Abstract
Lung cancer is a leading cause of cancer‐related deaths with an increasing incidence and poor prognoses. To further understand the regulatory mechanisms of lipidomic profiles in lung cancer subtypes, we measure the profiles of plasma lipidome between health and patients with lung cancer or among patients with squamous cell carcinomas, adenocarcinoma or small cell lung cancer and to correct lipidomic and genomic profiles of lipid‐associated enzymes and proteins by integrating the data of large‐scale genome screening. Our studies demonstrated that circulating levels of PS and lysoPS significantly increased, while lysoPE and PE decreased in patients with lung cancer. Our data indicate that lung cancer‐specific and subtype‐specific lipidomics in the circulation are important to understand mechanisms of systemic metabolisms and identify diagnostic biomarkers and therapeutic targets. The carbon atoms, dual bonds or isomerism in the lipid molecule may play important roles in lung cancer cell differentiations and development. This is the first try to integrate lipidomic data with lipid protein‐associated genomic expression among lung cancer subtypes as the part of clinical trans‐omics. We found that a large number of lipid protein‐associated genes significantly change among cancer subtypes, with correlations with altered species and spatial structures of lipid metabolites.
Keywords: biomarkers, lipidomics, lung cancer, subtypes
Lung cancer is a leading cause of cancer‐related deaths with an increasing incidence and poor prognoses, due to the lack of knowledge on heterogeneity complexity.1 Systems heterogeneity of lung cancer was described by integrating gene or protein expression, epigenetics, sequencing, transcription or interaction.2 Of those, there are a few studies on the heterogeneity of lipidomic profiles among lung diseases, which play an important role in the development, evolution, metabolism and mechanic function of lung. Marien et al3 investigated cancer and adjunct non‐malignant tissue contents of 91 phospholipid species in patients with small cell lung cancer (SCLC) and found that levels of SMs mainly reduced, while specific PIs elevated in cancer tissues. Reduced levels of PSs were correlated with some of elevated PE and PC species. Main changes were lipids with 40 or 42 carbon atoms in dual fatty acyl chains, with heterogeneity of phospholipids among cancer cells. Intratumour metabolic heterogeneity exists in cancer tissues, e.g renal cancer,4 contributing to the response of cancer cells to therapies. The human plasma lipidome is made up of thousands of ubiquitous lipid species to reflect systemic lipid metabolism and has the clinical relevance of diagnostics with the severity and risk of the disease. Although earlier studies implicated changes of lipid metabolism in tumour tissues,3, 5, 6 there is little known on systematic changes in lipidomic profiles in patients with lung cancer. The aim of our preliminary study is to explore the profiles of plasma lipidome between health and patients with lung cancer or among patients with squamous cell carcinomas (SCC), adenocarcinoma (ADC) or SCLC and to correct lipidomic and genomic profiles.
About 388 lipid molecules of plasma harvested from 8 health and 26 patients with SCC, ADC or SCLC were measured by MRM analysis performed with normal phase HPLC/MS. Patients with lung cancer as the first diagnosis and before any treatment were prospectively recruited at the initial clinic in Fudan University Zhongshan Hospital, without diabetes or other diseases. The study was approved by the Ethical Committee of Zhongshan Hospital. Total lipids were extracted from 200 μL plasma, performed with a modified method of Bligh & Dyer.7 Internal standard cocktails (Avanti Lipids Polar) were added at an amount of 10 μL to each sample, and lipid extracts were subjected to the normal‐phase silica liquid chromatography‐coupled triple‐quadrupole mass spectrometers (Qtrap® 4000 and 6500, Sciex, Framingham, MA, USA). Both negative and positive ESI modes were used, the Q‐Trap was operated in the MRM mode, and different precursor/product ion pairs were scanned. Each experiment was repeated thrice. MRM data were processed with MultiQuant™ software (AB Sciex), and peak area of each pair was used for further quantification. Lung cancer specificity was identified to compare the pooled group of all subtypes with the health. The subtype specificity was defined as the level significantly higher or lower than that in the health (>twofold and P < 0.05), but not in other subtypes.
