This review article focuses on the Notch3 gene subtype, summarizing its role in cancer, possible mechanisms of its action, and current cancer treatment strategies targeting Notch3 signaling.
Keywords: Notch3, Cancer, Tumor, CADASIL
Abstract
The Notch family is a highly conserved gene group that regulates cell‐cell interaction, embryogenesis, and tissue commitment. This review article focuses on the third Notch family subtype, Notch3. Regulation via Notch3 signaling was first implicated in vasculogenesis. However, more recent findings suggest that Notch3 signaling may play an important role in oncogenesis, tumor maintenance, and resistance to chemotherapy. Its role is mainly oncogenic, although in some cancers it appears to be tumor suppressive. Despite the wealth of published literature, it remains relatively underexplored and requires further research to shed more light on its role in cancer development, determine its tissue‐specific function, and elaborate novel treatment strategies. Herein we summarize the role of Notch3 in cancer, possible mechanisms of its action, and current cancer treatment strategies targeting Notch3 signaling.
Implications for Practice.
The Notch family is a highly conserved gene group that regulates cell‐cell interaction, embryogenesis, and tissue commitment. This review summarizes the existing data on the third subtype of the Notch family, Notch3. The role of Notch3 in different types of cancers is discussed, as well as implications of its modification and new strategies to affect Notch3 signaling activity.
Introduction
The Notch family was discovered one century ago; however, various functions of the gene products have not been elucidated until the last few decades. In mammals, there are four Notch receptors (Notch1, Notch2, Notch3, and Notch4) and five canonical ligands [1]. Three Delta‐like ligands (DLL1, DLL3, and DLL4) are homologues in the Drosophila Delta family, and two Jagged ligands (Jag1 and Jag2) are homologues in the Drosophila Serrate family [2]. The ligand proteins on a neighboring cell's surface bind to the extracellular domain of Notch and via in‐trans regulation induce a signaling cascade to control cell differentiation, proliferation, and many other processes.
All Notch proteins share a similar basic structure, including an extracellular domain (ECD) at the N‐terminus, a transmembrane domain (TMD), and the Notch intracellular domain (NICD) at the C‐terminus. The ECD of Notch includes 29–36 epidermal growth factor (EGF)‐like repeats that contain six residues of cysteine, which favor the formation of a stable loop‐like structure. The Notch ligands bind these EGF‐like repeats with high specificity. The TMD contains a conserved negative regulatory region, which consists of three Lin‐Notch repeats that form a complex with Fringe, a protein modulating the ligand preference of Notch, and one transmembrane region [3]. The NICDs of all Notch family proteins contain three conserved domains: C‐repeat/DRE binding factor 1 (CBF1)/Recombination Signal Binding Protein For Immunoglobulin Kappa J Region (RBP‐jκ)‐associated molecule (RAM), seven Ankyrin repeats (ANK), and proline‐glutamic acid‐serine‐threonine (PEST). Most proximal to the cell membrane is the RAM domain, which interacts with CBF1/Suppressor of Hairless/gene/protein required for Lin‐12 and glp‐1 signaling (Lag‐1) (CSL) to form a transcription complex. Both the ANK domain and the PEST sequence are located near the C‐terminus and are involved in regulating protein stability and degradation. Only Notch1 and Notch2 include a transactivation domain (TAD) between ANK and PEST [4].
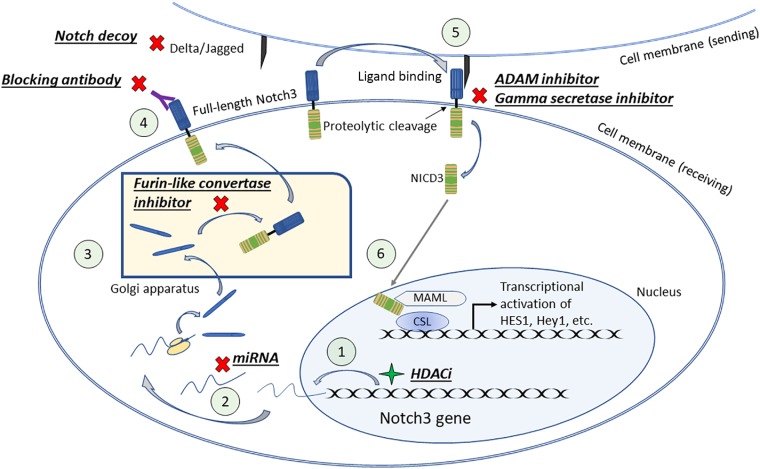
The Notch pathway leads to the expression of its target genes via a series of translocation, post‐translational modifications, and activation by its ligands. After translation, the Notch receptor is modified in the Golgi via proteolytic cleavage at site 1 (S1) by Furin‐like convertase. It is then transported to the cell surface as a heterodimer held together by noncovalent interactions. The Notch receptor on the signal‐receiving cell is activated by binding to a ligand on the cell surface of a neighboring signal‐sending cell. The binding causes a conformational change of the receptor to expose site 2 (S2) for cleavage by a disintegrin and metalloprotease (ADAM). Notch cleavage at S2 generates the membrane‐anchored Notch extracellular truncation (NEXT) fragment, a substrate for the γ‐secretase complex. γ‐secretase then cleaves the Notch transmembrane domain in the NEXT fragment progressively from site 3 (S3) to site 4 (S4), thereby releasing the NICD into the cytoplasm. NICD then enters the cell nucleus, where it associates with the DNA‐binding protein CSL. In the absence of NICD, CSL may associate with ubiquitous corepressor proteins and histone deacetylases (HDACs) to repress transcription of the target genes. However, in the event of NICD binding, allosteric change occurs in CSL that facilitates the displacement of transcriptional repressors. Subsequently, transcriptional coactivator Mastermind (MAM) recognizes the NICD/CSL interface, and the complex recruits additional coactivators mastermind‐like protein (MAML) and p300/P300/CBF‐associated factor (PCAF) to activate transcription. The aforementioned Notch pathway is depicted in Figure 1.
Notch signaling plays an important role in cell fate determination and organogenesis. Although the focus of this review is on Notch3, each of the other subtypes has been linked to human diseases. Missense mutations in the Notch1 gene causes bicuspid aortic valve formation, and its deletion is lethal during early stages of embryogenesis [5], [6]. It has also been shown to have both oncogenic and tumor suppressor roles in different types of cancers [7], [8]. Mutations in the Notch2 gene can cause Alagille and Hajdu‐Cheney syndromes, two autosomal‐dominant genetic conditions that affect skeletal, cardiovascular, neural, and gastrointestinal system development [9], [10]. An association between the mutation of the Notch4 gene and susceptibility to schizophrenia has been described [11].
The Notch3 gene, identified as the third mammalian Notch, was initially described in proliferating neuroepithelium [12]. Located on chromosome 19p13.12 (19: 15159632‐15200980), its 33 exons encode a single‐pass transmembrane heterodimer receptor protein composed of 2,321 amino acids. In canonical Notch3 signaling, Notch3 intracellular domain (NICD3) travels to the nucleus, where it binds to CSL to replace the transcription repressors HDAC1 and silencing mediator for retinoid or thyroid‐hormone receptors (SMRT) and recruits coactivators to activate transcription of Hey and Hes1, both of which are transcriptional regulators of the basic helix‐loop‐helix class.
A unique aspect of the Notch3 subtype is its restricted pattern of tissue distribution. Notch3 expression has been found in vascular smooth muscle, the central nervous system, and subsets of thymocytes [12], [13], [14]. Although the deletion of Notch1 and Notch2 is lethal to a developing embryo, deletion of Notch3 is not [5], [15]. Despite sharing a similar basic structure with Notch1 and 2, Notch3 displays a number of structural differences. The most apparent difference is a missing TAD, which could account for the weak transactivation activity of the NICD3 [4]. Slight differences are also evident in the Notch3 extracellular domain. It specifically lacks the Notch 1 and 2 equivalent of EGF repeat 21 and parts of EGF repeats 2 and 3 [12]. These differences could explain the differential ability of NICD3 to recruit coactivators or corepressors and undergo distinct conformational changes.
Changes in Notch activity have been associated with several benign and malignant diseases [7], [16], [17], [18], [19], [20], [21]. Although the primary function of Notch seems to be regulation of vasculogenesis, in which its dysregulation has been linked to a number of vasculopathies, it can also have tumor suppressive or oncogenic roles (Table 1).
Table 1. Oncogenic and tumor suppressive roles of Notch3.
Abbreviations: 5‐FU, fluorouracil; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; IL6, interleukin 6; MKP‐1, mitogen‐activated protein kinase phosphatase 1; NET, neuroendocrine tumor; NF‐κB, nuclear factor kappa‐light‐chain‐enhancer of activated B cells; NSCLC, non‐small cell lung cancer; p27kip1,; PDAC, pancreatic ductal adenocarcinoma; PEST, proline‐glutamic acid‐serine‐threonine; T‐ALL, T‐cell acute lymphoblastic leukemia; SCC, squamous cell cancer; siRNA, small interfering ribonucleic acid; STAT3, signal transducer and activator of transcription 3; T‐ALL, T‐cell acute lymphoblastic leukemia; TNBC, triple‐negative breast cancer.
