Abstract
Genetic studies have shown that FGF10/FGFR2 signaling is required for airway branching morphogenesis and FGF10 functions as a chemoattractant factor for distal epithelial cells during lung development. However, the detail downstream cellular and molecular mechanisms have not been fully characterized. Using live imaging of ex vivo cultured lungs, we found that tip airway epithelial progenitor cells migrate faster than cleft cells during airway bud formation and this migration process is controlled by FGFR2-mediated ERK1/2 signaling. Additionally, we found that airway progenitor cells that migrate faster tend to become distal airway progenitor cells. We identified that Anxa4 is a downstream target of ERK1/2 signaling. Anxa4−/− airway epithelial cells exhibit a “lag-behind” behavior and tend to stay at the stalk airways. Moreover, we found that Anxa4-overexpressing cells tend to migrate to the bud tips. Finally, we demonstrated that Anxa4 functions redundantly with Anxa1 and Anxa6 in regulating endoderm budding process. Our study demonstrates that ERK1/2/Anxa4 signaling plays a role in promoting the migration of airway epithelial progenitor cells to distal airway tips and ensuring their distal cell fate.
Introduction
The airways of mammalian lungs are generated via a process called branching morphogenesis. Fibroblast growth factor 10 (FGF10) and its receptor FGFR2b are known to be essential for airway branching morphogenesis1,2; both Fgf10-null and Fgfr2b-null mouse embryos have a lung agenesis phenotype3–5. Fgf10 is expressed in the distal mesenchyme, whereas Fgfr2b is uniformly expressed in the airway epithelium. As airway branching morphogenesis proceeds, Fgf10 is dynamically expressed in the distal mesenchyme prior to the appearance of each new airway bud6. FGF10 has been shown to induce lung endoderm bud expansion and budding in mesenchyme-free lung endoderm explant cultures6,7. It has also been shown in vitro that FGF10 acts as a chemoattractant factor for distal airway epithelium8,9. These findings have established an essential role of FGF10 in regulating the directional outgrowth of airway buds during branching morphogenesis. However, the underlying cellular and molecular mechanisms through which FGF10 regulates airway bud formation are not well understood.
It is now appreciated that airway branching morphogenesis requires epithelial-mesenchymal interactions. In response to growth factors that are expressed in the mesenchyme (e.g., FGF10), epithelial cells initiate several processes including cell proliferation and cell migration10,11. A previous study showed that inhibition of cell proliferation in cultured chicken lung explants did not block new bud formation12 and this observation is consistent with another study which reported that localized proliferation is not a triggering event for the initiation of bud outgrowth in cultured lung endoderm explants13. However, multiple lines of evidence have demonstrated that cell migration is required for branching morphogenesis in several organs. Specifically, in the Drosophila trachea system, Bnl/Btl (homologs of FGF/FGFR) signaling controls trachea cell migration and branching morphogenesis14. It has been demonstrated that MAPK-dependent collective cell migration drives the branching morphogenesis and tube elongation of mammary gland15,16. During renal branching morphogenesis, GDNF-Ret signaling is known to be essential for the competitive cell migration: Ret−/− cells exhibit a “lag-behind” behavior and contribute to trunks of ureteric bud17,18.
During airway branching morphogenesis, the airway epithelial progenitor cells in the distal tips differentiated into distinct cell lineages along its proximal-distal axis. The expression of Sox2 marks the proximal/stalk airway epithelial cell lineage whereas the expression of Sox9 and Id2 mark the distal airway epithelial cell lineage. Recent lineage-tracing studies using Id2CreER/+ mouse line show that airway epithelial progenitor cells in the distal bud tips are able to generate both distal and stalk airway epithelial cells at least up to E13.519. It has been showed that FGF10 signaling is essential for preventing distal airway progenitor cell from differentiating into stalk airway epithelial cell20,21. However, it is less clear how airway branching morphogenesis is orchestrated with the airway epithelial cell fate specification.
Annexin proteins are found in species ranging from fungi to higher vertebrates. They are a highly-conserved superfamily of proteins that bind with membrane phospholipids in a calcium-dependent manner and this binding links them to many membrane-related processes (e.g., basic membrane organization, membrane trafficking)22–24. Further, it has been shown that several members of the Annexin family are able to bind with and “bundle” F-actin filaments25–28. In vitro studies including both gain- and loss-of-function experiments have shown that Annexins play a role in promoting cell migration29,30. Despite the abundance and conservation of Annexins in most eukaryotic species, relatively little is known about the regulation of Annexin gene expression and little is known about the function of Annexin proteins during embryonic lung development.
Here, using a combination of live imaging, mouse genetics and lung endoderm culture system experiments, we found that tip airway epithelial progenitor cells migrate faster than cleft cells during airway bud formation. We identified Anxa4 (encoding Annexin A4) as a downstream target of ERK1/2 signaling and found that the expression level of Anxa4 is positively regulated by the activity of ERK1/2 signaling. We showed that Anxa4 is required for airway epithelial cell migration, both in vitro and in vivo. Furthermore, we found that Anxa4−/− epithelial cells exhibit a “lag-behind” behavior and tend to contribute to the stalk airways. In contrast, Anxa4-overexpressing cells tend to remain in bud tips during bud formation. We also found that Anxa4 functions redundantly with Anxa1 and Anxa6 in regulating the endoderm budding process. Our study establishes that FGF10-activated ERK1/2/Anxa4 signaling plays a role in promoting airway progenitor cell migration and ensuring their distal airway epithelial cell fate by regulating Anxa4 expression during airway bud formation.
Results
Airway progenitor cells that migrate faster tend to commit to distal airway cell fate
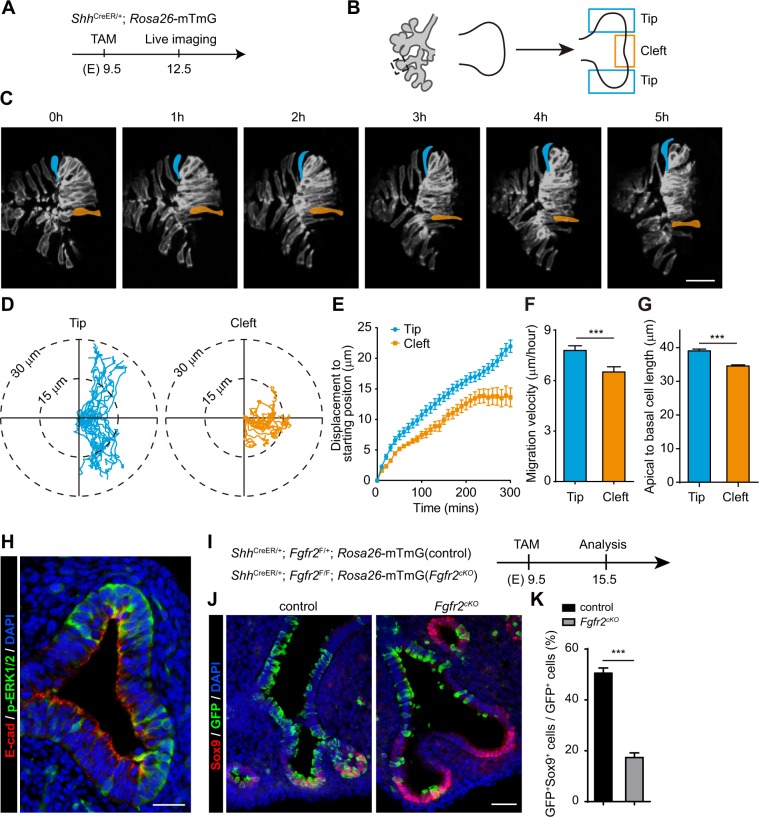
To investigate the cellular behaviors during airway bud formation, we conducted an ex vivo time-lapse imaging experiment with E12.5 lungs to monitor cell behaviors during airway bud formation. Pregnant females carrying ShhCreER/+; Rosa26-mTmG embryos were injected with one dose of tamoxifen at E9.5 (Fig. 1A). After a tamoxifen injection, membrane GFP was expressed in a mosaic manner in the airway epithelial cells of ShhCreER/+; Rosa26-mTmG lung, enabling tracing of individual airway epithelial cells. Airway epithelial progenitor cells at the tip of the first lateral branch in the right caudal lobe (RCd.L1) of lung at E12.5 were imaged (Fig. 1B). In wild type mice, this lateral branch undergoes a planar bifurcation that leads to the formation of two new buds31. The region between these two newly formed bud tips is referred to as the cleft (Fig. 1B). The RCd.L1 bud tip was imaged every 10 minutes for 5 hours. After five-hour live imaging, airway progenitor cells at the bud tip directly migrated out and formed a new bud tip, while airway progenitor cells at the cleft region migrated slower and formed the cleft (Fig. 1C and Supplementary Video 1).
