Abstract
Production of value-added compounds from a renewable aromatic polymer, lignin, has proven to be challenging. Chemical procedures, involving harsh reaction conditions, are costly and often result in nonselective degradation of lignin linkages. Therefore, enzymatic catalysis with selective cleavage of lignin bonds provides a sustainable option for lignin valorization. In this study, we describe the first functionally characterized fungal intracellular β-etherase from the wood-degrading white-rot basidiomycete Dichomitus squalens. This enzyme, Ds-GST1, from the glutathione-S-transferase superfamily selectively cleaved the β-O-4 aryl ether bond of a dimeric lignin model compound in a glutathione-dependent reaction. Ds-GST1 also demonstrated activity on polymeric synthetic lignin fractions, shown by a decrease in molecular weight distribution of the laccase-oxidized guaiacyl dehydrogenation polymer. In addition to a possible role of Ds-GST1 in intracellular catabolism of lignin-derived aromatic compounds, the cleavage of the most abundant linkages in lignin under mild reaction conditions makes this biocatalyst an attractive green alternative in biotechnological applications.
Keywords: Dichomitus squalens, White-rot fungi, Glutathione-S-transferase, β-Etherase, Lignin, β-O-4 linkage
Short abstract
Ds-GST1 is the first functionally characterized fungal β-etherase cleaving the β-O-4 aryl ether bond of a dimeric lignin model compound.
Introduction
Lignin is the largest renewable resource of aromatics with the production of approximately 100 million tons per year.1 Due to its recalcitrance imposed by, e.g., various C–C and C–O intermolecular bonds of which the arylglycerol β-O-4 aryl ether bond, accounting for 45–60% of the total linkages, is the most abundant,2 lignin is still largely considered as a side product of biorefineries. Other bonding patterns in native lignins include β-5 phenyl coumaran (6–12%), β-β′ pinoresinol (2–4%), 5-5′ biphenyl (9–22%), β-1 diaryl propane (1–9%) and 4-O-5′ diaryl ether (1–7%) linkage types, as well as dibenzodioxocin structure (5-5′-α, β-O-4′; 5–7%).3 Despite the structural hindrance, lignin holds a great potential as a sustainable feedstock for fuels, chemicals and materials.4
For the biorefinery concept to be fully sustainable and economically viable, all the lignocellulose fractions, including lignin, should be utilized. Currently, lignocellulosic biorefineries produce 60% excess lignin that could be used as a renewable feedstock.5 Development of new processes for the generation of lignin-derived value-added products is therefore required. Carbon fibers, plant-derived plastics and composites are examples of the successfully produced value-added materials originating from lignin.6 In addition to materials applications, some lignin fractions can as well be used for conversion to fuels such as syngas7 and chemicals like p-substituted benzyl alcohols or aldehydes.8
Efficient valorization requires depolymerization of lignin polymers to oligo- and monomeric aromatic compounds. Also, more uniform pools of desired aromatic products could be accomplished by selective enzymatic conversion of lignin. In this respect, microbial β-etherases catalyzing the reductive cleavage of β-O-4 bonds are promising candidates for biotechnological lignin applications. β-Etherases belong to the glutathione-S-transferase (GST; EC 2.5.1.18) protein superfamily, the representatives of which are ubiquitously present in both prokaryotes and eukaryotes.9 So far, functional β-etherases have been characterized only from α-proteobacteria Sphingobium sp. SYK-6,10Novosphingobium sp. PP1Y11 and Novosphingobium aromaticivorans,12 although putative β-etherase encoding genes are also widely detected in fungal genomes.13
Wood-decaying basidiomycete white-rot fungi, such as Dichomitus squalens, are the only organisms that efficiently degrade native lignin molecules.14,15 This is due to their ability to produce extracellular nonselective enzymes, e.g., lignin, manganese and versatile peroxidases, and laccases, which randomly attack lignin bonds. However, knowledge about the intracellular metabolism of lignin-derived compounds in white-rot fungi, including specific pathways and enzymes involved in the conversion processes, is scarce.16
We show here that an intracellular GST of D. squalens, Ds-GST1, selectively cleaves the β-O-4 bond of a dimeric lignin model compound and acts on a synthetic lignin dehydrogenation polymer. This is the first characterization of a functional intracellular β-etherase from a lignin-degrading basidiomycete fungus.