It was firstly reported that circulating levels of PS and lysoPS significantly increased, while lysoPE and PE decreased in patients with lung cancer. Our data demonstrated that the circulating levels of PE40s or lysoPE20s lipid species were significantly higher or lower in patients with lung cancer, respectively, of which about 28 lipid species co‐existed in all subtypes of lung cancer. It indicates that the systemic inflammation is in a high reactive condition, where cytokines may interact more with or cross local and systemic leucocytes or tumour cells.8 Lipidome comprehensive characterizations are dependent upon the number of carbon atoms, dual bonds or isomerism in the lipid molecule. For example, PS40:6, lysoPS22:6 or PS34:5 elevated >100‐fold, PS36:1 or 40:5, or lysoPS18:2 > 50, as well as PS34:1 or 34:2, lysoPS16:0 or 22:4 > 20, while d181So and lysoPE20:4(sn‐2) decreased >10‐fold, lysoPE20:3(sn‐2), 22:6(sn‐1) or 20:5(sn‐1) >5 and d171So, PS33:1, 33:2,35:4p or lysoPI22:0(sn‐1) >3. We further map the comprehensively lipidomic profiles and define subtype specificity of lung cancer, by comparing one subtype with healthy or other two subtypes. We noticed that 1 and 51 lipid species in SCC were significantly higher and lower, 34 and 6 in ADC or 5 and 4 in SCLC, respectively. Levels of PG or PI reduced mainly in SCC or ADC, while PS or PE elevated in ADC or SCLC, respectively (Table 1). Figure 1 demonstrates the heatmap of lipidomic profiles (Figure 1A), quantitative identification of main lipid elements, e.g PC, PS and PE (Figure 1B) and total levels of PC, PE, PS, SM and PI (Figure 1C) between health and lung cancer subtypes. The principal component analysis indicates the cover area (Figure 1D) and detailed distribution (Figure 1E) of lipidomic profiles in health and patients with various subtypes of lung cancer.
Table 1.
Up‐ or down‐regulated type‐special lipids in serum over twofold in adenocarcinoma (ADC), squamous cell carcinoma (SCC) or small cell lung cancer (SCLC)
| Lipid symbol | Fold | Lipid symbol | Fold |
|---|---|---|---|
| Up‐regulated type‐special lipids in SCC | |||
| PE 34:1 | 2.413927591 | ||
| Up‐regulated type‐special lipids in ADC | |||
| PS32:1 | 5.09765 | lysoPS15:1 | 11.06539345 |
| lysoPS20:2 | 17.472675 | lysoPS20:1 | 18.84013333 |
| PS30:1 | 19.00129722 | PS38:6 | 19.63504722 |
| lysoPS22:0 | 21.87011944 | PS36:3 | 26.31176111 |
| PS35:2 | 32.28302361 | PS35:1 | 33.0320804 |