Figure 1.
Airway progenitor cells that migrate faster tend to commit to distal airway cell fate. (A–C) Pregnant females carrying ShhCreER/+; Rosa26-mTmG embryos were treated with tamoxifen (TAM) at E9.5 and lungs were imaged at E12.5 (A). Bud tip of the first lateral branch in right caudal lobe (RCd.L1) was imaged. Finally, two new bud tips were formed while the middle cells between two bud tips formed the cleft (B). After five-hour imaging, the tip cells (one cell was highlighted by blue) directly moved out and formed a new bud tip, while the middle cells (one cell was highlighted by orange) moved slower and tended to lag behind (C). Scale bar: 50 μm. (D–G) Cell track plots of tip cells (24 cells) and cleft cells (17 cells) after aligning their starting positions (D), showing that tip cells can migrate longer than cleft cells (D,E); The migration velocity analysis showed that tip cells migrate faster than cleft cells (F); and the tip cells have longer apical-to-basal cell length (G). Data are presented as mean ± SEM; n = 3 live image samples; ***p < 0.001; Student’s t-test. (H) WT lung bud at E12.5 was analyzed by E-cadherin (red) and p-ERK1/2 (green) immunostaining. The bud tips showed higher p-ERK1/2 staining compared to cleft cells. Scale bar: 25 μm. (I–K) Pregnant females carrying ShhCreER/+; Fgfr2F/+; Rosa26-mTmG (control) and ShhCreER/+; Fgfr2F/F; Rosa26-mTmG (Fgfr2cKO) embryos were treated with tamoxifen (TAM) at E9.5 and lungs were sampled at E15.5 (I). Immunostaining using antibodies against GFP and Sox9 showed that less GFP+Sox9+ cells were detected in Fgfr2cKO lung as compared to control lung (J,K). Data are presented as mean ± SEM; ***p < 0.001; Student’s t-test. Scale bar: 25 μm.
To analyze cell migration of airway progenitor cells, we manually traced the positions of GFP cells at 31 time-points over a 5-hour-time-course live imaging by using Imaris software. Monitoring of the position data for each cell at each time points allowed us to directly analyze the migration tracks of cells and quantify both cell migration displacement and cell migration velocity (Fig. s1A). The cell migration displacement was calculated as the linear distance (in μm) between the current position of a given cell at a given time point and its starting position. The cell migration velocity was calculated as the ratio of cell migration distance (in μm) to migration time (in hours) (Fig. s1A,B). By analyzing migration tracks of airway progenitor cells at bud tip (tip cells) and airway progenitor cells at cleft region (cleft cells), we found that the tip cells migrated over longer distance and more directionally as compared to cleft cells (Fig. 1D). The average migration displacement over the 5 hours of the experiment was higher for tip cells as compared to cleft cells (Fig. 1E). Moreover, we found that the tip cells have higher migration velocity and longer apical-to-basal cell length as compared to cleft cells (Fig. 1F,G).
We next sought to identify the molecular mechanisms underlying our observation that tip cells migrate faster than cleft cells during planar bifurcation. Given that ERK1/2 signaling can be activated by FGF10/FGFR2 signaling and is essential for cell migration during development32,33, we hypothesized that FGF10/FGFR2 may regulate cell migration of tip cells via ERK1/2 signaling during airway bud formation. Indeed, the level of p-ERK1/2 was higher in cells of the two nascent bud tips than in cells of the cleft region during the planar bifurcation process, suggesting that ERK1/2 signaling may control tip cell migration (Fig. 1H).
Similar to lung, kidneys are highly branched organs and renal branching morphogenesis is known to requires GDNF and its receptor Ret34,35. It has been demonstrated that Ret is required for directed movement of progenitor cells during renal development and Ret−/− cells tend to lag behind and contribute to the renal trunks17. To investigate the effect of loss of FGFR2 signaling on cell migration, we conditionally knocked out Fgfr2 in airway epithelial cells at E9.5 using ShhCreER/+ mice and live imaged the RCd.L1 bud tips of ShhCreER/+; Fgfr2F/F; Rosa26-mTmG (Fgfr2cKO) lungs at E12.5 (Fig. s1C and Supplementary Video 2). We found that Fgfr2−/− cells showed shorter migration track and smaller migration displacement than control cells and that their migration velocity was much slower than control cells (Fig. s1D–F and Supplementary Video 2). Additionally, we found that some Fgfr2−/−;GFP+ cells underwent apoptosis (Fig. s1G), consistent with previous study that loss of Fgfr2 induces cell death36.
We next investigated the effect of loss of Fgfr2 on airway epithelial cell fate determination. Based on the patterns of gene expression, stalk and distal airway epithelial cells can be distinguished by the expression of Sox2 or Sox9. We quantified the ratio of GFP+Sox9+ cells to total GFP+ cells in either ShhCreER/+; Fgfr2F/+; Rosa26-mTmG (control) or ShhCreER/+; Fgfr2F/F; Rosa26-mTmG (Fgfr2cKO) lungs at E15.5 (Fig. 1I). We found that fewer GFP+ cells were present in distal airways of Fgfr2cKO lungs than in control lungs (Fig. 1J,K). Collectively, our findings indicate that Fgfr2 controls distal airway cell fate commitment by regulating ERK1/2-signaling-controlled cell migration.
ERK1/2 signaling regulates the expression of Anxa4
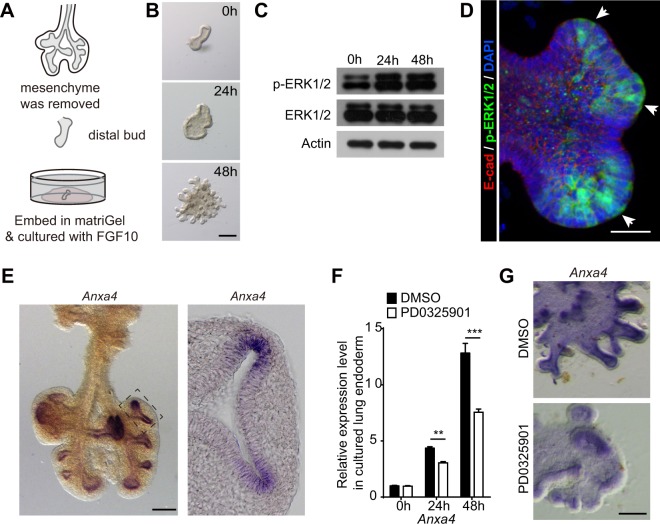
Seeking to identify downstream target(s) of ERK1/2 signaling in airway epithelial cells, we set up a mesenchyme-free lung endoderm culture system in which FGF10 is the only supplemented growth factor. Distal lung endoderm buds were dissected out from whole lungs at E11.5 and were subsequently cultured with serum-free medium supplemented only with FGF10 (800 ng/ml) (Fig. 2A). Based on our observation of both morphology and cell behavior, we defined the developmental process of the in vitro cultured endoderm explants into two stages: (i) within the initial 24 h of the culture period (“pre-budding stage”), the lung endoderm bud became sealed, grew and expanded into a cyst, progressing toward bud formation; (ii) from 24 h to 48 h (“budding stage”), the lung endoderm underwent branching and formed many buds at the cyst surfaces (Fig. 2B). We noted that the phosphorylation level of ERK1/2 was significantly increased at 24 h and at 48 h as compared to 0 h in the cultured lung endoderm explants (Fig. 2C). Similar to our finding that tip cells exhibit high p-ERK1/2 levels during planar bifurcation (Fig. 1H), we found that p-ERK1/2 levels were high in the bud tips of cultured lung endoderm explants (Fig. 2D).