Experimental section
Identification of Putative D. squalens β-Etherases
Putative β-etherase-encoding genes in the genome of D. squalens LYAD-421 SS1 (http://genome.jgi-psf.org/Dicsq1/Dicsq1.home.html) were identified by using Sphingobium sp. SYK-6 β-etherase LigE (GenBank accession: BAA02032) and three P. chrysosporium glutathione-S-transferase fungal specific class A (GSTFuA) amino acid sequences (JGI protein IDs: 5118, 5119, 5122) as queries in a BLASTP search (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins). In addition, “glutathione transferase” was used as a keyword for a search in the D. squalens genome portal. D. squalens protein sequences with Protein IDs 82843, 56311, 102613 and 61773, named as Ds-GST1, Ds-GST2, Ds-GST3 and Ds-GST4, respectively, showed the highest identity to P. chrysosporium GSTFuAs and LigE, and were selected for recombinant production in E. coli.
Microbial Strains and Cultivation Conditions
D. squalens FBCC312 was obtained from the HAMBI Fungal Biotechnology Culture Collection, University of Helsinki, Helsinki, Finland (fbcc@helsinki.fi), and maintained on 2% malt agar plates (2% (w/v) malt extract, 2% (w/v) agar agar). The fungus was cultivated on Norway spruce wood (Picea abies) cultures, from which total RNA was extracted and used for cDNA synthesis as described previously.17
E. coli DH5α and Rosetta(DE3)pLysS (Novagen) were used as cloning and production hosts, respectively. BL21(DE3)pLysS and Origami 2(DE3)pLysS strains (Novagen) were additionally tested as production hosts for Ds-GST4. Luria–Bertani medium (20 g/L tryptone (Lab M), 10 g/L yeast extract (Lab M), 20 g/L NaCl (Merck), pH 7.0) containing 50 μg/mL kanamycin (Sigma-Aldrich) was used for cultivation of DH5α, whereas for the production strains, 34 μg/mL chloramphenicol (Sigma-Aldrich) was also added. Materials and methods for cloning, heterologous expression and purification of Ds-GSTs are provided in the Supporting Information.
Enzymatic Cleavage of Dimers and Analysis by Liquid Chromatography
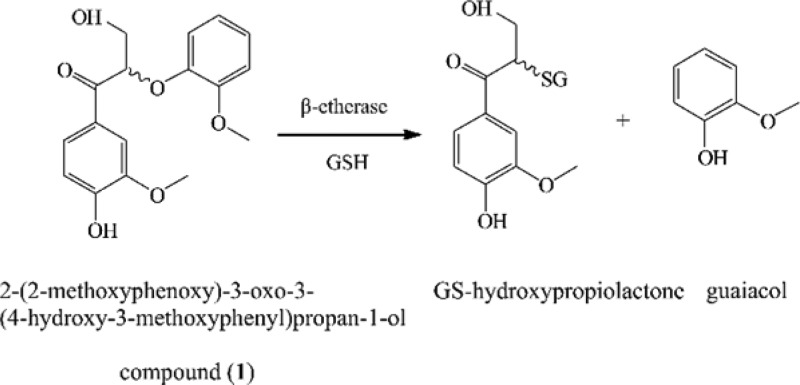
Materials and methods for the synthesis of lignin model compounds 2-(2-methoxyphenoxy)-3-oxo-3-(4-hydroxy-3-methoxyphenyl)propan-1-ol (1) (Figure 1A) and 2-(2-methoxyphenoxy)-3-(4-hydroxy-3-methoxyphenyl)propan-1,3-diol (2) (Figure 1B) and guaiacyl dehydrogenation polymer (G-DHP) are provided in the Supporting Information. The β-etherase activity of the purified Ds-GSTs and LigF was determined by following the degradation of (1) and release of guaiacol by ultrahigh performance liquid chromatography (UHPLC, Agilent 1290). The assay mixture (1 mL) contained 20 mM HEPES-buffer, pH 8.0, 2.5 mM (1), 6 mM reduced l-glutathione (GSH; Sigma-Aldrich), and 2.5 or 4 mg enzyme. Reactions were performed at 30 °C with constant mixing. Samples (100 μL) were taken prior to enzyme addition (zero point) and after 1, 24, and 72 h. Reactions were stopped with 25% (v/v) acetonitrile (Fischer Scientific) and stored at −20 °C. Precipitated proteins were removed by centrifugation (15 000g, 5 min), and the supernatant was filtered (Chromacol 4 mm Syringe Filter, 0.2 μm, Cellulose, Thermo Scientific) before UHPLC analysis. A negative control was prepared similarly as the reaction mixtures, but bacterial cell lysate containing empty plasmid was added instead of recombinant enzyme. Model compound (2) was treated as control substrate with Ds-GST1 and LigF.
