Abstract
Background
Behçet’s disease (BD) is a polygenic immune-mediated disorder characterized by a close association with the HLA-B*51 allele. The HLA region has a strong linkage disequilibrium (LD) and carries several genetic variants (e.g. MIC-A, TNF-α genes) identified as associated to BD because of their LD with HLA-B*51. In fact, the HLA-B*51 is inherited as part of extended HLA haplotypes which are well preserved in patients with BD. Sardinian population is highly differentiated from other Mediterranean populations because of a distinctive genetic structure with very highly preserved HLA haplotypes.
Patients and methods
In order to identify other genes of susceptibility to BD within the HLA region we investigated the distribution of human Allograft Inflammatory Factor-1 (AIF-1) gene variants among BD patients and healthy controls from Sardinia. Six (rs2736182; rs2259571; rs2269475; rs2857597; rs13195276; rs4711274) AIF-1 single nucleotide polymorphisms (SNPs) and related extended haplotypes have been investigated as well as their LD within the HLA region and with HLA-B*51. Overall, 64 BD patients, 43 HLA-B*51 positive healthy controls (HC) and 70 random HC were enrolled in the study.
Results
HLA-B*51 was the only allele with significantly higher frequency (pc = 0.0021) in BD patients (40.6%) than in HC (9.8%). The rs2259571T AIF-1 variant had a significantly reduced phenotypic, but not allelic frequency in BD patients (72.1%; pc = 0.014) compared to healthy population (91.3%). That was likely due to the LD between HLA-B*51 and rs2259571G (pc = 9E-5), even though the rs2259571G distribution did not significantly differ between BD patients and HC.
Conclusion
No significant difference in distribution of AIF-1 SNPs haplotypes was observed between BD patients and HC and between HLA-B*51 positive BD patients and HLA-B*51 positive HC. Taken together, these results suggest that AIF-1 gene is not associated with susceptibility to BD in Sardinia.
Introduction
Behçet’s disease (BD) is a chronic vasculitis characterized by recurrent oral ulcers, genital ulcers, ocular and skin manifestations with involvement of arteries and veins of all sizes. BD clusters in an area between latitudes 30° N and 45° N spanning from the far Eastern Asia to the Mediterranean basin [1,2]. Such a distinctive clustering seems related to geographical distribution of genetic susceptibility factors among general population [3,4]. As described in detail previously [5,6], several lines of evidence suggest that host genetic factors play a pivotal role in determining susceptibility to BD and its close association with the HLA-B*51 allele represents the clearest evidence of a genetic contribution to the disease. However, HLA-B*51 alone is neither necessary nor sufficient to BD development and other susceptibility genes, whose products are responsible for inflammatory and immune-mediated mechanisms, have been identified both outside and within the HLA region [6–8]. Genome wide association studies (GWAS) identified a strong association between BD and the HLA region comprehensive of HLA-B*51 within an extended haplotype [9]. These findings were suggestive of the presence of additional genes within the HLA region conferring susceptibility to BD. Actually, additional genes within the region such as other HLA class I alleles (e.g. B*15, B*57, A*26), the TNF-α and the MHC Class I chain-related gene A (MIC-A) have been associated to an increased risk of BD [10–12].
We previously pointed out that [13], in Sardinia, the BD-associated HLA-B*51:01 allele is inherited as part of a haplotype which is different from that characterizing the B*51:01-positive healthy controls. The HLA haplotype distribution in Sardinians, compared to other Mediterranean populations, is characterised by a small number of preserved and highly frequent haplotypes and by a very high number of rare haplotypes [14,15]. Therefore, the peculiar genetic background of Sardinians represents a valuable source for studying HLA-related genetic and epigenetic associations to BD [16].