Figure 2.
ERK1/2 signaling regulates the expression of Anxa4. (A) Diagram illustrating the method for mesenchyme-free lung endoderm culture. Whole lung at E11.5 was digested with trypsin on ice and then the mesenchyme was removed using fine forceps to get intact lung endoderm. The distal lung endoderm buds were dissected out, embedded in Matrigel and cultured in serum-free medium containing recombinant FGF10. (B) Images of cultured lung endoderm taken at different time points. There is no obvious bud formation at 24 h after culture. By 48 h, the lung endoderm starts budding and forms many buds. Scale bar: 200 μm. (C) Western blot of cultured lung endoderm explants at 0 h, 24 h and 48 h using antibodies against p-ERK1/2, total ERK1/2 and reference Actin. The phosphorylation level of ERK1/2 was increased significantly at both 24 h and 48 h. Full-length blots are presented in Supplementary Fig. s6. (D) Bud of cultured lung endoderm were analyzed by E-cadherin (red) and p-ERK1/2 (green) immunostaining. The bud tips (arrowhead) showed higher p-ERK1/2 staining compared to middle cells. Scale bar: 25 μm. (E) Whole-mount and section in situ hybridization of Anxa4 in WT lungs at E12.5. Anxa4 was highly expressed in the bud tip epithelial cells adjacent to FGF10 expressing site. Scale bar: 200 μm. (F,G) The expression level of Anxa4 in cultured lung endoderm explants at 0 h, 24 h and 48 h. The expression level of Anxa4 was increased significantly at both 24 h and 48 h as compared to 0 h and its expression level was inhibited by MEK inhibitor, PD0325901 (F). The whole mount in situ hybridization of Anxa4 confirmed that PD0325901 treatment could decrease the expression of Anxa4 (G). Data are presented as mean ± SEM, n = 3; **p < 0.01; ***p < 0.001; Student’s t-test. Scale bar: 100 μm.
Annexin proteins are highly-conserved calcium-dependent phospholipid binding proteins22,37 and several Annexins are known to bind with F-actin to regulate cell migration25,28,29. A previous study has shown that several Annexin family members may be downstream targets of FGF10 and may be involved in endoderm budding process38. Consistently, we here found that the expression levels of Anxa gene family members (encoding Annnexins) showed a steady increase from 0 h to 48 h in cultured lung endoderm explants (Fig. s2A). This increasing expression trend was positively correlated with increased budding activity in lung endoderm cultures, suggesting that Annexins may be downstream targets of FGF10 and, further, that these proteins may be involved in FGF10-induced lung endoderm budding process. However, only Anxa1, Anxa4 and Anxa6 showed significantly increased expression from 24 h to 48 h, the period during which lung endoderm budding occurs. Whole-mount RNA in situ hybridization experiments revealed that Anxa1 is highly expressed in the stalk airway epithelium and showed that Anxa4 is highly expressed in the distal airway epithelium; Anxa6 is expressed in both the epithelium and the mesenchyme (Figs 2E and s2B).
Given our finding that Anxa4 is highly expressed in the distal airway epithelium which also has high p-ERK1/2 level (Figs 1H and 2E), we next set out to investigate whether the expression of Anxa4 is regulated by ERK1/2 signaling. We cultured wild type lung endoderm explants with DMSO or PD0325901 (inhibitor of MEK, upstream of ERK1/2) at 0 h and then analyzed the expression levels of Anxa4 at 48 h using qPCR. We found that inhibition of ERK1/2 signaling decreased the expression of Anxa4; this inhibition also reduced the expression levels of Etv4 and Spry2, two well-known downstream targets of ERK1/2 signaling (Figs 2F and s2C). Whole-mount in situ analysis of Anxa4 in lung endoderm explants confirmed that the expression of Anxa4 is inhibited by treatment with PD0325901 (Fig. 2G). Taken together, these results establish that the expression of Anxa4 is regulated by ERK1/2 signaling.
Anxa4 promotes airway progenitor cell migration
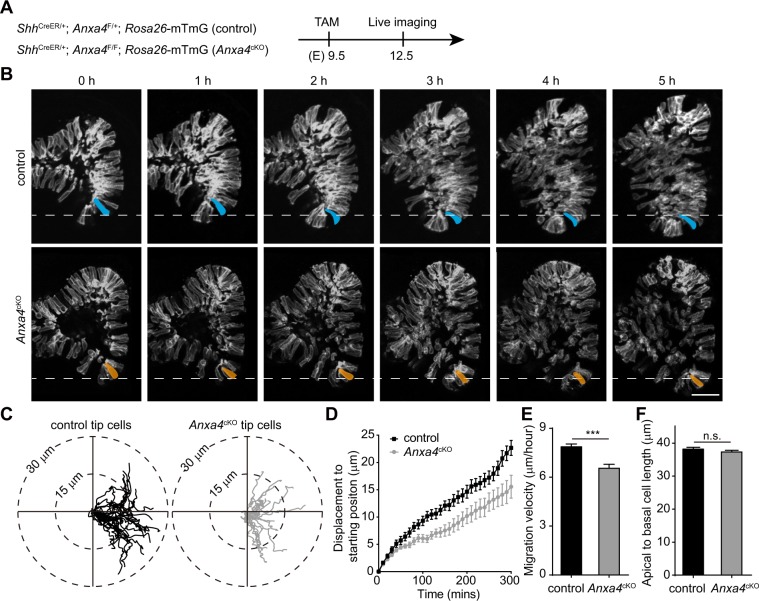
To gain insight into the function of Anxa4 during airway bud formation in vivo, we generated a Anxa4 flox mouse line (Fig. s3A). We next investigated the effect of the loss of Anxa4 on cell migration in vivo by generating pregnant female mice carrying ShhCreER/+; Anxa4F/+; Rosa26-mTmG (control) and ShhCreER/+; Anxa4F/F; Rosa26-mTmG (Anxa4cKO) embryos. After a tamoxifen injection at E9.5, we live-imaged the RCd.L1 bud tips of both control and Anxa4cKO lungs at E12.5 (Fig. 3A). By comparing the cell migration of Anxa4+/−;GFP+ cells in ShhCreER/+; Anxa4F/+; Rosa26-mTmG lungs with Anxa4+/+;GFP+ cells in ShhCreER/+; Rosa26-mTmG lungs, we found that Anxa4+/− cells and Anxa4+/+ cells shows no significant difference in cell migration displacement and migration velocity (data not shown), suggesting that loss of one copy of Anxa4 does not impair airway epithelial cell migration. We therefore used ShhCreER/+; Anxa4F/+; Rosa26-mTmG lung as control in the following experiments.
Figure 3.
