Abstract
Muscle proteins with ankyrin repeats (MARPs) ANKRD1 and ANKRD2 are titin-associated proteins with a putative role as transcriptional co-regulators in striated muscle, involved in the cellular response to mechanical, oxidative and metabolic stress. Since many aspects of the biology of MARPs, particularly exact mechanisms of their action, in striated muscle are still elusive, research in this field will benefit from novel animal model system. Here we investigated the MARPs found in zebrafish for protein structure, evolutionary conservation, spatiotemporal expression profiles and response to increased muscle activity. Ankrd1 and Ankrd2 show overall moderate conservation at the protein level, more pronounced in the region of ankyrin repeats, motifs indispensable for their function. The two zebrafish genes, ankrd1a and ankrd1b, counterparts of mammalian ANKRD1/Ankrd1, have different expression profiles during first seven days of development. Mild increase of ankrd1a transcript levels was detected at 72 hpf (1.74±0.24 fold increase relative to 24 hpf time point), while ankrd1b expression was markedly upregulated from 24 hpf onward and peaked at 72 hpf (92.18±36.95 fold increase relative to 24 hpf time point). Spatially, they exhibited non-overlapping expression patterns during skeletal muscle development in trunk (ankrd1a) and tail (ankrd1b) somites. Expression of ankrd2 was barely detectable. Zebrafish MARPs, expressed at a relatively low level in adult striated muscle, were found to be responsive to endurance exercise training consisting of two bouts of 3 hours of forced swimming daily, for five consecutive days. Three hours after the last exercise bout, ankrd1a expression increased in cardiac muscle (6.19±5.05 fold change), while ankrd1b and ankrd2 were upregulated in skeletal muscle (1.97±1.05 and 1.84±0.58 fold change, respectively). This study provides the foundation to establish zebrafish as a novel in vivo model for further investigation of MARPs function in striated muscle.
Introduction
The MARP family of stress responsive proteins is composed of three members: cardiac ankyrin repeat protein (ANKRD1/CARP), ankyrin repeat domain protein 2 (ANKRD2/ARPP) and diabetes related ankyrin repeat protein (ANKRD23/DARP) [1]. Their expression is mainly localized to cardiac and skeletal muscles, but to a different extent. In mammals, ANKRD1 and ANKRD2 proteins are predominantly expressed in cardiac and skeletal muscles, respectively [2–5], while DARP transcript is equally distributed between these tissues [6]. MARPs are implicated in a number of functions, ranging from mechanosensing to modulation of different signaling pathways and transcriptional regulation [1].
In striated muscle these proteins respond to various forms of mainly mechanical stress [7–14] which affect their expression level and cellular distribution. After prolonged stretch ANKRD1 and DARP proteins redistribute to the nucleus of fetal rat cardiac myocytes [15], while Ankrd2 gene expression is upregulated [10, 16]. Shuttling of ANKRD2 to the nucleus was observed in stressed mouse muscle fibers [17] and myofibers with damaged sarcomeres [18]. Accordingly, it is proposed that MARPs link the myofibrillar stress-related signaling pathways and muscle gene expression via stress-induced relocation from the cytoplasm to the nucleus [15]. ANKRD1 is involved in cardiomyocyte stress-response networks activated by myocardial infarction or pressure overload that leads to hypertrophy and heart failure [19]. It appears that ANKRD1 may have a more general role in mediating stress response, since its level is highly induced during the healing process of skin wounds in mice [20]. Overexpression of human ANKRD1 has been shown to improve several aspects of wound healing, including neoangiogenesis [21]. Apart from mechanical stress, oxidative stress was found to regulate intracellular localization of ANKRD2 [22], causing nuclear shuttling of overexpressed protein. Stress responsiveness of ANKRD2 is tightly related to its role in coordination of myogenic differentiation [23], but little is known about the function of ANKRD2 in mature muscle.
Altered expression of MARPs has been reported in various pathological conditions of the heart and skeletal muscle, indicating their clinical relevance [24–29]. Elevated expression level of ANKRD1 mRNA and protein was detected in patients with end-stage heart failure [29], as well as in dilated, hypertrophic and arrhythmogenic ventricular cardiomyopathies [28, 30–32]. Several studies have linked mutations in the ANKRD1 gene with cardiomyopathies [31–34]. ANKRD2 protein expression is altered in various skeletal muscle pathologies [5, 26, 27] and is likely associated with transition of muscle fiber types [16]. Recent findings suggest that ANKRD2 acts as a mediator of the pathological functions of the mutated LMNA gene in Emery-Dreifuss muscular dystrophy 2 (EDMD2). It was shown that mutated lamin A sequesters and mislocates ANKRD2 in the nucleus of EDMD2-affected human myotubes [35]. Although murine MARP proteins are not essential for development and function of cardiac and skeletal muscles, MARP triple knockout mice display mild changes in skeletal muscle sarcomere structure, as well as in performance, particularly in the case of eccentric contractions [36, 37].
To gain further insight into the functions of MARPs in developing and mature striated muscle, we studied these genes in zebrafish, an animal model system with well-known advantages in genetic manipulation and in vivo analysis [38]. Here we report characterization of zebrafish MARP genes and proteins: their structure, comparison to the mammalian orthologs, expression profiles and localization during early development. In addition, we find differential upregulation of MARPs gene expression in striated muscle of adult zebrafish following endurance exercise. This study provides a foundation for further functional characterization of the MARP proteins in zebrafish development and stress response.
Materials and methods
Fish
The zebrafish (Danio rerio) AB strain was maintained on a 14 h light/10 h dark cycle at 28.5°C. Embryos obtained from wild-type fish were visually examined for proper development [39] and collected at several time points post fertilization. For in situ hybridization (ISH) experiments embryos older than 24 hpf were treated with 0.003% 1-phenyl 2-thiourea (PTU, Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) to prevent pigmentation. Thirty two adult fish were used for exercise experiments and expression analysis. Zebrafish husbandry was performed under standard conditions in accordance with institutional and national ethical and animal welfare guidelines. Experiments were approved by the Veterinary Department, Darmstadt Regional Council, Germany and the Veterinary Directorate, Ministry of Agriculture, Forestry and Water Management, Republic of Serbia.
Exercise protocol
Adult zebrafish (8 months old) were exercised in a 5L glass beaker (external diameter 170 mm) with a 60x10 mm stir bar, filled with 4L of fish water and placed on a magnetic stirrer, similarly to the Spinning Task described by Blazina et al [40]. Maximum number of fish in one beaker was 10. In order to adapt to the new conditions fish were pre-exercised for two days, 3 hours per day. Stirrer speed was adjusted to generate a 2 cm deep vortex. On the third day the rotation speed was increased to generate a 10 cm deep vortex and experiment was continued if 8 good swimmers remained. Fish that were able to swim continuously, while avoiding the vortex, were subjected to a regime consisting of two sets of 3 hours swimming, with 1 hour resting and feeding in between, for 5 consecutive days. Organs were harvested 3 hours after the last exercise bout. Skeletal muscles were sampled individually or pooled by two, while two hearts were pooled in each sample. Experimental groups contained at least 6 animals. Movie showing zebrafish swimming during exercise is given as supplemental file (S1 Movie), while detailed protocol is available at dx.doi.org/10.17504/protocols.io.sa2eage.
RNA isolation and cDNA synthesis
Prior to RNA isolation embryos were mechanically dechorionated. Embryos and tissues of adult fish (axial skeletal muscles and whole hearts) were homogenized in Trizol (Life Technologies, Thermo Fisher Scientific, Waltham, Massachusetts, USA) using a Bullet Blender (Next Advance, Troy, New York, USA) or TissueLyser II (QIAGEN, Hilden, Germany). Total RNA from embryos was purified using RNeasy Mini Kit (Qiagen, Hilden, Germany), while RNA from adult tissues was isolated according to the standard manufacturer protocol for Trizol reagent. Isolated RNA was treated with Dnase I (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Concentration and purity of RNA were determined by spectrophotometry using a NanoDrop 2000c (Thermo Fisher Scientific, Waltham, Massachusetts, USA). RNAs with A260/A280 ratio of 1.8–2.0 were used for downstream applications. Random hexamer-primed cDNA was synthesized by reverse transcription from 500 ng (for embryos) or 2 μg (for adult tissues) of total RNA using SuperScript III First-Strand Synthesis System (Invitrogen, Thermo Fisher Scientific, Waltham, Massachusetts, USA), iScript cDNA Synthesis Kit (Bio-Rad, Hercules, California, USA) or High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA).
Quantitative real-time PCR (qPCR)
qPCR was performed in technical triplicate for each sample on a CFX Connect Real-Time Detection System (Bio-Rad, Hercules, California, USA) or 7500 Real Time PCR System (Thermo Fisher Scientific, Waltham, Massachusetts, USA), using DyNAmo ColorFlash SYBR Green Master Mix (Thermo Fisher Scientific, Waltham, Massachusetts, USA) or Hot FIREPol EvaGreen qPCR Mix Plus (Solis BioDyne, Tartu, Estonia), respectively. The transcript of 60S ribosomal protein L13a gene (rpl13a) served as an internal reference to normalize the mRNA levels in different samples. The rpl13a mRNA expression level was not affected by the exercise or at any stage during development. The primers are listed in Table 1. Reaction conditions were as follows: initial denaturation at 95°C for 10 min, 40 cycles of denaturation at 95°C for 15 s, annealing and elongation at 60°C for 20 s when SYBR Green was used, and annealing at 60°C for 32 s followed by 20 s of elongation at 72°C when EvaGreen chemistry was used. Amplification was followed by the melting curve/dissociation analysis. The qPCR data were analyzed using the 2(−ΔΔCt) method.
