Abstract
The gut microbial community (Gut microbiota) is known to impact metabolic functions as well as immune responses in our body. Diet plays an important role in determining the composition of the gut microbiota. Gut microbes help in assimilating dietary nutrients which are indigestible by humans. The metabolites produced by them not only modulate gastro-intestinal immunity, but also impact distal organs like lung and brain. Micro-aspiration of gut bacteria or movement of sensitized immune cells through lymph or bloodstream can also influence immune response of other organs. Dysbiosis in gut microbiota has been implicated in several lung diseases, including allergy, asthma and cystic fibrosis. The bi-directional cross-talk between gut and lung (termed as Gut-Lung axis) is best exemplified by intestinal disturbances observed in lung diseases. Some of the existing probiotics show beneficial effects on lung health. A deeper understanding of the gut microbiome which comprises of all the genetic material within the gut microbiota and its role in respiratory disorders is likely to help in designing appropriate probiotic cocktails for therapeutic applications.
Keywords: gut-lung axis, microbiome, lung immunity, gut microbiota, lung microbiota, diet, short chain fatty acids, SCFA
Introduction
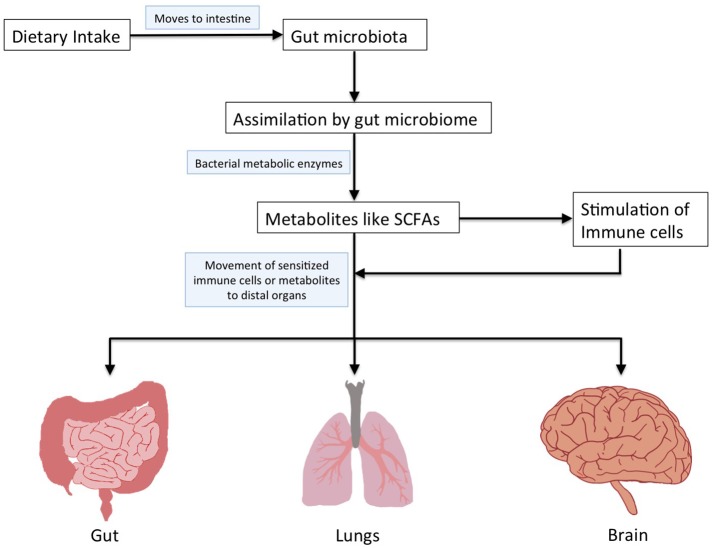
The human microbiota is constituted by the microbial communities residing in various body sites (gut, skin, mouth, airways, and vagina; Cho and Blaser, 2012). Majority of these microbes are present in the gastrointestinal tract (gut) and are important in maintaining our health (Shreiner et al., 2015). Dysbiosis within this homeostasis in the gut microbiota is associated with a multitude of health conditions affecting not only gut, but also distal organs like mouth, lung, brain, liver, vagina, etc., (Shreiner et al., 2015). The gut microbial community possesses enzymatic machinery for assimilating a variety of dietary nutrients leading to release of metabolites having multiple functions in the host (Albenberg and Wu, 2014; Figure 1). It is important to understand the influence of gut microbiota and metabolites produced by them on the functioning of various organs within the body (Figure 1). The gut microbiota affects other organs either by aspiration or produces metabolites that bring about changes when they reach other organs (Marsland et al., 2015; Bingula et al., 2017).
Figure 1.
Schematic representation to depict assimilation of dietary nutrients by gut microbiome and their impact on distal organs.
Scientific findings indicate a link between gut microbiota and lung immunity (Marsland et al., 2015; Bingula et al., 2017). In this review, we focus on the recent developments related to influence of diet on this “Gut-Lung axis” as well as discuss the impact of different types of diet on gut microbiota and how they influence lung immunity as well as lung diseases.
Diet and gut microbiota
A nutrient-rich diet augmented with dietary fibers is associated with a more diversified gut microbiota which is beneficial for the host (Simpson and Campbell, 2015; Xu and Knight, 2015). The metabolites produced by gut microbiome during assimilation of food significantly influence human health (Russell et al., 2013b; Joyce and Gahan, 2014; Sharon et al., 2014; Wu, 2016). The association between dysbiosis in gut microbiota and malnutrition (Gupta et al., 2011; Smith et al., 2013; Ghosh et al., 2014; De Clercq et al., 2016; Million et al., 2017) further demonstrates a link between diet and microbiota. Apart from intestinal disorders (diabetes, obesity, colon cancer, inflammatory bowel disease, etc.), changes in dietary patterns and their effects on gut microbiota are implicated in disorders of other organs like lung (asthma, COPD, etc.; Fujimura and Lynch, 2015; Panzer and Lynch, 2015) and brain (Alzheimers, depression, etc.; Dash et al., 2015; Köhler et al., 2016; Pistollato et al., 2016). The influence of dietary components on gut microbiota and associated physiological changes has been discussed in Table 1.
Table 1.
Influence of various types of dietary components on gut microbiome and associated physiological changes.
| Dietary component | Effect on gut bacteria | Metabolic changes by gut bacteria | Effects on health (Lung diseases which are associated with dietary intake have been highlighted in bold-italic font) | References (PMIDs) |
|---|---|---|---|---|
| PROTEINS | ||||
| Plant proteins (Whey and pea protein) |
|
Increased SCFA levels |
|
29311977 26463271 21276631 |
| Animal proteins |
|
Reduced SCFA levels |
|
29311977 29534465 20679230 22055893 28388917 |
| High protein/carbohydrate ratio |
|
Less fecal butyrate |
|
21389180 29534465 |
| CARBOHYDRATES | ||||
| Digestible carbohydrates (lactose) |
|
Increased SCFA levels |
|
22435727 28388917 22435727 |
| Non-digestible carbohydrates (Wheat bran) |
|
Increased SCFA levels |
|
26636660 29416529 |
| Non-digestible carbohydrates: Dietary Fiber (DF), Resistant starch (RS) and whole grain barley |
|
Increased fecal butyrate |
|
29042495 24440361 18063592 |
| Non-digestible carbohydrates Fructooligosaccharides (FOS), Galactooligosaccharides (GOS) and polydextrose |
|
Increased fecal butyrate |
|
23951074 25849971 27523186 |
| FATS | ||||
| Saturated fat (lard fat) |
|
Reduced SCFA levels |
|
26321659 28804483 28388917 |
| Unsaturated fat (fish oil) |
|
Increased fecal butyrate |
|
28388917 26784651 28951525 29215589 |
| SPECIFIC DIET REGIMES | ||||
| Western: High in animal protein and saturated and trans fat, Low in fiber |
|
Reduced SCFA levels |
|
29276171 26011307 28388917 |
| Gluten free |
|
Increased fecal butyrate |
|
27102333 25651995 |
| Mediterranean: Beneficial mono-unsaturated and poly-unsaturated fatty acids, high levels of polyphenols and other antioxidants, high fiber and low glycemic carbohydrates, more vegetables less animal protein |
|
Higher SCFA levels |
|
28789729 24987952 26416813 |
The role of diet in determining gut microbiota has been studied in different stages of life. Although the colonization of microbes in the gut begins at birth, their configuration keeps changing until a stable state is attained which is closer to the adult microbiota (Bergström et al., 2014; Backhed et al., 2015; Shukla et al., 2017). The composition of gut microbiota in early stages is determined by various factors, including mode of delivery and feeding, antibiotic exposure and surrounding environment. For example, the taxonomic composition of gut microbiota in breast-fed babies (higher Bifidobacteria, Lactobacilli, Staphylococci, and Streptococci) considerably differs from infants who are formula-fed (higher Bacteroides, Clostridia, and Proteobacteria; Timmerman et al., 2017). The gut microbiota in infants becomes stable after solid food diet is introduced, after which the composition is largely determined by dietary intake (Timmerman et al., 2017).
The gut microbiota of younger adults highly depends on their diet and lifestyle (Turnbaugh et al., 2009). As compared to individuals consuming Agrarian diet (low fat, higher plant products), those on Western diet (high fat and sugars) show significant decrease in Bacteroidetes (De Filippo et al., 2010). Children from rural Africa (having diet high in plant polysaccharide/protein and fiber; less animal protein) have higher levels of Actinobacteria and Bacteroidetes as against Firmicutes and Proteobacteria which occur in higher numbers in Western European children (diet protein, sugar, fat and low on fiber; De Filippo et al., 2010). The amount of short chain fatty acids (SCFAs) is about four times higher in African population with higher abundances of certain bacteria (Xylanibacter, Prevotella, Butyrivibrio, and Treponema) that are capable of digesting plant polysaccharides to yield SCFAs (De Filippo et al., 2010). The increased SCFA production by gut microbiome leads to lowering of pH in their gut, thereby reducing the pathogenic species like Escherichia coli and Enterobacteriaceae (Zimmer et al., 2012; Glick-Bauer and Yeh, 2014).