Human Allograft Inflammatory Factor-1 (AIF-1) is a 143 amino acid, 17 kDa, cytoplasmic calcium-binding protein, encoded within the HLA class III region on chromosome 6p21 which is densely clustered with genes involved in the inflammatory responses including TNF-α. Because its pro-inflammatory role, AIF-1 is involved in various inflammatory pathological processes such as allograft rejection, autoimmune diseases, inflammatory central nervous system injury. Several single-nucleotide polymorphisms (SNPs) have been identified in the AIF-1 gene as associated with autoimmune diseases [17,18]. Considering its position within the HLA region, between the TNF-α gene promoter and the HLA-B locus, and its pro-inflammatory activity we deemed interesting to study AIF-1 as a candidate gene for BD susceptibility.
The objective of this study was to investigate the association of selected AIF-1 SNPs with susceptibility to BD in Sardinian and their distribution within distinct HLA extended haplotypes harbouring the HLA-B*51 allele.
Materials and methods
Patients and controls
Overall, 64 unrelated consecutive Sardinian patients with BD referring to the Rheumatology Unit of Cagliari and classified according to the 1990 International Study Group criteria, were enrolled in this study between January 2014 and December 2016 (Table 1).
Table 1. Cumulative features of patients suffering from Behçet’s disease enrolled in this study.
| Features | N (%) |
|---|---|
| Gender | F/M = 2/1 |
| Age at diagnosis (mean ± SD) | 31.0 ± 9.7 |
| Oral ulcers | 64 (100) |
| Genital ulcers | 41 (64) |
| Cutaneous involvement | 36 (56.2) |
| Ocular involvement | 33 (51.4) |
| Neurological involvement | 6 (9.4) |
| Vascular involvement | 14 (21.8) |
| Musculoskeletal involvement | 21 (32.8) |
| Pathergy test positive | 7 (10.9) |
Unless otherwise specified, numbers are absolute values, number in brackets are percentage.
Overall, 43 consecutive HLA-B*51 positive and 70 unselected healthy bone marrow donors served as controls. All patients and controls, matched for gender, came from various areas of Sardinia and were representative of the islander population distribution. Both patients and controls gave their written informed consent to the study which protocol was specifically approved by the local ethics committee “Comitato Etico Indipendente AOU Cagliari” (n. 224/CE).
Genotyping
Peripheral blood from BD patients and HC was drawn in EDTA-containing vials and genomic DNA was extracted using the Nucleic Acid Extraction and Cell Separation Instrument (Manufacturer: DiaSorin Inc.). The amount of DNA was determined using the Qubit fluorometric quantitation that comprises the Qubit 3.0 Fluorometer and sensitive, specific Qubit quantitation DNA assay (Thermo Fisher Scientific). All patients and controls were genotyped for 6 different SNPs of the AIF-1 gene (Table 2) by the reverse sequence-specific oligonucleotide polymerase chain reaction (PCR) technique using TaqMan SNP Genotyping Assays from Life Technologies according to the manufacturer’s protocol.
Table 2. Features of the six SNPs of the AIF-1 gene typed.
| SNP ID | Location | Polymorphism | Molecular Consequences |
|---|---|---|---|
| rs2736182 | Chr.6: 31615535 | A/G, Transition Substitution | 2KB upstream variant, missense variant, 5’ UTR variant |
| rs2259571 | Chr.6: 31616050 | G/T;Transversion substitution | Intron variant 5’ UTR variant |
| rs2269475 | Chr.6: 31616154 | C/T, Transition Substitution | Intron variant, missense variant |
| rs2857597 | Chr.6: 31617223 | A/T, Transversion Substitution | 500B downstream variant |
| rs13195276 | Chr.6: 31616317 | C/T, Transition Substitution | intron variant, missense variant |
| rs4711274 | Chr.6: 31615389 | A/G, Transition Substitution | 2KB upstream variant, intron variant |
Source: Database of Short Genetic Variation (dbSNP)–NCBI- NIH.
Patients and controls were also typed for HLA-A, B, C, DRB1, DQA1 and DQB1 using commercial kits (HLA SSP kits; Biotest, Dreieich, Germany) in order to identify a different distribution of the AIF-1 SNPs in the extended HLA haplotypes.