| lysoPS16:0 | 35.35140556 | PS34:0 | 37.26924167 |
| PS34:3 | 50.05725958 | PS38:5 | 56.80440113 |
| PS38:2 | 59.53787133 | lysoPS22:4 | 61.52068609 |
| lysoPS20:3 | 63.70933056 | PS34:2 | 77.12018239 |
| lysoPS16:0 | 90.27303933 | lysoPS18:2 | 94.63135557 |
| PS36:0 | 100.2976925 | PS40:4 | 103.1282059 |
| PS38:4 | 110.1863088 | PS36:1 | 130.7878593 |
| PS40:5 | 141.3205679 | lysoPS22:6 | 144.0307609 |
| PS36:2 | 154.2687604 | PS38:3 | 170.3411736 |
| PS34:5 | 198.4490358 | PS40:6 | 237.5144428 |
| lysoPS18:1 | 271.9142996 | lysoPS20:4 | 422.4742472 |
| PS34:4 | 672.3756202 | lysoPS18:0 | 772.2299397 |
| Up‐regulated type‐special genes in SCLC | |||
| PC 40:1 | 2.452430888 | PC 40:4 | 3.035423914 |
| PE 38:1 | 6.919780093 | PE 42:7 | 8.224826755 |
| PE 40:3 | 12.80649768 | ||
| Down‐regulated type‐special lipids in ADC | |||
| PI 30:0 | 3.138308237 | PI 33:0 | 2.973606913 |
| PI 33:1e | 2.756489035 | PI 30:1 | 2.448825305 |
| PI 33:2e | 2.401033111 | PI 41:6 | 2.089884861 |
| Down‐regulated type‐special lipids in SCC | |||
| lysoPI 18:2 (sn‐1) | 3.174021852 | lysoPI 17:0 (sn‐1) | 2.686501872 |
| PG36:2 | 2.599471242 | PG35:2 | 2.599471242 |
| PG35:3 | 2.582298818 | PG35:1 | 2.573201161 |
| PG34:0 | 2.571442278 | PG39:7 | 2.57054263 |
| PG37:3 | 2.568963178 | PG35:4 | 2.560667263 |
| PG34:1 | 2.544927869 | PG36:0 | 2.540946865 |
| PG40:1 | 2.533367656 | PG37:6 | 2.519840973 |
| PG36:1 | 2.517071946 | PG33:1 | 2.509337261 |
| PG41:6 | 2.508824969 | lysoPG19:0 | 2.502973929 |
| PG35:5 | 2.498092401 | PG38:1 | 2.493572762 |
| PG37:4 | 2.478698123 | PG38:3 | 2.467495233 |
| PG37:2 | 2.466940759 | PG32:0 | 2.459511221 |
| PG37:5 | 2.457177588 | PG34:2 | 2.454550159 |
| lysoPG22:4 | 2.445162623 | lysoPG20:3 | 2.436735333 |
| lysoPG22:0 | 2.414907832 | PG40:4 | 2.412623442 |
| PG36:4 | 2.411241381 | lysoPG20:2 | 2.395150124 |
| PG33:2 | 2.394734033 | PG38:2 | 2.390172121 |
| PG40:5 | 2.37513548 | PG36:5 | 2.355426245 |
| PG38:4 | 2.350017951 | PG34:3 | 2.344849524 |
| lysoPG20:4 | 2.321610499 | PG31:0 | 2.318186638 |
| lysoPG20:5 | 2.302249424 | PG36:3 | 2.297141059 |
| lysoPG20:0 | 2.296203929 | lysoPG18:3 | 2.239909774 |
| lysoPG17:0 | 2.238864462 | PG40:8 | 2.236149498 |
| PG38:6 | 2.231677958 | PG38:5 | 2.21673939 |
| PG39:3 | 2.163513382 | PG40:6 | 2.162950362 |
| PG40:7 | 2.139169557 | ||
| Down‐regulated type‐special lipids in SCLC | |||
| lysoPC 20:5 | 3.679763534 | d171S1P | 2.622291711 |
| PE 35:5p | 2.377716405 | Cer120 | 2.227294449 |
Figure 1.