Notch3 and Vasculogenesis
Although Notch3 is almost undetectable in normal human microvascular endothelium, it is elevated in infantile hemangioma stem cells [22]. The importance of Notch3 in development of normal vasculature was suggested when Notch3‐null mice had decreased retinal neovascularization and increased Notch3 expression was shown in a hypoxic model [23]. Kaposi's sarcoma is a virus‐induced angioproliferative disease, and the protein that is necessary for virus reactivation has been shown to form a complex with RBP‐Jκ [24], [25]. Interestingly, the endothelial cells in lymphatic vessels infected with Kaposi's sarcoma herpesvirus had higher Notch3 receptor density compared with uninfected cells [26]. Additionally, it has been demonstrated that endothelial Jag1 can modulate Notch3 signaling in the perivascular smooth muscle layer with a proangiogenic effect [27].
Cerebral Autosomal‐Dominant Arteriopathy with Subcortical Infarcts and Leukoencephalopathy
Because of the distribution pattern of Notch3, abnormalities in this protein are linked primarily to vascular pathologies, such as small‐vessel disease in skin, coronary arteries, and the brain [21]. The first example of Notch3‐related disease was reported in 1993, when mutations in the extracellular domain were identified in patients with cerebral autosomal‐dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) [28]. Since then, over 70% of mutations in CADASIL have been reported to occur in EGF‐like repeats 3 and 4 of the Notch3 extracellular domain. Nearly all are missense mutations involving a cysteine residue, in which cysteine is replaced with another amino acid or vice versa. The odd number of cysteine residues are thought to lead to misfolding of the protein, ultimately affecting its trafficking, processing, ligand binding, and signal transduction. Three mechanisms that involve mutant Notch3 in the pathology of CADASIL have been explored:
Local accumulation of Notch3 extracellular domain in the vascular smooth muscle results from local aggregation and impaired endocytosis [14]. Accumulated mutant Notch3 prevents protein ubiquitination and degradation, which leads to the formation of granular osmiophilic material [29].
The mutations in the Notch3 extracellular domain lower its affinity to Furin and thereby reduce the frequency of S1 cleavage. The amount of the mutant Notch3 transported to the cell surface is reduced and subsequently results in intracellular aggregation [30].
Mutations in EGF repeats 10 and 11 may alter CBF1/RBP‐jκ interaction. Interestingly, there are reports that CADASIL mutant Notch3 receptors show no change in affinity for ligand binding and that some mutations may actually increase signaling [30].
Notch3 in Hypoxia and Inflammation
Chronic inflammatory conditions have been associated with development of cancer in the gastrointestinal tract [31]. The inflammatory process involves the interplay between proinflammatory cytokines (e.g., interleukin 6 [IL6]) and a variety of anti‐inflammatory mediators that may be produced by enzymes such as 15‐lipoxygenase‐1 (15‐LOX‐1). The constitutive activation of 15‐LOX‐1 in mice intestine decreased IL6 expression and signal transducer and activator of transcription 3 (STAT3) phosphorylation. Decreased IL6/STAT3 transcriptional activation resulted in reduced Notch3 expression and concomitant inhibition of colitis‐associated colorectal cancer development [32]. IL6‐mediated induction of Notch3 has also been reported to promote stem cell‐like features, spheroid formation, and fluorouracil (5‐FU) resistance [33].
The association between IL6 and Notch3 has been observed in many types of tumors other than those of the gastrointestinal tract. Epidermal growth factor receptor (EGFR)‐enriched basal breast tumors have been found to have high IL6 and Notch3 expression [34]. IL6 increases Notch3 through the microtubule associated protein kinase (MAPK) pathway that leads to mammosphere formation and increase of Carbonic anhydrase 9 (CA‐IX), an enzyme involved in survival during hypoxia [35]. Notch3 is activated during hypoxia in prostate cancer cells, and Notch3‐Hypoxia‐inducible factor 1‐alpha (HIF‐1α) interaction is necessary for induction of CA‐IX in breast carcinoma cells [36], [37]. Notch3‐HIF‐1α interaction has also been found to be important in hypoxia‐related radiation resistance in lung cancer [38].
Urogenital Cancers
One of the tumors that has been found to be associated with CADASIL syndrome is prostatic adenocarcinoma [39]. Prostate cancer has been reported to have upregulated Notch3 and its downstream target gene Hey1 [20]. Notch3 expression was found to be positively correlated with the Gleason score, and high expression of Notch3 was seen in prostate cancers with high metastatic potential [40], [41]. In addition, endothelial Jag1 can activate Notch3 signaling in prostate tumor, leading to cancer cell proliferation and dedifferentiation [42].
Notch3 was shown to contribute to drug resistance when Notch3 knockdown combined with cisplatin chemotherapy eradicated drug‐resistant urothelial cancer cell clones. Notch3 knockdown also reduced bladder cancer progression in vivo, an observation perhaps most suggestive that Notch3 could be a suitable chemotherapeutic target for treating urothelial cancer [43].
Notch3 expression was found to be positively correlated with the Gleason score, and high expression of Notch3 was seen in prostate cancers with high metastatic potential. In addition, endothelial Jag1 can activate Notch3 signaling in prostate tumor, leading to cancer cell proliferation and dedifferentiation.
Leukemia
Notch3 is expressed in the early stages of T‐cell development, but it normally becomes downregulated as cells transition to the double‐positive stage during maturation. Furthermore, Notch3 appears to be redundant for T‐cell development, whereas Notch1 is required [44]. Even so, Notch3 expression is found in the majority of T‐cell acute lymphoblastic leukemia (T‐ALL) cases. Although it is rarely mutated, Notch3 appears to play a significant role in the survival and proliferation of T‐ALL [45]. For example, when expressed in endothelial cells, DLL4, another Notch activator, promotes T‐ALL cell proliferation by activating Notch3 signaling [46].
Recently, it has been shown that prostate apoptosis response factor 4 and Thanatos‐associated domain‐containing apoptosis‐associated protein 1 cooperate to induce apoptosis in T‐ALL cells and that this is mediated through the increased expression of cell cycle and apoptosis regulator 1 (CCAR1). However, Notch3 binding to the promoter region of CCAR1 leads to a truncated form of the protein and attenuates the proapoptotic function of CCAR1 [47]. Increased Notch3 signaling also plays a role in both the aggressiveness and the survival of T‐ALL through the Ras/MAPK pathway. Although it does not appear to be a direct transcriptional target, the protein stability of mitogen‐activated protein kinase phosphatase 1 (MKP‐1) appears to be under the control of Notch3. Both Notch3 knockdown and γ‐secretase inhibitors reduced MKP‐1 levels, which led to impaired tumorigenicity and increased apoptosis [48].
Next to Ras/MAPK pathway, NF‐κB is perhaps one of the most commonly reported transcription factors interacting with the Notch signaling pathway. It has been shown that transgenic mice that express the Notch3 intracellular domain develop T‐cell lymphomas attributable to Notch3‐induced NF‐κB activation and that protein kinase C θ was a downstream target of Notch3 responsible for NF‐κB activation in T‐cell leukemia [49], [50].
As stated previously, Notch signaling can have either oncogenic or tumor suppressor roles. The silencing of tumor suppressor genes has been linked to the overexpression of some HDACs [51]. HDAC1 is involved not only in histone modifications but also as part of a corepressor complex in the Notch signaling [52]. However, both HDAC1 and p300 also directly interact with Notch3 via modulation of acetylation. The transcription factor p300 contains an acetyltransferase domain that promotes ubiquitination and subsequent degradation of Notch3, whereas HDAC1 can reverse this by deacetylation. Agents that inhibit HDAC favored p300 activity leading to hyperacetylation. T‐cell proliferation was enhanced in cells possessing a nonacetylable Notch3 mutant and was inhibited in cells possessing acetylable Notch3 after treatment with HDAC inhibitors (HDACi) [53]. Immunoprecipitation experiments showed that HDAC1 directly interacts with Notch3. In another study, HDACi did not change the levels of Notch1 protein or downstream gene expression in B‐cell acute lymphoblastic leukemia cells. However, cell proliferation was inhibited, cell cycle was blocked in the G1/S phase, and apoptosis was induced [54]. These results suggest that although Notch1 may be important in the initiation of leukemia, more work needs to be done to more fully appreciate the role Notch3 has in leukemic cell survival and disease progression.
Squamous Cell Carcinoma
Squamous cell carcinoma (SCC) has higher Notch3 expression compared with surrounding normal epithelial tissues. Examination of 74 pathologic specimens found that that Notch3 expression was higher in SCC compared with adjacent normal tissue [55]. Notch3 overexpression was seen in 18.2% of head and neck squamous cell cancers, in more than 92% of primary nasopharyngeal carcinomas, and in all epstein‐barr virus (EBV)‐positive tumor cell lines [56]. Cervical SCC had higher nuclear Notch3 immunoreactivity compared with normal cervical epithelium or cervical intraepithelial neoplasms [57]. Additionally, increased Notch3 mRNA expression was found in esophageal SCC cell lines compared with noncancerous squamous esophageal cell lines. This increased expression was associated with esophageal cancer cell aggressiveness and resistance to 5‐FU [58]. Knocking down Notch3 in nasopharyngeal carcinoma increased chemosensitivity to cisplatin [56].
Higher Notch3 expression was observed not only in SCC cells but also in cancer‐associated fibroblasts. Notch3 expression in normal human dermal fibroblasts was induced by direct cell‐cell contact with oral SCC cell lines. Additionally, microvessel density in oral SCC was associated with the expression of Notch3 in cancer‐associated fibroblasts [59].
