Here, we report how the analysis of viral genetic variation using next-generation sequencing (NGS) can be used as a tool to improve mumps virus diagnostics. Analysis of NGS data from recently circulating mumps virus isolates allowed optimization of the current mumps virus real-time reverse transcription-PCR (RT-PCR) by primer and probe modifications due to nucleotide variations.
KEYWORDS: mumps virus, real-time RT-PCR, next-generation sequencing, molecular homology modelling, genotype, vaccines
ABSTRACT
Here, we report how the analysis of viral genetic variation using next-generation sequencing (NGS) can be used as a tool to improve mumps virus diagnostics. Analysis of NGS data from recently circulating mumps virus isolates allowed optimization of the current mumps virus real-time reverse transcription-PCR (RT-PCR) by primer and probe modifications due to nucleotide variations. The modified assay showed a higher efficiency and sensitivity than the previously used CDC protocol for the detection of currently circulating mumps virus strains and could therefore offer better support for outbreak control. The NGS sequence data were also used to make predictions of changes in the hemagglutinin-neuraminidase protein structure that could explain possible immune escape mechanisms.
INTRODUCTION
The formerly common childhood disease mumps is caused by mumps virus (MuV), a member of the Rubulavirus genus in the Paramyxoviridae family. There are 12 different MuV genotypes (A to N, excluding E and M) recognized (1–3), which all belong to one serotype. A vaccine has been available since the 1960s, and the use of the MMR (mumps-measles-rubella) vaccine resulted in a more than 90% decline in mumps rates in developed countries (4). Nevertheless, in recent years, mumps outbreaks have been recorded from many countries and continents, including Canada, the United States, Europe, Russia, and Australia (5, 6). In January 2017, a mumps outbreak caused by a genotype G virus began in Auckland, New Zealand, resulting in more than 1,400 confirmed and probable cases to date (ESR KSC Outbreak Surveillance, 28 February 2018). Our lab is using a CDC-developed and WHO-recommended real-time reverse transcription-PCR (RT-PCR) targeting the nucleocapsid protein (NP) gene for the detection of MuVs in clinical samples and another CDC-developed RT-PCR targeting the highly variable small hydrophobic (SH) protein gene for genotyping of MuV strains via Sanger sequencing of the PCR amplicon (https://www.cdc.gov/mumps/lab/qa-lab-test-infect.html). Since the beginning of the outbreak, we have frequently observed MuV samples with reduced fluorescence intensity signals of the respective real-time RT-PCR amplification curves; this made them difficult to interpret and therefore a potential risk for missing positive samples. In this report, we present the importance of whole-genome sequencing of MuV to monitor for viral genome changes and their impact on primer and probe sequences used in real-time PCR protocols. A new primer and probe were designed and evaluated on archived clinical samples.
There is conflicting evidence about the cross-protection of genotype A vaccines to heterologous wild-type (WT) genotypes, since some studies have shown that the Jeryl Lynn mumps vaccine shows cross-protection for MuV genotype G based on neutralization experiments (7, 8), whereas other studies showed only weak cross-protection (9–11). New Zealand is currently using the Priorix MMR vaccine (GlaxoSmithKline), which contains the RIT4385 MuV derived from the major component JL5 (also called JL1) of the Jeryl Lynn genotype A MuV vaccine. Prior to 2017, the Jeryl Lynn vaccine Mumpsvax, propagated in chicken embryo cell culture, was the mumps component of the funded MMR II in New Zealand. Propagation in chicken embryo cell culture preferentially selects for the JL5 strain, which is identical to RIT4385. We used molecular homology modeling to further investigate if differences in hemagglutinin-neuraminidase epitopes between vaccine and WT strains might explain some degree of immune escape.
MATERIALS AND METHODS
Samples.
Forty-nine mumps RNA-positive samples (majority were buccal swabs and oral swabs, including throat swabs, and nasopharyngeal swabs in viral transport medium) collected between February and November 2017 were used for this study. Samples were stored at −80°C for further testing.