Table 1. List of primers used for in situ hybridization and quantitative PCR.
| amplicon | forward primer 5′ - 3′ | reverse primer 5′ - 3′ | size (bp) | |
|---|---|---|---|---|
| in situ hybridization | ||||
| ankrd1a | AGGGTGGGAGAAAGTGCTTGT | CAAATGCTGAAAAGTTGTTCATCTG | 902 | |
| ankrd1b | CTTCAAGCAACTGAAGTCCA | AATATGCAGGCTCATAATATCTCA | 466 | |
| ankrd2 | AGGCGTGAGATTGTTGATCTAGG | CTTTAGTGTCAAACTGCCACTGCT | 664 | |
| myod1 | CATTAACCCTCACTAAAGGGAATTCTACGACGACCCTTGCTT | TAATACGACTCACTATAGGGTTTCCAGCAGTGGATCAAAA | 902 | |
| quantitative PCR | ||||
| ankrd1a | GAAGGGTGGGAGAAAGTGCT | TTTGGCTTCAGTTCACTTGG | ||
| ankrd1b | CATCACAGGTGGAAACACAGA | CCGCTGAGAATGACTTCACC | ||
| ankrd2 | AGGGCATTACAGCCACTGAA | GTGCATCCCCAAGTGTTTGT | ||
| rpl13a | TCTGGAGGACTGTAAGAGGTATGC | AGACGCACAATCTTGAGAGCAG | ||
| tnni2b.2 | AGGTGGACAGAGTTAATTACATGG | TCAGATCCTCAATCTCTTTGTCAC | ||
| tmod4 | CGCAACAGATGCTGAAATGTG | TTTCACCACACTGTTGATGCC | ||
| casq1a | CTTCTTCAAGAGCAACAAATCC | GTTAATATCGTCTTCCCAGATCTC | ||
| casq1b | ATAACACAGAGAATCCTGACCT | CCAGATACTCTCAGCATCATCC | ||
| tgfb2l | CAGACACCTCCATATGCACAC | CACAGGTAAGGACAGTTCCC | ||
| mstnb | CATGGCCACAGAACCTGACC | CCGGTCTCAGATGAACCCAG | ||
| col8a2 | AGGGTGAGTTTGTAATCTTGTGAC | CGTACTTCATCTGAGGCATAGG | ||
| lamc3 | CTAAAGATGCCAAAGCCTCCT | GAAGAAACCATGTCCTCCTCTG | ||
| cpt1b | GCATTTCAGTTCACCGTCAC | AACACTGTTCTTAAAGCGGATGG | ||
| pfkma | TCATGTCAGCAAAGGTAAGATCAC | AGTCTGTGCCAATAGTCATGTC | ||
| pdk2b | GAATGAGCAACAGTTTGAAGGAG | AGAGTTTCCACAAATTCTGCGA | ||
| fbxo32 | CATTCAATCGCTTGGACTTCTG | TTGCTGATCATCGAGAACTTTCTG | ||
| dcn | AAATTCCACTTGATACCACTCTCC | CCAAGATGAGCGTTTGGAGAC | ||
| aplnrb | CATATTCTCTGATTCCCGTGCT | GAGCCAGGTTTCCAATGTAGAC | ||
| aplnra | TAATGACTCTGGGTGTGACTACTC | GTTGCCGATATAAACATCTGCC | ||
| cs | AGACCTCGTCCCTAAAGAACAG | CTCATTCCTCCATAAACCATGTCC | ||
| ppargc1a | ACCCAGGTATGACAGCTATGAG | CTCGCCTCTCCTCTATTGCT | ||
| igf1 | TTATTTCAGCAAACCGACAGGA | GTTGTGCTCGTAGAGATCGT | ||
| il6r | AGTGGATTTATAATGTGGACCCGA | CAGAAGGAGGATCTTGTCGAG | ||
| cxcl12b | TTCCAAGTCATTGCCAAGCTG | CTTTAGAGATTCTCCGCTGTCC | ||
| igfbp2a | CTAAACAGAGCCAGTGCCAG | CCACGATAGCCATTCACTGAC | ||
| casq2 | AACTTCCCATTGCTCATTCC | CTCGTCATCATTGGGTATCTC | ||
| sparc | GAACTACAACATGTACATCTTCCC | CGACATCCTGCTCTTTGATCC | ||
| gys1 | AGAGTCAAAGTGATCTTCCATCC | AAACAGCCAAACCCAGACAG | ||
| ctrb1 | GATACAATGCTCCCGATACTC | ACACGATACCAACCAAAGTC | ||
| col1a1a | GCTTCCAGTTCGAGTATGGC | GTGACACTGTATGTGAAGCGG | ||
| gpib | CGCTTTCTACCAGCTCATCC | CAACAGAATCTTGTGGTGAAGG | ||
| lpl | TCCATTATCAAGTGAAGGTCCA | GTTCAAAGTAGGCATAATGTAGGG | ||
| nppa | CCAAGCTCAAGAGCTTGCTG | CTGCTTCCTCTCGGTCTCTG | ||
| pkmb | CACACTCGGACCTGCTTCAC | ACGGACACTCTTGATGGTTTCAG | ||
| aldocb | GAACCGCCGTCTTTACCGTC | ACACCTTTGTCAACCTTGATTCCT | ||
| ldha | TGTTGGAATGGTAGGAATGGCTG | GCGGTCACACTGTAATCTTTATCC | ||
In situ hybridization
Templates for synthesis of ISH probes were generated by PCR amplification using cDNA of 72 hpf embryos. Primer sets are given in Table 1. Amplicons were cloned into the pGEM-T easy vector (Promega, Madison, Wisconsin, USA) and verified by sequencing. Labeled RNA probes were synthesized using mMESSAGE mMACHINE SP6 or T7 kits (Thermo Fisher Scientific, Waltham, Massachusetts, USA) and digoxigenin RNA labeling nucleotide mix (Roche, Basel, Switzerland), with linearized plasmids as templates. Probes were analyzed by agarose gel electrophoresis and purified using RNA Clean and Concentrator-5 kit (Zymo Research, Irvine, California, USA). Probe for myod1 was synthesized directly from PCR fragments amplified using primers containing T3 and T7 promoter sequences and cDNA of 48 hpf embryos. Whole mount ISH was performed according to the protocol of Thisse and Thisse [41]. Hybridization was carried out overnight at 67°C, with 300 ng of each probe. myod1 probe was used as positive control (S1 Fig). Whole-mount embryo imaging was performed on a Nikon SMZ25 stereomicroscope (Nikon, Tokyo, Japan).
Bioinformatics (protein sequence analysis, phylogeny and synteny)
The protein sequences of MARPs from different species were retrieved from the Ensembl database (http://www.ensembl.org/index.html). A list of accession numbers is shown in the S1 Table. Protein sequences were aligned using the Clustal Omega algorithm available on the EBI webserver [42]. Protein motifs for zebrafish, human and mouse MARPs were identified via the SMART database [43, 44]. PEST motifs and NLS sequences were identified using ePESTfind (http://emboss.bioinformatics.nl/cgi-bin/emboss/epestfind) and NLStradamus [45] web-based servers for sequence prediction, respectively. Analyses were conducted using the default parameters.
A phylogenetic tree was constructed using the maximum likelihood method of the PhyML algorithm (v3.0) [46], with bootstrapping value of 1000, via the ATGC webserver [47]. MARP protein sequences from zebrafish (Danio rerio), blind cave fish (Astyanax mexicanus), frog (Xenopus tropicalis), chicken (Gallus gallus), mouse (Mus musculus) and human (Homo sapiens) were analyzed. The genome assemblies are listed in S1 Table.
Synteny analysis of zebrafish and human genes was performed using Genomicus v87.01 genome browser synchronized with genomes from the Ensembl database [48].
Nomenclature
Zebrafish gene and protein symbols are written according to 2018 ZFIN zebrafish nomenclature conventions. The zebrafish protein symbol is the same as the gene symbol, but non-italic and the first letter is uppercase. Human and mouse gene and protein symbols are written in accordance with HUGO and MGI nomenclatures, respectively. Human protein and gene symbols are both uppercase, and gene symbol is italic. Mouse protein symbol is uppercase, while gene symbols are italicized, with first letter uppercase.
Statistical analysis
Developmental qPCR data were analyzed using one-way ANOVA followed by Tukey’s multiple comparison test. Expression levels in adult tissues were compared using the statistical t-test. Results were presented as mean ± SD, level of significance was P<0.05.
Results
Alignments, phylogenetic and synteny analysis of the zebrafish MARP genes
The Ensembl database contains entries for three MARP genes in the zebrafish genome: ankrd1a, ankrd1b and ankrd2. First two are paralog genes, counterparts of the mammalian ANKRD1/Ankrd1. No gene corresponding to ANKRD23/Ankrd23 was found in the zebrafish genome. The conserved structure of human, mouse and zebrafish MARP genes showing that all of them have nine exons is presented in Fig 1A.