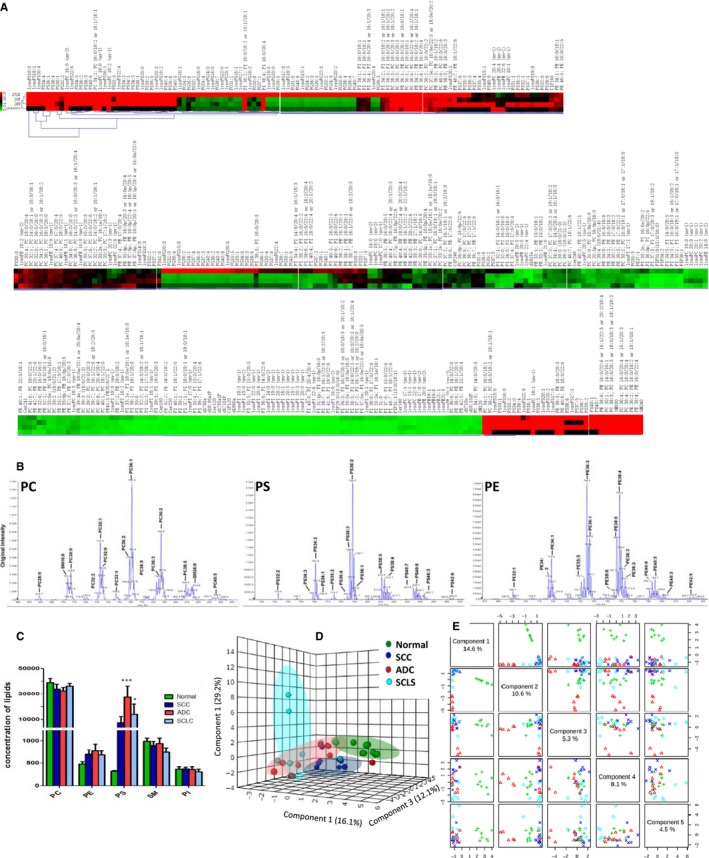
The Heatmap of Lipidomic Profiles (A) was Compared between the Health (as normal) and Patients with Squamous Cell Carcinomas (SCC), Adenocarcinoma (ADC) or Small Cell Lung Cancer (SCLC). The measurement quality of lipidomic profiles was shown by the precursor ion scan of phosphocholine (PC)‐, phosphatidylserine (PS)‐, phosphatidylethanolamine (PE)‐containing phospholipid species in plasma (B). The amounts (nmol lipid/mg DNA) of phospholipid head‐group classes, e.g PC, PE, PS, phosphatidylinositol (PI) or sphingomyelins (SM), were pooled and calculated among the health (as normal) or patients with SCC, ADC or SCLC, as presented in C (***<0.001). Three‐dimensional scatter plots of the principal component scores were illustrated in the validation set based on the weights from the discovery set (D). Component analysis of partial least squares‐discriminant analysis (PLS‐DA) by metabolism between the health (as normal) and patients with SCC, ADC or SCLC was scattered in E
Our results deliver the significant message that the heterogeneity of lipidomic profiles among lung cancer subtypes should be seriously considered and has the unique value of early diagnostics. For example, PI significantly increased with a decrease in PS in cancer tissues of patients with non‐small cell lung cancer,3 while there are no similar findings in plasma samples of patients with early lung cancer.9 Our present study demonstrated the elevation of PS in patients with lung cancer, as compared with healthy, rather than PI (Figure 1B). Altered PS species in the circulation can be valuable biomarkers to screen the risk population if those PS may be generated from lung cancer cells, or be an early lung cancer‐dependent pattern of lipid metabolism. The reduced amount of multi‐PS species within lung cancer cells can be due to the over‐consumed or over‐metabolized, to the over‐release of cytoplasm into the circulation or to the increased binding and activity of negatively charged lipids PS, PA and PG on lung cancer cell surfaces.10 The changed serum levels of phospholipids and their derivatives, e.g lysophosphatidylcholines and lysophosphatidylethanolamine, are considered as promising cancer markers.9, 11 On basis of previous finding that plasma level of the LPC reduced in patients with lung cancer and colorectal cancer,12 our results furthermore evidenced that lysoPG and lysoPI are lower in the plasma of patients with lung cancer than healthy individuals.