Anxa4 promotes airway epithelial progenitor cell migration. (A) Pregnant females carrying ShhCreER/+; Anxa4F/+; Rosa26-mTmG (control) and ShhCreER/+; Anxa4F/F; Rosa26-mTmG (Anxa4cKO) embryos were treated with tamoxifen (TAM) at E9.5 and lungs were imaged at E12.5. (B) Representative images of mosaic labeled control and Anxa4cKO airway bud formation. One tip cells in control and Anxa4cKO RCd.L1 buds are highlighted by blue and orange, respectively. Scale bar: 50 μm. (C–F) Cell track plots of tip cells in control lung (25 cells) and Anxa4cKO lungs (16 cells) after aligning their starting positions (C), showing that Anxa4cKO tip cells had shorter migration tracks and migration displacement than that of control tip cells (C,D); The migration velocity analysis showed that the migration velocity of Anxa4cKO cells was decreased significantly (E) and the apical-to-basal cell length of Anxa4cKO cells was similar to that of control cells (F). Data are presented as mean ± SEM; n = 3 samples per genotype; n.s., not significant; ***p < 0.001; Student’s t-test.
After live imaging the airway progenitor cells in the RCd.L1 bud tips of both control and Anxa4cKO lungs, we found that both control and Anxa4cKO lung buds are able to grow and to form two new buds (Fig. 3B and Supplementary Video 3 and 4). However, the migration tracks of tip cells of control and Anxa4cKO lungs revealed that the tip cells of Anxa4cKO lungs have shorter migration tracks and lesser displacement as compared to tip cells of control lungs (Fig. 3C,D). Analysis of cell migration velocity showed that tip cells of Anxa4cKO lungs migrate slower than did tip cells of control lungs (Fig. 3E). However, the apical-to-basal cell length of tip cells of Anxa4cKO lungs is similar to that of tip cells of control lungs (Fig. 3F), suggesting that loss of Anxa4 does not affect the tip cell shape change. These results demonstrate that Anxa4 is required for airway epithelial progenitor cell migration during airway bud formation.
Loss of Anxa4 negatively affects distal airway epithelial cell fate specification
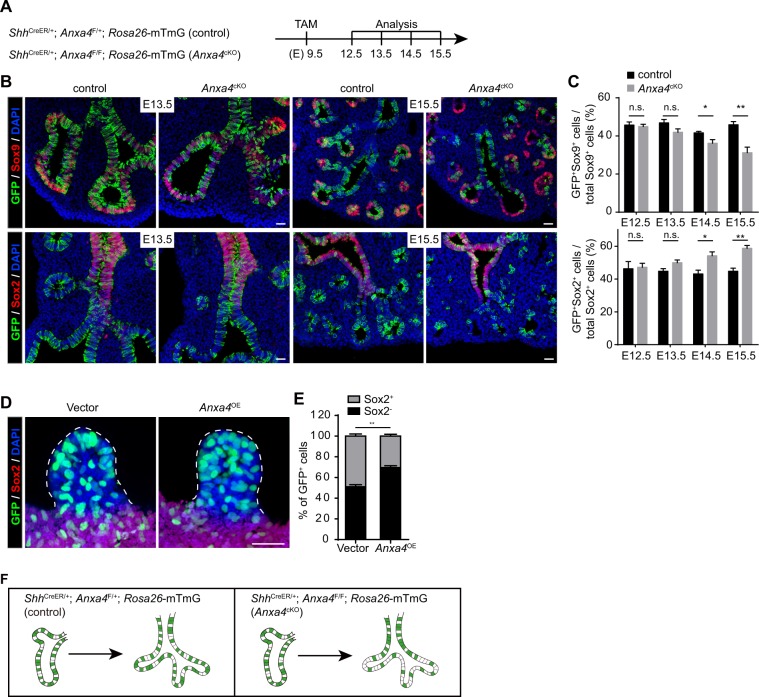
As Anxa4−/− airway progenitor cells migrate slower than wild type cells, we next assessed the ability of Anxa4−/− airway progenitor cells to become Sox9+ distal airway epithelial cells. Pregnant females carrying both control and Anxa4cKO embryos were injected with TAM at E9.5 and these embryos were sampled at different time points from E12.5 to E15.5 (Fig. 4A). Distal and stalk airway epithelial cells were distinguished based on the expression of Sox9 and Sox2, respectively. The proportions of GFP+ cells in the distal airways (Sox9+) of Anxa4cKO lungs decreased over time as compared to that of control lungs, while the proportions of GFP+ cells in the stalk airways (Sox2+) of Anxa4cKO lungs increased: at E12.5, the proportions of GFP+ cells in the distal airways and stalk airways of Anxa4cKO lungs were similar to that of control lungs; at E13.5, the proportion of GFP+ cells in the distal airways (Sox9+) of Anxa4cKO lungs slightly decreased as compared to that of control lungs; at E14.5 and E15.5, the proportion of GFP+ cells in the distal airways of AnxacKO lungs significantly decreased as compared to that of control lungs (Fig. 4B,C).
Figure 4.
Anxa4 promotes distal airway epithelial cell fate commitment. (A) Pregnant females carrying ShhCreER/+; Anxa4F/+; Rosa26-mTmG (control) and ShhCreER/+; Anxa4F/F; Rosa26-mTmG (Anxa4cKO) embryos were treated with tamoxifen (TAM) at E9.5 and lungs were analyzed at different time point from E12.5 to E15.5. (B,C) Lungs at different time points from E12.5 to E15.5 were double stained for antibodies against GFP (green) and Sox9 (red), or GFP (green) and Sox2 (red) (B). Note that the proportion of GFP+ cells in distal airways (Sox9+) of Anxa4cKO lungs decreased over time as compared to that of control lungs, while the proportion of GFP+ cells in stalk airways (Sox2+) of Anxa4cKO lungs increased over time (C). Data are presented as mean ± S.E.M, n = 4. n.s., not significant; *p < 0.05; **p < 0.01; Student’s t-test. Scale bar: 25 μm. (D,E) GFP (green) and Sox2 (red) staining in cultured whole lung endoderm explants infected with vector or Anxa4OE lentivirus (D). GFP is a reporter used to indicate lentivirus-infected epithelial cells. More GFP+ cells were located in the bud tips (Sox2−) in Anxa4OE lung endoderm explants than in control explants (E). Data are presented as mean ± S.E.M, n = 3. **p < 0.01; Student’s t-test. Scale bar: 25 μm. (F) A model illustrating the effect of loss of Anxa4 on epithelial cell behavior and cell fate. At E11.5, in both control and Anxa4cKO lung, mosaic labeled GFP cells are evenly distributed along the airways. Four days later (E15.5), in newly formed airways of control lung, GFP+ cells are still evenly distributed along the airways; however, in the newly formed airways of Anxa4cKO lung, GFP+ (Anxa4−/−) cells tend to lag behind and contribute to the stalk airways.
We next examined the effect of loss of Anxa4 in lung epithelial cells on cell proliferation and apoptosis using immunostaining against pH3 and Caspase3 in control and ShhCre/+; Anxa4F/F lungs. In ShhCre/+; Anxa4F/F lungs, Anxa4 was knocked out in all airway epithelial cells. The proliferation rate of airway epithelial cells did not differ significantly between littermate control and ShhCre/+; Anxa4F/F lungs (Fig. s4A,B). We did not detect Caspase3+ cells in either control or ShhCre/+; Anxa4F/F lungs (Fig. s4A). The proportions of GFP+ airway epithelial cells to the total airway epithelial cells were similar between control and Anxa4cKO lungs at both E11.5 and E15.5 (Fig. s4C–E), supporting that loss of Anxa4 doesn’t decrease cell proliferation or induce apoptosis. Transwell migration assays with isolated primary lung epithelial cells showed that the migration of Anxa4−/− epithelial cells toward FGF10 was significantly decreased as compared to control epithelial cells (Fig. s4F,G). These experiments demonstrated that loss of Anxa4 decreases distal cell fate commitment and that this decrease is most likely caused by decreased cell migration, as loss of Anxa4 does not affect cell proliferation or apoptosis.