Fig 1. Gene organization, protein sequence alignment and domain structure of MARP family members from different species.
(a) Exon-intron structure of human (Hs) ANKRD1 and ANKRD2 and their counterparts in mouse (Mm) and zebrafish (Dr). Exons (boxes) and introns (lines) are drawn to scale. White boxes indicate 5’ and 3’ UTRs. The numbers on the right indicate the length of the genomic region. (b) Amino acid sequence alignment of human, mouse and zebrafish proteins. (c) Schematic representation of the structural domains of human, mouse and zebrafish proteins. The predicted domains are indicated by colored boxes. Domain positions are listed in S2 Table.
The protein sequence alignment of human, mouse and zebrafish MARP orthologs shows substantial sequence similarities across species, particularly in the regions of ankyrin repeats (Fig 1B). BLAST comparison of zebrafish Ankrd1a, Ankrd1b and Ankrd2 with their human counterparts reveals 56%, 46% and 51% of identical amino acids (aa), respectively. Zebrafish Ankrd1a and Ankrd1b proteins show 47% identity. Zebrafish and mammalian MARP proteins also show similarities in other protein domains and motifs (Fig 1C). Ankyrin repeats, involved in protein-protein interactions, and PEST sequences, required for rapid intracellular proteolysis, are identified in all zebrafish MARPs. Protein oligomerization motif, coiled coils are predicted in Ankrd1a, but not in Ankrd1b and Ankrd2, while Ankrd1b lacks NLS. The position of these conserved functional domains within zebrafish MARP proteins is shown in S2 Table.
To investigate the evolutionary relationship, the phylogenetic tree based on Clustal Omega protein sequence alignment of MARP homologs was generated using the PhyML algorithm, with 1000 bootstraps (Fig 2). The tree topology segregates ANKRD1 and ANKRD2 homologs in two distinct groups. The Ankrd1b protein, present only in teleost, is closely related to Ankrd1a.
Fig 2. Phylogenetic tree of MARP homologs generated by PhyML algorithm.
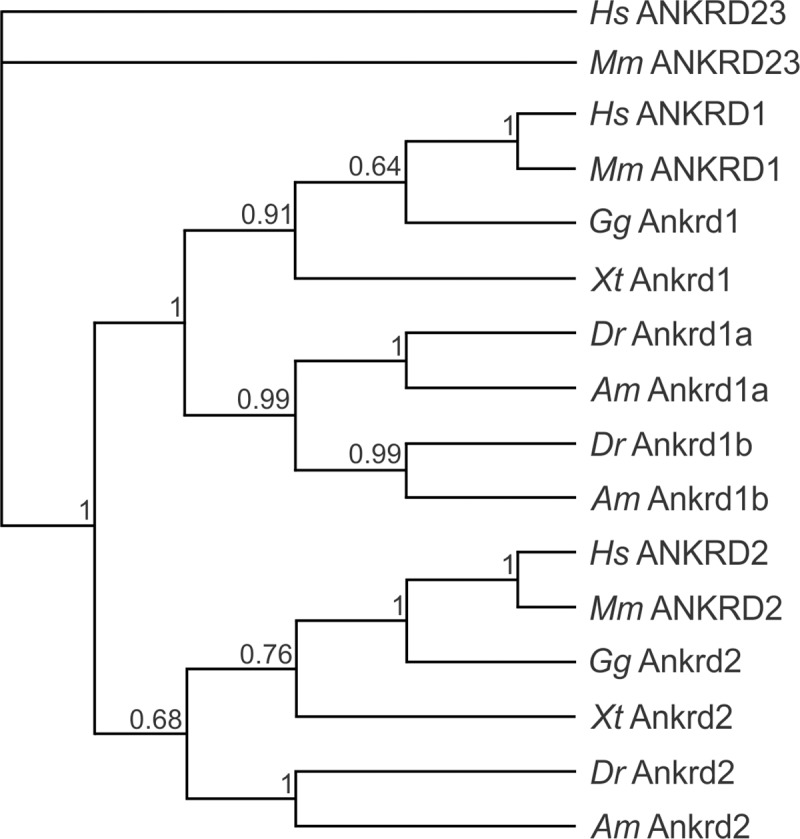
The GeneBank accession numbers of the sequences are listed in S1 Table. Species abbreviations: Hs, Homo sapiens; Mm, Mus musculus; Gg, Gallus gallus; Xt, Xenopus tropicalis; Dr, Danio rerio; Am, Astyanax mexicanus. Confidence of nodes is indicated by numbers on individual branches.
Syntenic analysis demonstrated that zebrafish MARP genes display a conserved genetic neighborhood with their human counterparts. Comparisons of chromosomal regions containing human ANKRD1 and zebrafish ankrd1a and ankrd1b genes show that despite the rearrangements at the macrosyntenic level, neighboring genes and their homologs have kept their relative location throughout evolution (Fig 3). Regarding ankrd2, neighboring genes remained close but their orientation is inverted in comparison to human orthologs.
Fig 3. Synteny comparisons between human (Hs) and zebrafish (Dr) gene loci.
ANKRD1/ankrd1a/ankrd1b (a) and ANKRD2/ankrd2 (b) genes, depicted in orange polygons, are presented with nearest neighbors (colored polygons, orthologs have the same color). The transcriptional orientation of the gene is indicated by the angled end of each polygon corresponding to the 3´ end. Image style was adapted from Genomicus.
Overall, zebrafish MARPs are similar to their mammalian counterparts in terms of gene organization, primary protein sequence and identified key domains.
Expression of the MARP genes in developing zebrafish
We analyzed the temporal and spatial expression of MARP transcripts in the developing zebrafish embryos by qPCR and whole mount ISH. qPCR was performed using four independent batches of twenty embryos or larvae at each developmental stage tested. In general, although very low, the expression of all MARPs was increasing up to the seventh day of development (Fig 4 and S3 Table). A mild increase of ankrd1a expression was observed at 72 hpf (1.74-fold increase relative to 24 hpf time point), after which the levels did not change significantly up to 168 hpf. The most prominent change was shown for ankrd1b expression, with average of 92.18-fold increase in the first 72 hpf. Levels of ankrd2 transcript were very low during first seven days of development and changes were not statistically significant. Among all MARP genes, ankrd2 was the least expressed, as confirmed by the analysis of qPCR amplicons by agarose gel electrophoresis (Fig 4 and S3 Table).
Fig 4. Quantification of zebrafish MARP transcripts during development.
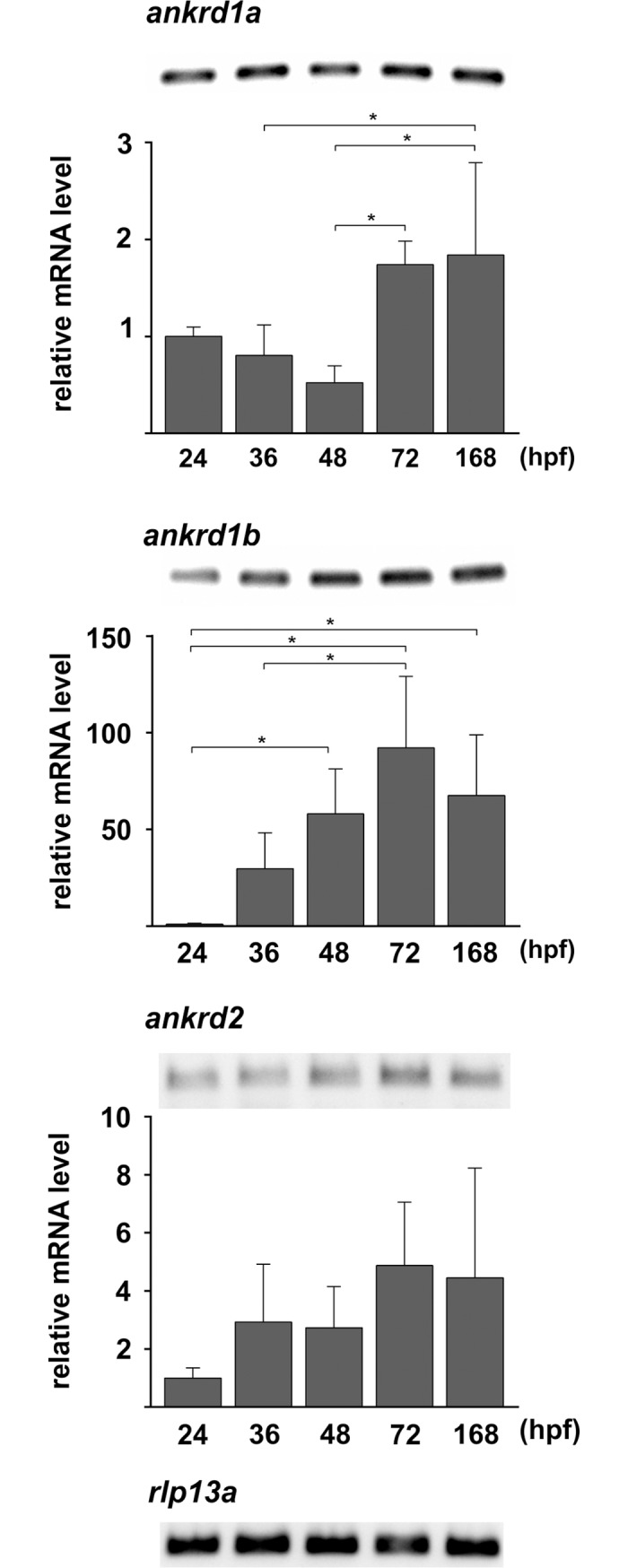
Expression levels of ankrd1a, ankrd1b and ankrd2 at indicated time points after fertilization were obtained using qPCR. The housekeeping gene rlp13a served as internal reference. Data (mean ± SD) are combined from four biological replicates and normalized to the 24 hpf time point. * denotes P<0.05 in comparison to control group (by one-way ANOVA/Tukey’s multiple comparison test). Agarose gels showing qPCR products for ankrd1a, ankrd1b, ankrd2 and rlp13a are also presented. Average Ct±SD values for MARP and reference (rpl13a) genes during zebrafish development at indicated time points are given in S3 Table.