The majority of published results suggest that high expression of Notch3 is associated with a poorer prognosis in SCC [60]. Patients with cervical SCC had a shorter overall survival if they had nuclear Notch3 expression compared with those who did not. Knocking down Notch3 had antiproliferative and apoptotic effect on cervical SCC, and the injection of γ‐secretase inhibitor in tumors caused tumor shrinkage in mice [57]. An increase of Notch3 expression in one third of oral SCC has been correlated with larger tumor size. Growth inhibition and induction of apoptosis were observed in nasopharyngeal carcinoma after Notch3 knockdown and was associated with a decrease in protein levels of CCND1, C‐MYC, NFKB1, BCL2, BCL‐XL, and SURVIVIN.
Breast Cancer
In breast cancer, Notch3 serves mainly as an oncogene with some exceptions. Notch3 causes mammary tumor development in transgenic mice [61]. In breast cancer cell lines, Notch3 signaling was constitutively active, and among other Notch receptors its activity was solely sufficient for promoting tumor growth both in vitro and in vivo [16]. The importance of Notch3 signaling among other Notch family members was also shown by a study that demonstrated that downregulation of Notch3 significantly suppressed proliferation and promoted apoptosis of the ErbB2‐negative tumor cell lines. This effect was not observed in the ErbB2‐positive tumor cells. In contrast, Notch1 siRNA did not suppress the proliferation of either ErbB2‐positive or ErbB2‐negative cell lines [62]. Notch3 activation was seen in a human inflammatory breast cancer xenograft model by RT‐quantitative reverse transcription polymerase chain reaction (qPCR), western blot, and immunohistochemistry [63]. Among Notch3 aberrations, amplifications are more common than mutations in triple‐negative breast cancer (TNBC), with mutations mostly involving the PEST domain (typically involved in the negative regulation of gene expression) [64], [65]. In TNBC samples, 34% of the tumors were positive for NICD3 compared with 4% positive for Notch1 intracellular domain (NICD1) [16]. However, the role of Notch3 in breast cancer is not fully resolved. One study suggested that induction of the Notch3 intracellular domain led to p27kip1 accumulation, resulting in cancer cell cycle arrest at the G0/G1 phase [66].
The role of Notch3 in epidermal‐to‐mesenchymal transition (EMT) of breast cancers is also controversial. While some investigators reported that Notch3 promotes tumor aggressiveness by inducing EMT, other investigators demonstrated that Notch3 actually opposes it. Notch3 was shown to be inhibiting EMT by upregulating the Hippo/Yap pathway [16], [67]. Another mechanism of EMT inhibition was reported to be the downregulation of Fos‐related antigen 1 (Fra1), which is an activator of EMT. This observation was made in Adriamycin‐resistant human breast cancer cells, in which Notch3 was found to be downregulated, whereas Fra1 was expressed at high levels [68]. On the other hand, in metastatic breast cancer cells, a decrease in Notch3 levels decreased levels of the EMT‐associated proteins Snail, fibronectin, and vimentin [69].
Notch3 appears to be promoting breast cancer metastasis. It is expressed at significantly higher levels in invasive cancer compared with ductal carcinoma in situ [70]. Truncated‐Fms Related Tyrosine Kinase 1 (FLT1) isoform associated with increased breast cancer invasiveness was upregulated by Notch1 and Notch3 [8]. Lymphovascular emboli of human inflammatory breast cancer have been found to express increased levels of Notch3. After transfection with NICD3, spheroid formation was detected in normal breast epithelial cells that was not seen after NICD1 transfection [71]. Selective increase of Notch3 expression in human breast cancer cell lines was seen after stimulation with transforming growth factor beta 1 (TGFβ1) secreted by bone marrow osteoblasts. Cancer cell colony formation was inhibited after anti‐TGFβ1 antibody treatment and Notch3 chemical inhibition. Notch3 expression was also shown to be associated with osteolytic bone lesions [69].
Another mechanism by which Notch3 can contribute to the aggressiveness of breast cancer is its involvement in cell metabolism. Notch3 activity is essential for the survival of hormone therapy‐resistant metastatic breast cancer. It was also shown to be promoting mitochondrial activity and exit from metabolic dormancy. Tamoxifen‐resistant cells showed reduced mitochondrial DNA copy number, mitochondrial antigen expression, and diminished oxidative phosphorylation. The reduction of mitochondrial activity was reversed by IL6 treatment that was mediated by Notch3 [72]. The essential role of Notch3 in reversal of tamoxifen‐induced mitochondrial toxicity was demonstrated when IL6 treatment failed to increase mitochondrial activity in Notch3 knockdown cells [72]. Several studies have reported that in metastatic breast cancer, functional Notch3 is essential for IL6 to maintain high levels of carbonic anhydrase, an enzyme associated with hypoxia survival and cancer invasiveness [35], [36].
Notch3 can increase the chemosensitivity in doxorubicin‐resistant breast cancer by negatively regulating Fra1. Fra1 has been shown to play an important role in EMT of colorectal cancer, ovarian cancer, glioma, and breast cancer cell lines [68], [73], [74], [75]. Fra1 was highly expressed in doxorubicin‐resistant breast cancer cells while Notch3 was underexpressed and this resistance was mediated by Fra1‐induced EMT. Adriamycin chemoresistance could be recreated in chemosensitive cells by knocking down Notch3 signaling [68]. Stromal cells can also contribute to chemoresistance by inducing Notch3 in breast cancer cells, an effect that is reversed with Notch3 siRNA or γ‐secretase inhibitor (GSI) treatment [76]. Another mechanism by which cancer cells resist treatment is tumor dormancy. Upregulation of the Notch3 pathway arrested the cell cycle in G0/G1 phase and induced Hes1, Mix1, and ΔNp63α proteins, which promote cellular quiescence [77].
Ovarian Cancer
Notch3 expression is significantly higher in ovarian cancer compared with normal ovaries or benign ovarian tumors [17]. More than twofold higher expression of Notch3 was detected in 63% of serous ovarian carcinomas compared with benign ovarian tissue [78]. Notch3 gene amplification was correlated with elevated Notch3 protein expression in high‐grade serous cancers. Knocking down Notch3 induced apoptosis and stopped cell proliferation only in cells with high Notch3 expression [79]. Both Notch3 siRNA and GSI treatment decreased cell survival and antiapoptotic proteins, such as cyclin D1, cyclin D3, and the B‐cell lymphoma (BCL) family, and increased antiproliferative and proapoptotic proteins, such as p21, p27, Bad, Bak, Bim, Bid, and Bax [80].
Notch3 expression was associated with higher tumor grade, lymph node (LN) and distant metastasis, and higher clinical stage in ovarian cancer [17], [78]. It is known that Notch3 promotes resistance to anoikis, a process involved in programed cell death after detachment from anchoring extracellular matrix [81]. Jag1 is a primary Notch ligand in ovarian carcinoma as well as in peritoneal mesothelial cells. Knocking down Jag1 in mesothelial cells inhibited cell proliferation and adhesion of ovarian cancer cells, suggesting that a Jag1‐Notch3 interaction promoted ovarian cancer dissemination within the peritoneal cavity [82].
High Notch3 expression is associated with tumor recurrence and poor survival [17], [78]. High Notch3 expression was associated with shorter overall survival and worse progression‐free survival in recurrent high‐grade serous carcinoma (HGSC) of the ovary. Furthermore, Notch3 expression was associated with recurrence of HGSC after chemotherapy. Induction of Notch3 increased the concentration that inhibits cell growth by 50% (IC50) for carboplatin in low‐grade serous carcinoma cell lines, and Notch3 knockdown sensitized ovarian carcinoma to chemotherapy [83]. Tumor chemosensitization after Notch3 knockdown was also shown by cisplatin and GSI combination treatment, which led to a synergistic effect on DNA damage and G2/M cell cycle arrest [84]. Additionally, a decrease in cell cycle markers, arrest in G2/M phase, and induction of apoptosis has been observed after Notch3 knockdown in combination with either platinum‐based or taxane chemotherapy [80], [85], [86]. Notch3 overexpression was associated with shorter progression‐free survival in patients with advanced stage ovarian cancer who were treated with taxanes or platinum‐based chemotherapeutic agents [86]. Treatment of paclitaxel‐resistant ovarian cancer cells with microRNA 150 (miR‐150) or overexpression of miR‐136 reduced Notch3 mRNA expression, reversed chemoresistance, and decreased stemness genes [87], [88]. Another mechanism of synergy between Notch3 inhibition and chemotherapy is that it decreases angiogenesis [80], [89]. Activation of Notch3 attenuates the apoptotic effect of carboplatin by inhibiting carboplatin‐induced extracellular signal‐regulated kinase (ERK) phosphorylation [85]. Although several studies reported that high Notch3 expression was associated with worse overall survival time, others had contradictory findings [90]. Additionally, high Notch3 expression has been correlated with better overall survival for all patients with ovarian cancer except with those who had serous cancer [90], [91].
Colorectal Cancer
Notch3 is commonly expressed at a significantly higher level in colorectal cancer (CRC) compared with normal tissue, with poorly differentiated tumors having higher expression levels than well‐differentiated tumors [92], [93]. Higher expression of Notch3 is associated with increased tumor growth rate, and knocking it down slows tumor growth [93]. Notch3 expression is significantly higher in micropapillary carcinomas compared with nonmicropapillary carcinomas (69 vs. 42%), with micropapillary carcinomas having more frequent lymphovascular invasion, higher tumor stage, and LN metastasis rate [94]. Thirty‐eight percent of stage II and III colorectal cancers overexpressed Notch3, which was correlated with higher tumor grades, higher rates of venous invasion and LN metastasis, and shorter recurrence‐free survival [92]. Notch3 was found to upregulate the exchange factor Asef, leading to increased cell migration and tumor invasiveness [95]. miR‐206, a microRNA that inhibits Notch3 translation, was found to activate apoptosis, induce cell cycle arrest, and block proliferation and migration when transfected into CRC cancer cell lines [96]. miR‐1 may be another microRNA regulating Notch3 with potential for treatment utility in CRC [95].