Nucleic acid extraction.
Nucleic acid extraction was performed using the NucliSENS easyMag platform (bioMérieux, France) and the generic 2.0.1 total nucleic acid protocol, as recommended by the manufacturer. Two hundred microliters of sample was extracted, and nucleic acid was eluted in 60 μl of elution buffer.
Real-time PCR.
Real-time RT-PCR was performed on an ABI 7500 thermocycler (Applied Biosystems, Thermo Fisher Scientific, New Zealand). The original CDC ABI AmpliTaq Gold protocol (https://www.cdc.gov/mumps/downloads/lab-rt-pcr-assay-detect.pdf) was replaced with the SuperScript III Platinum One-Step RT-PCR protocol (Invitrogen, Thermo Fisher Scientific); therefore, this assay is referred to as the CDC* protocol. The thermocycling parameters were 48°C for 20 min (SuperScript III RT step) and 95°C for 10 min (Platinum Taq polymerase activation), followed by 95°C for 15 s and 1 min at 60°C for amplification and acquisition of the fluorescence signal.
Modified real-time PCR.
New forward primer and probe sequences were designed from nucleoprotein (NP) sequence alignments and evaluated for improved performance on genotype G mumps virus. The primer was MUPCF (5′-GTATGACAGCKTACGACCAACCT-3′) and the probe was MUPCP (5′-FAM-CYGGRTCTGCWGATCGGCGAT-BHQ-3′; FAM, 6-carboxyfluorescein; BHQ, black hole quencher) (IDT, USA) with IUPAC-ambiguous 2-fold degenerated nucleotides (bold and underlined). Archived samples from the current genotype G outbreak were retested in parallel with the CDC* protocol and with our new method under the real-time RT-PCR conditions described previously.
Whole-genome sequencing.
Two MuV samples from the 2017 New Zealand outbreak were collected as follows: sample MuVi/NewPlymouth.NZL/30.17[G] from New Plymouth, North Island, was an oropharynx swab collected on 24 July 2017 from an unvaccinated 19-year-old case; and sample MuVi/Dunedin.NZL/32.17[G] from Dunedin, South Island, was collected from a site not stated on 11 August 2017 from a 21-year-old case mentioned to have received the MMR vaccine at ages 15 months and 11 years. These samples were isolated in Vero E6/hSLAM cells (passage 1). Cultured virus was first treated with 75 units of Benzonase (Sigma-Aldrich/Merck, St. Louis, MO, USA) for 2 h at 37°C to remove human and African green monkey nucleic acids, and capsid-protected viral nucleic acids were extracted from cell culture supernatants using the NucliSENS easyMag instrument (bioMérieux, France). Whole-genome sequence data of the two New Zealand mumps virus isolates (strain MuVi/NewPlymouth.NZL/30.17[G], GenBank accession no. MG765426; and strain MuVi/Dunedin.NZL/32.17[G], GenBank accession no. MG765427) were generated using Nextera XT library preparation and Illumina sequencing (San Diego, CA, USA). In brief, first-strand cDNA was synthesized with Transcriptor reverse transcriptase (Roche Diagnostics, Basel, Switzerland) for 1 h at 60°C using random-hexamer primers. Second-strand cDNA was generated using the NEBNext Ultra II nondirectional RNA second-strand synthesis module (NEB, Ipswich, MA, USA). The resulting double-stranded cDNA was purified using AMPure XP beads (Beckman Coulter, Brea, CA, USA) and converted into Illumina sequencing libraries using the Nextera XT library preparation kit. Sequencing was done on a MiSeq instrument (Illumina, San Diego, CA, USA) using v2 chemistry (2 × 150 bp). CLC Genomics Workbench version 9.5 (Qiagen, Hilden, Germany) was used for assembly, reference mapping, and sequence analysis.