It is well known that choosing preliminary candidate SNPs is critical for candidate gene association studies. The chosen SNPs were based on previously described associations in various immune-mediated diseases [17,18], as well as according to functional features deemed interesting by the authors.
Statistical analysis
Hardy–Weinberg equilibrium (HWE) was tested using the Chi-square test. To assess differences in the proportions of AIF-1 polymorphic alleles and disease associations in healthy controls (HC) versus BD patients, chi-square test or two-tailed Fisher’s exact test, for low frequency, were performed using MedCalc software (version 16.8.4, Mariakerke, Belgium). The strength of association was estimated by calculating the odds ratio (OR) with 95% Confidence Interval (95% CI). Under the assumption of independence, a value of p<0.05 was considered statistically significant where Bonferroni correction was applied for multiple comparisons to all novel associations, with a correction factor derived from the number of alleles examined; pc indicates where the Bonferroni correction was applied. The LD among the 6 SNPs of the AIF-1 gene and between single SNPs and HLA-B*51 was calculated using the HaploView 4.2 software.
Results
HLA-B*51 phenotype frequency was significantly higher (pc = 0.0021; OR = 6.2; 95%CI 2.5 to 15.8) in BD patients (40.6%) than in HC (9.8%). No other HLA class I and II alleles were independently associated with BD.
Six SNPs in AIF-1 were determined in 64 BD patients, 43 HLA-B*51 positive HC and 70 HC (Table 3).
Table 3. Phenotypic frequencies of the 6 AIF-1 SNPs determined in BD patients and healthy controls according to their HLA-B*51 status.
| SNP ID | Genotype/allele | BDn (%) | BD B*51+n (%) | HCn (%) | HC B*51+n (%) | BD vs. HCPc |
|---|---|---|---|---|---|---|
| rs2736182 | G | 64 (100) | 26 (100) | 70 (100) | 43 (100) | Ns |
| A | 5 (7.8) | 4 (15.4) | 5 (7.1) | 2 (4.6) | Ns | |
| G/G | 59 (92.2) | 22 (84.6) | 65 (92.8) | 41(95.3) | Ns | |
| A/A | 0 | 0 | 0 | 0 | Ns | |
| A/G | 5 (7.8) | 4 (15.4) | 5 (7.1) | 2 (4.6) | Ns | |
| rs2259571 | T | 44 (72.1) | 13 (56.5) | 63(91.3) | 27 (62.8) | 0.014 |
| G | 43 (70.5) | 18 (78.3) | 43(62.3) | 38 (88.4) | Ns | |
| T/T | 18 (29.5) | 5 (21.7) | 26 (37.7) | 5 (11.6) | Ns | |
| G/G | 17 (27.9) | 10 (43.5) | 6 (8.7) | 16 (37.2) | Ns | |
| T/G | 26 (42.6) | 8 (34.8) | 37 (53.6) | 22 (51.2) | Ns | |
| rs2269475 | C | 62 (100) | 25 (100) | 69 (100) | 43 (100) | Ns |
| T | 5 (8.1) | 3 (12.0) | 3 (4.3) | 4 (9.3) | Ns | |