The later stages of life have also been associated with changes in gut microbiota and might be attributed to factors like lifestyle, diet, immune strength, altered gut morphology and physiology, frequent infections, hospitalizations, medication, etc., (Vemuri et al., 2018). The diversity of gut microbiota reduces with aging, but the associated changes may vary across different geographies. Although it is unclear whether these changes are a cause or an effect of aging, the maintenance of gut homeostasis might be crucial for longevity. Apart from the taxonomic changes, aging has been associated with a reduced metabolic potential of microbiome, including less production of SCFAs (Kumar et al., 2016; Nagpal et al., 2018). These changes could be related to changes in dietary pattern with age. It has been speculated that the metabolites produced by gut microbiota might have an effect on longevity (Kumar et al., 2016; Nagpal et al., 2018).
The above-mentioned studies establish that differences in diet play an important role in determining the composition of the gut microbiota. The physiological impact of changes in gut microbiota with variation in dietary patterns is governed by differences in microbial metabolome profiles. In vitro and in vivo studies have shown that food components are utilized by gut bacteria to biosynthesize metabolites (like SCFAs from soluble fibers), which modulate the host immune system (Kosiewicz et al., 2011; Maslowski and Mackay, 2011; Tilg and Moschen, 2015; Tomkovich and Jobin, 2016). Thus, the influence of diet on intestinal microbial dynamics suggests “Diet-Microbiota-Immunity” link and delineates their role in maintenance of health (Kosiewicz et al., 2011; Maslowski and Mackay, 2011; Tilg and Moschen, 2015; Tomkovich and Jobin, 2016).
Roles of short chain fatty acids (SCFAs) produced by gut microbiota
SCFAs (acetate, propionate, and butyrate) promote recruitment as well as maturation of immune cells which provide protection against inflammatory response (Vinolo et al., 2011). They influence host metabolic activities by serving as a link between dietary fiber, commensal microbes and host (Ha et al., 2014; Kasubuchi et al., 2015; Yang and Duan, 2018). SCFAs produced in the gut are distributed systemically and are either utilized to provide energy or as signaling molecules (Morrison and Preston, 2016).
Energy and metabolism
A large part of SCFAs produced in the gut are utilized as energy source and can provide as high as ~10% of our daily caloric requirement (Bergman, 1990). Such roles of SCFAs thus indicate their association with metabolic disorders like obesity and diabetes (Cho et al., 2013; Chen et al., 2014; Ríos-Covián et al., 2016). In vivo and in vitro studies in mice have further shown that butyrate and propionate might induce hormone production by gut bacteria (Lin et al., 2012; Tolhurst et al., 2012). The SCFAs bind to G-protein coupled receptors (GPCRs; Den Besten et al., 2013; Sun et al., 2017) which sense metabolites and trigger signaling pathways leading to anti-inflammatory effects on the host immune responses (Sun et al., 2017).
Integrity of gut epithelium
The SCFAs in gut decrease the luminal pH, thereby curtailing growth of pathogenic microorganisms. Acetate production by Bifidobacteria has been observed to inhibit growth of enteropathogens in mice (Fukuda et al., 2011, 2012). Moreover, in vitro and in vivo studies have indicated that higher butyrate levels lead to increased mucin production which reduces bacterial adhesion and improves epithelial integrity (Jung et al., 2015; Jiminez et al., 2017).
Systemic immunity
Studies on mice have shown that butyrate production leads to an increase in the number of Treg cells. In vitro studies further established the role of butyrate in regulation of forkhead transcription factor FOXP3 (a lineage specification factor). Butyrate works as an inhibitor of histone deacetylases (HDAC) and promotes acetylation of histone H3 at the promoter of the gene encoding FOXP3 (Zeng and Chi, 2015; Kespohl et al., 2017; Supplementary Figure S1). SCFAs also influence the development of dendritic cells and inflammatory cytokines, thereby regulating the intestinal macrophages (Chauvistré et al., 2014; Supplementary Figure S1). Multiple roles of SCFAs in energy and metabolism support activation/differentiation of B cells as well as production of antibodies (IgM/IgA; Supplementary Figure S1; Kim, 2016; Kim et al., 2016).
Gut-lung axis
Dysbiosis in gut microbiota is associated with lung disorders and respiratory infections (Trompette et al., 2014; Shukla et al., 2017). For example, reduction in genus Bifidobacteria and increase in Clostridia in the intestine are associated with asthma in early life (Kalliomaki et al., 2001). Furthermore, murine studies show that depletion of certain species within the gut microbiota due to antibiotic intake influences lung diseases and allergic inflammation (Russell et al., 2013a; Dharmage et al., 2015; Metsälä et al., 2015). For instance, studies on mouse models indicate that removal of sensitive gut bacteria after administration of neomycin leads to an increase in the susceptibility to influenza virus infection in lungs (Ichinohe et al., 2011; Looft and Allen, 2012).
The changes in lung microbial community also influence the composition of gut microbiota. For example, influenza virus infection in the respiratory tract (in mice models) increases Enterobacteriaceae as well reduces Lactobacilli and Lactococci in the intestinal microbiota (Looft and Allen, 2012). The dysbiosis in lung microbiota upon administration of Lipopolysaccharide (LPS) in mice is accompanied by disturbances in their gut microbiota due to movement of bacteria from their lung into the bloodstream (Sze et al., 2014).
All the above-mentioned findings corroborate that gut and lung are intricately linked organs which influence each others' homeostasis.
Gut microbiota influence lung immune response
Perturbation of the normal gut microbiota may be associated with development of experimental asthma and other respiratory disorders, as demonstrated in animal models (Noverr et al., 2004). Evidence is increasing in support of a “common mucosal response,” which states that the effects of gut microbiota on the mucosal immunity may have an influence on the immune responses at distal mucosal sites, including that in lung (Mestecky, 1987; McGhee and Fujihashi, 2012; Hauptmann and Schaible, 2016; Date et al., 2017; Ipci et al., 2017). Further, gut bacterial cells as well as metabolites biosynthesized by them might stimulate the immune response in distal sites (Hauptmann and Schaible, 2016).
Migration of immune cells from gut to lung by common mucosal immune system (MIS)
The Mucosal Immune System (MIS) constitutes inductive as well as effector sites which are defined on the basis of their functional properties and anatomy. The immune cells migrate from these mucosal inductive to effector sites through the lymphatic system (McGhee and Fujihashi, 2012). This transfer of cells determines the immune response in different organs (gastro-intestinal tract, lung, etc.,). Mucosal inductive sites form mucosa-associated lymphoid tissue (MALT) that comprises of gut-associated lymphoid tissues (GALT) and nasopharyngeal-associated lymphoid tissues (NALT; McGhee and Fujihashi, 2012). The GALT consists of organized lymphoid tissues (mesenteric lymph nodes and Peyer patches) which work as inductive sites. It also possesses more diffusely scattered effector sites in the intestinal lamina and epithelium (Bekiaris et al., 2014; Brugman et al., 2015). The MALT is covered by microfold (M) cells which take up antigens present in the lumen of intestinal mucosa and transfer them to Dendritic cells (DCs) in the subepithelial regions (Zarzaur and Kudsk, 2001; Azzali, 2003; Cesta, 2006). These DCs carry antigens into inductive sites (Peyer's patch or mesenteric lymph nodes) where they bring about the initiation of mucosal T and B cell responses (McGhee and Fujihashi, 2012; Samuelson et al., 2015). The MALT possesses regions rich in T cells as well as areas populated by B cells harboring surface IgA-positive (sIgA+) B cells in significant numbers. Recent findings indicate that GALT-DCs can act as “antigen presenting cells” (APCs), thereby influencing differentiation and antibody secretion in B cells (Qi et al., 2006). These newly generated IgA-producing cells in inductive sites move out from GALT into the bloodstream and home to the intestinal effector site lamina propria where they act as defense against intestinal pathogens. The mucosal effector sites, including the lamina propria regions of the gastrointestinal tract, comprise of IgA-producing plasma cells and memory B and T cells. The CD4+ T helper cells (both CD4+ Th2 or CD4+ Th1 cells) support the development of IgA- producing plasma cells (McGhee and Fujihashi, 2012).
After stimulation by antigens (like gut pathogens) targeting the mucosa, plasma cells produce dimeric immunoglobulin A (IgA; with specificities toward antigens in mucosal tissues) which are secreted by epithelia expressing polymeric Ig receptor (IgR; Lycke and Bemark, 2012; Rios et al., 2016). The presence of commensals in gut is critical for controlling the induction and functioning of IgA. For example, IgA positive plasma cells are in lesser numbers in neonates and germ-free (GF) animals (Macpherson et al., 2005; Harris et al., 2006). The immunity and IgA production enhances after colonization by gut bacteria. The T and B cells, induced in Peyers patches, can move into the circulation and migrate to intestinal as well as extra-intestinal sites (including bronchial epithelium and lymphoid tissues; Matsuno et al., 2010). Such B-cells produce IgA which can be transported to the mucosal surface, thereby passing “immunological information” between different organs (Roth et al., 2014; Bingula et al., 2017). Thus, the lymph and/or bloodstream act as a link between the gut (where primary sensitization occurs) and the affected site on the lung (He et al., 2017; Figure 2).