Figure 1.
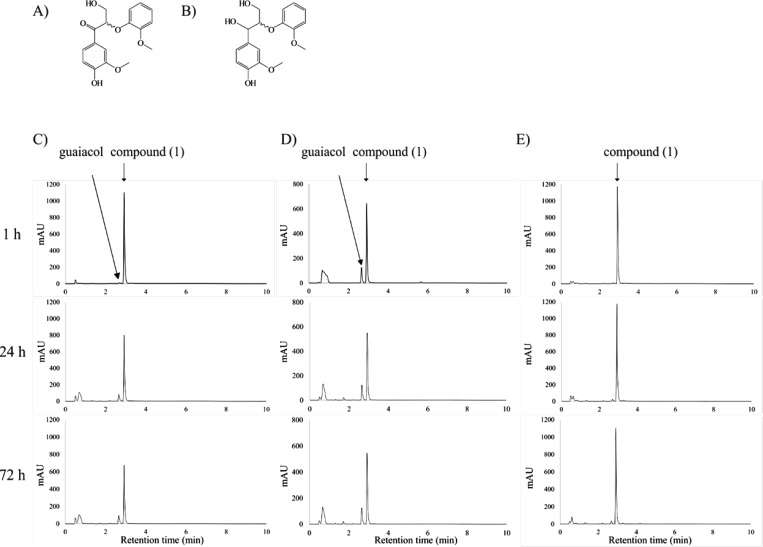
Lignin model compounds used in the study (A) compound 1, (B) compound 2. Release of guaiacol from compound 1 in the presence of GSH by (C) Ds-GST1, (D) positive control LigF and (E) negative control, after 1, 24 and 72 h as analyzed by UHPLC.
Three technical replicate reactions were run by UHPLC equipped with UV–vis diode array detector with reverse-phase Zorbax Eclipse XDB-C18 column (2.1 × 100 mm, 1.8 μm; Agilent) at 30 °C. The quantification was performed by using calibration curves of (1) and guaiacol (Sigma-Aldrich). Chromatographic separation was conducted in a binary gradient with water and acetonitrile (Fischer Scientific) as eluents, at a flow rate of 0.4 mL/min. The gradient was: 0–5 min from 10–90% acetonitrile; 5–6 min isocratic conditions 90% acetonitrile; 6–8 min from 90–10% acetonitrile. All compounds were detected at 280 nm.
Size Exclusion Chromatography Analysis of Enzymatically Treated G-DHP
The oxidized G-DHP (5 mg) was dissolved in 3 mL HEPES-buffered (20 mM, pH 8.5) 10% 1,4-dioxane and incubated overnight to ensure complete dissolution. Reaction mixtures containing oxidized G-DHP, 12 mM reduced GSH (Sigma-Aldrich) and 8 mg/mL Ds-GST1 or 4 mg/mL LigF were mixed (Eppendorf ThermoMixer C, 450 rpm) for 48 h at 25 °C, and acidified by two drops of 2 M HCl. Microcrystalline cellulose (200 mg, Fluka) was added to assist the handling of the material. Ethyl acetate (2 mL) was added on top of the reaction, vortexed and centrifuged (3,200 × g, 5 min, room temperature). The organic layer (low molecular weight fraction) was separated and the ethyl acetate extraction was repeated twice. The pellet of microcrystalline cellulose-bound G-DHP was extracted twice by adding 3 mL of 1,4-dioxane, mixed, incubated for 30 min at room temperature, and centrifuged (3,200g, 3 min, room temperature). Ethyl acetate and 1,4-dioxane extracts were evaporated to dryness. Acetylation for further analysis was conducted overnight by adding 0.5 mL pyridine (J.T. Baker) and acetic anhydride (Fluka).
The acetylated 1 mg/mL samples were dissolved in tetrahydrofuran (THF; Sigma-Aldrich) and filtered (0.20 μm Acrodisc GHP Membrane HPLC filters, Waters). The molecular weight distribution analysis was performed using Waters Acquity APC equipment with 10 μL injection. Acquity APC XT 200 2.5 and 45 1.7 μm columns (Waters) were used for separation at 30 °C with 0.8 mL/min THF elution. The detection was performed at 254 nm with UV and refractive index (RI) detector. The molar mass was calibrated using polystyrene standards (Scientific Polymer Products and Fluka Analytical). GPC Empower Software (Waters) was used for data processing to obtain Mn (number-average molecular weight) and Mw (weight-average molecular weight) as numerical output. The polydispersity PDI (Mw/Mn) was also calculated. The chromatograms are based on dioxane lignin extracts and were normalized against the highest peak in the chromatogram.