| C/C | 57 (91.9) | 22 (88.0) | 66 (95.6) | 39 (90.7) | Ns | |
| T/T | 0 | 0 | 0 | 0 | Ns | |
| C/T | 5 (8.1) | 3 (12.0) | 3 (4.3) | 4 (9.3) | Ns | |
| rs2857597 | T | 5 (7.8) | 3 (11.5) | 13 (18.6) | 4 (9.3) | Ns |
| A | 64 (100) | 26 (100) | 69 (98.6) | 43 (100) | Ns | |
| T/T | 0 | 0 | 1 (1.4) | 0 | Ns | |
| A/A | 59 (92.2) | 23 (88.5) | 57 (81.4) | 39 (90.7) | Ns | |
| A/T | 5 (7.8) | 3 (11.5) | 12 (17.1) | 4 (9.3) | Ns | |
| rs13195276 | C | 63 (100) | 25 (100) | 69 (100) | 43 (100) | Ns |
| T | 63(100) | 25 (100) | 69 (100) | 43 (100) | Ns | |
| C/C | 0 | 0 | 0 | 0 | Ns | |
| T/T | 0 | 0 | 0 | 0 | Ns | |
| C/T | 63 (100) | 25 (100) | 69 (100) | 43 (100) | Ns | |
| rs4711274 | G | 64 (100) | 26 (100) | 70 (100) | 39 (90.7) | Ns |
| A | 5 (7.8) | 3 (11.5) | 3 (4.3) | 0 | Ns | |
| G/G | 59 (92.2) | 23 (88.5) | 67 (95.7) | 39 (90.7) | Ns | |
| A/A | 0 | 0 | 0 | 0 | Ns | |
| A/G | 5 (7.8) | 3 (11.5) | 3 (4.3) | 0 | Ns |
Numbers are absolute values, number in brackets are percentages. ns: not significant
Five out of 6 AIF-1 SNPs (rs2736182; rs2269475; rs2857597; rs13195276; rs4711274) did not show different allelic and phenotypic distribution between patients and HC. Only the rs2259571 SNP had a significantly decreased phenotypic, but not allelic, frequency of the rs2259571T variant in BD patients (72.1%, Chi squared 9.31, pc = 0.014) compared to healthy population (91.3%) without a significantly different phenotypic distribution of the rs2259571G variant between BD patients and HC despite its LD with the HLA-B*51 (pc = 9E-5). Noteworthy, the rs2259571T phenotypic frequency distribution did not significantly differ between HLA-B*51 positive BD patients (56.5%) and HLA-B*51 positive HC (62.8%).
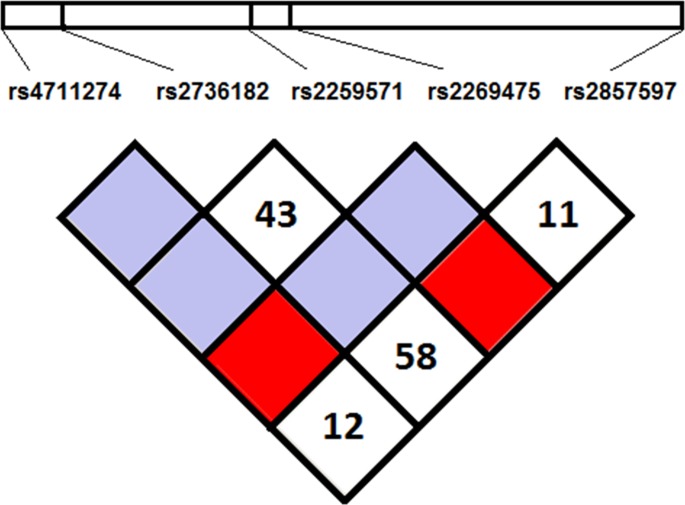
Analysing the distribution of AIF-1 SNPs haplotypes, no significantly different haplotype distribution between BD patients and HC was detected (Fig 1). The GGTCA (haplotype frequency 42.0% in BD and 48.0% in HC) and GGGCA (haplotype frequency 47.6% in BD and 35.5% in HC) were the most frequently detected AIF-1 SNP haplotypes. As effect of the LD between rs2259571G and HLA-B*51 the GGGCA was found at a higher frequency in HLA-B*51 positive subjects (56.3%) and the GGTCA was most frequently carried by HLA-B*51 negative subjects (52.9%) irrespective of the disease status.
Fig 1. Linkage disequilibrium plot of AIF-1 SNPs in HLA-B*51 positive and HLA-B*51 negative subjects irrespective of the disease status.