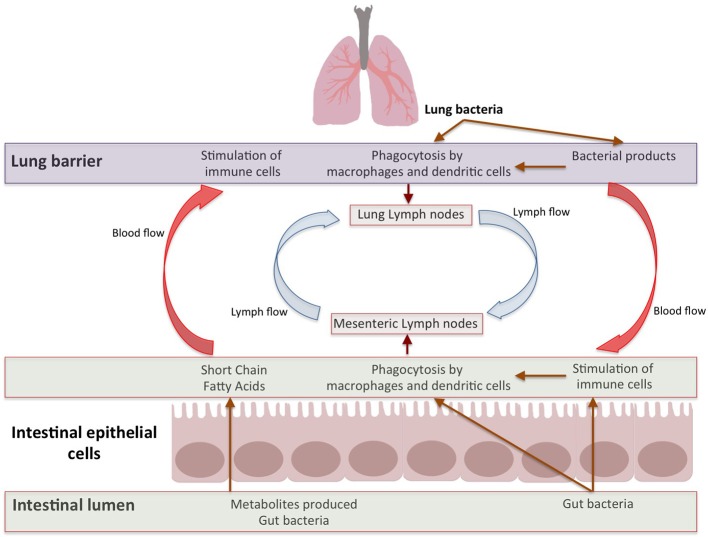
Figure 2.
Bidirectional Gut-Lung axis. The metabolites like SCFAs produced by gut bacteria move through blood stream to stimulate immune response in lung and different factors from lung effect gut immune response. Apart from metabolites, the immune cells induced by multitude of antigens move through lymphatic duct between both these organs which leads to modulation of immune response in both organs.
Gut bacterial cells or metabolites biosynthesized by them influence lung immunity
The immune cells which reside in the intestinal lamina propria and mesenteric lymph nodes can neutralize most of the translocating bacteria (Gross et al., 2015; Bingula et al., 2017). The surviving bacteria as well as the fragments of dead bacteria travel from the mesenteric lymphatic system and enter systemic circulation (Bingula et al., 2017). Further, several factors or metabolites (e.g., SCFAs) produced by gut microbiome enter systemic circulation via the intestinal lymphatic system. These bacterial fragments and metabolites can modulate lung immune response (Trompette et al., 2014; Marsland et al., 2015; Gray et al., 2017; Mcaleer and Kolls, 2018; Figure 2).
Homing receptors determine tissue specificity of immune cells
The immune cells (after leaving the bloodstream) destined to home into specific sites, interact with high endothelial venules (HEVs) on these effector sites (Matsuno et al., 2010). The adhesion mechanisms in mucosal effector sites involve interaction between mucosal addressin molecule MAdCAM-1 (expressed on HEVs in Peyers patches and Mesenteric Lymph nodes) and integrin α4β7 (expressed on lymphocytes; Matsuno et al., 2010; Supplementary Figure S2). The memory/effector lymphocytes further express tissue-specific chemokine receptors which contribute to their mucosal localization (Hart et al., 2010). The T-effector cells and IgA secreting plasmablasts destined to home in the small intestine show higher expression of CCR9 receptor which binds local ligand CCL25 (TECK; Matsuno et al., 2010). On the other hand, CCR10 is expressed by plasmablasts targeted toward large intestine where they interact with ligand CCL28 (MEC; Seong et al., 2017). The DCs in GALT possess capability to produce Retinoic Acid (RA) which induces the expression of these homing receptors (integrin α4β7 and CCR9) in T as well as B cells and thereby boost their migration to small intestine (Seong et al., 2017).
In contrast to the above mentioned specific interactions, certain non-tissue specific interactions allow homing of lymphocytes to multiple mucosal sites (Supplementary Figure S2). Interactions between PNad and L-selectin as well as VCAM-1 and α4β1 integrins are non-tissue specific (Sun et al., 2014). Further, the expression of certain chemokine receptors (CCR5, CXCR3, etc.) are also not tissue specific (Thomas et al., 2007). It is speculated that the immune cell homing in lungs might occur through non-tissue specific interactions (Mateer et al., 2015). However, no lung specific homing receptors are characterized till date. Further, pulmonary T-cells express these non-specific chemokine receptors.
The expression of the above-mentioned non-specific receptors is higher in the lymphocytes residing in intestinal regions during inflammation (IBD etc.; Ye et al., 2017). These gut immune cells might get into the systemic circulation and interact with lung tissues possessing ligands to bind these non-tissue specific receptors. This is likely to cause mis-homing of these lymphocytes in the lung tissues (Mateer et al., 2015). The T effector memory (TEM) cells formed during gut inflammation might use their L-selectin/CD62L to interact with PNAd ligands present on lung endothelial surface once they enter systemic circulation (Golubovskaya and Wu, 2016). Such mechanisms explain how gut lymphocytes (TEMs and IgA plasmablasts) can migrate to lung lymphoid tissues. Thus, lung and gut form part of the mucosal immune system and an inflammatory response in one of these organs may be mirrored in the other.
Lung bacteria influence intestinal immunity
Although the above-mentioned aspects account for the effect of gut microbiota on lung immunity, the reverse has been less explored. A number of studies in different animal models have established that the manifestations of pneumonia due to multi drug resistant Staphylococcus aureus or Pseudomonas aeruginosa, which originate in the lung, is likely to induce intestinal injury (Lobo et al., 2005; Perrone et al., 2012). Pneumonia caused by P. aeruginosa leads to a reduction in gut epithelial proliferation and blocks the M-phase of cell cycle (Coopersmith et al., 2003; He et al., 2017). Studies have shown that the lung microbiota is formed by the micro-aspiration of microbiota from oropharynx part of the upper respiratory tract (URT) to the lung (Bassis et al., 2015; Dickson et al., 2015, 2017). In addition, studies performed on C57/Bl6 mice show that dysbiosis in the airway microbiota by intra-tracheal single dose of lipopolysaccharide leads to the movement of lung bacteria into the bloodstream. This causes an increase in the bacterial load in intestine, thereby disrupting the gut microbial community (Sze et al., 2014). This dysbiosis could be due to the interaction between translocated lung immune cells and gut microbiota. Therefore, the gut–lung axis is a bidirectional loop which is stimulated by the changes in gut or lung immunity (Bingula et al., 2017; Budden et al., 2017; Figure 2).
Gut microbiota modulates development of allergic responses and cystic fibrosis
Low gut microbial diversity in the first month of life of an individual is correlated with the occurrence of asthma (Russell et al., 2012; Ranucci et al., 2017). The impairment of gut microbiota balance due to usage of antibiotics increases the risk of asthma (Dharmage et al., 2015; Metsälä et al., 2015). For example, microbial dysbiosis due to the use of macrolides in early life (which depletes Actinobacteria and increases abundances of Bacteroidetes and Proteobacteria) is correlated with an increased risk of asthma in 2–7 years old Finnish children (Korpela et al., 2016). Similarly, neonatal mice administered with vancomycin show significant changes in gut microbiota which is accompanied by an increase in allergic asthma as compared to control mice (Russell et al., 2012).
Canadian children at risk of asthma show decreased abundances of Lachnospira, Veillonella, Faecalibacterium, and Rothia in their gut (Arrieta et al., 2015). Further, inoculations of these bacteria in germ free mice show protection against airway inflammation and asthma as compared to mice which are given fecal transplantation without these bacteria (Arrieta et al., 2015). There exists an association between lower numbers of certain bacteria (Bifidobacteria, Akkermansia and Faecalibacterium) and higher risk of developing atopy/asthma in neonates (Fujimura et al., 2016). The genera Faecalibacterium and Lachnospira, which are reduced in asthmatic patients, comprise of strains that are capable of butyrate production utilizing pyruvate as substrate (Anand et al., 2016). Out of the four pathways which can produce butyrate, this butyrogenic pathway (from Pyruvate) is a marker for gut commensals (Anand et al., 2016). This reaffirms the importance of butyrate production by beneficial gut bacteria in maintaining the lung immune homeostasis.