Phylogenetic analysis.
A phylogenetic tree was generated from the nucleotide ClustalW alignment of available mumps virus whole-genome sequences. The tree was reconstructed in MEGA version 7.0.21 using a neighbor-joining method with Kimura 2-parameter model. The robustness of the internal branches was determined by 500 bootstrap replications.
Molecular homology modeling.
Amino acid sequences of the hemagglutinin-neuraminidase (HN) protein of strain MuVi/NewPlymouth.NZL/30.17[G] (GenBank accession no. MG765426, protein_id AUN87694) and NZ vaccine strain RIT4358[A] (GenBank accession no. FJ211584.1, protein_id ACN50023.1) were used for molecular homology modeling against SWISS-MODEL (http://swissmodel.expasy.org/) template libraries (12, 13). SWISS-MODEL generated predicted three-dimensional (3D) structures of the HN head domain based on crystallographic data of the mumps virus HN homodimer template PDB identification (ID) code 5b2c.1.A from vaccine strain Hoshino (genotype B) (14), with QMQE scores of 0.77 and 0.76 and QMEAN scores of −0.66 and −0.97, respectively. The obtained pdb files were superimposed using PyMOL version 2.0.7 (https://pymol.org/2/).
Accession number(s).
Sequences have been deposited in GenBank under accession numbers MG765426 and MG765427.
RESULTS
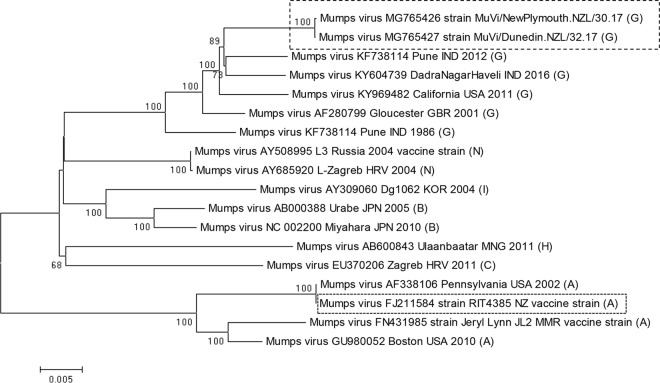
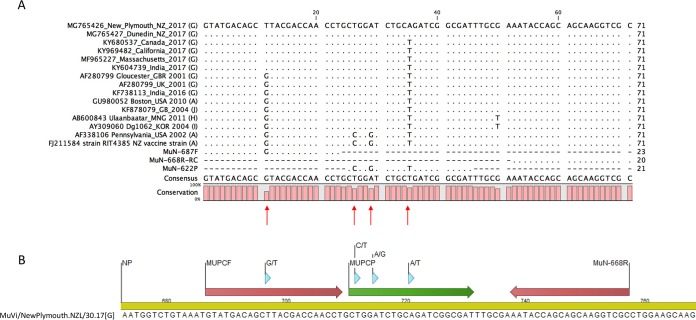
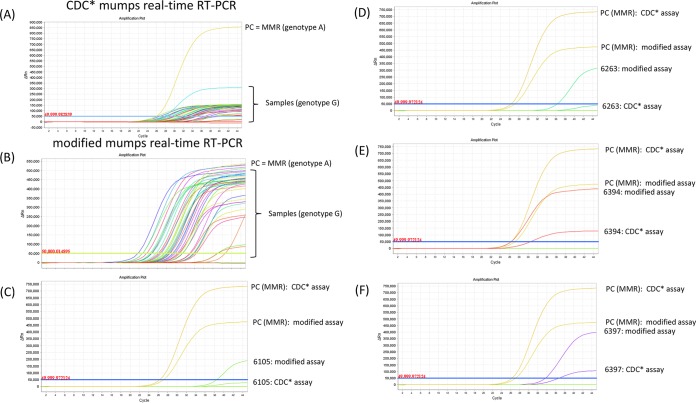
Genotyping of MuVs based on Sanger sequencing of the SH protein gene from the 2017 NZ outbreak revealed that they belong to genotype G, which is phylogenetically distant from genotype A viruses used in MMR vaccines (Fig. 1). We decided to produce whole-genome sequences via next-generation sequencing (NGS) of the circulating MuV strain to obtain the full NP and HN genes. Both MuV isolates from the North and South Island of New Zealand produced complete genome sequences, with average coverages of 250× for strain MuVi/NewPlymouth.NZL/30.17[G] and 280× for strain MuVi/Dunedin.NZL/32.17[G], respectively. The two sequences showed 99.97% sequence identity to each other. A pairwise comparison of hemagglutinin-neuraminidase (HN) protein sequences to a selection of genotype G and vaccine genotype A sequences showed 85.55% sequence identity of the NZ MuV strains to the HN protein sequence of the NZ vaccine strain RIT4385[A] (Table S1). The same low value was obtained from a MuV strain isolated from a vaccinated child in Gabon, Africa, in 2013 (GenBank accession no. KM597072). BLASTN analysis (NIH, NCBI) of the whole-genome sequences revealed that the NZ MuV strains show the closest sequence identity (98%, score 26969) to a genotype G strain from an outbreak in the United States in 2006 (GenBank accession no. JN012242), as well as to other genotype G strains involved in recent outbreaks in Croatia in 2005 (GenBank accession no. EU370207), Canada in 2009 (GenBank accession no. KY680537), the United States in 2016 (GenBank accession no. KY996512), and India in 2017 (GenBank accession no. KY604739). Analysis of the obtained