity of SS to DDR-targeting drugs was rather unexpected. To date, much of the focus on identifying tumour subtypes that might respond to targeted DDR targets has centred upon tumour subtypes such as high-grade serous ovarian cancers, where homologous recombination (HR) defects with expected genomic rearrangements exist (31). Although genomic instability has been reported in SS, this is generally restricted to adult patients and those with advanced disease; the majority of patients with SS have tumours that do not exhibit mutations or chromosomal alterations other than a pathognomonic SS18-SSX translocation (6, 45, 46). Interestingly, ATRi sensitivity has also been recently reported in models of Ewing’s sarcoma, a sarcoma of adolescent and young adult (AYA) age that is generally characterised by a single oncogenic fusion driver event (EWS-FLI translocations) (34). In our in vitro assays, SS cells appeared to have comparable ATRi sensitivity to EWS-FLI1 positive Ewing’s sarcoma tumour cells. This might suggest that assessing ATR inhibitor sensitivity in additional AYA and/or other translocation-associated sarcoma subtypes might also identify similar vulnerabilities.
At the mechanistic level, our data suggest that the expression of SS18-SSX fusion genes generates a relatively moderate form of replication fork stress that is enhanced by ATR inhibition (for example, Figure 3F-G). Our analysis of ATRi sensitivity in SS tumour cells suggests that this phenotype is dependent on a known mediator of replication fork stress, Cyclin E (40, 41), (Figure 4) but is not necessarily caused by an overt increase in Cyclin E expression, profound defects in either ATM, ATR or chromatin bound topoisomerase levels or a defect in RAD51 localisation to the site of DNA damage (Supplementary Figure 3). Another possibility might be that the expression of SS fusion genes and the subsequent effects on SWI/SNF function cause subtle yet critical differences in the chromatin structure of the genome that induces an increased impairment of replication fork progression and thus an enhanced reliance upon ATR (and sensitivity to ATRi); this model would be a form of induced essentiality (47).
Supplementary Material
Acknowledgements
We thank John Pollard and Philip Reaper (Vertex) for providing ATR inhibitors. Akira Kawai (National Cancer Center Hospital, Tokyo, Japan; SYO-1), Kazuyuki Itoh and Norifumi Naka (Osaka Medical Center for Cancer and Cardiovascular Diseases, Osaka, Japan; Yamato-SS and Aska-SS), and Cinzia Lanzi (Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, Italy; CME-1) for kindly providing the SS cell lines. We also thank Cigal Kadoch (Dana Farber/Harvard Cancer Center, Boston, USA) for useful discussions. This work was funded by a Cancer Research UK Programme Grant (Grant Number C347/A8363) to CJL. SEJ was supported by a Wellcome Trust Studentship. EDGF is supported by a Rubicon Fellowship (NWO 019.153LW.035). We acknowledge NIHR funding to the Royal Marsden Biomedical Research Centre.
Financial support:
This work was funded by a Cancer Research UK Programme Grant (Grant Number C347/A8363) to C.J. Lord. S.E. Jones was supported by a Wellcome Trust Studentship. E.D.G. Fleuren is supported by a Rubicon Fellowship (NWO 019.153LW.035). We acknowledge NIHR funding to the Royal Marsden Biomedical Research Centre.
Footnotes
Conflict of interest: SEJ, CTW, AA and CJL are inventors on patents describing the use of of DNA repair inhibitors and stand to gain from their use as part of the ICR “Rewards to Inventors” scheme.