The Haploview software automatically omitted rs13195276 SNP from the model because equally expressed in both populations. Values of the pair-wise D’ (multiply by 100) are shown in each white square. 1: rs4711274; 2: rs2736182; 3: rs2259571; 4: rs2269475; 6: rs2857597. Bright red blocks, D′ (normalized linkage disequilibrium measure or D) = 1.0, with logarithm of odds (LOD) score ≥ 2.0; white blocks, D′ < 1.0 with LOD < 2.0; blue blocks, D′ = 1.0 with LOD < 2.0. Numbers in blocks denote D′ values. The genomic organization is described above the LD plot. LOD was defined as log10(L1/L0), where L1 = likelihood of the data under linkage disequilibrium, and L0 = likelihood of the data under linkage equilibrium. D′ was calculated as follows: D′ = (D) divided by the theoretical maximum for the observed allele frequencies.
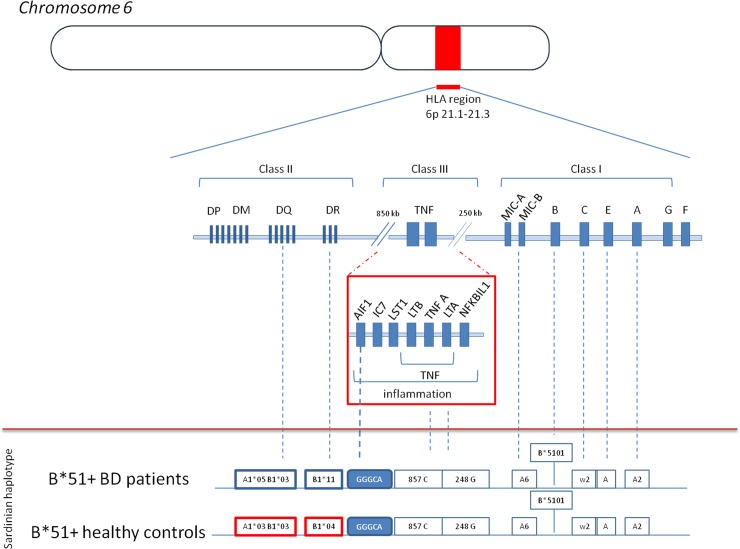
Finally, HLA-B*51 was harboured in two HLA-B-HLA-DR distinct haplotypes: B*51-DR*11 in 14/26 (53.8%) of HLA-B*51 positive BD patients and in 14/37 (37.8%) of HLA-B*51 positive HC (p = 0.231); and B*51-DR*4 in 1/26 (3.8%) of HLA-B*51 positive BD patients and in 14/37 (37.8%) of HLA-B*51 positive HC (p = 0.013; pc = 0.065 OR 0.06 95%CI 0.01–0.54) according to, but not fully confirming, the previous observation of a lack of association between B*51-DR*4 and BD susceptibility in Sardinians (13). No significant difference in the distribution of AIF-1 single or combined SNPs was observed between the B*51-DR*11 and B*51-DR*4 extended haplotypes (Fig 2).
Fig 2. Schematic representation of the HLA region and extended HLA haplotypes harbouring the HLA-B*51 and AIF-1 alleles in BD patients and healthy controls from Sardinia.
Discussion
The present study firstly reports on the association of six AIF-1 SNPs with BD susceptibility. The results showed no significant association between each investigated SNP or SNP haplotypes and BD in Sardinia. Despite of the LD between rs2259571G and HLA-B*51, the frequency of each SNP and the related GGGCA haplotype was not significantly increased in BD patients. As an effect of the LD with HLA-B*51, the rs2259571G was harboured in both B*51-DR*11 and B*51-DR*4 haplotypes, the latter having different distribution in BD patients and HC from Sardinia. These results suggest that polymorphisms of AIF-1 are not associated with the susceptibility to BD in the Sardinian population.