Hepatocellular Carcinoma
Immunohistochemical analysis of Notch3 expression in liver tissue found that 78% of hepatocellular carcinomas (HCCs) had abnormally elevated Notch3 levels, whereas it was undetectable in normal liver or tissue adjacent to HCCs with chronic hepatitis. In 95% of HCCs, Notch3 mRNA was elevated compared with matching cirrhotic tissue, and Notch3 mRNA expression was in parallel with Hes6 mRNA expression [97]. Among other Notch family members, Notch3 had the highest upregulation in HCC compared with normal liver [98]. High Notch3 expression in HCC was found to be associated with hepatitis B infection [99], [100].
Initial work suggested that Notch3 exerted its effect in HCC via the β‐catenin pathway. However, it was later concluded that cell proliferation and stemness in HCC was linked to Notch3 upregulation of Nanog, a transcription factor critical for embryonic stem cell self‐renewal. High expression of Notch3 is also associated with larger tumors and elevated α‐fetoprotein levels [98], [101], [102]. Notch3 helps maintain low differentiation and high migratory capacity in HCC [101], [102]. Elevated Notch3 expression correlate with more satellite lesions, tumor multifocality, and metastasis [98], [101]. High expression of Notch3, but not other subtypes, is associated with increased vascular invasion by HCC [100], [101]. Transient transfection of miR‐206 in HCC cells decreased Notch3, Hes1, BCL‐2, and MMP‐9, whereas p57 and Bax levels were upregulated. Additionally, miR‐206 transfection has been shown to decrease migration in HepG2 cells via inhibition of a Notch3/matrix metallopeptidase 9 (MMP‐9) pathway [96], [103]. This Notch3‐linked inhibition of HCC migration through decrease of MMP‐9 and MMP‐2 seems to be mediated by the ERK1/2 pathway [101].
Notch3 expression is associated with early recurrence and poor overall survival in HCC and was shown to oppose the antitumor effect of chemotherapy. Downregulation of Notch3 increased sensitivity to doxorubicin, cisplatin, and sorafenib [98], [99], [101], [102], [104], [105]. Inhibition of Notch3 enhanced the effect of sorafenib in vitro as well as in a mouse xenograft model and was associated with low levels of p21 [105]. A Notch3 knockdown sensitized HCC to the cytotoxic effect of doxorubicin and increased p53 mRNA, an effect not seen with Notch1 [104].
Notch3 has been found to have an oncogenic role in other liver tumors, such as cholangiocarcinoma (CCA). Tumor formation and progression was attenuated after genetic deletion of Notch3 in CCA cell lines. In CCA, Notch3 seemed to be affecting tumor progression via a noncanonical Phosphatidylinositol‐4,5‐Bisphosphate 3‐Kinase (PI3K)‐Akt pathway, independent of the RBPJ protein. The authors suggested that selectively targeting Notch3 via this noncanonical pathway could prevent toxicities that are known to be RBPJ dependent [106].
Pancreatic Ductal Adenocarcinoma
Notch3 has been reported to be overexpressed in pancreatic ductal adenocarcinoma (PDAC) and is associated with higher tumor grade, lymphovascular invasion, and rates of metastasis [107], [108], [109]. Both Notch3 and Hey1 overexpression were associated with vascular invasion and increased LN metastasis [108]. Notch3 promotes invasion by modifying E‐cadherin, MMP‐2, MMP‐9, and vascular endothelial growth factor (VEGF) expression, whereas knocking down Notch3 decreased invasiveness [107]. Notch3 also promoted angiogenesis by activating of VEGF‐A, MMP‐9, and Angiogenin‐1 [110]. Decreased Notch3 activity resulted in tumor cell apoptosis, cell cycle arrest, and the suppression of cell proliferation, invasion, and migration, each of which correlates with overexpression of miR‐613. Furthermore, downregulation of miR‐613 is associated with shorter overall survival, higher TNM grade, and poor differentiation of pancreatic cancer cells [111].
Notch3 expression has been correlated with shortened disease‐free and overall survival in pancreatic adenocarcinoma [108], [112]. Nuclear localization of Notch3 is not normally found in pancreatic ductal tissue, but it has been observed in 44% of PDAC and 100% of unresectable tumors and has a strong correlation to pAkt expression [109]. The link between Notch3 expression and the Akt pathway in PDAC was further supported when it was shown that knocking down Notch3 decreased PI3K/Akt activity and increased pancreatic cancer cell sensitivity to gemcitabine chemotherapy [113].
Neuroendocrine Cancers
Neuroendocrine tumors (NETs) include many types of neural crest‐derived tumors that secrete different biogenic amines or peptide hormones [114]. Notch signaling has been found to have a tumor suppressor role in NETs. Medullary thyroid carcinoma (MTC) is a neuroendocrine tumor that develops from calcitonin‐producing parafollicular C cells in the thyroid gland. Developing C cells express Achaete‐scute homolog 1 (ASCL‐1) at high levels, and the expression of ASCL‐1 is lost in mature C cells [115]. ASCL‐1 is a downstream target of Notch and is considered to be a neuroendocrine tumor marker [116]. Notch3 activation decreased neuroendocrine markers (chromogranin A, ASCL‐1) in MTC in a dose‐dependent manner. Induction of the Notch3 gene and increased protein activity was found to be associated with apoptosis and decreased MTC cell proliferation [117]. Notch1 has been shown to have tumor suppressive role in gastrointestinal carcinoids and pheochromocytoma; however, the role of Notch3 in these tumors is understudied and requires more research [118], [119], [120].
Lung Cancers
Notch3 plays an important role in lung development. Constitutive activation of Notch3 in mice causes an increase in undifferentiated epithelial cells, delayed lung maturation, and perinatal mortality [121]. It is overexpressed in about 40% of non‐small cell lung cancers (NSCLCs) [122], [123]. In genetically engineered mouse models, induced expression of Notch3 increased tumorigenicity in the lung [124]. Inhibiting Notch3 expression decreased malignant features both in vitro and in vivo via induced apoptosis and decreased cell proliferation, migration, and invasiveness [122], [123], [125]. The induction of apoptosis by Notch3 knockdown is likely mediated by Bim [126].
Clinically in NSCLC, Notch3 overexpression is associated with poorer disease‐free and overall survival, higher TNM stage, and increased LN metastasis rate [125], [127], [128]. Overexpression of Notch3 induces Zinc finger E‐box‐binding (ZEB‐1) activity, decreases E‐cadherin, and upregulates fibronectin, each contributing to EMT and tumor invasiveness. This increase in EMT was abolished by ZEB‐1 siRNA. Notch3 induced ZEB‐1 may promote EMT and bone metastasis in NSCLC through TGFβ; however, Notch3 expression itself may be under the control of Wnt Family Member 3A (WNT3a) [129], [130].
High levels of Notch3 may cause chemoresistance and promote tumor recurrence [124], [125]. Notch3 was shown to be involved in cancer stem cell formation after lung cancer treatment with the anti‐EGFR antibody erlotinib. Dramatic EGFR‐mutated lung cancer cell death was seen after erlotinib treatment with consequent formation of an aldehyde dehydrogenase (ALDH)‐positive stem cell‐like cell population [131]. ALDH positivity has been shown to be a marker of stem‐like cells that are dependent on Notch3 for tumor colony formation, and the role of notch signaling in cancer stem cell formation has been previously described [132], [133]. EGFR has been shown to decrease all subtypes of Notch proteins by protein ubiquitination [134]. Blocking EGFR led to Notch activation, which resulted in stem cell formation. This effect was abolished by selective blockage of Notch3 but not Notch1, underscoring the importance of Notch3 in stem cell formation and suggesting that combination treatment of erlotinib with a Notch inhibitor could prevent cancer stem cell formation [124], [131]. Although not specifically targeting cancer stem cells, a combination of erlotinib with Notch3 blockade may have a synergistic effect on the induction of Bim with consequent cancer cell apoptosis [126]. Another example of successful drug combination treatment against lung cancer is pharmacologic blockade of Notch and protein kinase Cι (PKCι). PKCι activates Notch3 expression, which causes asymmetric cell division leading to tumor initiation and maintenance. Treatment with gold salts selectively inhibit PKCι in combination with GSI and lead to enhanced inhibition of tumor growth compared with single drug therapy [135].
Notch3 inhibition also enhances the effectiveness of radiotherapy. Notch3 is increased during tissue hypoxia, a condition known to be a major cause of radioresistance [38], [136]. Suppression of HIF‐1α may prevent activation of the radiation‐induced Notch pathway. Simultaneous inhibition of both HIF‐1α and Notch had a synergistic effect on radiation‐induced lung cancer cytotoxicity [38].
Notch3 inhibition also enhances the effectiveness of radiotherapy. Notch3 is increased during tissue hypoxia, a condition known to be a major cause of radioresistance. Suppression of HIF‐1α may prevent activation of the radiation‐induced Notch pathway.