whole-genome data revealed one mismatch in the CDC-developed forward primer MuN-687F and three mismatches in the CDC-developed probe MuN-622P in the NP gene target region. A sequence alignment including the New Zealand genotype G viruses from the 2017 outbreak, as well as a selection of mumps virus genotype G, H, I, and J sequences from different geographical regions from 2001 until 2017 compared to the RIT4385 genotype A vaccine strain currently used in New Zealand and the CDC-developed primers and probe is shown in Fig. 2A. Some genotype G viruses contained one mismatch in the forward primer binding region, as seen in the New Zealand isolates. In addition, all genotype G viruses contained two mismatches in the 5′ end of the probe binding region, whereas only genotype G viruses from the New Zealand 2017 outbreak contained a third mismatch in the center of the probe binding region. A NCBI BLASTN search revealed that this third single nucleotide polymorphism (SNP) has not been detected in any other MuV sequence deposited in GenBank so far. We therefore developed a new forward primer and a new probe with IUPAC-ambiguous 2-fold degenerated nucleotides (see Materials and Methods, and Fig. 2B) and compared the two methods on 49 mumps genotype G-positive samples from the current outbreak plus Priorix genotype A positive control (Table S2). The modified MuV real-time RT-PCR showed much stronger fluorescence signals of the amplification curves and lower threshold cycle (CT) values (on average, 3.7 cycles lower) than the CDC* assay for the tested genotype G samples (Table S2 and Fig. 3). Other mumps genotypes could not be tested, because they were not available in our lab. Epidemiological data indicate that genotypes A, J, H, and I have been recently circulating in addition to genotype G, and bioinformatics analysis indicates that the modified primer and probe will detect them (see nucleotide alignment in Fig. 2A). In addition, the original CDC assay showed the specific detection of genotypes C, H, D, G, and A (https://www.cdc.gov/mumps/downloads/lab-rt-pcr-assay-detect.pdf). The amplification curve of the modified assay for the mumps virus (genotype A) in the MMR vaccine showed slightly reduced fluorescence intensity but a CT value similar to those of the CDC protocol (on average, 0.5 cycles higher). The negative-control samples showed no amplification. Since the modified positions contain degenerate nucleotides, the detection of other genotypes should not be impaired (Fig. 2A). A comparison of the HN amino acid sequences of the circulating NZ genotype G strain with the RIT4358 genotype A vaccine strain (Fig. 4) revealed several amino acid exchanges in regions considered to be epitopes or involved in immune escape (11). Superposition of the predicted HN structure of the vaccine strain onto that of the circulating G genotype strain identified several amino acids present in the epitope regions of the vaccine strain that mutated into smaller amino acids in the circulating G genotype (Fig. 4 and 5).