Feces and gut lavage samples from Cystic fibrosis (CF) patients show reduced microbial diversity as well as increased intestinal inflammatory markers (Bruzzese et al., 2014). As compared to healthy children, those suffering from CF have reduced abundances of beneficial bacteria like Eubacterium rectale, Bacteroides vulgatus, Bacteroides uniformis, Faecalibacterium prausnitzii, Bifidobacterium catenulatum, and Bifidobacterium adolescentis (Bruzzese et al., 2014). E. rectale and F. prausnitzii possess capability to produce butyrate using Pyruvate as substrate (Anand et al., 2016). Further, studies on pediatric fecal samples show that CF patients possess gastrointestinal environments which favor higher SCFA catabolism. The dysbiotic microbiomes in CF children display a decreased capacity for fatty acid biosynthesis and an increased potential for degrading SCFAs (including butyrate and propionate; Manor et al., 2016). Although, administration of probiotics to CF patients shows improvement in respiratory as well as gastro-intestinal clinical outcomes, the evidence has certain limitations (Ananthan et al., 2016; Anderson et al., 2017; Nikniaz et al., 2017). Meticulously designed random controlled trials with well-assessed clinical outcomes are required to ascertain the strains/species which can be used to alleviate lung diseases as well as the dosage required for desirable outcomes.
The reduction in the abundance of bacteria utilizing Pyruvate to produce butyrate in the above-mentioned diseases (asthma and CF) is in confirmation with a similar decrease observed in patients with allergic responses. This indicates the potential of utilizing this pathway (and bacteria possessing it) in early diagnosis of respiratory diseases. Further, these bacteria can find potential applications as probiotics for treatment of several lung diseases like allergy, COPD, asthma, etc. The influence of dietary intake on butyrate production highlights a link between nutrition and lung immunity.
Nutrition and lung function
A fiber rich diet changes not only the intestinal microbiota, but also affects the lung microbiota, indicating influence of nutrition on lung immunity (Trompette et al., 2014; Halnes et al., 2017). The dietary fiber increases SCFA levels in blood, thereby providing protection against allergic inflammation in lung (Trompette et al., 2014; Halnes et al., 2017). These findings emphasize the importance of diet and composition of gut microbiota in determining lung immune response.
Fiber rich diets show association with better lung function and lesser risk of lung disorders. A study based on 120,000 individuals who were followed up for around 12–16 years indicated that a healthy diet can lead to ~33% decline in chances of COPD (Varraso et al., 2015). The beneficial effect of fiber on lung function is clinically more significant in case of smokers, thereby indicating the potential of utilizing dietary modifications to tackle respiratory disorders. High fiber diet leads to reduction in mortality from respiratory disease (Varraso et al., 2015). A multitude of experimental interventions indicate the role of high fiber diets in modulating innate immunity, which can be corroborated by reduction in the levels of inflammatory markers (CRP and IL-6). Similarly, dietary fiber leads to a reduction in serum CRP by 20–30% from baseline (King et al., 2007).
Can probiotics be used for treating lung diseases?
There has been a growing interest in addressing the effect of probiotics on lung disorders such as asthma and COPD. Interestingly, studies on mice models indicate that Treg cells, which down-regulate the allergic response, can be induced by the administration of probiotic bacteria like Lactobacillus rhamnosus, Bifidobacterium lactis, and Bifidobacterium breve (Feleszko et al., 2007; Jang et al., 2012; Toh et al., 2012; Sagar et al., 2014). Similarly, administration of either Lactobacillus casei shirota or L. rhamnosus GG in Cystic Fibrosis patients leads to a reduction in exacerbation symptoms (Toh et al., 2012; West et al., 2017). Further, in vitro administration of L. rhamnosus and B. breve suppresses the pro-inflammatory mediators induced by exposure of macrophages to cigarette smoke. These findings may indicate importance of probiotics in treatment of cigarette smoke induced diseases like COPD (Mortaz et al., 2015).
Probiotics have shown promising results in improving inflammatory conditions (e.g., IBD) as well as regulating innate immunity using toll-like receptors and the corresponding signaling pathways (Toh et al., 2012; West et al., 2017). Further, they enhance barrier function in the intestine, thereby preventing antigens like lipopolysaccharide (LPS) from leaking through. Such beneficial roles of probiotics make them potential candidates for treatment of inflammatory diseases like IBD, allergy, COPD, asthma, etc.
Probiotics also show promising results in the field of lung oncology. Oral feeding of probiotic strain Lactobacillus acidophilus to mice lung cancer model undergoing cisplatin treatment shows reduction in tumor size and higher survival rate (Kelly et al., 2001; Gui et al., 2015). Further, gut microbiota has the potential to control lung cancer and has been attributed to the changes in antitumor immunity (Sivan et al., 2015). For example, when a mouse melanoma model is orally administered with Bifidobacterium cocktail (B. bifidum, B. longum, B. lactis, and B. breve), the tumor is observed to be controlled to an equal efficacy as that seen with PDL1 specific antibody therapy. This PDL1 specific antibody therapy has shown promise in the treatment of advanced Non-small Cell Lung Cancer (NSCLC; Sivan et al., 2015). The activated T-cells express programmed death protein-1 (PD-1) receptor that interacts with tumor-expressed ligands PD-L1/L2 and leads to an inhibition of T-cell activation, thereby promoting tumor cells escaping immunity. Antibodies against PD-1/PD-L1 prevent this interaction, thereby leading to T cell activation and thus acting as an immunotherapy against tumor cells (Valecha et al., 2017). On the other hand, Bifidobacterium cocktail treatment leads to an up regulation of 760 genes involved in multiple aspects of immune response like CD8+ T cell activation and co-stimulation, dendritic cell maturation, antigen processing presentation and type I interferon signaling (Sivan et al., 2015). Also, feeding Bacteroides fragilis to germ-free mice enhances the maturation of dendritic cells due to cross-reactivity between epitopes of this bacteria and the tumor (Vétizou et al., 2015). Interestingly, an improvement in advanced lung cancer patients is observed when Enterococcus hirae and Barnesiella intestine hominis are administered in combination with chemo-immunotherapy (Daillère et al., 2016). Both these strains can be considered as potential “oncomicrobiotics” for augmenting the efficacy of cancer treatments (Daillère et al., 2016). Probiotics show potential in modulating the immune response which can prove beneficial in handling a variety of lung diseases. Although, most of the studies on effect of probiotics have been performed with regards to lung cancer, the influence of gut microbiota on other lung diseases (discussed earlier) indicates promising opportunities for their treatment as well.
Despite the beneficial effects of probiotics on the aspects of human immune response that influence allergy and inflammation, a few inconsistencies pertaining to their efficacy in clinical trials have been observed (Vliagoftis et al., 2008; Elazab et al., 2013; Zuccotti et al., 2015). The subject population in these random controlled trials included children and adults with intermittent to mild persistent asthma. The durations of the studies ranged between 4 and 14 months. The microorganisms used included strains of lactic acid bacteria (L. casei, L. rhamnosus, and L. acidophilus, etc.,) as well as other genera including Enterococcus and Streptococcus. The placebo comprised of products having the same acidity and taste, but which lacked live microorganisms. The trials showed no significant differences in several laboratory parameters (forced expiratory volume in 1 s, blood eosinophil, serum total IgE, etc.) between the probiotic treated groups and the placebo treated individuals (Wheeler et al., 1997; Giovannini et al., 2007). Further, another trial indicated no considerable differences between the intervention and the placebo groups, in terms of clinical outcomes like life quality, number of asthma episodes and usage of medicine (Wheeler et al., 1997; Giovannini et al., 2007; Stockert et al., 2007). It is to be noted that these findings are based on a few clinical studies with smaller patient cohorts. The inconsistencies in the results of these studies make it difficult to conclude whether probiotics have beneficial roles in patients with respiratory disorders. Apart from recruitment of a heterogenous population, the design of these studies involved administration of different types and quantity of probiotics for variable durations. Further, symptom-based scoring systems used to evaluate the effects of interventions were also different. Thus, it is difficult to compare such studies in order to conclusively delineate the efficacy of probiotics in treatment of respiratory diseases.
Conclusion
The dietary intake of an individual determines the taxonomic constitution of his/her gut microbial community. The assimilation of dietary nutrients by gut microbiome leads to the production of metabolites which play significant roles in human health. Metabolites like SCFAs significantly influence the systemic immunity of not only gastro-intestinal system, but also other organs through lymphatic and circulatory systems. Thus, dysbiosis in gut microbiota affects not only gastrointestinal tract, but also influences the health of other distal organs. For example, the common mucosal system ensures that antigens introduced in gut are capable of eliciting an immune response in lung. Although the impact of changes in lung microbiota on the intestine has also been observed, an understanding of the system will require multiple mechanistic studies and clinical intervention trials. The increasing understanding of lung microbiota and its influence on the immune system might lead to more insights into these mechanisms.