Results and discussion
In this study, we show that an intracellular GSH-dependent β-etherase Ds-GST1 from the white-rot fungus D. squalens catalyzes selective cleavage of β-O-4 bond in a racemic β-O-4 aryl ether lignin model (1). Ds-GST1 is the first functionally characterized fungal β-etherase with known gene sequence, and it can be classified into the fungal specific GSTFuA class of GST enzymes. This is also the first study on aromatic metabolism of lignin-derived compounds in D. squalens, although its ability to degrade chlorophenoxyacetic acids by ether bond cleavage has previously suggested the presence of an intracellular etherase.18 So far, intracellular β-etherase enzymes have been described only from few bacterial species.10−12,19 Although extracellular enzyme with β-etherase activity has been purified from the ascomycete fungus Chaetomium sp. strain 2BW-1,20 the corresponding gene encoding the enzyme has not been so far identified.
We identified 42 putative GST encoding sequences in the D. squalens genome (Table S1), which is slightly higher than the 33 reported earlier.21 In line with a previous study,22 six of these sequences were classified as putative members of the fungal specific GSTFuA class. Ds-GST1, Ds-GST2, Ds-GST3 and Ds-GST4 with the highest homology (43%, 44%, 45% and 42%, respectively) to P. chrysosporium GSTFuAs were selected for heterologous expression in E. coli (Figure S1). Ds-GST1, Ds-GST2 and Ds-GST3 were produced as 32, 30 and 30 kDa proteins, respectively (Figure S2), corresponding well to their theoretical molecular masses (31.5, 30.0 and 30.8 kDa, respectively). Ds-GST4 could not be produced regardless of the production strain used.
β-Etherase activity of Ds-GSTs was tested toward the β-O-4 model (1). In UHPLC analysis, Ds-GST1 showed accumulation of guaiacol with a retention time of 2.65 min and reduction of (1) with a retention time of 2.92 min (Figure 1C). This was only detected in the presence of GSH, thus confirming Ds-GST1 as a GSH-dependent β-etherase similar to all characterized bacterial β-etherases.19,23 Ds-GST2 and Ds-GST3 were not able to catalyze cleavage of 1 as no guaiacol was detected. Ds-GST1 exhibited specific activity of 117 pmol of guaiacol/mg of enzyme/min and transformed 36% of the racemic 1, whereas the positive control LigF, with specific activity of 1813 pmol of guaiacol/mg of enzyme/min, was able to transform 50% of the racemic 1 (Figure 1D). Specific activities of both Ds-GST1 and LigF were low compared to the previous reports on LigF with similar substrates.12 However, this could be due to nonoptimal reaction conditions, such as temperature and pH used in our study. As substrate side chains have been shown to significantly influence bacterial β-etherase activity, p-hydroxy substituent of the β-O-4 model (1) may have reduced the activity of LigF.12 The previously used model substrates have exclusively been p-methoxy-substituted derivatives and therefore no direct comparison is possible. All bacterial β-etherases described so far exhibit stereospecificity. For example, LigF catalyzes stereospecific cleavage of β(S)-enantiomers, whereas LigE exhibits β(R)-stereospecificity.23 Because lignin is a racemic polymer, corresponding stereospecificities of multiple (iso)enzymes involved in the degradation of β-O-4 aryl ether linkages containing lignin-derived compounds are necessary. Increasing the concentration of Ds-GST1 from 2.5 to 4 mg/mL, resulted in 42% cleavage of 1 indicating that Ds-GST1 possesses stereospecific properties. This was confirmed by chiral HPLC analysis, showing Ds-GST1 is stereospecific toward β(S)-enantiomer (Figure S3). Although complete conversion was not achieved, 50% cleavage of the racemic model compound should be reached by optimizing reaction conditions. Therefore, pH and temperature optima for Ds-GST1 will be determined in the future.