FIG 1.
Phylogenetic comparison of mumps viruses from the NZ 2017 outbreak with a selection of available whole-genome reference sequences. A neighbor-joining phylogenetic tree was constructed after ClustalW alignment and inferred by bootstrapping for 500 replicates. Only bootstrap values of >50% are displayed.
FIG 2.
(A) Sequence alignment of the mumps virus real-time RT-PCR target region in the NP gene of a selection of genotype G, H, I, and J sequences from 2001 until 2017 in comparison to genotype A strains and the CDC-developed primers and probe. The two analyzed NZ genotype G isolates are depicted on the top of the alignment. The CDC forward primer (MuN-687F), reverse primer (MuN-668R), and probe (MuN-622P) are at the bottom of the alignment. The letters in parentheses indicate the genotype. (B) Primer and probe modifications in relation to NP target sequence of strain MuVi/NewPlymouth.NZL/30.17[G].
FIG 3.
Comparison of the CDC*-developed MuV real-time PCR (A) with the modified assay (B) on a selection of positive patient samples from the current NZ genotype G outbreak. PC, MMR positive control (genotype G); NC, negative control. Samples, genotype G. (C and D) Samples with high CT values that could have been missed using the CDC* protocol. (E and F) Samples with low CT values that still perform better with the modified assay.
FIG 4.
Sequence alignment of the HN amino acid sequences of vaccine strain RIT4358 and NZ genotype G strain MuVi/NewPlymouth.NZL/30.17[G] (which shows the amino acid sequence identical to that of strain MuVi/Dunedin.NZL/32. 17[G]). The color coding represents the features in Fig. 5.
FIG 5.
Molecular homology modeling of the homodimeric head domains of mumps HN proteins using SWISS-MODEL and superposition of predicted 3D structures via PyMOL. (A) The turquoise dotted mesh shows the HN surface of the NZ genotype G strain MuVi/NewPlymouth.NZL/30.17[G]. (B) The orange sticks show the HN structure of the vaccine strain RIT4385[A], which is superimposed onto the NZ genotype G strain in panels C and D. Mutated amino acids are shown as spheres. Vaccine strain amino acids protruding from the genotype G surface are depicted in magenta and labeled, and internal amino acid exchanges are depicted in blue color (not labeled).
DISCUSSION
This publication shows the importance of monitoring viral genetic variation and its impact on diagnostic assays. Whole-genome sequencing can be used as a tool to optimize molecular targets of real-time PCR assays but in addition can provide further insight into outbreak investigations, viral evolution, and possible mechanisms of immune escape. Based on the obtained NZ MuV whole-genome sequences, the introduction of one 2-fold degenerate nucleotide in the forward primer and three 2-fold degenerate nucleotides in the probe sequence enhanced the fluorescence intensity and clinical sensitivity of the real-time RT-PCR. This modified assay showed better performance on clinical samples for the currently circulating genotype G MuV, leading to CT values which were on average 3.7 cycles lower (corresponding to approximately 11% increase in clinical sensitivity) than the CT values of the corresponding CDC* real-time RT-PCR. A change in 3 to 4 cycles is equivalent to a 10-fold increase in sensitivity; therefore, samples at the lower limit of detection could be missed by the CDC* protocol (Fig. 3C and D; further assay performance characteristics can be found in the supplemental material). This is especially critical if the timing of the sample taken is not optimal, causing low concentrations of MuV. Since isolation of positive patients is crucial for containment of the outbreak, the modified MuV real-time RT-PCR might support outbreak control and has therefore been distributed to other NZ labs involved in MuV diagnostics. It should be noted that even though the modified assay shows an improvement to the previous CDC* assay, both target the same region of the NP gene. The design of a diagnostic real-time RT-PCR suitable to detect all mumps virus genotypes equally well may rely on the selection of a different, more conserved region of the NP gene or other parts of the genome (3). The same modified assay could have been developed just based on Sanger sequencing of the real-time PCR target region. However, the advantage of NGS lies in the generation of comprehensive genome information, which could support further optimization of the real-time PCR target depending on the modified assay's performance. In addition, WGS data allowed us to analyze parts of the genome that are involved in immune recognition.