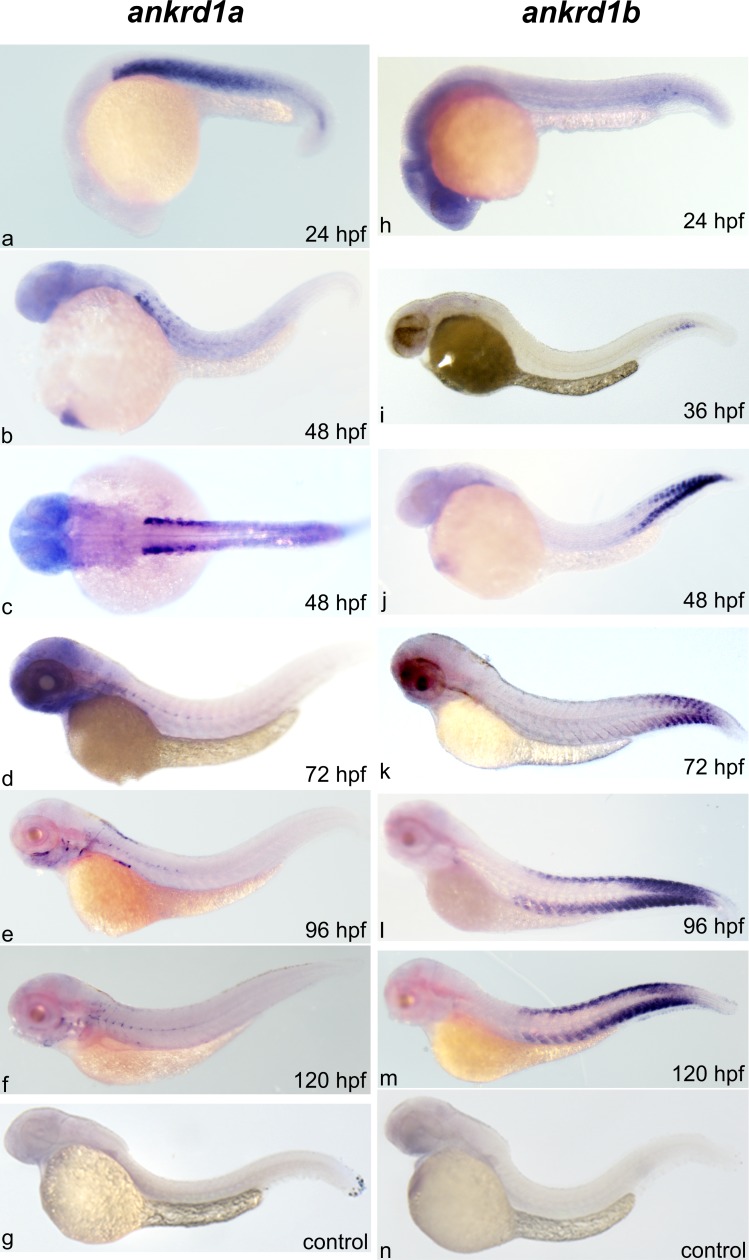
As demonstrated by ISH, ankrd1a expression preceded that of the ankrd1b (Fig 5). First transcripts of ankrd1a were observed at 24 hpf, in the ventral part of the developing somites. At later stages expression of ankrd1a was more spatially restricted, concentrated in the apex of the chevron-shaped somites. Conversely, ankrd1b expression was first observed at 36 hpf in the tail somites. As development progresses, ankrd1b gene expression expands caudally, being more pronounced in the most ventral and dorsal parts of the somites. We were not able to detect any signal for ankrd2 even after prolonged staining, consistent with qPCR results.
Fig 5. Spatiotemporal expression of zebrafish ankrd1a and ankrd1b during development, from 24 to 120 hpf.
Representative images of whole mount ISH using probes detecting ankrd1a (a-f) and ankrd1b (h-m) transcripts at designated time points. Control staining for ankrd1a (g) and ankrd1b (n) was performed in 48 hpf embryos. Lateral and one dorsal (c) views are shown, anterior to the left.
In conclusion, during development, ankrd1a and ankrd1b are expressed in somites, in a non-overlapping pattern, and at relatively low levels.
Endurance exercise differentially upregulates expression of the MARP genes in adult zebrafish heart and skeletal muscle
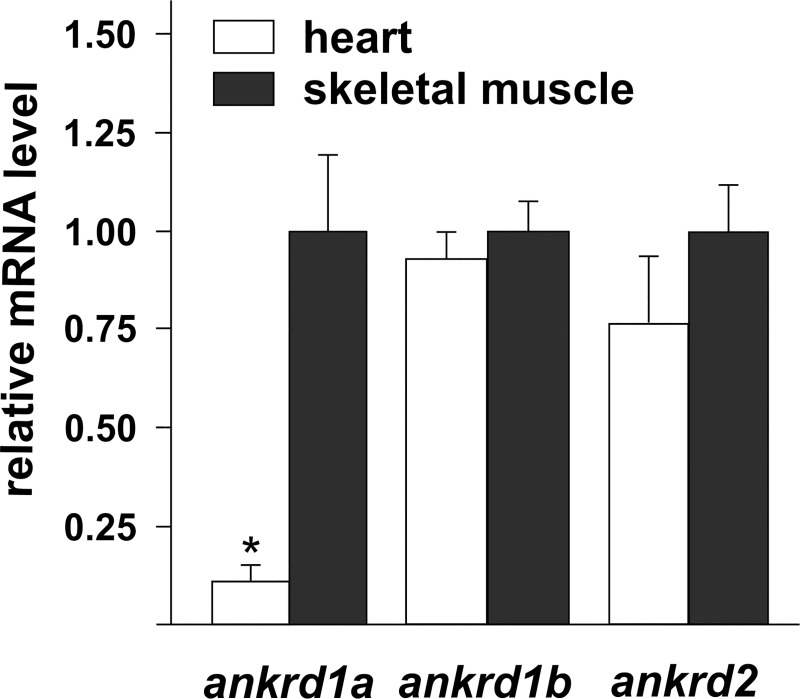
Since human ANKRD1 and ANKRD2 genes are differentially expressed in cardiac and skeletal muscles, we investigated relative expression of zebrafish genes in these two organs in adult animals. Under basal conditions, all MARP genes are expressed at low levels (S4 Table), ankrd1a being the most abundant. There is more ankrd1a transcript in the skeletal muscle, compared to the heart, while ankrd1b and ankrd2 show no significant difference in distribution between analyzed tissues (Fig 6).
Fig 6. Expression of the zebrafish MARP genes in adult heart and skeletal muscle.
Quantification of ankrd1a, ankrd1b and ankrd2 expression was done by qPCR, using mRNA isolated from the hearts of 8 fish, pooled in 4 groups and the skeletal muscles of 4 fish. Relative level of transcripts in adult heart is normalized to the transcript level in the skeletal muscle, set as 1. Bars represent the mean ± SD. * denotes P<0.05 in comparison to control group (by t-test). Average Ct±SD values for MARP and reference (rpl13a) genes in adult zebrafish heart and skeletal muscle are given in S4 Table.
In order to analyze the responsiveness of zebrafish ankrd1a, ankrd1b and ankrd2 genes to increased muscle activity we quantified their mRNA levels in whole hearts and skeletal muscle after one week of endurance exercise. To validate that employed exercise protocol is able to cause changes in cardiac and skeletal muscle gene expression comparable to those observed after tunnel swimming, we measured expression of exercise-responsive genes listed in S5 Table. Majority of these genes were selected based on their reported altered expression in zebrafish after tunnel swimming of varying duration [49–52]. Additionally, several genes from mammalian model organisms and humans were included in the analysis [53–55]. Among the tested genes, four in the heart (col1a1a, lpl, gys1 and ctrb1) and four in skeletal muscle (ppargc1a, aplnra, aplnrb and igf1) showed significant change in expression after exercise (Fig 7). These results recapitulate known effects of swim tunnel exercise and support the expected activation of muscles.
Fig 7. Effects of swimming exercise on heart and skeletal muscle genes expression.
mRNA expression levels of chymotrypsinogen B1 (ctrb1), glycogen synthase 1 (gys1), lipoprotein lipase (lpl) and type I collagen, alpha 1a (col1a1a) in heart, and apelin receptor a (aplnra), apelin receptor b (aplnrb), peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (ppargc1a) and insulin-like growth factor 1 (igf1) in skeletal muscle of exercised fish expressed as a fold change over non-exercised controls set to 1. Bars represent the mean ± SD. * denotes P<0.05 and ** denotes P<0.005 in comparison to control group (by t-test). Fold change ± SD values for all tested cardiac and skeletal muscle genes of trained and control adult zebrafish are given in S5 Table.