Unlike NSCLCs, small cell lung cancer (SCLC) was reported not to express Notch [137], [138]. Classical SCLC shows neuroendocrine differentiation, and Notch has been reported to be tumor suppressive in most of the neuroendocrine tumors [117], [139], [140]. The role of Notch3 signaling in SCLC is controversial and largely unexplored. The lower expression of Notch3 in SCLC compared with normal lung tissue might suggest that Notch3 has tumor suppressive role in this type of cancer [141]. This is further supported by the finding that activation of Notch signaling in SCLC‐induced cell‐cycle arrest at G1 phase was associated with p21waf1/cip1 and p27kip1 induction [142]. However, these findings are somewhat contradicted by studies showing that inactivation of Notch 2 and Notch3 by monoclonal antibody decreases SCLC growth and that Notch3 expression levels do not correlate with SCLC cell proliferation [138], [143].
Connective Tissue Tumors
Notch3 is upregulated in osteosarcoma and chondrosarcoma, with high expression in hypercellular areas of chondrosarcoma suggesting that it plays a role in regulating stemness in this tumor type [144]. Notch3 is overexpressed in rhabdomyosarcoma cell lines compared with normal myoblasts and has been shown to induce proliferation via a Hes1‐dependent mechanism [145]. Downregulation of Notch3 in vitro resulted in increased differentiation of alveolar rhabdomyosarcoma cells and a reduction in cellular proliferation [145], [146]. It has been suggested that osteosarcoma maintains an undifferentiated state by depressing of Notch3 and its downstream target Hes5 [147].
Conclusion
Notch3 dysregulation has been associated with a wide variety of cancers. It has been shown to affect tumor aggressiveness, maintenance, and resistance to chemotherapy. Notch genes have a significant role in directing tissue commitment and cell differentiation. Activation of Notch pathways lead to increased expression of various downstream targets, some of which include transcriptional regulators. Depending on the tissue type, Notch‐regulated transcriptional targets may conceivably be either oncogenes or tumor suppressors. Conflicting data within specific cancer types further suggest that the full role of Notch is not yet known. For example, it has been shown that in breast cancer, Notch3 can act as oncogene but may also cause cell cycle arrest and prevent cellular proliferation [66], [77].
Targeted therapies can be used to modify Notch3 levels in the cells (Fig. 1). Gamma secretase inhibitors have been used to suppress Notch activity in cancers with high Notch expression, but they are not specific to Notch subtypes. Additional putative targets inhibiting Notch enzymatic processing would include Furin‐like convertase and ADAM inhibitors. To increase the specificity and avoid side effects of pan‐Notch blockade, one study used monoclonal antibodies (tarextumab) to block Notch2/Notch3. MicroRNAs targeting specific Notch subtypes have been used for inhibition, including miR‐136 and miR‐206 for Notch3. Additionally, targeting the Notch/Jagged or Notch/Delta interaction is another route for potential therapeutic development. In cases in which Notch3 is responsible for regulating a tumor suppressor gene, it would be beneficial to induce Notch3 expression rather than inhibit it. Several histone deacetylase inhibitors have been shown to increase Notch expression in a variety of neuroendocrine tumor cells in which Notch plays a tumor suppressive role. These histone deacetylase inhibitors were shown to increase NICD3, inhibit tumor growth, induce apoptosis, and decrease tumor marker secretion. The exact mechanism by which HDACi increase Notch expression is unknown.
Figure 1.
The Notch pathway, showing therapeutic points for targeting Notch3. (1) HDAC inhibitors can enhance Notch3 gene transcription. (2) Notch3 mRNA translation can be inhibited by miRNA. (3) Furin‐like convertase inhibitors can inhibit Notch3 protein folding in the Golgi apparatus. (4) Blocking antibodies can be used to block Notch3 extracellular domain and prevent its interaction with the ligand. (5) ADAM and gamma‐secretase inhibitors can be used to prevent Notch3 protein cleavage and release of Notch3 intracellular domain. (6) Notch3 is translocated to the nucleus, it binds to transcription factors and modifies target gene transcription.
Abbreviations: ADAM, disintegrin and metalloprotease; CSL, CBF1/Suppressor of Hairless/Lag‐1; HDACi, histone deacetylase inhibitor; MAML, mastermind‐like protein 1; miRNA, microRNA; NICD3, Notch3 intracellular domain.
Modifying Notch3 activity based on Notch3 expression in tumors has been shown to have significant anticancer effect. Unfortunately, there is no clinically available laboratory test that would help us determine the level of Notch3 expression in a patient's tumor. Conceivably, immunohistochemical staining could be used on biopsy or resection specimens to determine the level of Notch3 expression and guide treatment with Notch pathway inhibitors. A less invasive method of indirectly determining Notch3 expression could be measuring serum chromogranin A (CgA) levels. CgA expression has been shown to be inhibited by the Notch pathway and has the benefit of being an established clinical test. For example, neuroendocrine tumors are known to have low Notch expression and conversely high CgA levels that decrease when treated with Notch‐inducing drugs. However, this is purely theoretical, as to our knowledge, no study has correlated Notch expression to serum CgA in a clinical setting. Additional research is required to determine if this approach or others will be clinically relevant in the wide array of diseases associated with Notch3 dysregulation.
Author Contributions
Conception/design: J. Bart Rose
Manuscript writing: Zviadi Aburjania, Samuel Jang, Jason Whitt, Renata Jaskula‐Stzul, Herbert Chen, J. Bart Rose
Final approval of manuscript: J. Bart Rose
Disclosures
The authors indicated no financial relationships.
References
- 1. Lai EC. Notch signaling: Control of cell communication and cell fate. Development 2004;131:965–973. [DOI] [PubMed] [Google Scholar]
- 2. Wang W, Struhl G. Distinct roles for Mind bomb, Neuralized and Epsin in mediating DSL endocytosis and signaling in Drosophila. Development 2005;132:2883–2894. [DOI] [PubMed] [Google Scholar]
- 3. Ju BG, Jeong S, Bae E et al. Fringe forms a complex with Notch. Nature 2000;405:191–195. [DOI] [PubMed] [Google Scholar]
- 4. Beatus P, Lundkvist J, Oberg C et al. The origin of the ankyrin repeat region in Notch intracellular domains is critical for regulation of HES promoter activity. Mech Dev 2001;104:3–20. [DOI] [PubMed] [Google Scholar]
- 5. Hamada Y, Kadokawa Y, Okabe M et al. Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development 1999;126:3415–3424. [DOI] [PubMed] [Google Scholar]
- 6. McKellar SH, Tester DJ, Yagubyan M et al. Novel NOTCH1 mutations in patients with bicuspid aortic valve disease and thoracic aortic aneurysms. J Thorac Cardiovasc Surg 2007;134:290–296. [DOI] [PubMed] [Google Scholar]
- 7. Yu XM, Jaskula‐Sztul R, Georgen MR et al. Notch1 signaling regulates the aggressiveness of differentiated thyroid cancer and inhibits SERPINE1 expression. Clin Cancer Res 2016;22:3582–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mezquita B, Mezquita J, Barrot C et al. A truncated‐Flt1 isoform of breast cancer cells is upregulated by Notch and downregulated by retinoic acid. J Cell Biochem 2014;115:52–61. [DOI] [PubMed] [Google Scholar]
- 9. Alagille D, Odièvre M, Gautier M et al. Hepatic ductular hypoplasia associated with characteristic facies, vertebral malformations, retarded physical, mental, and sexual development, and cardiac murmur. J Pediatr 1975;86:63–71. [DOI] [PubMed] [Google Scholar]
- 10. Iwaya T, Taniguchi K, Watanabe J et al. Hajdu‐Cheney syndrome. Arch Orthop Trauma Surg 1979;95:293–302. [DOI] [PubMed] [Google Scholar]
- 11. Shayevitz C, Cohen OS, Faraone SV et al. A re‐review of the association between the NOTCH4 locus and schizophrenia. Am J Med Genet B Neuropsychiatr Genet 2012;159B:477–483. [DOI] [PubMed] [Google Scholar]
- 12. Lardelli M, Dahlstrand J, Lendahl U. The novel Notch homologue mouse Notch 3 lacks specific epidermal growth factor‐repeats and is expressed in proliferating neuroepithelium. Mech Dev 1994;46:123–136. [DOI] [PubMed] [Google Scholar]
- 13. Anastasi E, Campese AF, Bellavia D et al. Expression of activated Notch3 in transgenic mice enhances generation of T regulatory cells and protects against experimental autoimmune diabetes. J Immunol 2003;171:4504–4511. [DOI] [PubMed] [Google Scholar]
- 14. Joutel A, Andreux F, Gaulis S et al. The ectodomain of the Notch3 receptor accumulates within the cerebrovasculature of CADASIL patients. J Clin Invest 2000;105:597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krebs LT, Xue Y, Norton CR et al. Characterization of Notch3‐deficient mice: Normal embryonic development and absence of genetic interactions with a Notch1 mutation. Genesis 2003;37:139–143. [DOI] [PubMed] [Google Scholar]
- 16. Choy L, Hagenbeek TJ, Solon M et al. Constitutive NOTCH3 signaling promotes the growth of basal breast cancers. Cancer Res 2017;77:1439–1452. [DOI] [PubMed] [Google Scholar]