The majority of NZ mumps cases in the current outbreak are age 15 to 29 years, and the New Zealand Ministry of Health (MoH) estimates that approximately 40% of New Zealanders born between 1990 and 2005 may not be fully vaccinated and may therefore be vulnerable to measles, mumps, and rubella outbreaks. Prior to 2005, immunization rates were not recorded nationally, resulting in people with unknown vaccination history. In addition, the MuV component is the weakest component of the MMR vaccine, with an efficacy of 88% (range, 31 to 95%) after two doses (https://www.cdc.gov/mumps/vaccination.html). MuV genotyping performed in our lab revealed that the circulating MuV belongs to genotype G, which is phylogenetically distinct from genotype A vaccine viruses (Fig. 1). The genetic distance between genotype A vaccine strains and non-genotype A wild strains has been proposed to be a determinant factor in vaccine failure (5). Nevertheless, many countries use MuV vaccines based on Jeryl Lynn genotype A strains, since these strains have never shown any virologically proven postvaccine-associated aseptic meningitis cases (15–17) and are therefore considered to be safer than other mumps vaccines, including L-Zagreb (genotype N) or Urabe AM-9 (genotype B), which on the other hand might be more immunogenic. In the past years, genotype G viruses were responsible for several MuV outbreaks in North America, Europe, and Asia, and the number of reports of mumps outbreaks in fully vaccinated populations is increasing (10, 18). Although in NZ, several mumps cases could be attributed to members of Pacific Island communities that have only received MR (measles and rubella) vaccination, a reasonable proportion (44%) of infections also appeared in people having received two doses of MMR immunization (source, Institute of Environmental Science and Research [ESR] EpiSurv, unpublished data). New Zealand has not seen such a pronounced mumps outbreak in previous years. This might indicate that genetic drift in G genotype viruses has contributed to the extent of the 2017 mumps outbreak. Since the HN protein of MuV is the major target of neutralizing antibodies (8), the low sequence identity to the vaccine strain displayed in Table S1 could indicate a lower immunological response. The alignment of the HN proteins of RIT4385 vaccine strain and circulating NZ genotype G strain (Fig. 4) shows several amino acid substitutions in predicted epitope regions or regions that seem to be involved in immune escape (11). All of these amino acid substitutions resulted in smaller amino acids introduced in the immunological important regions of HN in the genotype G virus compared to the genotype A vaccine strain (Ile279Thr, Ile287Val, Leu336Ser, Glu356Asp, Tyr442Ser, and Gln444Pro). Substitutions against bigger amino acids (Ser293Thr and Ser372Asp) were only seen in regions not implicated in immune recognition. In addition, one substitution (His464Asp) causes an additional predicted N-linked glycosylation site in the NZ genotype G virus that is not present in the vaccine strain. Glycans attached to these N-linked glycosylation sites could reduce the accessibility of antibodies to epitopes and may therefore facilitate immune escape (19). Based on the recently published crystal structure of the HN head domain of mumps virus (14), we generated a molecular homology modeling to compare the HN proteins of the vaccine strain and circulating G genotype strain. The overlay of the molecular homology models