To further investigate the role of Anxa4 in cell fate commitment, we generated an Anxa4-overexpression (Anxa4OE) lentivirus carrying a nuclear H2B-GFP reporter and then co-cultured lung endoderm explants with this lentivirus to overexpress Anxa4. After 48 h co-culture, we did whole-mount immunostaining using antibodies against GFP and Sox2. Our immunostaining results showed that more GFP+ cells remained in the endoderm bud tips (Sox2−) of Anxa4OE endoderm explants as compared to vector-lentivirus infected endoderm explants, while fewer GFP+ cells were detected in the proximal endoderm (Sox2+) of Anxa4OE endoderm explants (Fig. 4D,E). Taken together, our findings show that Anxa4 controls cell migration and promotes distal airway epithelial cell fate specification (Fig. 4F).
Anxa4 functions redundantly with Anxa1 and Anxa6 in regulating endoderm budding process
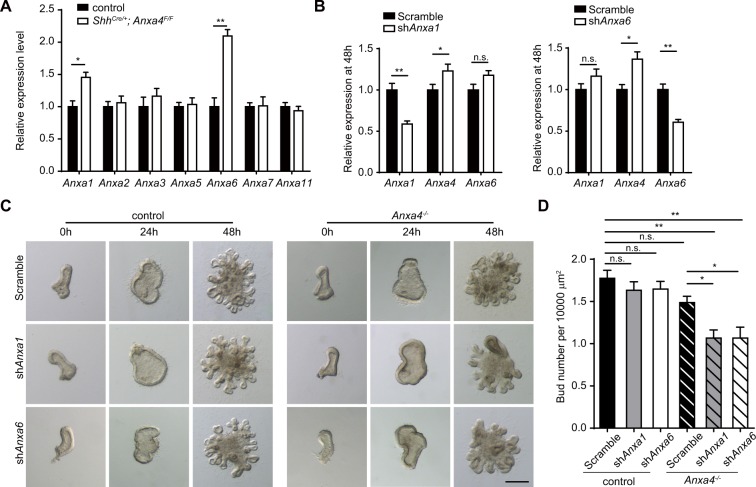
We next used ShhCre/+ mouse line to knockout Anxa4 in all lung epithelial cells to investigate the effect of the loss of Anxa4 on airway branching morphogenesis and found that neither the branching pattern nor the tube shape of ShhCre/+; Anxa4F/F mouse lungs differed significantly from their somite-matched littermate controls (Fig. s3B). We therefore hypothesized that the normal lung development phenotype that we observed in ShhCre/+; Anxa4F/F mice may result from functional redundancy among the Annexins22,24. Similar to a previous study that reported increased expression of other Anxa genes in Anxa1−/− embryos39, we found that the expression levels of Anxa1 and Anxa6 were increased in ShhCre/+; Anxa4F/F mutant lungs (Fig. 5A). It was also notable that the expression levels of Anxa1 and Anxa6 increased significantly between 24 h and 48 h in lung endoderm culture (Fig. s2A).
Figure 5.
Anxa4 functions redundantly with Anxa1 and Anxa6 in regulating endoderm budding process. (A) Relative expression level of Anxa1, Anxa2, Anxa3, Anxa5, Anxa6, Anxa7 and Anxa11 in control and ShhCre/+; Anxa4F/F lungs. (B) The relative expression level of Anxa1, Anxa4 and Anxa6 at 48 h in cultured control lung endoderm treated with Scramble shRNA, or shAnxa1, or shAnxa6. The knockdown efficiency of shAnxa1 or shAnxa6 in cultured lung endoderm explants at 48 h was about 40%. Data are presented as mean ± S.E.M, n = 3. ***p < 0.001; Student’s t-test. (C) Representative images of cultured control or Anxa4−/− lung endoderm explants treated with Scramble shRNA, shAnxa1, or shAnxa6. Scale bar: 200 μm. (D) Quantification of bud number of different lung endoderm groups in (B). Data are presented as mean ± S.E.M, n = 12. n.s., not significant; *p < 0.05; ***p < 0.001; Student’s t-test.
We therefore knocked down either Anxa1 or Anxa6 in control and Anxa4−/− endoderm explants with lentivirus carrying shRNA targeting Anxa1 or Anxa6. The RNAi knockdown efficiency for shAnxa1 and for shAnxa6 in the lung endoderm explants was about 40% at 48 h (Fig. 5B). Note that a slight increase in the Anxa4 expression level was detected in Anxa1 and Anxa6 knock-down endoderm explants (Fig. 5B). In control lung endoderm explants, knockdown of Anxa1 or Anxa6 by lentivirus-mediated RNAi did not impair their budding process (Fig. 5C,D). We also co-cultured Anxa4−/− endoderm explants with Scramble shRNA lentivirus and found that Anxa4−/− endoderm explants showed slight (not significant) decreases in the number of buds as compared with control endoderm explants (Fig. 5C,D). However, in the Anxa4−/− endoderm explants, knockdown of Anxa1 or Anxa6 resulted in significantly fewer buds as compared to Scramble-shRNA-treated control or Scramble-shRNA-treated Anxa4−/− lung endoderm explants (Fig. 5C,D). Immunostaining against pH3 and Caspase3 showed that loss of Anxa1, or Anxa4, or Anxa6 did not impair the relative proportions of pH3+ and Caspase3+ cells in these lung endoderm explants (Fig. s5A,B), suggesting that the impaired endodermal budding process was not caused by impaired cell proliferation or by apoptosis. These experiments indicate that Anxa1, Anxa4 and Anxa6 function redundantly in regulating lung endoderm budding process.
Discussion
Here, by combining live imaging, mouse genetics and mesenchyme-free lung endoderm culture system, we found that airway epithelial progenitor cells that migrate faster are more likely to become Sox9+ distal airway epithelial cells. This process is controlled by FGFR2-mediated ERK1/2 signaling. Using the lung endoderm culture system, we identified Anxa4 as a downstream target of ERK1/2 signaling. We further demonstrated, both in vitro and in vivo, that Anxa4 promotes airway epithelial cell migration. Loss of Anxa4 decreases airway epithelial cell migration during bud formation, while overexpression of Anxa4 promotes cell migration. We also found that Anxa4−/− epithelial cells tend to exhibit a lag-behind behavior and contribute to stalk airways in vivo, suggesting that Anxa4 plays a role in promoting distal cell fate commitment. This lag behind behavior is most likely caused by decreased cell migration, as loss of Anxa4 does not affect cell proliferation or apoptosis.
Attempts have been made to investigate the underlying mechanisms of branching morphogenesis at cellular level over the past decades. Studies combining fluorescent reporters, mouse genetics and live imaging have revealed the dynamics and kinematics of branching morphogenesis in a variety of model organs. These studies have shown that a variety of cellular behaviors, including local proliferation, cell migration, cell invasion, apical constriction, can contribute to branching morphogenesis in different contexts12,14,16,18. FGF10/FGFR2 signaling is essential for cell proliferation and cell migration during lung development6,8,36. Studies have shown that localized cell proliferation is not required for the initiation of bud formation12,13. Other studies have shown that ERK1/2-signaling-controlled cell migration is involved in lung endoderm budding and renal branching morphogenesis17,40. In the present study, we found that cell migration is involved in airway bud formation and that tip cells migrate faster than cleft cells. Further, changes in cell shape are known to accompany cell migration41, we found here that tip cells are more elongated than cleft cells, a result suggesting that we are here observing active cell migration.
A previous study used microarrays to profile the transcriptomes of cultured mesenchyme-free lung endoderm explants with the goal of identifying downstream targets of FGF10/FGFR2 signaling in the lung epithelial cells during branching morphogenesis38. This microarray study implicated approximately 200 genes, including several members of the Anxa family, in the initial stages of bud formation. Consistently, we here found that the expression levels of Anxa family members are increased over time in lung endoderm culture and identified Anxa4 as a downstream target of ERK1/2 signaling. We also showed that loss of Anxa4 impairs airway epithelial cell migration, without affecting cell proliferation.