- 17. Liu Z, Yun R, Yu X et al. Overexpression of Notch3 and pS6 is associated with poor prognosis in human ovarian epithelial cancer. Mediators Inflamm 2016;2016:5953498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pelullo M, Quaranta R, Talora C et al. Notch3/Jagged1 circuitry reinforces notch signaling and sustains T‐ALL. Neoplasia 2014;16:1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Somnay YR, Yu XM, Lloyd RV et al. Notch3 expression correlates with thyroid cancer differentiation, induces apoptosis, and predicts disease prognosis. Cancer 2017;123:769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pedrosa AR, Graça JL, Carvalho S et al. Notch signaling dynamics in the adult healthy prostate and in prostatic tumor development. Prostate 2016;76:80–96. [DOI] [PubMed] [Google Scholar]
- 21. Joutel A, Monet‐Leprêtre M, Gosele C et al. Cerebrovascular dysfunction and microcirculation rarefaction precede white matter lesions in a mouse genetic model of cerebral ischemic small vessel disease. J Clin Invest 2010;120:433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu JK, Adepoju O, De Silva D et al. A switch in Notch gene expression parallels stem cell to endothelial transition in infantile hemangioma. Angiogenesis 2010;13:15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu H, Zhang W, Kennard S et al. Notch3 is critical for proper angiogenesis and mural cell investment. Circ Res 2010;107:860–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DeCotiis JL, Lukac DM. KSHV and the role of Notch receptor dysregulation in disease progression. Pathogens 2017;6:pii.E34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goncalves PH, Ziegelbauer J, Uldrick TS et al. Kaposi sarcoma herpesvirus‐associated cancers and related diseases. Curr Opin HIV AIDS 2017;12:47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu R, Li X, Tulpule A et al. KSHV‐induced notch components render endothelial and mural cell characteristics and cell survival. Blood 2010;115:887–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu H, Kennard S, Lilly B. NOTCH3 expression is induced in mural cells through an autoregulatory loop that requires endothelial‐expressed JAGGED1. Circ Res 2009;104:466–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Joutel A, Corpechot C, Ducros A et al. Notch3 mutations in CADASIL, a hereditary adult‐onset condition causing stroke and dementia. Nature 1996;383:707–710. [DOI] [PubMed] [Google Scholar]
- 29. Yamamoto Y, Craggs LJ, Watanabe A et al. Brain microvascular accumulation and distribution of the NOTCH3 ectodomain and granular osmiophilic material in CADASIL. J Neuropathol Exp Neurol 2013;72:416–431. [DOI] [PubMed] [Google Scholar]
- 30. Haritunians T, Chow T, De Lange RP et al. Functional analysis of a recurrent missense mutation in Notch3 in CADASIL. J Neurol Neurosurg Psychiatry 2005;76:1242–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pastille E, Frede A, McSorley HJ et al. Intestinal helminth infection drives carcinogenesis in colitis‐associated colon cancer. PLoS Pathog 2017;13:e1006649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mao F, Xu M, Zuo X et al. 15‐Lipoxygenase‐1 suppression of colitis‐associated colon cancer through inhibition of the IL‐6/STAT3 signaling pathway. FASEB J. 2015;29:2359–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ying J, Tsujii M, Kondo J et al. The effectiveness of an anti‐human IL‐6 receptor monoclonal antibody combined with chemotherapy to target colon cancer stem‐like cells. Int J Oncol 2015;46:1551–1559. [DOI] [PubMed] [Google Scholar]
- 34. Farnie G, Clarke RB, Spence K et al. Novel cell culture technique for primary ductal carcinoma in situ: Role of Notch and epidermal growth factor receptor signaling pathways. J Natl Cancer Inst 2007;99:616–627. [DOI] [PubMed] [Google Scholar]
- 35. Sansone P, Storci G, Tavolari S et al. IL‐6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest 2007;117:3988–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shareef MM, Udayakumar TS, Sinha VK et al. Interaction of HIF‐1α and Notch3 is required for the expression of carbonic anhydrase 9 in breast carcinoma cells. Genes Cancer 2013;4:513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meunier A, Flores AN, McDermott N et al. Hypoxia regulates Notch‐3 mRNA and receptor activation in prostate cancer cells. Heliyon 2016;2:e00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ikezawa Y, Sakakibara‐Konishi J, Mizugaki H et al. Inhibition of Notch and HIF enhances the antitumor effect of radiation in Notch expressing lung cancer. Int J Clin Oncol 2017;22:59–69. [DOI] [PubMed] [Google Scholar]
- 39. Hassan WA, Udaka N, Ueda A et al. Neoplastic lesions in CADASIL syndrome: Report of an autopsied Japanese case. Int J Clin Exp Pathol 2015;8:7533–7539. [PMC free article] [PubMed] [Google Scholar]
- 40. Ross AE, Marchionni L, Vuica‐Ross M et al. Gene expression pathways of high grade localized prostate cancer. Prostate 2011;71:1568–1577. [DOI] [PubMed] [Google Scholar]
- 41. Danza G, Serio Di C, Ambrosio MR et al. Notch3 is activated by chronic hypoxia and contributes to the progression of human prostate cancer. Int J Cancer 2013;133:2577–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pedrosa AR, Trindade A, Carvalho C et al. Endothelial Jagged1 promotes solid tumor growth through both pro‐angiogenic and angiocrine functions. Oncotarget 2015;6:24404–24423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang H, Liu L, Liu C et al. Notch3 overexpression enhances progression and chemoresistance of urothelial carcinoma. Oncotarget 2017;8:34362–34373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Waegemans E, Van de Walle I, De Medts J et al. Notch3 activation is sufficient but not required for inducing human T‐lineage specification. J Immunol 2014;193:5997–6004. [DOI] [PubMed] [Google Scholar]
- 45. Lee SH, Jeong EG, Yoo NJ et al. Mutational analysis of NOTCH1, 2, 3 and 4 genes in common solid cancers and acute leukemias. APMIS 2007;115:1357–1363. [DOI] [PubMed] [Google Scholar]
- 46. Indraccolo S, Minuzzo S, Masiero M et al. Cross‐talk between tumor and endothelial cells involving the Notch3‐Dll4 interaction marks escape from tumor dormancy. Cancer Res 2009;69:1314–1323. [DOI] [PubMed] [Google Scholar]
- 47. Lu C, Li JY, Ge Z et al. Par‐4/THAP1 complex and Notch3 competitively regulated pre‐mRNA splicing of CCAR1 and affected inversely the survival of T‐cell acute lymphoblastic leukemia cells. Oncogene 2013;32:5602–5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Masiero M, Minuzzo S, Pusceddu I et al. Notch3‐mediated regulation of MKP‐1 levels promotes survival of T acute lymphoblastic leukemia cells. Leukemia 2011;25:588–598. [DOI] [PubMed] [Google Scholar]
- 49. Felli MP, Vacca A, Calce A et al. PKC theta mediates pre‐TCR signaling and contributes to Notch3‐induced T‐cell leukemia. Oncogene 2005;24:992–1000. [DOI] [PubMed] [Google Scholar]
- 50. Bellavia D, Campese AF, Alesse E et al. Constitutive activation of NF‐kappaB and T‐cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J 2000;19:3337–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene 2007;26:5420–5432. [DOI] [PubMed] [Google Scholar]
- 52. Kao HY, Ordentlich P, Koyano‐Nakagawa N et al. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev 1998;12:2269–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Palermo R, Checquolo S, Giovenco A et al. Acetylation controls Notch3 stability and function in T‐cell leukemia. Oncogene 2012;31:3807–3817. [DOI] [PubMed] [Google Scholar]
- 54. Shao N, Ma D, Wang J et al. Notch1 signaling is irresponsible to the anti‐leukemic effect of HDACis in B‐ALL Nalm‐6 cells. Ann Hematol 2013;92:33–39. [DOI] [PubMed] [Google Scholar]
- 55. Zhang TH, Liu HC, Zhu LJ et al. Activation of Notch signaling in human tongue carcinoma. J Oral Pathol Med 2011;40:37–45. [DOI] [PubMed] [Google Scholar]
- 56. Man CH, Wei‐Man Lun S, Wai‐Ying Hui J et al. Inhibition of NOTCH3 signalling significantly enhances sensitivity to cisplatin in EBV‐associated nasopharyngeal carcinoma. J Pathol 2012;226:471–481. [DOI] [PubMed] [Google Scholar]
- 57. Yeasmin S, Nakayama K, Rahman MT et al. Expression of nuclear Notch3 in cervical squamous cell carcinomas and its association with adverse clinical outcomes. Gynecol Oncol 2010;117:409–416. [DOI] [PubMed] [Google Scholar]
- 58. Liu J, Fan H, Ma Y et al. Notch1 is a 5‐fluorouracil resistant and poor survival marker in human esophagus squamous cell carcinomas. PLoS One 2013;8:e56141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kayamori K, Katsube K, Sakamoto K et al. NOTCH3 Is induced in cancer‐associated fibroblasts and promotes angiogenesis in oral squamous cell carcinoma. PLoS One 2016;11:e0154112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Krikelis D, Kotoula V, Bobos M et al. Protein and mRNA expression of notch pathway components in operable tumors of patients with laryngeal cancer. Anticancer Res 2014;34:6495–6503. [PubMed] [Google Scholar]
- 61. Hu C, Diévart A, Lupien M et al. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am J Pathol 2006;168:973–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yamaguchi N, Oyama T, Ito E et al. NOTCH3 signaling pathway plays crucial roles in the proliferation of ErbB2‐negative human breast cancer cells. Cancer Res 2008;68:1881–1888. [DOI] [PubMed] [Google Scholar]