With 2, no guaiacol was detected, indicating that Ds-GST1 is active only on compounds with a keto group at the Cα position similar to all bacterial β-etherases known so far.10,12,24 However, in natural lignins, hydroxyl groups are present in Cα positions. In Sphingobium sp. SYK-6, several NAD+-dependent Cα-dehydrogenases, i.e., LigD, LigL, LigN and LigO, first oxidize these hydroxyl groups to the corresponding keto groups,24−26 after which β-etherases can cleave the β-O-4 bond. Enzymes with a similar function remain to be identified in D. squalens.
Most of the previous studies have used β-O-4 containing lignin model dimers to identify putative β-etherases.11,19,24,27,28 However, this does not necessarily imply activity of the enzymes on more complex lignin polymers, which is a crucial aspect in biotechnological processing of lignin into value-added products. Therefore, Ds-GST1 was also tested on a synthetic G-DHP lignin polymer, which was oxidized by laccase-MeS system to provide Cα-carbonyl functional groups. As a result, a clear shift in the molecular weight distributions (decrease of the Mw and Mn) of G-DHP was detected (Figure 2), confirming the ability of Ds-GST1 to act on lignin polymeric fractions. In addition, also LigF changed the molecular weight distribution of the laccase-oxidized G-DHP (Figure 2), thus indicating its ability to act on polymeric lignin. This is in line with the cleavage of the β-aryl ether linked fluorogen from synthetic DHP lignin that has been reported for bacterial β-etherases from Sphingobium sp. SYK-6, Novosphingobium sp. PP1Y and N. aromaticivorans DSM 12444 without12 and with the NAD+-dependent Cα-dehydrogenase LigD.29
Figure 2.
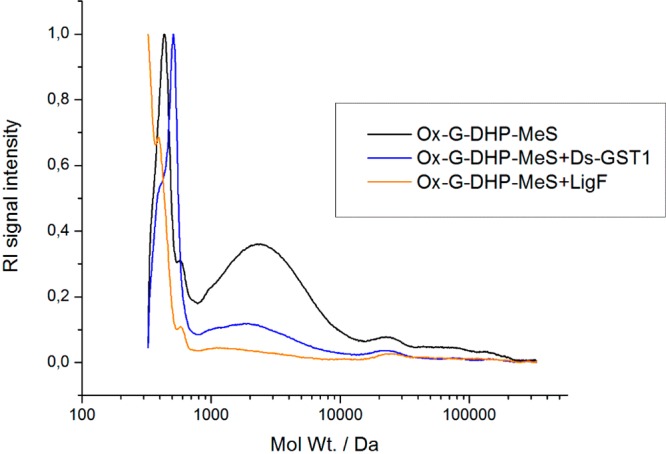
Size exclusion chromatography analysis showing the molecular weight distribution of oxidized synthetic lignin (Ox-G-DHP-MeS) after incubation with D. squalens GST1 (Ds-GST1). Sphingobium sp. SYK-6 LigF was used as a positive control. Ox-G-DHP-MeS, nontreated sample; Ox-G-DHP-MeS+Ds-GST1, sample treated with Ds-GST1; Ox-G-DHP-MeS+LigF, sample treated with LigF. The chromatograms are presented using a relative scale and were normalized against the highest peak in the chromatogram.
No change in molecular weight distribution has been detected when softwood and hardwood alkali kraft lignin, and bagasse organosolv lignin were used as substrates for the mixture of LigD, LigF and LigG.27 This could be due to the low abundance of β-O-4 linkages in technical lignins, as well as inhibition caused by sulfides and residual solvents from organosolv process.27 However, in the recent study the activity of LigE and N. aromaticivorans LigF-NA on laccase-oxidized OrganoCat beech wood lignin was reported.30 This multistep biocatalytic lignin depolymerization process employed also a fourth enzyme, LigG-TD from Thiobacillus denitrificans, which is a glutathione lyase. LigG-TD catalyzes a GSH-dependent thioether cleavage of glutathione adduct formed by LigF and LigE. As a result, low-molecular mass aromatic compounds (coniferylaldehyde, other guaiacyl and syringyl units) were detected together with larger lignin fractions. Similar multienzyme cascade conversion with five Sphingobium sp. SYK-6 enzymes (LigD, LigL, LigF, LigE and LigG) on lignin model compound GGE (1-(4-hydroxy-3-methoxyphenyl)-2-(2-methoxyphenoxy)propane-1,3-diol has been previously reported,31 where instead of laccase, LigD and LigL were utilized for the oxidation of Cα-hydroxyl groups. This resulted in almost complete conversion of GGE to 3-hydroxy-1-(4-hydroxy-3-methoxyphenyl)-propan-1-one. Our results demonstrate that D. squalens Ds-GST1, belonging to the GSTFuA class of GSTs, possesses β-etherase activity, thus suggesting a role in intracellular catabolism of lignin-derived aromatic compounds. In addition, the activity of Ds-GST1 on polymeric lignin model highlights its potential in selective conversion of lignin into value-added aromatic compounds. β-Etherases, including Ds-GST1, hold a great potential for future development of selective methods for lignin depolymerization. By catalyzing this chemically demanding reaction under mild conditions and in highly selective manner,32 these enzymes are both eco-friendly and industrially favorable. However, future work should address limiting factors such as regeneration of GSH cofactor and preparation of corresponding substrates with oxidized Cα-hydroxyl groups, to make the process industrially applicable.