Mice deficient in Anxa1, Anxa2, Anxa5, or Anxa6 have been generated and used to evaluate the physiological roles of Annexins39,42–44. However, all of the mice strains that lack a single Annexin are viable and exhibit normal development. It has been shown that the expression levels of other members of the Annexin family are altered in the tissues of Anxa1−/− mice, suggesting the existence of reciprocal regulation between Annexin family members and of functional redundancy among Annexins39. Future studies that achieve the deletion of multiple Annexin genes will help to elucidate the precise functions of particular Annexin genes in embryonic lung branching morphogenesis.
Materials and Methods
Mice
The ShhCre/+ 45, ShhCreER/+ 45, Rosa26-mTmG46 and Fgfr2F/F 47 mice have been described previously. All mice experiments were performed in accordance with the guidelines for the use and care of laboratory animals of the National Institute of Biological Sciences, Beijing. The experimental protocol was approved by the National Institute of Biological Sciences, Beijing (protocol number NIBS2012M0017). Mice were housed under standard environmental conditions (20–22 °C, 12–12 hr light–dark cycle) and provided food and water ad libitum. Animals were anesthetized by using Pentobarbital sodium before sacrifice. For live imaging experiments, pregnant females carrying ShhCreER/+; Rosa26-mTmG or ShhCreER/+; Anxa4F/F; Rosa26-mTmG or ShhCreER/+; Fgfr2F/F; Rosa26-mTmG were injected with one dose (30 μg/g) of Tamoxifen at E9.5. For the cell fate lineage tracing experiments, pregnant females were injected with one dose (75 μg/g) of Tamoxifen at E9.5.
Generation and genotyping of Anxa4flox mice
A conditional, replacement-type targeting vector was produced by inserting one LoxP site 210 bp upstream of Anxa4 exon 3. A fragment containing a Loxp-flanked neo-cassette was cloned into a site 244 bp downstream of Anxa4 exon 3. The targeting construct was linearized and electroporated into C57/BL6 ES cells and was then selected with G418 on embryonic fibroblast feeder cells. Recombinant clones containing a floxed Anxa4 gene were identified by PCR using primers P1 (GGTGAACCATCTCTCGTCCTAAGCTCG) and P2 (CTGCTAACACATTCTCCCATCCGTCAC). The targeted clones were injected into C57BL/6J blastocysts, yielding 3 lines of chimeric mice that transmitted the Anxa4flox allele through the germ line.
Time-lapse imaging of embryonic lung explants and imaging analysis
The procedures for ex vivo time-lapse imaging have been described previously48. Briefly, lungs at E12.5 were dissected out and immediately embedded in 0.4% low melting-point agarose (Lonza) dissolved in the culture medium (1% insulin-transferrin-selenium +10 μM vitamin C and 1% penicillin/streptomycin + BGJb media). the culture dish with embedded lung explants were firstly cultured in a cell incubator (5% CO2, 37 °C) for 1 h, then were carried out and placed on a 37 °C heated platform for time-lapse images imaging. Time-lapse imaging was taken with a two-photon microscope (FV1000, Olympus) using a 25× water immersion objective. Imaging stacks of 512 × 512 pixels × 25 optical sections (xyzt sampling: 0.994 × 0.994 × 5 μm × 10 min) were acquired every 10 min for 5 h. For live imaging analysis, the images were opened by Imaris software, cells in the bud tip or cleft were distinguished by the end time point of live imaging: if the cells were in the two newly formed bud tips at the end time point, then we defined them as “tip cells”; if the cells were between the two newly formed bud tips at the end time point, then we defined them as “cleft cells”. We traced the xy positions of GFP+ tip cells and cleft cells at all time-points. All these xy position data were used for analyzed in cell migration trace plot, cell displacement to starting position and cell migration velocity.
E-cadherin whole-mount staining
Lungs were dissected out from mouse embryos fixed with 4% PFA in PBS for 1 h at 4 °C. To facilitate the analysis of branching morphogenesis, whole lungs were stained with E-cadherin to visualize all airway epithelial cells. Whole-mount immunostaining was performed as previously described31.
Lung endoderm isolation and culture
Lung endoderm isolation was performed according to Weaver et al.49. Briefly, lung explants were dissected from ICR mice at E11.5 in HBSS, washed three times in Tyrode-Ringer’s solution and then incubated in a pancreatin-trypsin solution for 5 min on ice. Lung endoderm explants were isolated from the mesenchyme using tungsten needles in DMEM/F12 media with 10% fetal bovine serum. The distal lung endoderm buds were cut off and embedded in 50% growth factor reduced Matrigel (Corning) and cultured in DMEM/F12 media supplemented with 800 ng/ml human recombinant FGF10 (Sino Biological). In some experiments, a MEK inhibitor (PD0325901, 1 μM, Selleck) was added at 1 h after culture initiation. In shRNA knockdown or Anxa4-overexpressing experiments, the lung endoderm was co-cultured with lentivirus added in 50% Matrigel and culture media at the time of initial culturing.
Lentivirus production and concentration
Lentiviral shRNA plasmids corresponding to Anxa1, a4 and a6 were obtained from Sigma (Mission TRC-Mm 1.5). The corresponding TRC IDs are: Anxa1 (TRCN0000109725, TRCN0000109728), Anxa4 (TRCN0000110706, TRCN0000110709), Anxa6 (TRCN0000110650, TRCN0000110653), hAnxa4 (TRCN0000310693, TRCN0000056278). For Anxa4-overexpressing lentivirus, Anxa4 was cloned into modified pCDH-CMV-MCS-PGK-H2BGFP plasmid. Lentivirus was produced by co-transfecting HEK293ft cells with a transfer vector together with the packaging vector (psPAX2) and the VSV-G envelope protein vector (pMD2.G). The supernatant containing the virus was harvested 48 h after transfection and was filtered through a 0.45 μm filter. The virus was concentrated by centrifugation at 70,000 × g for 2 h at 20 °C. The titer of the viral supernatant was determined using serial dilutions to infect NIH 3T3 cells.
Quantitative real-time PCR
Total RNA from lung endoderm or whole lung samples was extracted using Trizol reagent (Ambion) and Direct-Zol RNA Miniprep kits (ZYMO research). RNA was reverse transcribed into first-strand complementary DNA using HiScript II Q RT SuperMix (Vazyme). The quantitative real-time PCR analysis was performed on a CFX96 Tough instrument (Bio-Rad) using KAPA SYBR FAST qPCR Master Mix (KAPA). The sequences of the primers that were used in the real-time PCR analysis are: Anxa1-F: AGCTTTCCTCATCTTCGCAG; Anxa1-R: TGGCACACTTCACGATGG; Anxa2-F: GTCTACTGTCCACGAAATCCTG; Anxa2-R: ACTCCTTTGGTCTTGACTGC; Anxa3-F: CTCAGATCCTCTATAATGCTGGTG; Anxa3-R: TGCTGTCCTCAATGTCCTTC; Anxa4-F: GAACTGTATGAGGCTGGAGAG; Anxa4-R: TGCTCTGTTCAATGTCCTTCTG; Anxa5-F: CGAATAGAGACCCTGATACTGC; Anxa5-R: ACTGCGTGTCCCAAAGATG; Anxa6-F: AGAGTGTCAAGAACAAGCCTC; Anxa6-R: TTCTCAATGAATTCCCTCCGG; Anxa7-F: TCAGATACCTCGGGACATTTTG; Anxa7-R: GATTCATCCGTTCCCAGTCTC; Anxa11-F: AATGCCTCAAGAACACCCC; Anxa11-R: CATCCGCTTATACTCTGCTCG; GAPDH-F: AAGGTCGGTGTGAACGGATTTGG; GAPDH-R: CGTTGAATTTGCCGTGAGTGGAG.