The beneficial effect of pre- and probiotics have been observed in several lung ailments, although the results are inconsistent. These inconsistencies may arise because of dosage of the pre/pro-biotic administered as well as the strains used as probiotics. It is to be noted that probiotics usually comprise of a few strains, which might not be able to function in certain dysbiotic communities. Further, the efficacy of different strains might differ with changes in environmental states and geographical location. These strains might show better efficacy in the presence of other bacterial species or strains which coexist in a natural environment. Therefore, in order to delineate benefits of such treatments in lung diseases, an understanding of functional interactions within the entire microbial community is necessary. Further, understanding the metabolic potential of each strain and its capability to survive in conditions of interest also needs to be assessed. Our analyses on gut metagenomes in asthma, allergy and CF patients show decreased abundances of pyruvate utilizing butyrate-producing bacteria. This finding emphasizes the potential of utilizing the (Pyruvate -> Butyrate) pathway for diagnosis and assessment of lung diseases. In addition, our observation can be utilized for designing potential probiotic cocktails for treatment of lung disorders. Thus, the information based on the interactions amongst host genetics, diet and microbiota can be potentially used for designing personalized precision approaches for prevention as well as treatment of lung disorders.
Author contributions
SA and SM designed the review and drafted the manuscript. Both authors approved the final version of the manuscript for submission.
Conflict of interest statement
SM and SA are employed by company Tata Consultancy Services Ltd., (TCS) and are part of TCS Research. Both authors declare no competing interests. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02147/full#supplementary-material
References
- Albenberg L. G., Wu G. D. (2014). Diet and the intestinal microbiome: associations, functions, and implications for health and disease. Gastroenterology 146, 1564–1572. 10.1053/j.gastro.2014.01.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand S., Kaur H., Mande S. S. (2016). Comparative in silico analysis of butyrate production pathways in gut commensals and pathogens. Front. Microbiol. 7:1945. 10.3389/fmicb.2016.01945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthan A., Balasubramanian H., Rao S., Patole S. (2016). Probiotic supplementation in children with cystic fibrosis-a systematic review. Eur. J. Pediatr. 175, 1255–1266. 10.1007/s00431-016-2769-8 [DOI] [PubMed] [Google Scholar]
- Anderson J. L., Miles C., Tierney A. C. (2017). Effect of probiotics on respiratory, gastrointestinal and nutritional outcomes in patients with cystic fibrosis: a systematic review. J. Cyst. Fibros. 16, 186–197. 10.1016/j.jcf.2016.09.004 [DOI] [PubMed] [Google Scholar]
- Arrieta M. C., Stiemsma L. T., Dimitriu P. A., Thorson L., Russell S., Yurist-Doutsch S., et al. (2015). Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 7, 307ra152. 10.1126/scitranslmed.aab2271 [DOI] [PubMed] [Google Scholar]
- Azzali G. (2003). Structure, lymphatic vascularization and lymphocyte migration in mucosa-associated lymphoid tissue. Immunol. Rev. 195, 178–189. 10.1034/j.1600-065X.2003.00072.x [DOI] [PubMed] [Google Scholar]
- Backhed F., Roswall J., Peng Y., Feng Q., Jia H., Kovatcheva-Datchary P., et al. (2015). Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703. 10.1016/j.chom.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Bassis C. M., Erb-Downward J. R., Dickson R. P., Freeman C. M., Schmidt T. M., Young V. B., et al. (2015). Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio 6, e00037. 10.1128/mBio.00037-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekiaris V., Persson E. K., Agace W. W. (2014). Intestinal dendritic cells in the regulation of mucosal immunity. Immunol. Rev. 260, 86–101. 10.1111/imr.12194 [DOI] [PubMed] [Google Scholar]
- Bergman E. N. (1990). Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70, 567–590. 10.1152/physrev.1990.70.2.567 [DOI] [PubMed] [Google Scholar]
- Bergström A., Skov T. H., Bahl M. I., Roager H. M., Christensen L. B., Ejlerskov K. T., et al. (2014). Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl. Environ. Microbiol. 80, 2889–2900. 10.1128/AEM.00342-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingula R., Filaire M., Radosevic-Robin N., Bey M., Berthon J. Y., Bernalier-Donadille A., et al. (2017). Desired turbulence? gut-lung axis, immunity, and lung cancer. J. Oncol. 2017, 5035371. 10.1155/2017/5035371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugman S., Perdijk O., Van Neerven R. J., Savelkoul H. F. (2015). Mucosal immune development in early life: setting the stage. Arch. Immunol. Ther. Exp. (Warsz). 63, 251–268. 10.1007/s00005-015-0329-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzese E., Callegari M. L., Raia V., Viscovo S., Scotto R., Ferrari S., et al. (2014). Disrupted intestinal microbiota and intestinal inflammation in children with cystic fibrosis and its restoration with Lactobacillus GG: a randomised clinical trial. PLoS ONE 9:e87796. 10.1371/journal.pone.0087796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budden K. F., Gellatly S. L., Wood D. L., Cooper M. A., Morrison M., Hugenholtz P., et al. (2017). Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 15, 55–63. 10.1038/nrmicro.2016.142 [DOI] [PubMed] [Google Scholar]
- Cesta M. F. (2006). Normal structure, function, and histology of mucosa-associated lymphoid tissue. Toxicol. Pathol. 34, 599–608. 10.1080/01926230600865531 [DOI] [PubMed] [Google Scholar]
- Chauvistré H., Küstermann C., Rehage N., Klisch T., Mitzka S., Felker P., et al. (2014). Dendritic cell development requires histone deacetylase activity. Eur. J. Immunol. 44, 2478–2488. 10.1002/eji.201344150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., He X., Huang J. (2014). Diet effects in gut microbiome and obesity. J. Food Sci. 79, R442–451. 10.1111/1750-3841.12397 [DOI] [PubMed] [Google Scholar]
- Cho I., Blaser M. J. (2012). The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270. 10.1038/nrg3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. S., Qi L., Fahey G. C., Jr., Klurfeld D. M. (2013). Consumption of cereal fiber, mixtures of whole grains and bran, and whole grains and risk reduction in type 2 diabetes, obesity, and cardiovascular disease. Am. J. Clin. Nutr. 98, 594–619. 10.3945/ajcn.113.067629 [DOI] [PubMed] [Google Scholar]
- Coopersmith C. M., Stromberg P. E., Davis C. G., Dunne W. M., Amiot D. M., Karl I. E., et al. (2003). Sepsis from Pseudomonas aeruginosa pneumonia decreases intestinal proliferation and induces gut epithelial cell cycle arrest. Crit. Care Med. 31, 1630–1637. 10.1097/01.CCM.0000055385.29232.11 [DOI] [PubMed] [Google Scholar]
- Daillère R., Vétizou M., Waldschmitt N., Yamazaki T., Isnard C., Poirier-Colame V., et al. (2016). Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity 45, 931–943. 10.1016/j.immuni.2016.09.009 [DOI] [PubMed] [Google Scholar]
- Dash S., Clarke G., Berk M., Jacka F. N. (2015). The gut microbiome and diet in psychiatry: focus on depression. Curr. Opin. Psychiatry 28, 1–6. 10.1097/YCO.0000000000000117 [DOI] [PubMed] [Google Scholar]
- Date Y., Ebisawa M., Fukuda S., Shima H., Obata Y., Takahashi D., et al. (2017). NALT M cells are important for immune induction for the common mucosal immune system. Int. Immunol. 29, 471–478. 10.1093/intimm/dxx064 [DOI] [PubMed] [Google Scholar]
- De Clercq N. C., Groen A. K., Romijn J. A., Nieuwdorp M. (2016). Gut microbiota in obesity and undernutrition. Adv. Nutr. 7, 1080–1089. 10.3945/an.116.012914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C., Cavalieri D., Di Paola M., Ramazzotti M., Poullet J. B., Massart S., et al. (2010). Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U.S.A. 107, 14691–14696. 10.1073/pnas.1005963107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Besten G., Van Eunen K., Groen A. K., Venema K., Reijngoud D. J., Bakker B. M. (2013). The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 54, 2325–2340. 10.1194/jlr.R036012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmage S. C., Lodge C. J., Lowe A. J., Allen K. J. (2015). Antibiotics and risk of asthma: a debate that is set to continue. Clin. Exp. Allergy 45, 6–8. 10.1111/cea.12424 [DOI] [PubMed] [Google Scholar]