References
- 1.Nielsen TO, Poulin NM, Ladanyi M. Synovial sarcoma: recent discoveries as a roadmap to new avenues for therapy. Cancer Discov. 2015;5:124–34. doi: 10.1158/2159-8290.CD-14-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Graaf WT, Blay JY, Chawla SP, Kim DW, Bui-Nguyen B, Casali PG, et al. Pazopanib for metastatic soft-tissue sarcoma (PALETTE): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet. 2012;379:1879–86. doi: 10.1016/S0140-6736(12)60651-5. [DOI] [PubMed] [Google Scholar]
- 3.Sultan I, Rodriguez-Galindo C, Saab R, Yasir S, Casanova M, Ferrari A. Comparing children and adults with synovial sarcoma in the Surveillance, Epidemiology, and End Results program, 1983 to 2005: an analysis of 1268 patients. Cancer. 2009;115:3537–47. doi: 10.1002/cncr.24424. [DOI] [PubMed] [Google Scholar]
- 4.Clark J, Rocques PJ, Crew AJ, Gill S, Shipley J, Chan AM, et al. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet. 1994;7:502–8. doi: 10.1038/ng0894-502. [DOI] [PubMed] [Google Scholar]
- 5.Amary MF, Berisha F, Bernardi Fdel C, Herbert A, James M, Reis-Filho JS, et al. Detection of SS18-SSX fusion transcripts in formalin-fixed paraffinembedded neoplasms: analysis of conventional RT-PCR, qRT-PCR and dual color FISH as diagnostic tools for synovial sarcoma. Mod Pathol. 2007;20:482–96. doi: 10.1038/modpathol.3800761. [DOI] [PubMed] [Google Scholar]
- 6.Vlenterie M, Hillebrandt-Roeffen MH, Flucke UE, Groenen PJ, Tops BB, Kamping EJ, et al. Next generation sequencing in synovial sarcoma reveals novel gene mutations. Oncotarget. 2015;6:34680–90. doi: 10.18632/oncotarget.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell. 2013;153:71–85. doi: 10.1016/j.cell.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su L, Sampaio AV, Jones KB, Pacheco M, Goytain A, Lin S, et al. Deconstruction of the SS18-SSX fusion oncoprotein complex: insights into disease etiology and therapeutics. Cancer Cell. 2012;21:333–47. doi: 10.1016/j.ccr.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia CB, Shaffer CM, Alfaro MP, Smith AL, Sun J, Zhao Z, et al. Reprogramming of mesenchymal stem cells by the synovial sarcoma-associated oncogene SYT-SSX2. Oncogene. 2012;31:2323–34. doi: 10.1038/onc.2011.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagai M, Tanaka S, Tsuda M, Endo S, Kato H, Sonobe H, et al. Analysis of transforming activity of human synovial sarcoma-associated chimeric protein SYT-SSX1 bound to chromatin remodeling factor hBRM/hSNF2 alpha. Proc Natl Acad Sci U S A. 2001;98:3843–8. doi: 10.1073/pnas.061036798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trautmann M, Sievers E, Aretz S, Kindler D, Michels S, Friedrichs N, et al. SS18-SSX fusion protein-induced Wnt/beta-catenin signaling is a therapeutic target in synovial sarcoma. Oncogene. 2014;33:5006–16. doi: 10.1038/onc.2013.443. [DOI] [PubMed] [Google Scholar]
- 12.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–92. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 13.Smith-Roe SL, Nakamura J, Holley D, Chastain PD, 2nd, Rosson GB, Simpson DA, et al. SWI/SNF complexes are required for full activation of the DNA-damage response. Oncotarget. 2015;6:732–45. doi: 10.18632/oncotarget.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen J, Peng Y, Wei L, Zhang W, Yang L, Lan L, et al. ARID1A Deficiency Impairs the DNA Damage Checkpoint and Sensitizes Cells to PARP Inhibitors. Cancer Discov. 2015;5:752–67. doi: 10.1158/2159-8290.CD-14-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito J, Asano N, Kawai A, Yoshida A. The diagnostic utility of reduced immunohistochemical expression of SMARCB1 in synovial sarcomas: a validation study. Hum Pathol. 2015 doi: 10.1016/j.humpath.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 17.Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017;355:1152–58. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karnitz LM, Zou L. Molecular Pathways: Targeting ATR in Cancer Therapy. Clin Cancer Res. 2015;21:4780–5. doi: 10.1158/1078-0432.CCR-15-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller RE, Brough R, Bajrami I, Williamson CT, McDade S, Campbell J, et al. Synthetic Lethal Targeting of ARID1A-Mutant Ovarian Clear Cell Tumors with Dasatinib. Mol Cancer Ther. 2016;15:1472–84. doi: 10.1158/1535-7163.MCT-15-0554. [DOI] [PubMed] [Google Scholar]
- 20.Campbell J, Ryan CJ, Brough R, Bajrami I, Pemberton HN, Chong IY, et al. Large-Scale Profiling of Kinase Dependencies in Cancer Cell Lines. Cell Rep. 2016;14:2490–501. doi: 10.1016/j.celrep.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boutros M, Bras LP, Huber W. Analysis of cell-based RNAi screens. Genome Biol. 2006;7:R66. doi: 10.1186/gb-2006-7-7-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lord CJ, McDonald S, Swift S, Turner NC, Ashworth A. A high-throughput RNA interference screen for DNA repair determinants of PARP inhibitor sensitivity. DNA Repair (Amst) 2008;7:2010–9. doi: 10.1016/j.dnarep.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 23.Prichard MN, Shipman C., Jr A three-dimensional model to analyze drug-drug interactions. Antiviral Res. 1990;14:181–205. doi: 10.1016/0166-3542(90)90001-n. [DOI] [PubMed] [Google Scholar]