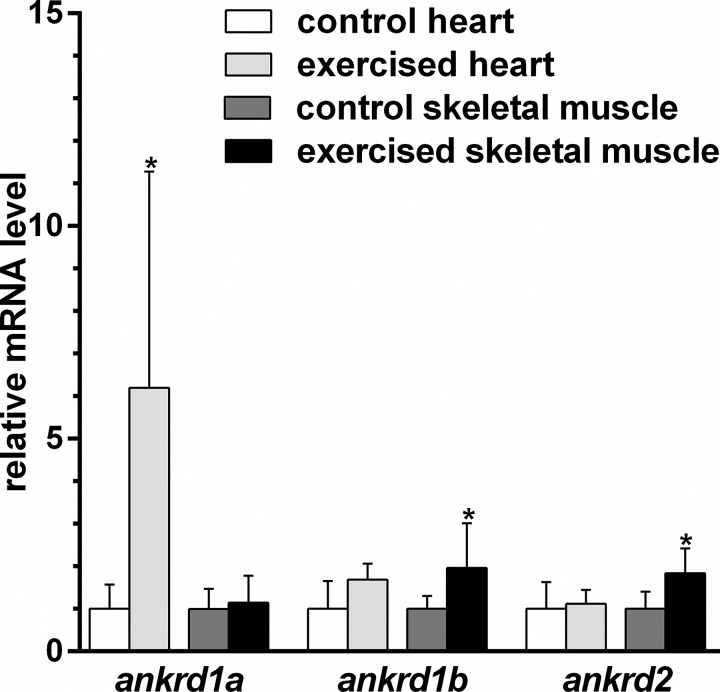
Endurance exercise caused significant increase in ankrd1a mRNA level (fold change 6.19 ± 5.08 compared to non-exercised control fish) in adult hearts (Fig 8). No change was observed for ankrd1b and ankrd2 transcripts levels in the heart. However, a slight upregulation of ankrd1b and ankrd2 expression in skeletal muscles was detected (1.97±1.05 and 1.84±0.58, respectively) in exercised animals. These results indicate that, like in mammals, zebrafish MARPs are responsive to increased load imposed on striated muscle.
Fig 8. Zebrafish MARP genes are differentially upregulated in adult heart and in skeletal muscle after endurance exercise.
Adult fish were subjected to endurance exercise for one week. Relative expression of MARP genes in zebrafish heart and skeletal muscle is expressed as fold change to non-exercised controls. Bars represent the mean ± SD. * denotes P<0.05 in comparison to control group (by t-test).
Discussion
In this study we investigated protein structure, evolutionary conservation, spatiotemporal expression profiles and responsiveness to increased muscle activity of zebrafish MARP genes ankrd1a, ankrd1b and ankrd2. As suggested by the phylogenetic tree topology, the former two are paralog genes, orthologous to mammalian ANKRD1/Ankrd1, most probably originating from a duplication event in the teleost lineage. This notion is further supported by the synteny analysis which shows similarities shared by the two loci in regard to the genomic context.
The primary sequences of MARP proteins in all compared taxa are modestly conserved, with identical residue percentage being around 50% in all pairwise comparisons. Protein sequences of the two zebrafish paralogs, Ankrd1a and Ankrd1b, show more divergence in the N-terminal region, with the former being more similar to its orthologs in other species. Despite showing 50% aa identity to human and mouse orthologs, there are notable differences in the N-terminal region of the zebrafish Ankrd2 protein.
Most of the protein sequence identity is shared between the ankyrin repeats, a motif all MARPs have in common, which is indispensable for their interactions with other proteins [1]. The coiled-coil domain, essential for homo- and heterodimerization of MARP proteins in antiparallel fashion [56, 57], is present in Ankrd1a, but missing from Ankrd1b and Ankrd2. The NLS is predicted in Ankrd1a and Ankrd2, but not in Ankrd1b. The identified structural similarities of zebrafish MARP family members to their relatively distant mammalian counterparts suggests that their function is conserved. Further experimental work is needed to characterize zebrafish MARPs at the protein level in cardiac and skeletal muscle, including their relative amounts and subcellular distribution.
Profiles of basal expression of ankrd1a, ankrd1b and ankrd2 genes in developing and adult zebrafish mostly differ from those observed for their mammalian counterparts. During cardiogenesis, murine Ankrd1 transcript and protein are expressed specifically in the myocardium and slightly stronger in the atrium than in the ventricle [3, 58]. Human ANKRD1 was found to be strongly expressed in the fetal heart, diffusely distributed throughout the atria and ventricles, while ANKRD2 was detectable at trace levels [4, 59]. During the first five days of zebrafish development, none of the MARP transcripts were detected in the heart by in situ hybridization, indicating no or very low expression, undetectable by this method. Expression of mouse fetal Ankrd2 transcript is restricted to skeletal muscle [10], in contrast to Ankrd1, which is not detected in this organ during development [3]. In humans, both ANKRD1 and ANKRD2 were detected in fetal skeletal muscle [4]. Zebrafish ankrd1a and ankrd1b transcripts were only found in developing axial muscles. Absence of ankrd2 expression suggests no important role during early zebrafish development. Our results are in accordance with the data in the EMBO Expression Atlas on ankrd1a and ankrd1b expression [60]. Gene expression analysis by qPCR revealed that ankrd1b mRNA levels increased during development, peaking at 72 hpf. Similarly, ankrd1a expression was detected during embryonic stages, observing a significant increase in larvae at 72 and 168 hpf. Interestingly, our ISH analysis shows that at larval stages the number of ankrd1a expressing cells is reduced. A possible explanation is that, despite being present in lower number than at earlier stages, these cells express higher levels of ankrd1a. On the other hand, contribution of other larval cells expressing low amounts of ankrd1a, not detectable by ISH staining, cannot be excluded. Differential spatial expression patterns suggest that ankrd1a and ankrd1b expression may be regulated by trunk and tail muscle specific regulators. Development of zebrafish trunk and tail muscle domains is controlled by different mechanisms. Trunk domains are established via Nodal signaling, whereas the tail domain requires BMP during early development [61]. A spatial expression pattern similar to that of ankrd1a and ankrd1b was observed for myosin heavy chain isoforms coded by fmyhc1.2 and fmyhc2.1 whose differential expression in trunk and tail, respectively, is coordinated by retinoic acid and Wnt signaling [62]. Use of gene knockout, overexpressing and reporter zebrafish lines will help in deciphering regulatory mechanisms of ankrd1a and ankrd1b expression and their distinct functions during zebrafish development.
Expression of MARPs in adult mammalian skeletal muscle and the heart is well documented [3–5, 59, 63]. In humans, ANKRD1 and ANKRD2 were detected mostly in the heart and skeletal muscles, respectively. Contrary to human homologs, adult zebrafish MARP genes have low expression under basal conditions. While ankrd1b and ankrd2 transcripts are equally distributed between heart and skeletal muscles, ankrd1a is preferentially expressed in skeletal muscle, similarly to avian ANKRD1 gene [64]. Low levels of ankrd1a and ankrd1b expression in the adult zebrafish heart, detected by qPCR, is in line with results obtained during transcriptome analysis of zebrafish genes homologous to dilated cardiomyopathy-associated human genes [65].
Mammalian MARPs are known to be upregulated by various stress stimuli. Endurance exercise and eccentric contractions [9, 11, 13, 66], hypertrophic overload of skeletal muscle [67], chronic immobilization of leg muscles in a stretched position [10, 16], submaximal exhaustive exercise [14] and fatiguing jumping exercise [12] all increase expression of mammalian ANKRD1/Ankrd1 and/or ANKRD2/Ankrd2. Here we demonstrated evolutionary conservation of MARPs responsiveness to endurance exercise which differentially upregulated ankrd1a in heart and ankrd1b and ankrd2 in skeletal muscle. Differential response of ankrd1a and ankrd1b paralogs to endurance exercise suggests their non-redundant functions. Generally, after gene duplication, one of the paralog genes is often lost from the genome due to redundancy [68], while if duplicated genes acquire non-redundant functions, both are likely to be retained [69]. In the case of ankrd1a and ankrd1b it is possible that a functional specialization in the cardiac and skeletal muscle occurred.
Alterations in expression of cardiac and skeletal muscle genes after exercise were mostly studied in fish subjected to linear tunnel swimming [49–51, 70, 71] and their transcriptomic response to endurance exercise was determined by microarray analysis [49, 51]. The exercise method used in this study is similar to published set-ups with varying swimming conditions, employed to study motor coordination [40] and swim performance [72]. To demonstrate that our swimming protocol caused the activation of striated muscle in adult zebrafish, we analyzed the expression of selected exercise responsive genes from different functional categories: muscle growth and development, muscle contraction, extracellular matrix, protein synthesis and degradation, metabolism, and myokines [49–51]. Recapitulation of known aspects of tunnel swimming training suggests that exercise method used in this study is a valuable tool for investigating muscle response to increased load in zebrafish, as an affordable alternative to costly swim tunnels. It is worth noting that exercise as short as the one used in this study induces the expression of MARPs, demonstrating their early responsiveness in zebrafish striated muscle to increased activity. Since response of mammalian MARP genes vary depending on the type, intensity and duration of the exercise and recovery period, expression studies after various forms and regimes of exercise are needed. The zebrafish model provides a tool to asses MARPs function in stressed striated muscle by virtue of the protocol we used, which is adjustable to facilitate various exercise regimes.