Immunostaining and western blotting
Whole lung was dissected out and fixed in 4% PFA in PBS for 4 hours at 4 °C. Cultured lung endoderm explants were fixed with 4% PFA in PBS for 20 min at RT. After fixation, samples were washed twice with PBS, immersed in 30% sucrose and then embedded in OCT for cryosectioning. The primary and secondary antibodies and dilutions used were as follows: Rabbit anti-pH3 (1:250, 06–570, Millipore), Chicken anti-GFP (1:500, ab13970, Abcam), Rat anti-E-cadherin (1:100, 13–1900, Invitrogen), Rabbit anti-Caspase3 (#9664 s, CST), Rabbit anti-pERK1/2 (4370 s, CST), Goat anti-Sox2 (1:100, sc-17320, Santa Cruz); Alexa Fluor 488-Donkey anti-goat (Jackson Immuno Research), Alexa Fluor Cy3-Donkey anti-rabbit (Jackson Immuno Research), Alexa Fluor 488-Donkey anti-chicken (Jackson Immuno Research), Alexa Fluor 568-Donkey anti-rat (Jackson Immuno Research). Immunofluorescence images were taken by Lecia TCS LSI confocal. For western blotting, lungs were lysed on ice in RIPA buffer with a protease inhibitor cocktail (Roche) and PhosSTOP (Roche). The proteins were resolved via 4–20% gradient SDS-PAGE. Proteins were transferred to a PVDF membrane and incubated overnight at 4 °C and were then incubated with corresponding secondary antibody: HRP-Donkey anti-Rabbit (Jackson Immuno Research) and HRP-Donkey anti-mouse (Jackson Immuno Research), at RT for 1 h. Finally, proteins were visualized using LumiGLO chemiluminescent substrate (CST). The primary antibodies and dilutions used were as follows: rabbit anti-p-ERK/12 (1:2500, #4370, CST); rabbit anti-ERK/12 (1:1000, #9102, CST); rabbit anti-Anxa4 (1:1000, 10087–1-AP, ProteinTech); mouse anti-b-Actin (1:2500, A2228, Sigma).
Whole-mount in situ hybridization
Lungs were dissected out quickly and fixed with 4% PFA in PBS for 1 h at 4 °C, washed three times with DEPC-PBS and then dehydrated through a graded methanol series in DEPC-PBS (25%, 50%, 75%, 100%, 100%) and stored at −20 °C. Whole-mount in situ hybridization was performed as described previously50. The DNA templates used for DIG-labeled probe synthesis were generated by PCR with the primers containing either the T7 or the T3 promoter. The primer sequences are: Anxa1 5′ primer: AATTAACCCTCACTAAAGGCCCTACCCTTCCTTCAATGTATC; Anxa1 3′ primer: TAATACGACTCACTATAGGCACAGAGCCACCAGGATTT; Anxa4 5′ primer: AATTAACCCTCACTAAAGGCAGAGGAACTTCTGCAGGTG; Anxa4 3′ primer: TAATACGACTCACTATAGGCAGTACTCGCTGGAACATGAA; Anxa6 5′ primer: AATTAACCCTCACTAAAGGCAGGAAGATGCCCAGGAAATAG; Anxa6 3′ primer: TAATACGACTCACTATAGGCTCAAGTCAGGCAGGGTTATG. PCR products were purified and used as templates for probe labeling. DIG-labeled RNA probes were generated using the appropriate RNA polymerase, as described in the manufacturer’s manual (Roche). The quality of labeled RNA probes was assessed by agarose gel electrophoresis. The RNA density was quantified by the ImageJ software.
Isolation of primary lung epithelial cells and Transwell migration assays
The procedures for primary lung epithelial cell isolation have been described previously51. Briefly, after removing the heart and trachea, E14.5 lung lobes were minced into very small pieces and digested in DMEM media with neutral protease (Worthington) and DNase I (Roche) at 37 °C for 20 min. After stopping digestion and passing cells through 40 μm filter, we next centrifuged samples for 10 min at 350 × g, re-suspended the cell pellet in RBC lysis buffer (BD Biosciences) to remove red blood cells and centrifuged samples again for 10 min at 350 × g. The cell pellet was suspended with DMEM/F12 plus 10% FBS and cultured in 35 mm dish for 1.5 hours. The supernatant was collected and centrifuged for 3 min at 350 × g to obtain purified primary lung epithelial cells, which were suspended with DMEM/F12 plus 1% FBS and quantified (cell number) using a hemocytometer. For the Transwell® migration assays, 5 × 104 cells in 200 μl were seeded into the upper well of the Transwell® apparatus (6.5 mm diameter, 8 μm pore size, Corning Costar); 600 μl of DMEM/F12 medium supplemented with 400 ng/ml human recombinant FGF10 was added into the lower chamber. Following incubation for 48 hours at 37 °C to allow cell migration, cells were fixed with 4% PFA and perform crystal violet staining.
Electronic supplementary material
Author Contributions
K.J. and N.T. conceived and designed research; K.J. and Z.T. performed experiments; K.J. and N.T. performed data analysis and interpretation; F.W. generated the Anxa4 flox mouse line; J.L. provided technical support; K.J. and N.T. wrote the paper. All authors reviewed this manuscript.
Data Availability
All data generated and analyzed during this study are either included in this publication and the Supplementary Information, or available from the corresponding author upon reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-32494-z.
References
- 1.Cardoso WV, Lu J. Regulation of early lung morphogenesis: questions, facts and controversies. Development. 2006;133:1611–1624. doi: 10.1242/dev.02310. [DOI] [PubMed] [Google Scholar]
- 2.Morrisey EE, Hogan BL. Preparing for the first breath: genetic and cellular mechanisms in lung development. Developmental cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Min H, et al. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes & development. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sekine K, et al. Fgf10 is essential for limb and lung formation. Nature genetics. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- 5.Arman E, Haffner-Krausz R, Gorivodsky M, Lonai P. Fgfr2 is required for limb outgrowth and lung-branching morphogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:11895–11899. doi: 10.1073/pnas.96.21.11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- 7.Goetz R, Mohammadi M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nature reviews. Molecular cell biology. 2013;14:166–180. doi: 10.1038/nrm3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park WY, Miranda B, Lebeche D, Hashimoto G, Cardoso WV. FGF-10 is a chemotactic factor for distal epithelial buds during lung development. Developmental biology. 1998;201:125–134. doi: 10.1006/dbio.1998.8994. [DOI] [PubMed] [Google Scholar]
- 9.Weaver M, Dunn NR, Hogan BL. Bmp4 and Fgf10 play opposing roles during lung bud morphogenesis. Development. 2000;127:2695–2704. doi: 10.1242/dev.127.12.2695. [DOI] [PubMed] [Google Scholar]
- 10.Ochoa-Espinosa A., Affolter M. Branching Morphogenesis: From Cells to Organs and Back. Cold Spring Harbor Perspectives in Biology. 2012;4(10):a008243–a008243. doi: 10.1101/cshperspect.a008243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Affolter M, Zeller R, Caussinus E. Tissue remodelling through branching morphogenesis. Nature reviews. Molecular cell biology. 2009;10:831–842. doi: 10.1038/nrm2797. [DOI] [PubMed] [Google Scholar]
- 12.Kim HY, Varner VD, Nelson CM. Apical constriction initiates new bud formation during monopodial branching of the embryonic chicken lung. Development. 2013;140:3146–3155. doi: 10.1242/dev.093682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nogawa H, Morita K, Cardoso WV. Bud formation precedes the appearance of differential cell proliferation during branching morphogenesis of mouse lung epithelium in vitro. Developmental dynamics: an official publication of the American Association of Anatomists. 1998;213:228–235. doi: 10.1002/(SICI)1097-0177. [DOI] [PubMed] [Google Scholar]
- 14.Ghabrial AS, Krasnow MA. Social interactions among epithelial cells during tracheal branching morphogenesis. Nature. 2006;441:746–749. doi: 10.1038/nature04829. [DOI] [PubMed] [Google Scholar]