- Dickson R. P., Erb-Downward J. R., Freeman C. M., Mccloskey L., Beck J. M., Huffnagle G. B., et al. (2015). Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann. Am. Thorac. Soc. 12, 821–830. 10.1513/AnnalsATS.201501-029OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. P., Erb-Downward J. R., Freeman C. M., Mccloskey L., Falkowski N. R., Huffnagle G. B., et al. (2017). Bacterial topography of the healthy human lower respiratory tract. MBio 8, e02287-16. 10.1128/mBio.02287-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elazab N., Mendy A., Gasana J., Vieira E. R., Quizon A., Forno E. (2013). Probiotic administration in early life, atopy, and asthma: a meta-analysis of clinical trials. Pediatrics 132, e666–e676. 10.1542/peds.2013-0246 [DOI] [PubMed] [Google Scholar]
- Feleszko W., Jaworska J., Rha R. D., Steinhausen S., Avagyan A., Jaudszus A., et al. (2007). Probiotic-induced suppression of allergic sensitization and airway inflammation is associated with an increase of T regulatory-dependent mechanisms in a murine model of asthma. Clin. Exp. Allergy 37, 498–505. 10.1111/j.1365-2222.2006.02629.x [DOI] [PubMed] [Google Scholar]
- Fujimura K. E., Lynch S. V. (2015). Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe 17, 592–602. 10.1016/j.chom.2015.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K. E., Sitarik A. R., Havstad S., Lin D. L., Levan S., Fadrosh D., et al. (2016). Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 22, 1187–1191. 10.1038/nm.4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S., Toh H., Hase K., Oshima K., Nakanishi Y., Yoshimura K., et al. (2011). Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547. 10.1038/nature09646 [DOI] [PubMed] [Google Scholar]
- Fukuda S., Toh H., Taylor T. D., Ohno H., Hattori M. (2012). Acetate-producing bifidobacteria protect the host from enteropathogenic infection via carbohydrate transporters. Gut Microbes 3, 449–454. 10.4161/gmic.21214 [DOI] [PubMed] [Google Scholar]
- Ghosh T. S., Gupta S. S., Bhattacharya T., Yadav D., Barik A., Chowdhury A., et al. (2014). Gut microbiomes of Indian children of varying nutritional status. PLoS ONE 9:e95547. 10.1371/journal.pone.0095547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannini M., Agostoni C., Riva E., Salvini F., Ruscitto A., Zuccotti G. V., et al. (2007). A randomized prospective double blind controlled trial on effects of long-term consumption of fermented milk containing Lactobacillus casei in pre-school children with allergic asthma and/or rhinitis. Pediatr. Res. 62, 215–220. 10.1203/PDR.0b013e3180a76d94 [DOI] [PubMed] [Google Scholar]
- Glick-Bauer M., Yeh M. C. (2014). The health advantage of a vegan diet: exploring the gut microbiota connection. Nutrients 6, 4822–4838. 10.3390/nu6114822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovskaya V., Wu L. (2016). Different subsets of T cells, memory, effector functions, and CAR-T immunotherapy. Cancers (Basel) 8, E36. 10.3390/cancers8030036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray L. E., O'hely M., Ranganathan S., Sly P. D., Vuillermin P. (2017). The maternal diet, gut bacteria, and bacterial metabolites during pregnancy influence offspring asthma. Front. Immunol. 8:365. 10.3389/fimmu.2017.00365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross M., Salame T. M., Jung S. (2015). Guardians of the gut-murine intestinal macrophages and dendritic cells. Front. Immunol. 6:254. 10.3389/fimmu.2015.00254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui Q. F., Lu H. F., Zhang C. X., Xu Z. R., Yang Y. H. (2015). Well-balanced commensal microbiota contributes to anti-cancer response in a lung cancer mouse model. Genet. Mol. Res. 14, 5642–5651. 10.4238/2015.May.25.16 [DOI] [PubMed] [Google Scholar]
- Gupta S. S., Mohammed M. H., Ghosh T. S., Kanungo S., Nair G. B., Mande S. S. (2011). Metagenome of the gut of a malnourished child. Gut Pathog. 3, 7. 10.1186/1757-4749-3-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha C. W., Lam Y. Y., Holmes A. J. (2014). Mechanistic links between gut microbial community dynamics, microbial functions and metabolic health. World J. Gastroenterol. 20, 16498–16517. 10.3748/wjg.v20.i44.16498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halnes I., Baines K. J., Berthon B. S., Macdonald-Wicks L. K., Gibson P. G., Wood L. G. (2017). Soluble fibre meal challenge reduces airway inflammation and expression of GPR43 and GPR41 in asthma. Nutrients 9, E57. 10.3390/nu9010057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris N. L., Spoerri I., Schopfer J. F., Nembrini C., Merky P., Massacand J., et al. (2006). Mechanisms of neonatal mucosal antibody protection. J. Immunol. 177, 6256–6262. 10.4049/jimmunol.177.9.6256 [DOI] [PubMed] [Google Scholar]
- Hart A. L., Ng S. C., Mann E., Al-Hassi H. O., Bernardo D., Knight S. C. (2010). Homing of immune cells: role in homeostasis and intestinal inflammation. Inflamm. Bowel Dis. 16, 1969–1977. 10.1002/ibd.21304 [DOI] [PubMed] [Google Scholar]
- Hauptmann M., Schaible U. E. (2016). Linking microbiota and respiratory disease. FEBS Lett. 590, 3721–3738. 10.1002/1873-3468.12421 [DOI] [PubMed] [Google Scholar]
- He Y., Wen Q., Yao F., Xu D., Huang Y., Wang J. (2017). Gut-lung axis: the microbial contributions and clinical implications. Crit. Rev. Microbiol. 43, 81–95. 10.1080/1040841X.2016.1176988 [DOI] [PubMed] [Google Scholar]
- Ichinohe T., Pang I. K., Kumamoto Y., Peaper D. R., Ho J. H., Murray T. S., et al. (2011). Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. U.S.A. 108, 5354–5359. 10.1073/pnas.1019378108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipci K., Altintoprak N., Muluk N. B., Senturk M., Cingi C. (2017). The possible mechanisms of the human microbiome in allergic diseases. Eur. Arch. Otorhinolaryngol. 274, 617–626. 10.1007/s00405-016-4058-6 [DOI] [PubMed] [Google Scholar]
- Jang S. O., Kim H. J., Kim Y. J., Kang M. J., Kwon J. W., Seo J. H., et al. (2012). Asthma prevention by lactobacillus rhamnosus in a mouse model is associated with CD4(+)CD25(+)Foxp3(+) T Cells. Allergy Asthma Immunol. Res. 4, 150–156. 10.4168/aair.2012.4.3.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiminez J. A., Uwiera T. C., Abbott D. W., Uwiera R. R. E., Inglis G. D. (2017). Butyrate supplementation at high concentrations alters enteric bacterial communities and reduces intestinal inflammation in mice infected with citrobacter rodentium. mSphere 2, e00243-17. 10.1128/mSphere.00243-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce S. A., Gahan C. G. (2014). The gut microbiota and the metabolic health of the host. Curr. Opin. Gastroenterol. 30, 120–127. 10.1097/MOG.0000000000000039 [DOI] [PubMed] [Google Scholar]
- Jung T. H., Park J. H., Jeon W. M., Han K. S. (2015). Butyrate modulates bacterial adherence on LS174T human colorectal cells by stimulating mucin secretion and MAPK signaling pathway. Nutr. Res. Pract. 9, 343–349. 10.4162/nrp.2015.9.4.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalliomaki M., Kirjavainen P., Eerola E., Kero P., Salminen S., Isolauri E. (2001). Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J. Allergy Clin. Immunol. 107, 129–134. 10.1067/mai.2001.111237 [DOI] [PubMed] [Google Scholar]
- Kasubuchi M., Hasegawa S., Hiramatsu T., Ichimura A., Kimura I. (2015). Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients 7, 2839–2849. 10.3390/nu7042839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly K., Crowley J., Bunn P. A., Jr., Presant C. A., Grevstad P. K., Moinpour C. M., et al. (2001). Randomized phase III trial of paclitaxel plus carboplatin versus vinorelbine plus cisplatin in the treatment of patients with advanced non-small cell lung cancer: a Southwest Oncology Group trial. J. Clin. Oncol. 19, 3210–3218. 10.1200/JCO.2001.19.13.3210 [DOI] [PubMed] [Google Scholar]
- Kespohl M., Vachharajani N., Luu M., Harb H., Pautz S., Wolff S., et al. (2017). The microbial metabolite butyrate induces expression of Th1-associated factors in CD4(+) T Cells. Front. Immunol. 8:1036. 10.3389/fimmu.2017.01036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. H. (2016). B cell-helping functions of gut microbial metabolites. Microb Cell 3, 529–531. 10.15698/mic2016.10.536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Qie Y., Park J., Kim C. H. (2016). Gut microbial metabolites fuel host antibody responses. Cell Host Microbe 20, 202–214. 10.1016/j.chom.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King D. E., Egan B. M., Woolson R. F., Mainous A. G., III., Al-Solaiman Y., et al. (2007). Effect of a high-fiber diet vs. a fiber-supplemented diet on C-reactive protein level. Arch. Intern. Med. 167, 502–506. 10.1001/archinte.167.5.502 [DOI] [PubMed] [Google Scholar]