BD is considered as a complex polygenic disorder with a mixed autoinflammatory and autoimmune pathogenesis [19]. GWAS identified several susceptibility loci associated with BD susceptibility [9,20–23], but always confirmed the major role of the HLA region and especially of the HLA-B*51 allele in BD susceptibility [9,20–23]. Nevertheless, the highest contribution of HLA-B*51 to the overall BD genetic susceptibility was estimated to be only 19% [24]. GWAS also confirmed a strong LD in the HLA region of BD patients, mainly due to the fact that HLA-B*51 was found almost exclusively on a single extended haplotype [9]. In Sardinia, two distinct extended haplotypes harbouring HLA-B*51:01 were identified: A2-Cw2-B*5101-DRB1*11-DQA1*05-DQB1*03, which marks the B*51 positive patients with BD in Sardinia, and A2-Cw2-B*5101-DRB1*04-DQA1*03-DQB1*03, which is significantly more frequent in Sardinian HC than in BD [13]. Considering the high LD in the HLA region, it is conceivable that genes besides HLA-B*51, somewhat involved in the innate and adaptive immune responses and inherited as part of distinct HLA-B*51:01 haplotypes, may play a role in BD susceptibility.
Because of its position within the HLA class III region, between the HLA-B and HLA-DR loci, and its pro-inflammatory effect, the AIF-1 gene was deemed as a possible determinant of genetic susceptibility to BD. In humans, in fact, AIF-1 influences the immune system at several key points and boosts the expression of inflammatory mediators such as cytokines (IL6, TNF-α), chemokines, inducible nitric oxide synthase and promotes inflammatory cell proliferation and migration [25]. Moreover, AIF-1 is involved in some model of autoimmune diseases such as experimental autoimmune uveitis, encephalomyelitis and neuritis [26,27]. The role of AIF-1 in rheumatoid arthritis and systemic sclerosis has been investigated and the rs2269475 SNP was found associated with an increased risk of developing both diseases [28,29]. Although preliminary data pointed to a possible role of AIF-1 in BD susceptibility, we did not find any suggestion for this in our study population.
To the best of our knowledge, this was the first study investigating the role of AIF-1 in BD. With the aim to elucidate the genetic basis of BD we set a candidate gene case-control association study and we tested six different AIF-1 SNPs. Major strengths are represented by the peculiar genetic background of Sardinians and by the enrolment of two different control populations allowing to identify a different distribution of AIF-1 in patients and in controls but also in HLA-B*51 carriers versus other subjects. A major limitation is related to sample size, therefore caution is advised when interpreting results as they may be related to the small size of the population under study.
In conclusion, the present study does not support the hypothesis that a genetically determined regulation of AIF-1 expression or change in protein structure may predispose to the development of BD in Sardinian patients. Further, larger studies are required to confirm our findings in other populations.
Supporting information
Genotyping results for AIF1 are reported here.
(XLSX)
Acknowledgments
Authors would like to thank all patients and controls participating in this study.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Ohno S, Ohguchi M, Hirose S, Matsuda H, Wakisaka A, Aizawa M. Close association of HLA-Bw51 with Behçet's disease. Arch Ophthalmol. 1982;100:1455–8. [DOI] [PubMed] [Google Scholar]
- 2.Verity DH, Marr JE, Ohno S, Wallace GR, Stanford MR. Behçet's disease, the Silk Road and HLA-B51: historical and geographical perspectives. Tissue Antigens. 1999;54:213–20. [DOI] [PubMed] [Google Scholar]