- 15.Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Developmental cell. 2008;14:570–581. doi: 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huebner RJ, Neumann NM, Ewald AJ. Mammary epithelial tubes elongate through MAPK-dependent coordination of cell migration. Development. 2016;143:983–993. doi: 10.1242/dev.127944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riccio P, Cebrian C, Zong H, Hippenmeyer S, Costantini F. Ret and Etv4 Promote Directed Movements of Progenitor Cells during Renal Branching Morphogenesis. PLoS biology. 2016;14:e1002382. doi: 10.1371/journal.pbio.1002382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chi X, et al. Ret-dependent cell rearrangements in the Wolffian duct epithelium initiate ureteric bud morphogenesis. Developmental cell. 2009;17:199–209. doi: 10.1016/j.devcel.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rawlins EL, Clark CP, Xue Y, Hogan BL. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development. 2009;136:3741–3745. doi: 10.1242/dev.037317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mucenski ML, et al. beta-Catenin is required for specification of proximal/distal cell fate during lung morphogenesis. The Journal of biological chemistry. 2003;278:40231–40238. doi: 10.1074/jbc.M305892200. [DOI] [PubMed] [Google Scholar]
- 21.Volckaert T, et al. Localized Fgf10 expression is not required for lung branching morphogenesis but prevents differentiation of epithelial progenitors. Development. 2013;140:3731–3742. doi: 10.1242/dev.096560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerke V, Moss SE. Annexins: from structure to function. Physiological reviews. 2002;82:331–371. doi: 10.1152/physrev.00030.2001. [DOI] [PubMed] [Google Scholar]
- 23.Rescher U, Gerke V. Annexins–unique membrane binding proteins with diverse functions. Journal of cell science. 2004;117:2631–2639. doi: 10.1242/jcs.01245. [DOI] [PubMed] [Google Scholar]
- 24.Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nature reviews. Molecular cell biology. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- 25.Patel DM, Ahmad SF, Weiss DG, Gerke V, Kuznetsov SA. Annexin A1 is a new functional linker between actin filaments and phagosomes during phagocytosis. Journal of cell science. 2011;124:578–588. doi: 10.1242/jcs.076208. [DOI] [PubMed] [Google Scholar]
- 26.Gabel M, et al. Annexin A2-dependent actin bundling promotes secretory granule docking to the plasma membrane and exocytosis. The Journal of cell biology. 2015;210:785–800. doi: 10.1083/jcb.201412030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Babiychuk EB, Palstra RJ, Schaller J, Kampfer U, Draeger A. Annexin VI participates in the formation of a reversible, membrane-cytoskeleton complex in smooth muscle cells. The Journal of biological chemistry. 1999;274:35191–35195. doi: 10.1074/jbc.274.49.35191. [DOI] [PubMed] [Google Scholar]
- 28.Hayes MJ, Rescher U, Gerke V, Moss SE. Annexin-actin interactions. Traffic. 2004;5:571–576. doi: 10.1111/j.1600-0854.2004.00210.x. [DOI] [PubMed] [Google Scholar]
- 29.Mogami T, et al. Annexin A4 is involved in proliferation, chemo-resistance and migration and invasion in ovarian clear cell adenocarcinoma cells. PloS one. 2013;8:e80359. doi: 10.1371/journal.pone.0080359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Melero A, et al. Annexin A6 and Late Endosomal Cholesterol Modulate Integrin Recycling and Cell Migration. The Journal of biological chemistry. 2016;291:1320–1335. doi: 10.1074/jbc.M115.683557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metzger RJ, Klein OD, Martin GR, Krasnow MA. The branching programme of mouse lung development. Nature. 2008;453:745–750. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nature reviews. Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 33.Li J, et al. The Strength of Mechanical Forces Determines the Differentiation of Alveolar Epithelial Cells. Developmental cell. 2018;44:297–312 e295. doi: 10.1016/j.devcel.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 34.Costantini F, Shakya R. GDNF/Ret signaling and the development of the kidney. BioEssays: news and reviews in molecular, cellular and developmental biology. 2006;28:117–127. doi: 10.1002/bies.20357. [DOI] [PubMed] [Google Scholar]
- 35.Costantini F. Renal branching morphogenesis: concepts, questions and recent advances. Differentiation; research in biological diversity. 2006;74:402–421. doi: 10.1111/j.1432-0436.2006.00106.x. [DOI] [PubMed] [Google Scholar]
- 36.Abler LL, Mansour SL, Sun X. Conditional gene inactivation reveals roles for Fgf10 and Fgfr2 in establishing a normal pattern of epithelial branching in the mouse lung. Developmental dynamics: an official publication of the American Association of Anatomists. 2009;238:1999–2013. doi: 10.1002/dvdy.22032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lizarbe MA, Barrasa JI, Olmo N, Gavilanes F, Turnay J. Annexin-phospholipid interactions. Functional implications. International journal of molecular sciences. 2013;14:2652–2683. doi: 10.3390/ijms14022652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu J, Izvolsky KI, Qian J, Cardoso WV. Identification of FGF10 targets in the embryonic lung epithelium during bud morphogenesis. The Journal of biological chemistry. 2005;280:4834–4841. doi: 10.1074/jbc.M410714200. [DOI] [PubMed] [Google Scholar]
- 39.Hannon R, et al. Aberrant inflammation and resistance to glucocorticoids in annexin 1−/− mouse. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2003;17:253–255. doi: 10.1096/fj.02-0239fje. [DOI] [PubMed] [Google Scholar]
- 40.Xing Y, Wang R, Li C, Minoo P. PTEN regulates lung endodermal morphogenesis through MEK/ERK pathway. Developmental biology. 2015;408:56–65. doi: 10.1016/j.ydbio.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. The Journal of cell biology. 2010;188:11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brachvogel B, et al. Annexin A5 is not essential for skeletal development. Molecular and cellular biology. 2003;23:2907–2913. doi: 10.1128/MCB.23.8.2907-2913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ling Q, et al. Annexin II regulates fibrin homeostasis and neoangiogenesis in vivo. The Journal of clinical investigation. 2004;113:38–48. doi: 10.1172/JCI19684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hawkins TE, Roes J, Rees D, Monkhouse J, Moss SE. Immunological development and cardiovascular function are normal in annexin VI null mutant mice. Molecular and cellular biology. 1999;19:8028–8032. doi: 10.1128/MCB.19.12.8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harfe BD, et al. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 46.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 47.Yu K, et al. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–3074. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- 48.Tang Z, et al. Mechanical Forces Program the Orientation of Cell Division during Airway Tube Morphogenesis. Developmental cell. 2018;44:313–325 e315. doi: 10.1016/j.devcel.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 49.Weaver M, Batts L, Hogan BL. Tissue interactions pattern the mesenchyme of the embryonic mouse lung. Developmental biology. 2003;258:169–184. doi: 10.1016/S0012-1606(03)00117-9. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y, Jiang H, Crawford HC, Hogan BL. Role for ETS domain transcription factors Pea3/Erm in mouse lung development. Developmental biology. 2003;261:10–24. doi: 10.1016/S0012-1606(03)00359-2. [DOI] [PubMed] [Google Scholar]
- 51.Lebeche D, Malpel S, Cardoso WV. Fibroblast growth factor interactions in the developing lung. Mechanisms of development. 1999;86:125–136. doi: 10.1016/S0925-4773(99)00124-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and analyzed during this study are either included in this publication and the Supplementary Information, or available from the corresponding author upon reasonable request.