- 24.Schwab RA, Niedzwiedz W. Visualization of DNA replication in the vertebrate model system DT40 using the DNA fiber technique. J Vis Exp. 2011:e3255. doi: 10.3791/3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barham W, Frump AL, Sherrill TP, Garcia CB, Saito-Diaz K, VanSaun MN, et al. Targeting the Wnt pathway in synovial sarcoma models. Cancer Discov. 2013;3:1286–301. doi: 10.1158/2159-8290.CD-13-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vijayakumar S, Liu G, Rus IA, Yao S, Chen Y, Akiri G, et al. High-frequency canonical Wnt activation in multiple sarcoma subtypes drives proliferation through a TCF/beta-catenin target gene, CDC25A. Cancer Cell. 2011;19:601–12. doi: 10.1016/j.ccr.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de la Roche M, Worm J, Bienz M. The function of BCL9 in Wnt/beta-catenin signaling and colorectal cancer cells. BMC Cancer. 2008;8:199. doi: 10.1186/1471-2407-8-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fry AM, O'Regan L, Sabir SR, Bayliss R. Cell cycle regulation by the NEK family of protein kinases. J Cell Sci. 2012;125:4423–33. doi: 10.1242/jcs.111195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sonamoto R, Kii I, Koike Y, Sumida Y, Kato-Sumida T, Okuno Y, et al. Identification of a DYRK1A Inhibitor that Induces Degradation of the Target Kinase using Co-chaperone CDC37 fused with Luciferase nanoKAZ. Sci Rep. 2015;5:12728. doi: 10.1038/srep12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouwman P, Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat Rev Cancer. 2012;12:587–98. doi: 10.1038/nrc3342. [DOI] [PubMed] [Google Scholar]
- 32.Williamson CT, Miller R, Pemberton HN, Jones SE, Campbell J, Konde A, et al. ATR inhibitors as a synthetic lethal therapy for tumours deficient in ARID1A. Nat Commun. 2016;7:13837. doi: 10.1038/ncomms13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol. 2011;7:428–30. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 34.Nieto-Soler M, Morgado-Palacin I, Lafarga V, Lecona E, Murga M, Callen E, et al. Efficacy of ATR inhibitors as single agents in Ewing sarcoma. Oncotarget. 2016;7:58759–67. doi: 10.18632/oncotarget.11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Llona-Minguez S, Hoglund A, Jacques SA, Koolmeister T, Helleday T. Chemical strategies for development of ATR inhibitors. Expert Rev Mol Med. 2014;16:e10. doi: 10.1017/erm.2014.10. [DOI] [PubMed] [Google Scholar]
- 36.Charrier JD, Durrant SJ, Golec JM, Kay DP, Knegtel RM, MacCormick S, et al. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J Med Chem. 2011;54:2320–30. doi: 10.1021/jm101488z. [DOI] [PubMed] [Google Scholar]
- 37.Mohni KN, Kavanaugh GM, Cortez D. ATR pathway inhibition is synthetically lethal in cancer cells with ERCC1 deficiency. Cancer Res. 2014;74:2835–45. doi: 10.1158/0008-5472.CAN-13-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nat Rev Genet. 2015;16:583–97. doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- 39.Schoppy DW, Ragland RL, Gilad O, Shastri N, Peters AA, Murga M, et al. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. J Clin Invest. 2012;122:241–52. doi: 10.1172/JCI58928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones RM, Mortusewicz O, Afzal I, Lorvellec M, Garcia P, Helleday T, et al. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene. 2013;32:3744–53. doi: 10.1038/onc.2012.387. [DOI] [PubMed] [Google Scholar]
- 41.Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol. 2011;18:721–7. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim H, George E, Ragland R, Rafial S, Zhang R, Krepler C, et al. Targeting the ATR/CHK1 Axis with PARP Inhibition Results in Tumor Regression in BRCA-Mutant Ovarian Cancer Models. Clin Cancer Res. 2017;23:3097–108. doi: 10.1158/1078-0432.CCR-16-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohni KN, Thompson PS, Luzwick JW, Glick GG, Pendleton CS, Lehmann BD, et al. A Synthetic Lethal Screen Identifies DNA Repair Pathways that Sensitize Cancer Cells to Combined ATR Inhibition and Cisplatin Treatments. PLoS One. 2015;10 doi: 10.1371/journal.pone.0125482. e0125482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yazinski SA, Comaills V, Buisson R, Genois MM, Nguyen HD, Ho CK, et al. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev. 2017;31:318–32. doi: 10.1101/gad.290957.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lagarde P, Przybyl J, Brulard C, Perot G, Pierron G, Delattre O, et al. Chromosome instability accounts for reverse metastatic outcomes of pediatric and adult synovial sarcomas. J Clin Oncol. 2013;31:608–15. doi: 10.1200/JCO.2012.46.0147. [DOI] [PubMed] [Google Scholar]
- 46.Przybyl J, Sciot R, Wozniak A, Schoffski P, Vanspauwen V, Samson I, et al. Metastatic potential is determined early in synovial sarcoma development and reflected by tumor molecular features. Int J Biochem Cell Biol. 2014;53:505–13. doi: 10.1016/j.biocel.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 47.Tischler J, Lehner B, Fraser AG. Evolutionary plasticity of genetic interaction networks. Nat Genet. 2008;40:390–1. doi: 10.1038/ng.114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.