Differential upregulation of zebrafish MARP genes in cardiac and skeletal muscles after one week of endurance exercise training suggests their possible muscle-type specific role in physiological remodeling and may be a reflection of differences between mechanisms of cardiac and skeletal muscles adaptation to increased workload. It was already demonstrated that endurance exercise differentially stimulates development of heart and axial muscle in zebrafish [70]. The phenotype of axial muscles was shifted towards a slow aerobic, while heart muscle gained a faster phenotype, but does not become more aerobic.
Despite differences in primary structure, gene number and expression patterns between zebrafish and mammalian MARPs, their responsiveness to increased muscle activity is encouraging for including zebrafish as a model organism for further functional studies of these genes in mature muscles. It is worth mentioning that ankrd1a has been identified in recent studies as an early response gene in regeneration of injured zebrafish heart [73–75]. These and our data point to remodeling of skeletal muscles and cardiac regeneration as processes in which the role of zebrafish MARPs warrants further investigation.
Conclusions
The expression profiles of ankrd1a and ankrd1b indicate an active role in the development of somites, while upregulation of gene expression of all zebrafish MARPs after relatively short endurance exercise suggests that their function in response to increased activity in striated muscles is conserved. This initial study provides a foundation from which the zebrafish could be established as a novel model organism for further functional studies of MARPs in mature striated muscle.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(TIF)
(AVI)
Acknowledgments
We are grateful to Michelle Collins for providing the control ISH probe and help with optimization of ISH protocol.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the Ministry of Education, Science and Technological Development of the Republic of Serbia (Project No. 173008 to DR), bilateral project funded by the German Academic Exchange Service and the Ministry of Education, Science and Technological Development of the Republic of Serbia (Project-ID 451-03-01766/2014-09/3 to SK and 57140778 to DYRS) and the Max Planck Society (DYRS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kojic S, Radojkovic D, Faulkner G. Muscle ankyrin repeat proteins: their role in striated muscle function in health and disease. Crit Rev Clin Lab Sci. 2011;48(5–6):269–94. Epub 2011/12/22. 10.3109/10408363.2011.643857 . [DOI] [PubMed] [Google Scholar]
- 2.Wette SG, Smith HK, Lamb GD, Murphy RM. Characterization of muscle ankyrin repeat proteins in human skeletal muscle. Am J Physiol Cell Physiol. 2017;313(3):C327–c39. Epub 2017/06/16. 10.1152/ajpcell.00077.2017 ; PubMed Central PMCID: PMCPMC5625093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zou Y, Evans S, Chen J, Kuo HC, Harvey RP, Chien KR. CARP, a cardiac ankyrin repeat protein, is downstream in the Nkx2-5 homeobox gene pathway. Development. 1997;124(4):793–804. Epub 1997/02/01. . [DOI] [PubMed] [Google Scholar]
- 4.Ishiguro N, Baba T, Ishida T, Takeuchi K, Osaki M, Araki N, et al. Carp, a cardiac ankyrin-repeated protein, and its new homologue, Arpp, are differentially expressed in heart, skeletal muscle, and rhabdomyosarcomas. Am J Pathol. 2002;160(5):1767–78. Epub 2002/05/10. 10.1016/S0002-9440(10)61123-6 ; PubMed Central PMCID: PMCPMC1850855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pallavicini A, Kojic S, Bean C, Vainzof M, Salamon M, Ievolella C, et al. Characterization of human skeletal muscle Ankrd2. Biochem Biophys Res Commun. 2001;285(2):378–86. Epub 2001/07/11. 10.1006/bbrc.2001.5131 . [DOI] [PubMed] [Google Scholar]
- 6.Ikeda K, Emoto N, Matsuo M, Yokoyama M. Molecular identification and characterization of a novel nuclear protein whose expression is up-regulated in insulin-resistant animals. J Biol Chem. 2003;278(6):3514–20. Epub 2002/11/29. 10.1074/jbc.M204563200 . [DOI] [PubMed] [Google Scholar]
- 7.Chen YW, Nader GA, Baar KR, Fedele MJ, Hoffman EP, Esser KA. Response of rat muscle to acute resistance exercise defined by transcriptional and translational profiling. J Physiol. 2002;545(Pt 1):27–41. Epub 2002/11/16. 10.1113/jphysiol.2002.021220 ; PubMed Central PMCID: PMCPMC2290672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsukamoto Y, Senda T, Nakano T, Nakada C, Hida T, Ishiguro N, et al. Arpp, a new homolog of carp, is preferentially expressed in type 1 skeletal muscle fibers and is markedly induced by denervation. Lab Invest. 2002;82(5):645–55. Epub 2002/05/11. . [DOI] [PubMed] [Google Scholar]
- 9.Barash IA, Mathew L, Ryan AF, Chen J, Lieber RL. Rapid muscle-specific gene expression changes after a single bout of eccentric contractions in the mouse. Am J Physiol Cell Physiol. 2004;286(2):C355–64. Epub 2003/10/17. 10.1152/ajpcell.00211.2003 . [DOI] [PubMed] [Google Scholar]
- 10.Kemp TJ, Sadusky TJ, Saltisi F, Carey N, Moss J, Yang SY, et al. Identification of Ankrd2, a novel skeletal muscle gene coding for a stretch-responsive ankyrin-repeat protein. Genomics. 2000;66(3):229–41. Epub 2000/06/30. 10.1006/geno.2000.6213 . [DOI] [PubMed] [Google Scholar]
- 11.Mohamed JS, Lopez MA, Cox GA, Boriek AM. Anisotropic regulation of Ankrd2 gene expression in skeletal muscle by mechanical stretch. Faseb j. 2010;24(9):3330–40. Epub 2010/05/06. 10.1096/fj.10-158386 ; PubMed Central PMCID: PMCPMC2923360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehti M, Kivela R, Komi P, Komulainen J, Kainulainen H, Kyrolainen H. Effects of fatiguing jumping exercise on mRNA expression of titin-complex proteins and calpains. J Appl Physiol (1985). 2009;106(4):1419–24. Epub 2009/01/20. 10.1152/japplphysiol.90660.2008 . [DOI] [PubMed] [Google Scholar]
- 13.Lehti TM, Silvennoinen M, Kivela R, Kainulainen H, Komulainen J. Effects of streptozotocin-induced diabetes and physical training on gene expression of titin-based stretch-sensing complexes in mouse striated muscle. Am J Physiol Endocrinol Metab. 2007;292(2):E533–42. Epub 2006/09/28. 10.1152/ajpendo.00229.2006 . [DOI] [PubMed] [Google Scholar]
- 14.Koskinen SOA, Kyrolainen H, Flink R, Selanne HP, Gagnon SS, Ahtiainen JP, et al. Human skeletal muscle type 1 fibre distribution and response of stress-sensing proteins along the titin molecule after submaximal exhaustive exercise. Histochem Cell Biol. 2017;148(5):545–55. Epub 2017/07/18. 10.1007/s00418-017-1595-z . [DOI] [PubMed] [Google Scholar]
- 15.Miller MK, Bang ML, Witt CC, Labeit D, Trombitas C, Watanabe K, et al. The muscle ankyrin repeat proteins: CARP, ankrd2/Arpp and DARP as a family of titin filament-based stress response molecules. J Mol Biol. 2003;333(5):951–64. Epub 2003/10/30. . [DOI] [PubMed] [Google Scholar]
- 16.McKoy G, Hou Y, Yang SY, Vega Avelaira D, Degens H, Goldspink G, et al. Expression of Ankrd2 in fast and slow muscles and its response to stretch are consistent with a role in slow muscle function. J Appl Physiol (1985). 2005;98(6):2337–43; discussion 20. Epub 2005/01/29. 10.1152/japplphysiol.01046.2004 . [DOI] [PubMed] [Google Scholar]
- 17.Ojima K, Kawabata Y, Nakao H, Nakao K, Doi N, Kitamura F, et al. Dynamic distribution of muscle-specific calpain in mice has a key role in physical-stress adaptation and is impaired in muscular dystrophy. The Journal of Clinical Investigation. 2010;120(8):2672–83. 10.1172/JCI40658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsukamoto Y, Hijiya N, Yano S, Yokoyama S, Nakada C, Uchida T, et al. Arpp/Ankrd2, a member of the muscle ankyrin repeat proteins (MARPs), translocates from the I-band to the nucleus after muscle injury. Histochem Cell Biol. 2008;129(1):55–64. Epub 2007/10/11. 10.1007/s00418-007-0348-9 . [DOI] [PubMed] [Google Scholar]
- 19.Mikhailov AT, Torrado M. The enigmatic role of the ankyrin repeat domain 1 gene in heart development and disease. Int J Dev Biol. 2008;52(7):811–21. 10.1387/ijdb.082655am . [DOI] [PubMed] [Google Scholar]