Acknowledgments
This research was supported by the European Commission Marie Curie ITN network SuBiCat FP7 (grant no: 607044) (MM), FP7 project OPTIBIOCAT (grant no: 613868) (AD), Horizon 2020 project FALCON (grant no: 720918) (PN) and the Academy of Finland (grant no: 297847) (MM, JK). The authors would like to acknowledge Dr. Francisco J. Ruiz-Dueñas, CIB-CSIC, Madrid, Spain, for providing the plasmids and E. coli strains. Ms. Janetta Salin is thanked for technical assistance.
Glossary
Abbreviations
- G-DHP
guaiacyl dehydrogenation polymer
- GSH
reduced glutathione
- GST
glutathione-S-transferase
- GSTFuA
glutathione-S-transferase fungal specific class A
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- MeS
methyl syringate
- THF
tetrahydrofuran
- UHPLC
ultrahigh performance liquid chromatography
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssuschemeng.7b03619.
Cloning, heterologous expression and purification of enzymes; lignin model compounds synthesis; chiral chromatography; Ds-GSTs alignment with homologues from Sphingobium sp. SYK-6 (LigE and LigF), and P. chrysosporium (Pc-FuA1); SDS-PAGE analysis; chiral chromatography of lignin model (1), LigF and Ds-GST1 enzymatic reactions; putative GST encoding sequences of D. squalens (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Rinaldi R.; Jastrzebski R.; Clough M. T.; Ralph J.; Kennema M.; Bruijnincx P. C. A.; Weckhuysen B. M. Paving the way for lignin valorisation: recent advances in bioengineering, biorefining and catalysis. Angew. Chem., Int. Ed. 2016, 55, 8164–8215. 10.1002/anie.201510351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler E. Lignin chemistry—past, present and future. Wood Sci. Technol. 1977, 11, 169–218. 10.1007/BF00365615. [DOI] [Google Scholar]
- Balakshin M. Y.; Capanema E. A.; Chang H.. Recent advances in the isolation and analysis of lignins and lignin–carbohydrate complexes. In Characterization of Lignocellulosic Materials; Hu T. Q., Ed.; Blackwell Publishing: Oxford, U. K., 2008; pp 148–170. [Google Scholar]
- Li C.; Zhao X.; Wang A.; Huber G. W.; Zhang T. Catalytic transformation of lignin for the production of chemicals and fuels. Chem. Rev. 2015, 115, 11559–11624. 10.1021/acs.chemrev.5b00155. [DOI] [PubMed] [Google Scholar]
- Sannigrahi P.; Ragauskas A. Characterization of fermentation residues from the production of bio-ethanol from lignocellulosic feedstocks. J. Biobased Mater. Bioenergy 2011, 5, 514–519. 10.1166/jbmb.2011.1170. [DOI] [Google Scholar]
- Ragauskas A. J.; Beckham G. T.; Biddy M. J.; Chandra R.; Chen F.; Davis M. F.; Davison B. H.; Dixon R. A.; Gilna P.; Keller M.; Langan P.; Naskar A. K.; Saddler J. N.; Tschaplinski T. J.; Tuskan G. A.; Wyman C. E. Lignin valorization: improving lignin processing in the biorefinery. Science 2014, 344, 1246843. 10.1126/science.1246843. [DOI] [PubMed] [Google Scholar]
- Abdelaziz O. Y.; Brink D. P.; Prothmann J.; Ravi K.; Sun M.; García-Hidalgo J.; Sandahl M.; Hulteberg C. P.; Turner C.; Lidén G.; Gorwa-Grauslund M. F. Biological valorization of low molecular weight lignin. Biotechnol. Adv. 2016, 34, 1318–1346. 10.1016/j.biotechadv.2016.10.001. [DOI] [PubMed] [Google Scholar]
- Zakzeski J.; Bruijnincx P. C.; Jongerius A. L.; Weckhuysen B. M. The catalytic valorization of lignin for the production of renewable chemicals. Chem. Rev. 2010, 110, 3552–3599. 10.1021/cr900354u. [DOI] [PubMed] [Google Scholar]