- 3.Piga M, Mathieu A. The origin of Behçet's disease geoepidemiology: possible role of a dual microbial-driven genetic selection. Clin Exp Rheumatol. 2014;32(S84):S123–9. [PubMed] [Google Scholar]
- 4.Ozturk O, Arikan S, Bahadir A, Atalay A, Atalay EO. Genetic origin of Behçet's disease population in Denizli, Turkey; population genetics data analysis; historical demography and geographical perspectives based on β-globin gene cluster haplotype variation. Genes Immun. 2017;18:28–32. 10.1038/gene.2016.46 [DOI] [PubMed] [Google Scholar]
- 5.de Menthon M, Lavalley MP, Maldini C, Guillevin L, Mahr A. HLA-B51/B5 and the risk of Behçet's disease: a systematic review and meta-analysis of case-control genetic association studies. Arthritis Rheum. 2009;61:1287–96. 10.1002/art.24642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piga M, Mathieu A. Genetic susceptibility to Behçet's disease: role of genes belonging to the MHC region. Rheumatology (Oxford). 2011;50:299–310. [DOI] [PubMed] [Google Scholar]
- 7.Direskeneli H. Autoimmunity vs autoinflammation in Behçet’s disease: do we oversimplify a complex disorder? Rheumatology 2006;45:1461–5. 10.1093/rheumatology/kel329 [DOI] [PubMed] [Google Scholar]
- 8.Kirino Y, Bertsias G, Ishigatsubo Y, Mizuki N, Tugal-Tutkun I, Seyahi E, et al. Genome-wide association analysis identifies new susceptibility loci for Behçet's disease and epistasis between HLA-B*51 and ERAP1. Nat Genet. 2013;45:202–7. 10.1038/ng.2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Remmers EF, Cosan F, Kirino Y, Ombrello MJ, Abaci N, Satorius C, et al. Genome-wide association study identifies variants in the MHC class I, IL10, and IL23R-IL12RB2 regions associated with Behçet's disease. Nat Genet. 2010;42:698–702. 10.1038/ng.625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ombrello MJ, Kirino Y, de Bakker PIW, Gül A, Kastner DL, Remmers EF. Behçet disease-associated MHC class I residues implicate antigen binding and regulation of cell-mediated cytotoxicity. PNAS 2014. 111 (24) 8867–8872. 10.1073/pnas.1406575111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Liao D, Yang L, Hou S. Association between Functional MICA-TM and Behçet's Disease: A Systematic Review and Meta-analysis. Sci Rep. 2016. February 15;6:21033 10.1038/srep21033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmad T, Wallace GR, James T, Neville M, Bunce M, Mulcahy-Hawes K, et al. Mapping the HLA association in Behçet’s disease: a role for tumor necrosis factor polymorphisms? Arthritis Rheum 2003;48:807–13. 10.1002/art.10815 [DOI] [PubMed] [Google Scholar]
- 13.Piga M, Paladini F, Lai S, Erre G, Passiu G, Carcassi C, et al. Genetics of Behçet's disease in Sardinia: two distinct extended HLA haplotypes harbour the B*51 allele in the normal population and in patients. Clin Exp Rheumatol. 2012;30(S72):51–56. [PubMed] [Google Scholar]
- 14.Contu L, Arras M, Carcassi C, La Nasa G, Mulargia M: HLA structure of the Sardinian population: a haplotype study of 551 families. Tissue Antigens 1992;40:165–74. [DOI] [PubMed] [Google Scholar]
- 15.Ll Cavalli-Sforza, Menozzi P, Piazza A: The history and geography of human genes. Abridged ed Princeton, N.J.; Chichester: Princeton University Press, 1996; 1994. [Google Scholar]
- 16.Erre GL, Piga M, Carru C, Angius A, Carcangiu L Piras M, et al. Global microRNA profiling of peripheral blood mononuclear cells in patients with Behçet's disease. Clin Exp Rheumatol. 2015. Nov-Dec;33(S94):S72–9. [PubMed] [Google Scholar]