- Köhler C. A., Maes M., Slyepchenko A., Berk M., Solmi M., Lanctôt K. L., et al. (2016). The gut-brain axis, including the microbiome, leaky gut and bacterial translocation: mechanisms and pathophysiological role in Alzheimer's disease. Curr. Pharm. Des. 22, 6152–6166. 10.2174/1381612822666160907093807 [DOI] [PubMed] [Google Scholar]
- Korpela K., Salonen A., Virta L. J., Kekkonen R. A., Forslund K., Bork P., et al. (2016). Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat. Commun. 7:10410. 10.1038/ncomms10410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosiewicz M. M., Zirnheld A. L., Alard P. (2011). Gut microbiota, immunity, and disease: a complex relationship. Front. Microbiol. 2:180. 10.3389/fmicb.2011.00180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Babaei P., Ji B., Nielsen J. (2016). Human gut microbiota and healthy aging: recent developments and future prospective. Nutr. Healthy Aging 4, 3–16. 10.3233/NHA-150002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. V., Frassetto A., Kowalik E. J., Jr., Nawrocki A. R., Lu M. M., et al. (2012). Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS ONE 7:e35240. 10.1371/journal.pone.0035240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo S. M., Orrico S. R., Queiroz M. M., Cunrath G. S., Chibeni G. S., Contrin L. M., et al. (2005). Pneumonia-induced sepsis and gut injury: effects of a poly-(ADP-ribose) polymerase inhibitor. J. Surg. Res. 129, 292–297. 10.1016/j.jss.2005.05.018 [DOI] [PubMed] [Google Scholar]
- Looft T., Allen H. K. (2012). Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes 3, 463–467. 10.4161/gmic.21288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycke N. Y., Bemark M. (2012). The role of Peyer's patches in synchronizing gut IgA responses. Front. Immunol. 3:329. 10.3389/fimmu.2012.00329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson A. J., Geuking M. B., Mccoy K. D. (2005). Immune responses that adapt the intestinal mucosa to commensal intestinal bacteria. Immunology 115, 153–162. 10.1111/j.1365-2567.2005.02159.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor O., Levy R., Pope C. E., Hayden H. S., Brittnacher M. J., Carr R., et al. (2016). Metagenomic evidence for taxonomic dysbiosis and functional imbalance in the gastrointestinal tracts of children with cystic fibrosis. Sci. Rep. 6, 22493. 10.1038/srep22493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsland B. J., Trompette A., Gollwitzer E. S. (2015). The gut-lung axis in respiratory disease. Ann. Am. Thorac. Soc. 12, S150–156. 10.1513/AnnalsATS.201503-133AW [DOI] [PubMed] [Google Scholar]
- Maslowski K. M., Mackay C. R. (2011). Diet, gut microbiota and immune responses. Nat. Immunol. 12, 5–9. 10.1038/ni0111-5 [DOI] [PubMed] [Google Scholar]
- Mateer S. W., Maltby S., Marks E., Foster P. S., Horvat J. C., Hansbro P. M., et al. (2015). Potential mechanisms regulating pulmonary pathology in inflammatory bowel disease. J. Leukoc. Biol. 98, 727–737. 10.1189/jlb.3RU1114-563R [DOI] [PubMed] [Google Scholar]
- Matsuno K., Ueta H., Shu Z., Xue-Dong X., Sawanobori Y., Kitazawa Y., et al. (2010). The microstructure of secondary lymphoid organs that support immune cell trafficking. Arch. Histol. Cytol. 73, 1–21. 10.1679/aohc.73.1 [DOI] [PubMed] [Google Scholar]
- Mcaleer J. P., Kolls J. K. (2018). Contributions of the intestinal microbiome in lung immunity. Eur. J. Immunol 48, 39–49. 10.1002/eji.201646721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. R., Fujihashi K. (2012). Inside the mucosal immune system. PLoS Biol. 10:e1001397. 10.1371/journal.pbio.1001397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestecky J. (1987). The common mucosal immune system and current strategies for induction of immune responses in external secretions. J. Clin. Immunol. 7, 265–276. 10.1007/BF00915547 [DOI] [PubMed] [Google Scholar]
- Metsälä J., Lundqvist A., Virta L. J., Kaila M., Gissler M., Virtanen S. M. (2015). Prenatal and post-natal exposure to antibiotics and risk of asthma in childhood. Clin. Exp. Allergy 45, 137–145. 10.1111/cea.12356 [DOI] [PubMed] [Google Scholar]
- Million M., Diallo A., Raoult D. (2017). Gut microbiota and malnutrition. Microb. Pathog. 106, 127–138. 10.1016/j.micpath.2016.02.003 [DOI] [PubMed] [Google Scholar]
- Morrison D. J., Preston T. (2016). Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7, 189–200. 10.1080/19490976.2015.1134082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortaz E., Adcock I. M., Ricciardolo F. L., Varahram M., Jamaati H., Velayati A. A., et al. (2015). Anti-inflammatory effects of lactobacillus rahmnosus and bifidobacterium breve on cigarette smoke activated human macrophages. PLoS ONE 10:e0136455. 10.1371/journal.pone.0136455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal R., Mainali R., Ahmadi S., Wang S., Singh R., Kavanagh K., et al. (2018). Gut microbiome and aging: physiological and mechanistic insights. Nutr. Healthy Aging 4, 267–285. 10.3233/NHA-170030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikniaz Z., Nikniaz L., Bilan N., Somi M. H., Faramarzi E. (2017). Does probiotic supplementation affect pulmonary exacerbation and intestinal inflammation in cystic fibrosis: a systematic review of randomized clinical trials. World J. Pediatr. 13, 307–313. 10.1007/s12519-017-0033-6 [DOI] [PubMed] [Google Scholar]
- Noverr M. C., Noggle R. M., Toews G. B., Huffnagle G. B. (2004). Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect. Immun. 72, 4996–5003. 10.1128/IAI.72.9.4996-5003.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzer A. R., Lynch S. V. (2015). Influence and effect of the human microbiome in allergy and asthma. Curr. Opin. Rheumatol. 27, 373–380. 10.1097/BOR.0000000000000191 [DOI] [PubMed] [Google Scholar]
- Perrone E. E., Jung E., Breed E., Dominguez J. A., Liang Z., Clark A. T., et al. (2012). Mechanisms of methicillin-resistant Staphylococcus aureus pneumonia-induced intestinal epithelial apoptosis. Shock 38, 68–75. 10.1097/SHK.0b013e318259abdb [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistollato F., Sumalla Cano S., Elio I., Masias Vergara M., Giampieri F., Battino M. (2016). Role of gut microbiota and nutrients in amyloid formation and pathogenesis of Alzheimer disease. Nutr. Rev. 74, 624–634. 10.1093/nutrit/nuw023 [DOI] [PubMed] [Google Scholar]
- Qi H., Egen J. G., Huang A. Y., Germain R. N. (2006). Extrafollicular activation of lymph node B cells by antigen-bearing dendritic cells. Science 312, 1672–1676. 10.1126/science.1125703 [DOI] [PubMed] [Google Scholar]
- Ranucci G., Buccigrossi V., De Freitas M. B., Guarino A., Giannattasio A. (2017). Early-life intestine microbiota and lung health in children. J. Immunol. Res. 2017:8450496 10.1155/2017/8450496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios D., Wood M. B., Li J., Chassaing B., Gewirtz A. T., Williams I. R. (2016). Antigen sampling by intestinal M cells is the principal pathway initiating mucosal IgA production to commensal enteric bacteria. Mucosal Immunol. 9, 907–916. 10.1038/mi.2015.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos-Covián D., Ruas-Madiedo P., Margolles A., Gueimonde M., De Los Reyes-Gavilán C. G., Salazar N. (2016). Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 7:185. 10.3389/fmicb.2016.00185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth K., Oehme L., Zehentmeier S., Zhang Y., Niesner R., Hauser A. E. (2014). Tracking plasma cell differentiation and survival. Cytometry A 85, 15–24. 10.1002/cyto.a.22355 [DOI] [PubMed] [Google Scholar]
- Russell S. L., Gold M. J., Hartmann M., Willing B. P., Thorson L., Wlodarska M., et al. (2012). Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 13, 440–447. 10.1038/embor.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S. L., Gold M. J., Willing B. P., Thorson L., Mcnagny K. M., Finlay B. B. (2013a). Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes 4, 158–164. 10.4161/gmic.23567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell W. R., Hoyles L., Flint H. J., Dumas M. E. (2013b). Colonic bacterial metabolites and human health. Curr. Opin. Microbiol. 16, 246–254. 10.1016/j.mib.2013.07.002 [DOI] [PubMed] [Google Scholar]