Lipid synthesis and its metabolites were suggested as a metabolic liability and potential biomarkers of lung cancer.13 Lipidome varied between the health and patients with lung cancer or among subtypes implies the existence of lipidomic heterogeneity, which may be caused by local and systemic cell metabolism and inflammation. To further understand the regulatory mechanisms of lipidomic profiles in lung cancer subtypes, we mined the genomic profiles of lipid‐associated enzymes and proteins from the global database integrated the data of large‐scale genome screening and other large mutation databases, as reported previously.14 We found that a large number of lipid protein‐associated genes significantly change among cancer subtypes, with correlations with altered species and spatial structures of lipid metabolites, like ACOT, ACSL, PKD gene families. Most of the long‐chain fatty acids (carbon atoms > 40) were up‐regulated in lung cancer. The over‐expression of fatty acid synthase (FASN) occurs in cancers, leading to the over‐production of endogenous fatty acids. Acyl‐CoA synthetases (ACSLs) enzymes, which convert free long‐chain fatty acids into fatty acyl‐CoA esters, also participate in the metabolic reprogramming of cancer cells. Recent study showed ACSL3 is the critical element for the oncogenic capacity of mutant KRAS in lung cancer.15
A number of genes and proteins are suggested as molecular biomarkers or therapeutic targets, which may contribute to the transit process from chronic obstructive pulmonary diseases to lung cancer.16 It is questioned whether lipid species or lipidomic profiles can be identified as stable biomarkers to monitor such transit process. It is also important to define the consistence and correlation of lipidomic profiles with genomic and proteomic ones. Our previous studies demonstrate that it is important and valuable to compare genomics and proteomics, while more important to integrate genomics and proteomics with clinical phenomes.17, 18, 19, 20 It will be a new challenge to merge or fuse lipidomics with clinical phenomics as the part of clinical trans‐omics.21 Thus, our data indicate that lung cancer‐specific and subtype‐specific lipidomics in the circulation are important to understand mechanisms of systemic metabolisms and identify diagnostic biomarkers and therapeutic targets. However, the roles of carbon atoms, dual bonds or isomerism in the lipid molecule in lung cancer cell differentiations should be further explored in future studies.
CONFLICT OF INTEREST
The authors confirm that there are no conflicts of interest.
ACKNOWLEDGEMENTS
The work was supported by Zhongshan Distinguished Professor Grant (XDW), The National Nature Science Foundation of China (91230204, 81270099, 81320108001, 81270131, 81300010, 81700008), The Shanghai Committee of Science and Technology (12JC1402200, 12431900207, 11410708600, 14431905100), Operation funding of Shanghai Institute of Clinical Bioinformatics, Ministry of Education for Academic Special Science and Research Foundation for PhD Education (20130071110043), and National Key Research and Development Program (2016YFC0902400, 2017YFSF090207).
Lv J, Gao D, Zhang Y, Wu D, Shen L, Wang X. Heterogeneity of lipidomic profiles among lung cancer subtypes of patients. J Cell Mol Med. 2018;22:5155–5159. 10.1111/jcmm.13782
Jiapei Lv, Danyan Gao, and Yong Zhang are contribute to this work equally
REFERENCES
- 1. Wang DC, Wang X. Tomorrow's genome medicine in lung cancer. Semin Cancer Biol. 2017;42:39‐43. [DOI] [PubMed] [Google Scholar]
- 2. Wang DC, Wang X. Systems heterogeneity: an integrative way to understand cancer heterogeneity. Semin Cell Dev Biol. 2017;64:1‐4. [DOI] [PubMed] [Google Scholar]