- 17.Nishimura M, Obayashi H, Mizuta I, Hara H, Adachi T, Ohta M, et al. TNF, TNF receptor type 1, and allograft inflammatory factor-1 gene polymorphisms in Japanese patients with type 1 diabetes. Hum Immunol. 2003. February;64(2):302–9. [DOI] [PubMed] [Google Scholar]
- 18.Pawlik A, Kurzawski M, Szczepanik T, Dziedziejko V, Safranow K, Borowiec-Chłopek Z, et al. Association of allograft inflammatory factor-1 gene polymorphism with rheumatoid arthritis. Tissue Antigens. 2008. August;72(2):171–5. 10.1111/j.1399-0039.2008.01086.x [DOI] [PubMed] [Google Scholar]
- 19.Grateau G, Hentgen V, Stojanovic KS, Jéru I, Amselem S, Steichen O. How should we approach classification of autoinflammatory diseases? Nat Rev Rheumatol. 2013;9:624–9. 10.1038/nrrheum.2013.101 [DOI] [PubMed] [Google Scholar]
- 20.Mizuki N, Meguro A, Ota M, Ohno S, Shiota T, Kawagoe T, et al. Genome-wide association studies identify IL23R-IL12RB2 and IL10 as Behçet's disease susceptibility loci. Nat Genet. 2010. August;42(8):703–6. 10.1038/ng.624 Epub 2010 Jul 11. [DOI] [PubMed] [Google Scholar]
- 21.Takeuchi M, Mizuki N, Meguro A, Ombrello MJ, Kirino Y, Satorius C, et al. Dense genotyping of immune-related loci implicates host responses to microbial exposure in Behçet's disease susceptibility. Nat Genet. 2017;49:438–443. 10.1038/ng.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes T, Coit P, Adler A, Yilmaz V, Aksu K, Düzgün N, et al. Identification of multiple independent susceptibility loci in the HLA region in Behçet's disease. Nat Genet. 2013. March;45(3):319–24. 10.1038/ng.2551 Epub 2013 Feb 10. [DOI] [PubMed] [Google Scholar]
- 23.Kirino Y, Bertsias G, Ishigatsubo Y, Mizuki N, Tugal-Tutkun I, Seyahi E et al. Genome-wide association analysis identifies new susceptibility loci for Behçet's disease and epistasis between HLA-B*51 and ERAP1. Nat Genet. 2013;45:202–7. 10.1038/ng.2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gul A, Hajeer AH, Worthington J, Barrett JH, Ollier WE, Silman AJ. Evidence for linkage of the HLA-B locus in Behçet’s disease, obtained using the transmission disequilibrium test. Arthritis Rheum 2001;44:239–40. [DOI] [PubMed] [Google Scholar]
- 25.Zhao YY, Yan DJ, Chen ZW. Role of AIF-1 in the regulation of inflammatory activation and diverse disease processes. Cell Immunol. 2013;284:75–83. 10.1016/j.cellimm.2013.07.008 [DOI] [PubMed] [Google Scholar]
- 26.Schluesener HJ, Seid K, Meyermann R. Effects of autoantigen and dexamethasone treatment on expression of endothelialmonocyte activating polypeptide II and allograft-inflammatory factor-1 by activated macrophages and microglial cells in lesions of experimental autoimmune encephalomyelitis, neuritis and uveitis. Acta Neuropathol. (Berl.), 1999;97:119–126. [DOI] [PubMed] [Google Scholar]
- 27.Storch MK, Weissert R, Steffer A, Birnbacher R, Wallstrom E, Dahlman I, et al. MHC gene related effects on microglia and macrophages in experimental autoimmune encephalomyelitis determine the extent of axonal injury. Brain Pathol. 2002;12:287–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alkassab F, Gourh P, Tan FK, McNearney T, Fischbach M, Ahn C, et al. An allograft inflammatory factor 1 (AIF1) single nucleotide polymorphism (SNP) is associated with anticentromere antibody positive systemic sclerosis. Rheumatology (Oxford). 2007;46:1248–51. [DOI] [PubMed] [Google Scholar]
- 29.Pawlik A, Kurzawski M, Szczepanik T, Dziedziejko V, Safranow K, Borowiec-Chłopek Z, et al. Association of allograft inflammatory factor-1 gene polymorphism with rheumatoid arthritis. Tissue Antigens. 2008;72:171–5. 10.1111/j.1399-0039.2008.01086.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genotyping results for AIF1 are reported here.
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.