- Sheehan D.; Meade G.; Foley V. M.; Dowd C. A. Structure, function and evolution of glutathione transferases: Implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 2001, 360, 1–16. 10.1042/0264-6021:3600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai E.; Katayama Y.; Kawai S.; Nishikawa S.; Yamasaki M.; Morohoshi N. Cloning and sequencing of the gene for a Pseudomonas paucimobilis enzyme that cleaves β-aryl ether. J. Bacteriol. 1991, 173, 7950–7955. 10.1128/jb.173.24.7950-7955.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall D. L.; Ralph J.; Donohue T. J.; Noguera D. R. A group of sequence-related sphingomonad enzymes catalyzes cleavage of β-aryl ether linkages in lignin β-guaiacyl and β-syringyl ether dimers. Environ. Sci. Technol. 2014, 48, 12454–12463. 10.1021/es503886d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picart P.; Muller C.; Mottweiler J.; Wiermans L.; Bolm C.; Domínguez de Maria P.; Schallmey A. From gene towards selective biomass valorization: bacterial β-etherases with catalytic activity on lignin-like polymers. ChemSusChem 2014, 7, 3164–3171. 10.1002/cssc.201402465. [DOI] [PubMed] [Google Scholar]
- Mathieu Y.; Gelhaye E.; Dumarcay S.; Gerardin P.; Harvengt L.; Buee M. Selection and validation of enzymatic activities as functional markers in wood biotechnology and fungal ecology. J. Microbiol. Methods 2013, 92, 157–163. 10.1016/j.mimet.2012.11.017. [DOI] [PubMed] [Google Scholar]
- Mäkelä M. R.; Hildén K. S.; de Vries R. P.. Degradation and modification of plant biomass by fungi. In Fungal Genomics; Nowrousian M., Ed.; The Mycota Book Series; Springer-Verlag: Berlin, 2014; pp 175–208. [Google Scholar]
- Rytioja J.; Hildén K.; Yuzon J.; Hatakka A.; de Vries R. P.; Mäkelä M. R. Plant-polysaccharide-degrading enzymes from basidiomycetes. Microbiol. Mol. Biol. Rev. 2014, 78, 614–649. 10.1128/MMBR.00035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä M. R.; Marinović M.; Nousiainen P.; Liwanag A. J. M.; Benoit I.; Sipilä J.; Hatakka A.; de Vries R. P.; Hildén K. S. Aromatic metabolism of filamentous fungi in relation to the presence of aromatic compounds in plant biomass. Adv. Appl. Microbiol. 2015, 91, 63–137. 10.1016/bs.aambs.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Rytioja J.; Hildén K.; Hatakka A.; Mäkelä M. R. Transcriptional analysis of selected cellulose-acting enzymes encoding genes of the white-rot fungus Dichomitus squalens on spruce wood and microcrystalline cellulose. Fungal Genet. Biol. 2014, 72, 91–98. 10.1016/j.fgb.2013.12.008. [DOI] [PubMed] [Google Scholar]
- Reddy G. V.; Joshi D. K.; Gold K. M. Degradation of chlorophenoxyacetic acids by the lignin-degrading fungus Dichomitus squalens. Microbiology 1997, 143, 2353–2360. 10.1099/00221287-143-7-2353. [DOI] [PubMed] [Google Scholar]
- Masai E.; Katayama Y.; Kubota S.; Kawai S.; Yamasaki M.; Morohoshi N. A bacterial enzyme degrading the model lignin compound β-etherase is a member of the glutathione-S-transferase superfamily. FEBS Lett. 1993, 323, 135–140. 10.1016/0014-5793(93)81465-C. [DOI] [PubMed] [Google Scholar]
- Otsuka Y.; Sonoki T.; Ikeda S.; Kajita S.; Nakamura M.; Katayama Y. Detection and characterization of a novel extracellular fungal enzyme that catalyzes the specific and hydrolytic cleavage of lignin guaiacylglycerol β-aryl ether linkages. Eur. J. Biochem. 2003, 270, 2353–2362. 10.1046/j.1432-1033.2003.03545.x. [DOI] [PubMed] [Google Scholar]