for the RIT4385 vaccine strain and the circulating NZ genotype G strain (Fig. 5C and D) clearly identify those big amino acids present in the vaccine strain to protrude the surface of the genotype G strain (marked in magenta), especially in the predicted epitope regions. Therefore, antibodies directed against such epitopes might not be able to recognize the altered, flatter epitopes in the genotype G virus (Fig. 5A). This observation is in agreement with the immunoinformatic analysis of Homan and Bremel (20), who showed that the Jeryl Lynn JL5 major strain/RIT4385 is an outlier in all measures of immunomic features, which distinguish it from other mumps virus strains (20). Computational comparison of HN proteins of different MuV outbreak strains could yield further insights into the amino acid substitutions involved in mounting immune escape. It has been suggested that a combination of differences in antigenic epitopes and waning immunity by the absence of encountering wild-type mumps virus boosters, especially in antibodies directed against conserved epitopes, could lead to vaccine-induced immunity dropping below a threshold required for protection against heterologous MuV strains in vaccinated individuals (21). The immunological pressure could be increased by introducing a third dose of the MMR vaccine, which could booster the levels of antibodies directed against conserved epitopes, and which has been shown to be effective in mumps outbreak control (22). Alternatively, a polyvalent vaccine covering several genotypes, like suggested by May et al. (21), could be beneficial.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by institutional funds from Canterbury Health Laboratories.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00405-18.
REFERENCES
- 1.Echevarria JE, Castellanos A, Sanz JC, Martinez de Aragon MV, Peña Rey I, Mosquera M, de Ory F, Royuela E. 2010. Mumps virus genotyping: basis and known circulating genotypes. Open Vaccine J 3:37–41. doi: 10.2174/1875035401003020037. [DOI] [Google Scholar]
- 2.WHO. 2012. Mumps virus nomenclature update: 2012. Wkly Epidemiol Rec. 87:217–24. [PubMed] [Google Scholar]
- 3.Jin L, Örvell C, Myers R, Rota PA, Nakayama T, Forcic D, Hiebert J, Brown KE. 2015. Genomic diversity of mumps virus and global distribution of the 12 genotypes. Rev Med Virol 25:85–101. doi: 10.1002/rmv.1819. [DOI] [PubMed] [Google Scholar]
- 4.WHO. 2007. Mumps virus vaccines. Wkly Epidemiol Rec. 82:51–60. [PubMed] [Google Scholar]
- 5.Kaaijk P, van der Zeijst B, Boog M, Hoitink C. 2008. Increased mumps incidence in the Netherlands: review on the possible role of vaccine strain and genotype. Euro Surveill 13:pii=18914. doi: 10.2807/ese.13.26.18914-en. [DOI] [PubMed] [Google Scholar]
- 6.Bangor-Jones RD, Dowse GK, Giele CM, van Buynder PG, Hodge MM, Whitty MM. 2009. A prolonged mumps outbreak among highly vaccinated Aboriginal people in the Kimberley region of Western Australia. Med J Aust 191:398–401. [DOI] [PubMed] [Google Scholar]
- 7.Rubin SA, Qi L, Audet SA, Sullivan B, Carbone KM, Bellini WJ, Rota PA, Sirota L, Beeler J. 2008. Antibody induced by immunization with the Jeryl Lynn mumps vaccine strain effectively neutralizes a heterologous wild-type mumps virus associated with a large outbreak. J Infect Dis 198:508–15. doi: 10.1086/590115. [DOI] [PubMed] [Google Scholar]