- 63. Fernandez SV, Robertson FM, Pei J et al. Inflammatory breast cancer (IBC): Clues for targeted therapies. Breast Cancer Res Treat 2013;140:23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang K, Zhang Q, Li D et al. PEST domain mutations in Notch receptors comprise an oncogenic driver segment in triple‐negative breast cancer sensitive to a γ‐secretase inhibitor. Clin Cancer Res 2015;21:1487–1496. [DOI] [PubMed] [Google Scholar]
- 65. Kawazu M, Kojima S, Ueno T et al. Integrative analysis of genomic alterations in triple‐negative breast cancer in association with homologous recombination deficiency. PLoS Genet 2017;13:e1006853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chen CF, Dou XW, Liang YK et al. Notch3 overexpression causes arrest of cell cycle progression by inducing Cdh1 expression in human breast cancer cells. Cell Cycle 2016;15:432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang X, Liu X, Luo J et al. Notch3 inhibits epithelial‐mesenchymal transition by activating Kibra‐mediated Hippo/YAP signaling in breast cancer epithelial cells. Oncogenesis 2016;5:e269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gu X, Lu C, He D et al. Notch3 negatively regulates chemoresistance in breast cancers. Tumour Biol 2016. [Epub ahead of print]. 10.1007/s13277‐016‐5412‐4. [DOI] [PubMed] [Google Scholar]
- 69. Zhang Z, Wang H, Ikeda S et al. Notch3 in human breast cancer cell lines regulates osteoblast‐cancer cell interactions and osteolytic bone metastasis. Am J Pathol 2010;177:1459–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Doebar SC, Sieuwerts AM, de Weerd V et al. Gene expression differences between ductal carcinoma in situ with and without progression to invasive breast cancer. Am J Pathol 2017;187:1648–1655. [DOI] [PubMed] [Google Scholar]
- 71. Xiao Y, Ye Y, Zou X et al. The lymphovascular embolus of inflammatory breast cancer exhibits a Notch 3 addiction. Oncogene 2011;30:287–300. [DOI] [PubMed] [Google Scholar]
- 72. Sansone P, Ceccarelli C, Berishaj M et al. Self‐renewal of CD133(hi) cells by IL6/Notch3 signalling regulates endocrine resistance in metastatic breast cancer. Nat Commun 2016;7:10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Antony J, Tan TZ, Kelly Z et al. The GAS6‐AXL signaling network is a mesenchymal (Mes) molecular subtype‐specific therapeutic target for ovarian cancer. Sci Signal 2016;9:ra97. [DOI] [PubMed] [Google Scholar]
- 74. Liu H, Ren G, Wang T et al. Aberrantly expressed Fra‐1 by IL‐6/STAT3 transactivation promotes colorectal cancer aggressiveness through epithelial‐mesenchymal transition. Carcinogenesis 2015;36:459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang L, Liu H, Mu X et al. Dysregulation of Fra1 expression by Wnt/β‐catenin signalling promotes glioma aggressiveness through epithelial‐mesenchymal transition. Biosci Rep 2017;37:pii.BSR20160643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Boelens MC, Wu TJ, Nabet BY et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell 2014;159:499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kent S, Hutchinson J, Balboni A et al. ΔNp63α promotes cellular quiescence via induction and activation of Notch3. Cell Cycle 2011;10:3111–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jung SG, Kwon YD, Song JA et al. Prognostic significance of Notch 3 gene expression in ovarian serous carcinoma. Cancer Sci 2010;101:1977–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hu W, Liu T, Ivan C et al. Notch3 pathway alterations in ovarian cancer. Cancer Res 2014;74:3282–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kang H, Jeong JY, Song JY et al. Notch3‐specific inhibition using siRNA knockdown or GSI sensitizes paclitaxel‐resistant ovarian cancer cells. Mol Carcinog 2016;55:1196–1209. [DOI] [PubMed] [Google Scholar]
- 81. Brown CW, Brodsky AS, Freiman RN. Notch3 overexpression promotes anoikis resistance in epithelial ovarian cancer via upregulation of COL4A2. Mol Cancer Res 2015;13:78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Choi JH, Park JT, Davidson B et al. Jagged‐1 and Notch3 juxtacrine loop regulates ovarian tumor growth and adhesion. Cancer Res 2008;68:5716–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Park JT, Chen X, Tropè CG et al. Notch3 overexpression is related to the recurrence of ovarian cancer and confers resistance to carboplatin. Am J Pathol 2010;177:1087–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. McAuliffe SM, Morgan SL, Wyant GA et al. Targeting Notch, a key pathway for ovarian cancer stem cells, sensitizes tumors to platinum therapy. Proc Natl Acad Sci USA 2012;109:E2939–E2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gupta N, Xu Z, El‐Sehemy A et al. Notch3 induces epithelial‐mesenchymal transition and attenuates carboplatin‐induced apoptosis in ovarian cancer cells. Gynecol Oncol 2013;130:200–206. [DOI] [PubMed] [Google Scholar]
- 86. Rahman MT, Nakayama K, Rahman M et al. Notch3 overexpression as potential therapeutic target in advanced stage chemoresistant ovarian cancer. Am J Clin Pathol 2012;138:535–544. [DOI] [PubMed] [Google Scholar]
- 87. Kim TH, Jeong JY, Park JY et al. miR‐150 enhances apoptotic and anti‐tumor effects of paclitaxel in paclitaxel‐resistant ovarian cancer cells by targeting Notch3. Oncotarget 2017;8:72788–72800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Jeong JY, Kang H, Kim TH et al. MicroRNA‐136 inhibits cancer stem cell activity and enhances the anti‐tumor effect of paclitaxel against chemoresistant ovarian cancer cells by targeting Notch3. Cancer Lett 2017;386:168–178. [DOI] [PubMed] [Google Scholar]
- 89. Shah MM, Zerlin M, Li BY et al. The role of Notch and gamma‐secretase inhibition in an ovarian cancer model. Anticancer Res 2013;33:801–808. [PMC free article] [PubMed] [Google Scholar]
- 90. Chen C, Wang X, Huang S et al. Prognostic roles of Notch receptor mRNA expression in human ovarian cancer. Oncotarget 2017;8:32731–32740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhou X, Teng L, Wang M. Distinct prognostic values of four‐Notch‐receptor mRNA expression in ovarian cancer. Tumour Biol 2016;37:6979–6985. [DOI] [PubMed] [Google Scholar]
- 92. Ozawa T, Kazama S, Akiyoshi T et al. Nuclear Notch3 expression is associated with tumor recurrence in patients with stage II and III colorectal cancer. Ann Surg Oncol 2014;21:2650–2658. [DOI] [PubMed] [Google Scholar]
- 93. Serafin V, Persano L, Moserle L et al. Notch3 signalling promotes tumour growth in colorectal cancer. J Pathol 2011;224:448–460. [DOI] [PubMed] [Google Scholar]
- 94. Lee HJ, Eom DW, Kang GH et al. Colorectal micropapillary carcinomas are associated with poor prognosis and enriched in markers of stem cells. Mod Pathol 2013;26:1123–1131. [DOI] [PubMed] [Google Scholar]
- 95. Furukawa S, Kawasaki Y, Miyamoto M et al. The miR‐1‐NOTCH3‐Asef pathway is important for colorectal tumor cell migration. PLoS One 2013;8:e80609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wang XW, Xi XQ, Wu J et al. MicroRNA‐206 attenuates tumor proliferation and migration involving the downregulation of NOTCH3 in colorectal cancer. Oncol Rep 2015;33:1402–1410. [DOI] [PubMed] [Google Scholar]
- 97. Gramantieri L, Giovannini C, Lanzi A et al. Aberrant Notch3 and Notch4 expression in human hepatocellular carcinoma. Liver Int 2007;27:997–1007. [DOI] [PubMed] [Google Scholar]
- 98. Hu L, Xue F, Shao M et al. Aberrant expression of Notch3 predicts poor survival for hepatocellular carcinomas. Biosci Trends 2013;7:152–156. [PubMed] [Google Scholar]
- 99. Yu T, Han C, Zhu G et al. Prognostic value of Notch receptors in postsurgical patients with hepatitis B virus‐related hepatocellular carcinoma. Cancer Med 2017;6:1587–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Yang SL, Ren QG, Zhang T et al. Hepatitis B virus X protein and hypoxia‐inducible factor‐1α stimulate Notch gene expression in liver cancer cells. Oncol Rep 2017;37:348–356. [DOI] [PubMed] [Google Scholar]
- 101. Zhou L, Zhang N, Song W et al. The significance of Notch1 compared with Notch3 in high metastasis and poor overall survival in hepatocellular carcinoma. PLoS One 2013;8:e57382. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 102. Zhang Q, Lu C, Fang T et al. Notch3 functions as a regulator of cell self‐renewal by interacting with the β‐catenin pathway in hepatocellular carcinoma. Oncotarget 2015;6:3669–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Liu W, Xu C, Wan H et al. MicroRNA‐206 overexpression promotes apoptosis, induces cell cycle arrest and inhibits the migration of human hepatocellular carcinoma HepG2 cells. Int J Mol Med 2014;34:420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Giovannini C, Gramantieri L, Chieco P et al. Selective ablation of Notch3 in HCC enhances doxorubicin's death promoting effect by a p53 dependent mechanism. J Hepatol 2009;50:969–979. [DOI] [PubMed] [Google Scholar]
- 105. Giovannini C, Baglioni M, Baron Toaldo M et al. Notch3 inhibition enhances sorafenib cytotoxic efficacy by promoting GSK3b phosphorylation and p21 down‐regulation in hepatocellular carcinoma. Oncotarget 2013;4:1618–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Guest RV, Boulter L, Dwyer BJ et al. Notch3 drives development and progression of cholangiocarcinoma. Proc Natl Acad Sci USA 2016;113:12250–12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhou JX, Zhou L, Li QJ et al. Association between high levels of Notch3 expression and high invasion and poor overall survival rates in pancreatic ductal adenocarcinoma. Oncol Rep 2016;36:2893–2901. [DOI] [PubMed] [Google Scholar]