- Morel M.; Meux E.; Mathieu Y.; Thuillier A.; Chibani K.; Harvengt L.; Jacquot J.; Gelhaye E. Xenomic networks variability and adaptation traits in wood decaying fungi. Microb. Biotechnol. 2013, 6, 248–263. 10.1111/1751-7915.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu Y.; Prosper P.; Favier F.; Harvengt L.; Didierjean C.; Jacquot J. P.; Morel-Rouhier M.; Gelhaye E. Diversification of fungal specific class A glutathione transferases in saprotrophic fungi. PLoS One 2013, 8, e80298. 10.1371/journal.pone.0080298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall D. L.; Kim H.; Lu F.; Donohue T. J.; Noguera D. R.; Ralph J. Stereochemical features of glutathione-dependent enzymes in the Sphingobium sp. strain SYK-6 β-aryl etherase pathway. J. Biol. Chem. 2014, 289, 8656–8667. 10.1074/jbc.M113.536250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai E.; Katayama Y.; Nishikawa S.; Yamasaki M.; Morohoshi N.; Haraguchi T. Detection and localization of a new enzyme catalyzing the β-aryl ether cleavage in the soil bacterium (Pseudomonas paucimobilis SYK-6). FEBS Lett. 1989, 249, 348–352. 10.1016/0014-5793(89)80656-8. [DOI] [PubMed] [Google Scholar]
- Masai E.; Ichimura A.; Sato Y.; Miyauchi K.; Katayama Y.; Fukuda M. Roles of the enantioselective glutathione S-transferases in cleavage of β-aryl ether. J. Bacteriol. 2003, 185, 1768–1775. 10.1128/JB.185.6.1768-1775.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y.; Moriuchi H.; Hishiyama S.; Otsuka Y.; Oshima K.; Kasai D.; Nakamura M.; Ohara S.; Katayama Y.; Fukuda M.; Masai E. Identification of three alcohol dehydrogenase genes involved in the stereospecific catabolism of arylglycerol-β-aryl ether by Sphingobium sp. strain SYK-6. Appl. Environ. Microbiol. 2009, 75, 5195–5201. 10.1128/AEM.00880-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter J.; Strittmatter H.; Wiemann L. O.; Schieder D.; Sieber V. Enzymatic cleavage of lignin β-O-4 aryl ether bonds via net internal hydrogen transfer. Green Chem. 2013, 15, 1373–1381. 10.1039/c3gc40295a. [DOI] [Google Scholar]
- Tanamura K.; Abe T.; Kamimura N.; Kasai D.; Hishiyama S.; Otsuka Y.; Nakamura M.; Kajita S.; Katayama Y.; Fukuda M.; Masai E. Characterization of the third glutathione S-transferase gene involved in enantioselective cleavage of the β-aryl ether by Sphingobium sp. strain SYK-6. Biosci., Biotechnol., Biochem. 2011, 75, 2404–2407. 10.1271/bbb.110525. [DOI] [PubMed] [Google Scholar]
- Sonoki T.; Iimura Y.; Masai E.; Kajita S.; Katayama Y. Specific degradation of β-aryl ether linkage in synthetic lignin (dehydrogenative polymerizate) by bacterial enzymes of Sphingomonas paucimobilis SYK-6 produced in recombinant Escherichia coli. J. Wood Sci. 2002, 48, 429–433. 10.1007/BF00770705. [DOI] [Google Scholar]
- Picart P.; Liu H.; Grande P. M.; Anders N.; Zhu L.; Klankermayer J.; Leitner W.; Domínguez de Maria P.; Schwaneberg U.; Schallmey A. Multi-step biocatalytic depolymerization of lignin. Appl. Microbiol. Biotechnol. 2017, 101, 6277–6287. 10.1007/s00253-017-8360-z. [DOI] [PubMed] [Google Scholar]
- Rosini E.; Allegretti C.; Melis R.; Cerioli L.; Conti G.; Pollegioni L.; D’Arrigo P. Cascade enzymatic cleavage of the small β-O-4 linkage in a lignin model compound. Catal. Sci. Technol. 2016, 6, 2195–2205. 10.1039/C5CY01591J. [DOI] [Google Scholar]
- Picart P.; Domínguez de María P.; Schallmey A. From gene to biorefinery: microbial β-etherases as promising biocatalysts for lignin valorization. Front. Microbiol. 2015, 6, 916. 10.3389/fmicb.2015.00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.