- 8.Rubin SA, Link MA, Sauder CJ, Zhang C, Ngo L, Rima BK, Duprex WP. 2012. Recent mumps outbreaks in vaccinated populations: no evidence of immune escape. J Virol 86:615–20. doi: 10.1128/JVI.06125-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gouma S, Ten Hulscher HI, Schurink-van't Klooster TM, de Melker HE, Boland GJ, Kaaijk P, van Els CACM, Koopmans MPG, van Binnendijk RS. 2016. Mumps-specific cross-neutralization by MMR vaccine-induced antibodies predicts protection against mumps virus infection. Vaccine 34:4166–4171. doi: 10.1016/j.vaccine.2016.06.063. [DOI] [PubMed] [Google Scholar]
- 10.Šantak M, Lang-Balija M, Ivancic-Jelecki J, Kosutic-Gulija T, Ljubin-Sternak S, Forcic D. 2013. Antigenic differences between vaccine and circulating wild-type mumps viruses decrease neutralization capacity of vaccine-induced antibodies. Epidemiol Infect 141:1298–1309. doi: 10.1017/S0950268812001896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaidya SR, Dvivedi GM, Jadhav SM. 2016. Cross-neutralization between three mumps viruses & mapping of haemagglutinin-neuraminidase (HN) epitopes. Indian J Med Res 143:37–42. doi: 10.4103/0971-5916.178587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 13.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubota M, Takeuchi K, Watanabe S, Ohno S, Matsuoka R, Kohda D, Nakakita SI, Hiramatsu H, Suzuki Y, Nakayama T, Terada T, Shimizu K, Shimizu N, Shiroishi M, Yanagi Y, Hashiguchi T. 2016. Trisaccharide containing α2,3-linked sialic acid is a receptor for mumps virus. Proc Natl Acad Sci U S A 113:11579–11584. doi: 10.1073/pnas.1608383113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ki M, Park T, Yi SG, Oh JK, Choi B. 2003. Risk analysis of aseptic meningitis after measles-mumps-rubella vaccination in Korean children by using a case-crossover design. Am J Epidemiol 157:158–165. doi: 10.1093/aje/kwf167. [DOI] [PubMed] [Google Scholar]
- 16.Miller E, Andrews N, Stowe J, Grant A, Waight P, Taylor B. 2007. Risks of convulsion and aseptic meningitis following measles-mumps-rubella vaccination in the United Kingdom. Am J Epidemiol 165:704–9. doi: 10.1093/aje/kwk045. [DOI] [PubMed] [Google Scholar]
- 17.WHO. 2007. Global Advisory Committee on Vaccine Safety, 29–30 November 2006 Wkly Epidemiol Rec 82:18–24. [PubMed] [Google Scholar]
- 18.Peltola H, Kulkarni PS, Kapre SV, Paunio M, Jadhav SS, Dhere RM. 2007. Mumps outbreaks in Canada and the United States: time for new thinking on mumps vaccines. Clin Infect Dis 45:459–66. doi: 10.1086/520028. [DOI] [PubMed] [Google Scholar]
- 19.Cole KS, Steckbeck JD, Rowles JL, Desrosiers RC, Montelaro RC. 2004. Removal of N-linked glycosylation sites in the V1 region of simian immunodeficiency virus gp120 results in redirection of B-cell responses to V3. J Virol 78:1525–39. doi: 10.1128/JVI.78.3.1525-1539.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homan EJ, Bremel RD. 2014. Are cases of mumps in vaccinated patients attributable to mismatches in both vaccine T-cell and B-cell epitopes?: an immunoinformatic analysis. Hum Vaccin Immunother 10:290–300. doi: 10.4161/hv.27139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.May M, Rieder CA, Rowe RJ. 2018. Emergent lineages of mumps virus suggest the need for a polyvalent vaccine. Int J Infect Dis 66:1–4. doi: 10.1016/j.ijid.2017.09.024. [DOI] [PubMed] [Google Scholar]
- 22.Cardemil CV, Dahl RM, James L, Wannemuehler K, Gary HE, Shah M, Marin M, Riley J, Feikin DR, Patel M, Quinlisk P. 2017. Effectiveness of a third dose of MMR vaccine for mumps outbreak control. N Engl J Med 377:947–956. doi: 10.1056/NEJMoa1703309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.