- 108. Mann CD, Bastianpillai C, Neal CP et al. Notch3 and HEY‐1 as prognostic biomarkers in pancreatic adenocarcinoma. PLoS One 2012;7:e51119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Doucas H, Mann CD, Sutton CD et al. Expression of nuclear Notch3 in pancreatic adenocarcinomas is associated with adverse clinical features, and correlates with the expression of STAT3 and phosphorylated Akt. J Surg Oncol 2008;97:63–68. [DOI] [PubMed] [Google Scholar]
- 110. Tang J, Zhu Y, Xie K et al. The role of the AMOP domain in MUC4/Y‐promoted tumour angiogenesis and metastasis in pancreatic cancer. J Exp Clin Cancer Res 2016;35:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Cai H, Yao J, An Y et al. LncRNA HOTAIR acts a competing endogenous RNA to control the expression of notch3 via sponging miR‐613 in pancreatic cancer. Oncotarget 2017;8:32905–32917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Eto K, Kawakami H, Kuwatani M et al. Human equilibrative nucleoside transporter 1 and Notch3 can predict gemcitabine effects in patients with unresectable pancreatic cancer. Br J Cancer 2013;108:1488–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Yao J, Qian C. Inhibition of Notch3 enhances sensitivity to gemcitabine in pancreatic cancer through an inactivation of PI3K/Akt‐dependent pathway. Med Oncol 2010;27:1017–1022. [DOI] [PubMed] [Google Scholar]
- 114. Ramage JK, Davies AH, Ardill J et al. Guidelines for the management of gastroenteropancreatic neuroendocrine (including carcinoid) tumours. Gut 2005;54(suppl 4):iv1–iv16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Lanigan TM, DeRaad SK, Russo AF. Requirement of the MASH‐1 transcription factor for neuroendocrine differentiation of thyroid C cells. J Neurobiol 1998;34:126–134. [PubMed] [Google Scholar]
- 116. Pitt SC, Chen H, Kunnimalaiyaan M. Inhibition of phosphatidylinositol 3‐kinase/Akt signaling suppresses tumor cell proliferation and neuroendocrine marker expression in GI carcinoid tumors. Ann Surg Oncol 2009;16:2936–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Jaskula‐Sztul R, Eide J, Tesfazghi S et al. Tumor‐suppressor role of Notch3 in medullary thyroid carcinoma revealed by genetic and pharmacological induction. Mol Cancer Ther 2015;14:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wyche TP, Dammalapati A, Cho H et al. Thiocoraline activates the Notch pathway in carcinoids and reduces tumor progression in vivo. Cancer Gene Ther 2014;21:518–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Mohammed TA, Holen KD, Jaskula‐Sztul R et al. A pilot phase II study of valproic acid for treatment of low‐grade neuroendocrine carcinoma. The Oncologist 2011;16835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Adler JT, Hottinger DG, Kunnimalaiyaan M et al. Histone deacetylase inhibitors upregulate Notch‐1 and inhibit growth in pheochromocytoma cells. Surgery 2008;144:956–61; discussion 961–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Dang TP, Eichenberger S, Gonzalez A et al. Constitutive activation of Notch3 inhibits terminal epithelial differentiation in lungs of transgenic mice. Oncogene 2003;22:1988–1997. [DOI] [PubMed] [Google Scholar]
- 122. Konishi J, Kawaguchi KS, Vo H et al. Gamma‐secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res 2007;67:8051–8057. [DOI] [PubMed] [Google Scholar]
- 123. Haruki N, Kawaguchi KS, Eichenberger S et al. Dominant‐negative Notch3 receptor inhibits mitogen‐activated protein kinase pathway and the growth of human lung cancers. Cancer Res 2005;65:3555–3561. [DOI] [PubMed] [Google Scholar]
- 124. Zheng Y, de la Cruz CC, Sayles LC et al. A rare population of CD24(+)ITGB4(+)Notch(hi) cells drives tumor propagation in NSCLC and requires Notch3 for self‐renewal. Cancer Cell 2013;24:59–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Shi C, Qian J, Ma M et al. Notch 3 protein, not its gene polymorphism, is associated with the chemotherapy response and prognosis of advanced NSCLC patients. Cell Physiol Biochem 2014;34:743–752. [DOI] [PubMed] [Google Scholar]
- 126. Konishi J, Yi F, Chen X et al. Notch3 cooperates with the EGFR pathway to modulate apoptosis through the induction of bim. Oncogene 2010;29:589–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Chen CY, Chen YY, Hsieh MS et al. Expression of notch gene and its impact on survival of patients with resectable non‐small cell lung cancer. J Cancer 2017;8:1292–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Xiong J, Zhang X, Chen X et al. Prognostic roles of mRNA expression of notch receptors in non‐small cell lung cancer. Oncotarget 2017;8:13157–13165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Li C, Song G, Zhang S et al. Wnt3a increases the metastatic potential of non‐small cell lung cancer cells in vitro in part via its upregulation of Notch3. Oncol Rep 2015;33:1207–1214. [DOI] [PubMed] [Google Scholar]
- 130. Liu L, Chen X, Wang Y et al. Notch3 is important for TGF‐β‐induced epithelial‐mesenchymal transition in non‐small cell lung cancer bone metastasis by regulating ZEB‐1. Cancer Gene Ther 2014;21:364–372. [DOI] [PubMed] [Google Scholar]
- 131. Arasada RR, Amann JM, Rahman MA et al. EGFR blockade enriches for lung cancer stem‐like cells through Notch3‐dependent signaling. Cancer Res 2014;74:5572–5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Wang Z, Li Y, Banerjee S et al. H. Emerging role of Notch in stem cells and cancer. Cancer Lett 2009;279:8–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Sullivan JP, Spinola M, Dodge M et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Res 2010;70:9937–9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Aguirre A, Rubio ME, Gallo V. Notch and EGFR pathway interaction regulates neural stem cell number and self‐renewal. Nature 2010;467:323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Ali SA, Justilien V, Jamieson L et al. Protein kinase Cι drives a NOTCH3‐dependent stem‐like phenotype in mutant KRAS lung adenocarcinoma. Cancer Cell 2016;29:367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Dewhirst MW, Cao Y, Moeller B. Cycling hypoxia and free radicals regulate angiogenesis and radiotherapy response. Nat Rev Cancer 2008;8:425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Fujino K, Motooka Y, Hassan WA et al. Insulinoma‐associated protein 1 Is a crucial regulator of neuroendocrine differentiation in lung cancer. Am J Pathol 2015;185:3164–3177. [DOI] [PubMed] [Google Scholar]
- 138. Hassan WA, Yoshida R, Kudoh S et al. Evaluation of role of Notch3 signaling pathway in human lung cancer cells. J Cancer Res Clin Oncol 2016;142:981–993. [DOI] [PubMed] [Google Scholar]
- 139. Jang S, Janssen A, Aburjania Z et al. Histone deacetylase inhibitor thailandepsin‐A activates Notch signaling and suppresses neuroendocrine cancer cell growth in vivo. Oncotarget 2017;8:70828–70840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Gazdar AF, Carbone DP. The Biology and Molecular Genetics of Lung Cancer. Austin, TX: R.G. Landes; 1994. [Google Scholar]
- 141. Zhou M, Jin WY, Fan ZW et al. Analysis of the expression of the Notch3 receptor protein in adult lung cancer. Oncol Lett 2013;5:499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Sriuranpong V, Borges MW, Ravi RK et al. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res 2001;61:3200–3205. [PubMed] [Google Scholar]
- 143. Yen WC, Fischer MM, Axelrod F et al. Targeting Notch signaling with a Notch2/Notch3 antagonist (tarextumab) inhibits tumor growth and decreases tumor‐initiating cell frequency. Clin Cancer Res 2015;21:2084–2095. [DOI] [PubMed] [Google Scholar]
- 144. Siar CH, Ha KO, Aung LO et al. Immunolocalization of notch signaling protein molecules in a maxillary chondrosarcoma and its recurrent tumor. Eur J Med Res 2010;15:456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. De Salvo M, Raimondi L, Vella S et al. Hyper‐activation of Notch3 amplifies the proliferative potential of rhabdomyosarcoma cells. PLoS One 2014;9:e96238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Raimondi L, Ciarapica R, De Salvo M et al. Inhibition of Notch3 signalling induces rhabdomyosarcoma cell differentiation promoting p38 phosphorylation and p21(Cip1) expression and hampers tumour cell growth in vitro and in vivo. Cell Death Differ 2012;19:871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Ongaro A, Pellati A, Bagheri L et al. Characterization of notch signaling during osteogenic differentiation in human osteosarcoma cell line MG63. J Cell Physiol 2016;231:2652–2663. [DOI] [PubMed] [Google Scholar]
- 148. Xie T, Li Y, Li SL et al. Astragaloside IV enhances cisplatin chemosensitivity in human colorectal cancer via regulating NOTCH3. Oncol Res 2016;24:447–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Suliman MA, Zhang Z, Na H et al. Niclosamide inhibits colon cancer progression through downregulation of the Notch pathway and upregulation of the tumor suppressor miR‐200 family. Int J Mol Med 2016;38:776–784. [DOI] [PMC free article] [PubMed] [Google Scholar]