- 20.Samaras SE, Almodovar-Garcia K, Wu N, Yu F, Davidson JM. Global deletion of Ankrd1 results in a wound-healing phenotype associated with dermal fibroblast dysfunction. Am J Pathol. 2015;185(1):96–109. Epub 2014/12/03. 10.1016/j.ajpath.2014.09.018 ; PubMed Central PMCID: PMCPMC4278243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi Y, Reitmaier B, Regenbogen J, Slowey RM, Opalenik SR, Wolf E, et al. CARP, a cardiac ankyrin repeat protein, is up-regulated during wound healing and induces angiogenesis in experimental granulation tissue. Am J Pathol. 2005;166(1):303–12. Epub 2005/01/06. 10.1016/S0002-9440(10)62254-7 ; PubMed Central PMCID: PMCPMC1602297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cenni V, Bavelloni A, Beretti F, Tagliavini F, Manzoli L, Lattanzi G, et al. Ankrd2/ARPP is a novel Akt2 specific substrate and regulates myogenic differentiation upon cellular exposure to H(2)O(2). Mol Biol Cell. 2011;22(16):2946–56. Epub 2011/07/09. 10.1091/mbc.E10-11-0928 ; PubMed Central PMCID: PMCPMC3154889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bean C, Facchinello N, Faulkner G, Lanfranchi G. The effects of Ankrd2 alteration indicate its involvement in cell cycle regulation during muscle differentiation. Biochim Biophys Acta. 2008;1783(6):1023–35. 10.1016/j.bbamcr.2008.01.027 . [DOI] [PubMed] [Google Scholar]
- 24.Nakada C, Oka A, Nonaka I, Sato K, Mori S, Ito H, et al. Cardiac ankyrin repeat protein is preferentially induced in atrophic myofibers of congenital myopathy and spinal muscular atrophy. Pathol Int. 2003;53(10):653–8. Epub 2003/10/01. . [DOI] [PubMed] [Google Scholar]
- 25.Nakada C, Tsukamoto Y, Oka A, Nonaka I, Takeda S, Sato K, et al. Cardiac-restricted ankyrin-repeated protein is differentially induced in duchenne and congenital muscular dystrophy. Lab Invest. 2003;83(5):711–9. Epub 2003/05/15. . [DOI] [PubMed] [Google Scholar]
- 26.Nakada C, Tsukamoto Y, Oka A, Nonaka I, Sato K, Mori S, et al. Altered expression of ARPP protein in skeletal muscles of patients with muscular dystrophy, congenital myopathy and spinal muscular atrophy. Pathobiology. 2004;71(1):43–51. Epub 2003/10/14. 10.1159/000072961 . [DOI] [PubMed] [Google Scholar]
- 27.Nakamura K, Nakada C, Takeuchi K, Osaki M, Shomori K, Kato S, et al. Altered expression of cardiac ankyrin repeat protein and its homologue, ankyrin repeat protein with PEST and proline-rich region, in atrophic muscles in amyotrophic lateral sclerosis. Pathobiology. 2002;70(4):197–203. Epub 2003/04/08. doi: 69329. 10.1159/000069329 . [DOI] [PubMed] [Google Scholar]
- 28.Nagueh SF, Shah G, Wu Y, Torre-Amione G, King NM, Lahmers S, et al. Altered titin expression, myocardial stiffness, and left ventricular function in patients with dilated cardiomyopathy. Circulation. 2004;110(2):155–62. Epub 2004/07/09. 10.1161/01.CIR.0000135591.37759.AF . [DOI] [PubMed] [Google Scholar]
- 29.Zolk O, Frohme M, Maurer A, Kluxen FW, Hentsch B, Zubakov D, et al. Cardiac ankyrin repeat protein, a negative regulator of cardiac gene expression, is augmented in human heart failure. Biochem Biophys Res Commun. 2002;293(5):1377–82. Epub 2002/06/11. 10.1016/S0006-291X(02)00387-X . [DOI] [PubMed] [Google Scholar]
- 30.Wei YJ, Cui CJ, Huang YX, Zhang XL, Zhang H, Hu SS. Upregulated expression of cardiac ankyrin repeat protein in human failing hearts due to arrhythmogenic right ventricular cardiomyopathy. Eur J Heart Fail. 2009;11(6):559–66. Epub 2009/04/11. 10.1093/eurjhf/hfp049 . [DOI] [PubMed] [Google Scholar]
- 31.Arimura T, Bos JM, Sato A, Kubo T, Okamoto H, Nishi H, et al. Cardiac ankyrin repeat protein gene (ANKRD1) mutations in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2009;54(4):334–42. Epub 2009/07/18. 10.1016/j.jacc.2008.12.082 . [DOI] [PubMed] [Google Scholar]
- 32.Crocini C, Arimura T, Reischmann S, Eder A, Braren I, Hansen A, et al. Impact of ANKRD1 mutations associated with hypertrophic cardiomyopathy on contraction parameters of engineered heart tissue. Basic Res Cardiol. 2013;108(3):349 Epub 2013/04/11. 10.1007/s00395-013-0349-x . [DOI] [PubMed] [Google Scholar]
- 33.Moulik M, Vatta M, Witt SH, Arola AM, Murphy RT, McKenna WJ, et al. ANKRD1, the gene encoding cardiac ankyrin repeat protein, is a novel dilated cardiomyopathy gene. J Am Coll Cardiol. 2009;54(4):325–33. Epub 2009/07/18. 10.1016/j.jacc.2009.02.076 ; PubMed Central PMCID: PMCPMC2915893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duboscq-Bidot L, Charron P, Ruppert V, Fauchier L, Richter A, Tavazzi L, et al. Mutations in the ANKRD1 gene encoding CARP are responsible for human dilated cardiomyopathy. Eur Heart J. 2009;30(17):2128–36. Epub 2009/06/16. 10.1093/eurheartj/ehp225 . [DOI] [PubMed] [Google Scholar]
- 35.Angori S, Capanni C, Faulkner G, Bean C, Boriani G, Lattanzi G, et al. Emery-Dreifuss Muscular Dystrophy-Associated Mutant Forms of Lamin A Recruit the Stress Responsive Protein Ankrd2 into the Nucleus, Affecting the Cellular Response to Oxidative Stress. Cell Physiol Biochem. 2017;42(1):169–84. Epub 2017/05/23. 10.1159/000477309 . [DOI] [PubMed] [Google Scholar]
- 36.Barash IA, Bang ML, Mathew L, Greaser ML, Chen J, Lieber RL. Structural and regulatory roles of muscle ankyrin repeat protein family in skeletal muscle. Am J Physiol Cell Physiol. 2007;293(1):C218–27. Epub 2007/03/30. 10.1152/ajpcell.00055.2007 . [DOI] [PubMed] [Google Scholar]
- 37.Bang ML, Gu Y, Dalton ND, Peterson KL, Chien KR, Chen J. The muscle ankyrin repeat proteins CARP, Ankrd2, and DARP are not essential for normal cardiac development and function at basal conditions and in response to pressure overload. PLoS One. 2014;9(4):e93638 Epub 2014/04/17. 10.1371/journal.pone.0093638 ; PubMed Central PMCID: PMCPMC3988038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gut P, Reischauer S, Stainier DYR, Arnaout R. LITTLE FISH, BIG DATA: ZEBRAFISH AS A MODEL FOR CARDIOVASCULAR AND METABOLIC DISEASE. Physiol Rev. 2017;97(3):889–938. Epub 2017/05/05. 10.1152/physrev.00038.2016 ; PubMed Central PMCID: PMCPMC5817164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. Epub 1995/07/01. 10.1002/aja.1002030302 . [DOI] [PubMed] [Google Scholar]
- 40.Blazina AR, Vianna MR, Lara DR. The spinning task: a new protocol to easily assess motor coordination and resistance in zebrafish. Zebrafish. 2013;10(4):480–5. Epub 2013/09/21. 10.1089/zeb.2012.0860 . [DOI] [PubMed] [Google Scholar]
- 41.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3(1):59–69. Epub 2008/01/15. 10.1038/nprot.2007.514 . [DOI] [PubMed] [Google Scholar]
- 42.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539 Epub 2011/10/13. 10.1038/msb.2011.75 ; PubMed Central PMCID: PMCPMC3261699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95(11):5857–64. Epub 1998/05/30. ; PubMed Central PMCID: PMCPMC34487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43(Database issue):D257–60. Epub 2014/10/11. 10.1093/nar/gku949 ; PubMed Central PMCID: PMCPMC4384020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics. 2009;10:202 Epub 2009/07/01. 10.1186/1471-2105-10-202 ; PubMed Central PMCID: PMCPMC2711084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. Epub 2003/10/08. . [DOI] [PubMed] [Google Scholar]
- 47.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–21. Epub 2010/06/09. 10.1093/sysbio/syq010 . [DOI] [PubMed] [Google Scholar]
- 48.Louis A, Muffato M, Roest Crollius H. Genomicus: five genome browsers for comparative genomics in eukaryota. Nucleic Acids Res. 2013;41(Database issue):D700–5. Epub 2012/11/30. 10.1093/nar/gks1156 ; PubMed Central PMCID: PMCPMC3531091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palstra AP, Rovira M, Rizo-Roca D, Torrella JR, Spaink HP, Planas JV. Swimming-induced exercise promotes hypertrophy and vascularization of fast skeletal muscle fibres and activation of myogenic and angiogenic transcriptional programs in adult zebrafish. BMC Genomics. 2014;15:1136 Epub 2014/12/19. 10.1186/1471-2164-15-1136 ; PubMed Central PMCID: PMCPMC4378002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rovira M, Arrey G, Planas JV. Exercise-Induced Hypertrophic and Oxidative Signaling Pathways and Myokine Expression in Fast Muscle of Adult Zebrafish. Front Physiol. 2017;8:1063 Epub 2018/01/13. 10.3389/fphys.2017.01063 ; PubMed Central PMCID: PMCPMC5741866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rovira M. Skeletal muscle and cardiac adaptations to swimming-induced exercise in adult zebrafish. PhD Thesis. The University of Barcelona. 2016. Available from: https://www.tesisenred.net/bitstream/handle/10803/402623/MRiB_PhD_THESIS.pdf?sequence=1&isAllowed=y.