- 3. Marien E, Meister M, Muley T, et al. Non‐small cell lung cancer is characterized by dramatic changes in phospholipid profiles. Int J Cancer. 2015;137:1539‐1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Okegawa T, Morimoto M, Nishizawa S, et al. Intratumor heterogeneity in primary kidney cancer revealed by metabolic profiling of multiple spatially separated samples within tumors. EBioMedicine. 2017;19:31‐38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hansen AS, Cattoglio C, Darzacq X, Tjian R. Recent evidence that TADs and chromatin loops are dynamic structures. Nucleus. 2018;9:20‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goto T, Terada N, Inoue T, et al. The expression profile of phosphatidylinositol in high spatial resolution imaging mass spectrometry as a potential biomarker for prostate cancer. PLoS ONE. 2014;9:e90242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911‐917. [DOI] [PubMed] [Google Scholar]
- 8. Oyler‐Yaniv J, Oyler‐Yaniv A, Shakiba M, et al. Catch and release of cytokines mediated by tumor phosphatidylserine converts transient exposure into long‐lived inflammation. Mol Cell. 2017;66:635‐647. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ros‐Mazurczyk M, Jelonek K, Marczyk M, et al. Serum lipid profile discriminates patients with early lung cancer from healthy controls. Lung Cancer. 2017;112:69‐74. [DOI] [PubMed] [Google Scholar]
- 10. Desai TJ, Udugamasooriya DG. A comprehensive lipid binding and activity validation of a cancer‐specific peptide‐peptoid hybrid PPS1. Biochem Biophys Res Comm. 2017;486:545‐550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yu Z, Chen H, Ai J, et al. Global lipidomics identified plasma lipids as novel biomarkers for early detection of lung cancer. Oncotarget. 2017;8:107899‐107906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoh C, Boocock D, Marczylo T, et al. Pilot study of oral silibinin, a putative chemopreventive agent, in colorectal cancer patients: silibinin levels in plasma, colorectum, and liver and their pharmacodynamic consequences. Clin Cancer Res. 2006;12:2944‐2950. [DOI] [PubMed] [Google Scholar]
- 13. Merino Salvador M, Gomez de Cedron M, Moreno Rubio J, et al. Lipid metabolism and lung cancer. Crit Rev Oncol Hematol. 2017;112:31‐40. [DOI] [PubMed] [Google Scholar]
- 14. Lu J, Wang W, Xu M, Li Y, Chen C, Wang X. A global view of regulatory networks in lung cancer: an approach to understand homogeneity and heterogeneity. Semin Cancer Biol. 2017;42:31‐38. [DOI] [PubMed] [Google Scholar]
- 15. Padanad MS, Konstantinidou G, Venkateswaran N, et al. Fatty acid oxidation mediated by Acyl‐CoA synthetase long chain 3 is required for mutant KRAS lung tumorigenesis. Cell Rep. 2016;16:1614‐1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X. New biomarkers and therapeutics can be discovered during COPD‐lung cancer transition. Cell Biol Toxicol. 2016;32(5):359‐361. [DOI] [PubMed] [Google Scholar]
- 17. Shi L, Zhu B, Xu M, Wang X. Selection of AECOPD‐specific immunomodulatory biomarkers by integrating genomics and proteomics with clinical informatics. Cell Biol Toxicol. 2018;34(2):109‐123. [DOI] [PubMed] [Google Scholar]
- 18. Chen C, Shi L, Li Y, Wang X, Yang S. Disease‐specific dynamic biomarkers selected by integrating inflammatory mediators with clinical informatics in ARDS patients with severe pneumonia. Cell Biol Toxicol. 2016;32(3):169‐184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen H, Wang Y, Bai C, Wang X. Alterations of serum inflammatory biomarkers in the healthy and chronic obstructive pulmonary disease patients with or without acute exacerbation. J Cell Mol Med. 2012;16(6):1286‐1297.21883889 [Google Scholar]
- 20. Gu J, Wang X. New future of cell biology and toxicology: thinking deeper. Cell Biol Toxicol. 2016;32(1):1‐3. [DOI] [PubMed] [Google Scholar]
- 21. Wang X. Clinical trans‐omics: an integration of clinical phenomics with multiomics. Cell Biol Toxicol. 2018;34:163‐166. [DOI] [PubMed] [Google Scholar]