- Sagar S., Morgan M. E., Chen S., Vos A. P., Garssen J., Van Bergenhenegouwen J., et al. (2014). Bifidobacterium breve and Lactobacillus rhamnosus treatment is as effective as budesonide at reducing inflammation in a murine model for chronic asthma. Respir. Res. 15, 46. 10.1186/1465-9921-15-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson D. R., Welsh D. A., Shellito J. E. (2015). Regulation of lung immunity and host defense by the intestinal microbiota. Front. Microbiol. 6:1085. 10.3389/fmicb.2015.01085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong Y., Lazarus N. H., Sutherland L., Habtezion A., Abramson T., He X. S., et al. (2017). Trafficking receptor signatures define blood plasmablasts responding to tissue-specific immune challenge. JCI Insight 2:e90233. 10.1172/jci.insight.90233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G., Garg N., Debelius J., Knight R., Dorrestein P. C., Mazmanian S. K. (2014). Specialized metabolites from the microbiome in health and disease. Cell Metab. 20, 719–730. 10.1016/j.cmet.2014.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shreiner A. B., Kao J. Y., Young V. B. (2015). The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 31, 69–75. 10.1097/MOG.0000000000000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S. D., Budden K. F., Neal R., Hansbro P. M. (2017). Microbiome effects on immunity, health and disease in the lung. Clin. Transl. Immunol. 6:e133. 10.1038/cti.2017.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson H. L., Campbell B. J. (2015). Review article: dietary fibre-microbiota interactions. Aliment. Pharmacol. Ther. 42, 158–179. 10.1111/apt.13248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan A., Corrales L., Hubert N., Williams J. B., Aquino-Michaels K., Earley Z. M., et al. (2015). Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089. 10.1126/science.aac4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. I., Yatsunenko T., Manary M. J., Trehan I., Mkakosya R., Cheng J., et al. (2013). Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339, 548–554. 10.1126/science.1229000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockert K., Schneider B., Porenta G., Rath R., Nissel H., Eichler I. (2007). Laser acupuncture and probiotics in school age children with asthma: a randomized, placebo-controlled pilot study of therapy guided by principles of Traditional Chinese Medicine. Pediatr. Allergy Immunol. 18, 160–166. 10.1111/j.1399-3038.2006.00493.x [DOI] [PubMed] [Google Scholar]
- Sun H., Liu J., Zheng Y., Pan Y., Zhang K., Chen J. (2014). Distinct chemokine signaling regulates integrin ligand specificity to dictate tissue-specific lymphocyte homing. Dev. Cell. 30, 61–70. 10.1016/j.devcel.2014.05.002 [DOI] [PubMed] [Google Scholar]
- Sun M., Wu W., Liu Z., Cong Y. (2017). Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J. Gastroenterol. 52, 1–8. 10.1007/s00535-016-1242-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze M. A., Tsuruta M., Yang S. W., Oh Y., Man S. F., Hogg J. C., et al. (2014). Changes in the bacterial microbiota in gut, blood, and lungs following acute LPS instillation into mice lungs. PLoS ONE 9:e111228. 10.1371/journal.pone.0111228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S. Y., Banerji A., Medoff B. D., Lilly C. M., Luster A. D. (2007). Multiple chemokine receptors, including CCR6 and CXCR3, regulate antigen-induced T cell homing to the human asthmatic airway. J. Immunol. 179, 1901–1912. 10.4049/jimmunol.179.3.1901 [DOI] [PubMed] [Google Scholar]
- Tilg H., Moschen A. R. (2015). Food, immunity, and the microbiome. Gastroenterology 148, 1107–1119. 10.1053/j.gastro.2014.12.036 [DOI] [PubMed] [Google Scholar]
- Timmerman H. M., Rutten N., Boekhorst J., Saulnier D. M., Kortman G. A. M, Contractor N., et al. (2017). Intestinal colonisation patterns in breastfed and formula-fed infants during the first 12 weeks of life reveal sequential microbiota signatures. Sci. Rep. 7:8327. 10.1038/s41598-017-08268-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh Z. Q., Anzela A., Tang M. L., Licciardi P. V. (2012). Probiotic therapy as a novel approach for allergic disease. Front. Pharmacol. 3:171. 10.3389/fphar.2012.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhurst G., Heffron H., Lam Y. S., Parker H. E., Habib A. M., Diakogiannaki E., et al. (2012). Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes 61, 364–371. 10.2337/db11-1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkovich S., Jobin C. (2016). Microbiota and host immune responses: a love-hate relationship. Immunology 147, 1–10. 10.1111/imm.12538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompette A., Gollwitzer E. S., Yadava K., Sichelstiel A. K., Sprenger N., Ngom-Bru C., et al. (2014). Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20, 159–166. 10.1038/nm.3444 [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ridaura V. K., Faith J. J., Rey F. E., Knight R., Gordon J. I. (2009). The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1, 6ra14. 10.1126/scitranslmed.3000322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valecha G. K., Vennepureddy A., Ibrahim U., Safa F., Samra B., Atallah J. P. (2017). Anti-PD-1/PD-L1 antibodies in non-small cell lung cancer: the era of immunotherapy. Expert. Rev. Anticancer Ther. 17, 47–59. 10.1080/14737140.2017.1259574 [DOI] [PubMed] [Google Scholar]
- Varraso R., Chiuve S. E., Fung T. T., Barr R. G., Hu F. B., Willett W. C., et al. (2015). Alternate healthy eating index 2010 and risk of chronic obstructive pulmonary disease among US women and men: prospective study. BMJ 350:h286. 10.1136/bmj.h286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri R., Gundamaraju R., Shastri M. D., Shukla S. D., Kalpurath K., Ball M., et al. (2018). Gut microbial changes, interactions, and their implications on human lifecycle: an ageing perspective. Biomed. Res. Int. 2018:4178607 10.1155/2018/4178607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vétizou M., Pitt J. M., Daillère R., Lepage P., Waldschmitt N., Flament C., et al. (2015). Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084. 10.1126/science.aad1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinolo M. A., Rodrigues H. G., Nachbar R. T., Curi R. (2011). Regulation of inflammation by short chain fatty acids. Nutrients 3, 858–876. 10.3390/nu3100858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vliagoftis H., Kouranos V. D., Betsi G. I., Falagas M. E. (2008). Probiotics for the treatment of allergic rhinitis and asthma: systematic review of randomized controlled trials. Ann. Allergy Asthma Immunol. 101, 570–579. 10.1016/S1081-1206(10)60219-0 [DOI] [PubMed] [Google Scholar]
- West C. E., Dzidic M., Prescott S. L., Jenmalm M. C. (2017). Bugging allergy; role of pre-, pro- and synbiotics in allergy prevention. Allergol Int. 66, 529–538. 10.1016/j.alit.2017.08.001 [DOI] [PubMed] [Google Scholar]
- Wheeler J. G., Shema S. J., Bogle M. L., Shirrell M. A., Burks A. W., Pittler A., et al. (1997). Immune and clinical impact of Lactobacillus acidophilus on asthma. Ann. Allergy Asthma Immunol. 79, 229–233. 10.1016/S1081-1206(10)63007-4 [DOI] [PubMed] [Google Scholar]
- Wu G. D. (2016). The gut microbiome, its metabolome, and their relationship to health and disease. Nestle Nutr. Inst. Workshop Ser. 84, 103–110. 10.1159/000436993 [DOI] [PubMed] [Google Scholar]
- Xu Z., Knight R. (2015). Dietary effects on human gut microbiome diversity. Br. J. Nutr. 113, S1–5. 10.1017/S0007114514004127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Duan Z. (2018). The local defender and functional mediator: gut microbiome. Digestion 97, 137–145. 10.1159/000484687 [DOI] [PubMed] [Google Scholar]
- Ye X., Liu S., Hu M., Song Y., Huang H., Zhong Y. (2017). CCR5 expression in inflammatory bowel disease and its correlation with inflammatory cells and β-arrestin2 expression. Scand. J. Gastroenterol. 52, 551–557. 10.1080/00365521.2017.1281435 [DOI] [PubMed] [Google Scholar]
- Zarzaur B. L., Kudsk K. A. (2001). The mucosa-associated lymphoid tissue structure, function, and derangements. Shock 15, 411–420. 10.1097/00024382-200115060-00001 [DOI] [PubMed] [Google Scholar]
- Zeng H., Chi H. (2015). Metabolic control of regulatory T cell development and function. Trends Immunol. 36, 3–12. 10.1016/j.it.2014.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer J., Lange B., Frick J. S., Sauer H., Zimmermann K., Schwiertz A., et al. (2012). A vegan or vegetarian diet substantially alters the human colonic faecal microbiota. Eur. J. Clin. Nutr. 66, 53–60. 10.1038/ejcn.2011.141 [DOI] [PubMed] [Google Scholar]
- Zuccotti G., Meneghin F., Aceti A., Barone G., Callegari M. L., Di Mauro A., et al. (2015). Probiotics for prevention of atopic diseases in infants: systematic review and meta-analysis. Allergy 70, 1356–1371. 10.1111/all.12700 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.