- 52.LeMoine CM, Craig PM, Dhekney K, Kim JJ, McClelland GB. Temporal and spatial patterns of gene expression in skeletal muscles in response to swim training in adult zebrafish (Danio rerio). J Comp Physiol B. 2010;180(1):151–60. Epub 2009/08/21. 10.1007/s00360-009-0398-5 . [DOI] [PubMed] [Google Scholar]
- 53.Stuewe SR, Gwirtz PA, Agarwal N, Mallet RT. Exercise training enhances glycolytic and oxidative enzymes in canine ventricular myocardium. J Mol Cell Cardiol. 2000;32(6):903–13. Epub 2000/07/11. 10.1006/jmcc.2000.1131 . [DOI] [PubMed] [Google Scholar]
- 54.Ibanez J, Gauquelin G, Desplanches D, Qiu HY, Dalmaz Y, Fareh J, et al. Atrial natriuretic peptide response to endurance physical training in the rat. Eur J Appl Physiol Occup Physiol. 1990;60(4):265–70. Epub 1990/01/01. . [DOI] [PubMed] [Google Scholar]
- 55.Kanda K, Sugama K, Sakuma J, Kawakami Y, Suzuki K. Evaluation of serum leaking enzymes and investigation into new biomarkers for exercise-induced muscle damage. Exerc Immunol Rev. 2014;20:39–54. Epub 2014/07/01. . [PubMed] [Google Scholar]
- 56.Witt SH, Labeit D, Granzier H, Labeit S, Witt CC. Dimerization of the cardiac ankyrin protein CARP: implications for MARP titin-based signaling. J Muscle Res Cell Motil. 2005;26(6–8):401–8. Epub 2006/02/02. 10.1007/s10974-005-9022-9 . [DOI] [PubMed] [Google Scholar]
- 57.Lun AS, Chen J, Lange S. Probing muscle ankyrin-repeat protein (MARP) structure and function. Anat Rec (Hoboken). 2014;297(9):1615–29. Epub 2014/08/16. 10.1002/ar.22968 ; PubMed Central PMCID: PMCPMC4135402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jeyaseelan R, Poizat C, Baker RK, Abdishoo S, Isterabadi LB, Lyons GE, et al. A novel cardiac-restricted target for doxorubicin. CARP, a nuclear modulator of gene expression in cardiac progenitor cells and cardiomyocytes. J Biol Chem. 1997;272(36):22800–8. Epub 1997/09/05. . [DOI] [PubMed] [Google Scholar]
- 59.Moriyama M, Tsukamoto Y, Fujiwara M, Kondo G, Nakada C, Baba T, et al. Identification of a novel human ankyrin-repeated protein homologous to CARP. Biochem Biophys Res Commun. 2001;285(3):715–23. Epub 2001/07/17. 10.1006/bbrc.2001.5216 . [DOI] [PubMed] [Google Scholar]
- 60.Petryszak R, Keays M, Tang YA, Fonseca NA, Barrera E, Burdett T, et al. Expression Atlas update—an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 2016;44(D1):D746–52. Epub 2015/10/21. 10.1093/nar/gkv1045 ; PubMed Central PMCID: PMCPMC4702781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szeto DP, Kimelman D. The regulation of mesodermal progenitor cell commitment to somitogenesis subdivides the zebrafish body musculature into distinct domains. Genes Dev. 2006;20(14):1923–32. Epub 2006/07/19. 10.1101/gad.1435306 ; PubMed Central PMCID: PMCPMC1522088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nord H, Burguiere AC, Muck J, Nord C, Ahlgren U, von Hofsten J. Differential regulation of myosin heavy chains defines new muscle domains in zebrafish. Mol Biol Cell. 2014;25(8):1384–95. Epub 2014/02/14. 10.1091/mbc.E13-08-0486 ; PubMed Central PMCID: PMCPMC3983002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jasnic-Savovic J, Nestorovic A, Savic S, Karasek S, Vitulo N, Valle G, et al. Profiling of skeletal muscle Ankrd2 protein in human cardiac tissue and neonatal rat cardiomyocytes. Histochem Cell Biol. 2015;143(6):583–97. Epub 2015/01/15. 10.1007/s00418-015-1307-5 . [DOI] [PubMed] [Google Scholar]
- 64.Ma G, Wang H, Gu X, Li W, Zhang X, Cui L, et al. CARP, a myostatin-downregulated gene in CFM Cells, is a novel essential positive regulator of myogenesis. Int J Biol Sci. 2014;10(3):309–20. Epub 2014/03/20. 10.7150/ijbs.7475 ; PubMed Central PMCID: PMCPMC3957086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shih YH, Zhang Y, Ding Y, Ross CA, Li H, Olson TM, et al. Cardiac transcriptome and dilated cardiomyopathy genes in zebrafish. Circ Cardiovasc Genet. 2015;8(2):261–9. Epub 2015/01/15. 10.1161/CIRCGENETICS.114.000702 ; PubMed Central PMCID: PMCPMC4406804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hentzen ER, Lahey M, Peters D, Mathew L, Barash IA, Friden J, et al. Stress-dependent and -independent expression of the myogenic regulatory factors and the MARP genes after eccentric contractions in rats. J Physiol. 2006;570(Pt 1):157–67. Epub 2005/10/22. 10.1113/jphysiol.2005.093005 ; PubMed Central PMCID: PMCPMC1464283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carson JA, Nettleton D, Reecy JM. Differential gene expression in the rat soleus muscle during early work overload-induced hypertrophy. Faseb j. 2002;16(2):207–9. Epub 2001/12/18. 10.1096/fj.01-0544fje . [DOI] [PubMed] [Google Scholar]
- 68.Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3(11):827–37. Epub 2002/11/05. 10.1038/nrg928 . [DOI] [PubMed] [Google Scholar]
- 69.Force A, Lynch M, Pickett FB, Amores A, Yan Y-l, Postlethwait J. Preservation of Duplicate Genes by Complementary, Degenerative Mutations. Genetics. 1999;151(4):1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van der Meulen T, Schipper H, van den Boogaart JG, Huising MO, Kranenbarg S, van Leeuwen JL. Endurance exercise differentially stimulates heart and axial muscle development in zebrafish (Danio rerio). Am J Physiol Regul Integr Comp Physiol. 2006;291(4):R1040–8. Epub 2006/09/13. 10.1152/ajpregu.00116.2006 . [DOI] [PubMed] [Google Scholar]
- 71.McClelland GB, Craig PM, Dhekney K, Dipardo S. Temperature- and exercise-induced gene expression and metabolic enzyme changes in skeletal muscle of adult zebrafish (Danio rerio). J Physiol. 2006;577(Pt 2):739–51. Epub 2006/09/23. 10.1113/jphysiol.2006.119032 ; PubMed Central PMCID: PMCPMC1890438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Usui T, Noble DWA, O'Dea RE, Fangmeier ML, Lagisz M, Hesselson D, et al. The French press: a repeatable and high-throughput approach to exercising zebrafish (Danio rerio). PeerJ. 2018;6:e4292 Epub 2018/01/27. 10.7717/peerj.4292 ; PubMed Central PMCID: PMCPMC5775754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goldman JA, Kuzu G, Lee N, Karasik J, Gemberling M, Foglia MJ, et al. Resolving Heart Regeneration by Replacement Histone Profiling. Dev Cell. 2017;40(4):392–404.e5. Epub 2017/03/02. 10.1016/j.devcel.2017.01.013 ; PubMed Central PMCID: PMCPMC5367476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu CC, Kruse F, Vasudevarao MD, Junker JP, Zebrowski DC, Fischer K, et al. Spatially Resolved Genome-wide Transcriptional Profiling Identifies BMP Signaling as Essential Regulator of Zebrafish Cardiomyocyte Regeneration. Dev Cell. 2016;36(1):36–49. Epub 2016/01/11. 10.1016/j.devcel.2015.12.010 . [DOI] [PubMed] [Google Scholar]
- 75.Lai SL, Marin-Juez R, Moura PL, Kuenne C, Lai JKH, Tsedeke AT, et al. Reciprocal analyses in zebrafish and medaka reveal that harnessing the immune response promotes cardiac regeneration. Elife. 2017;6 Epub 2017/06/21. 10.7554/eLife.25605 ; PubMed Central PMCID: PMCPMC5498136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(TIF)
(AVI)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.