Significance
Phagocytosis and subsequent destruction of pathogens when the phagosomes in which they reside are fused with lysosomes are pillars of the eukaryotic innate immune defense. Consequently, evading trafficking to phagolysosomes is a fundamental survival strategy of most intracellular pathogens that replicate inside eukaryotic host cells. The obligatory intracellular bacterium Ehrlichia chaffeensis also avoids routing to host-cell phagolysosomes, but in a unique way: Ehrlichia secretes a protein, Ehrlichia translocated factor-2 (Etf-2), that has a Tre2-Bub2-Cdc16 (TBC)-like motif lacking RAB-GTPase–activating protein (GAP) activity. Etf-2 binds RAB5 on Ehrlichia inclusions and interferes with the engagement of RAB5-specific GAP with RAB5, thereby maintaining RAB5 in a GTP-bound active form on bacterial inclusions. Etf-2 is a unique example of a RAB-associated regulatory protein with a TBC-like motif lacking RABGAP activity.
Keywords: Ehrlichia chaffeensis, T4SS effector, TBC motif, Rab5, RABGAP5
Abstract
Ehrlichia chaffeensis, an obligatory intracellular bacterium, infects monocytes/macrophages by sequestering a regulator of endosomal traffic, the small GTPase RAB5, on its membrane-bound inclusions to avoid routing to host-cell phagolysosomes. How RAB5 is sequestered on ehrlichial inclusions is poorly understood, however. We found that native Ehrlichia translocated factor-2 (Etf-2), a previously predicted effector of the Ehrlichia type IV secretion system, and recombinant Etf-2 (cloned into the Ehrlichia genome) are secreted into the host-cell cytoplasm and localize to ehrlichial inclusions. Ectopically expressed Etf-2–GFP also localized to inclusions and membranes of early endosomes marked with RAB5 and interacted with GTP-bound RAB5 but not with a GDP-bound RAB5. Etf-2, although lacking a RAB GTPase-activating protein (GAP) Tre2-Bub2-Cdc16 (TBC) domain, contains two conserved TBC domain motifs, namely an Arg finger and a Gln finger, and site-directed mutagenesis revealed that both Arg188 and Gln245 are required for Etf-2 localization to early endosomes. The yeast two-hybrid assay and microscale thermophoresis revealed that Etf-2 binds tightly to GTP-bound RAB5 but not to GDP-bound RAB5. However, Etf-2 lacks RAB5-specific GAP activity. Etf-2 localized to bead-containing phagosomes as well as endosomes containing beads coated with the C-terminal fragment of EtpE (entry-triggering protein of Ehrlichia), an Ehrlichia outer-membrane invasin, and significantly delayed RAB5 dissociation from and RAB7 localization to phagosomes/endosomes and RABGAP5 localization to endosomes. Thus, binding of Etf-2 to RAB5-GTP appears to delay RAB5 inactivation by impeding RABGAP5 localization to endosomes. This suggests a unique mechanism by which RAB5 is sequestered on ehrlichial inclusions to benefit bacterial survival and replication.
For intracellular pathogens of eukaryotic cells, evading destruction in lysosomes following entry into permissive host cells is an essential step for successful colonization. Well-known strategies for this are (i) rapid diversion from endosomes/phagosomes to organellar compartments that do not fuse with lysosomes, such as the endoplasmic reticulum (ER) (utilized by Brucella and Legionella) (1–3) or exocytic Golgi compartments (Chlamydia) (4); (ii) blocking vacuolar-type H+ ATPase acquisition to the bacterial compartment so that lysosomal enzymes cannot be activated (Mycobacterium) (5); (iii) blocking maturation of RAB7-containing parasitophorus vacuoles to the degradative compartment (Salmonella) (6, 7); (iv) resistance to lysosomal acidification and enzymes (Coxiella and Yersinia) (8, 9); and (v) escape from endosomes/phagosomes/vacuoles into the cytosol (Listeria, Rickettsia, Orientia, and Shigella) (10–13).
Ehrlichia chaffeensis, an obligatory intracellular bacterium in the order Rickettsiales, replicates within human monocytes and macrophages and causes severe flu-like symptoms accompanied by hematologic abnormalities and hepatitis (14). The disease is called “human monocytic ehrlichiosis,” one of the most prevalent, life-threatening, emerging tick-borne diseases in the United States (15, 16). The parasitophorous vacuole (inclusion) where Ehrlichia replicates has early endosome-like characteristics, including the presence of transferrin, transferrin receptor, vacuolar-type H+-ATPase, and the small GTPase RAB5 and its effectors EEA1 (early-endosome antigen 1), PIK3C3/VPS34 (human phosphatidylinositol 3-kinase, catalytic subunit type 3/mammalian homolog of yeast vacuolar protein sorting 34), and Rabankyrin-5, but inclusions lack late endosomal or lysosomal markers or NADPH oxidase components (17–19). Within inclusions, Ehrlichia acquires all nutrients for its reproduction to yield numerous mature infectious forms. Little is known, however, about how Ehrlichia creates this intracellular haven.
RABs cycle between activated GTP-bound and inactivated GDP-bound states (20, 21). RAB5-GTP localizes to the endosomal membrane, whereas RAB5-GDP is mostly cytosolic and is associated with the RAB GDP dissociation inhibitor (22). RAB5 effectors bind to RAB5-GTP but not to RAB5-GDP (23), and thus a key determinant of RAB5 function is the duration of the GTP-bound state (24). The localization of RAB5-GTP to phagosomes, which have engulfed diverse bacteria, parasites, viruses, or latex beads, is generally transient (∼30 min), as the phagosomes mature to late endosomes and fuse with lysosomes or the phagosomal traffic is altered to form various membrane-bound vacuoles containing intracellular pathogens. However, endogenous and ectopically expressed RAB5 and its effectors localize persistently (>3 d) to the inclusion membrane harboring replicating Ehrlichia (18, 19), indicating that GTP-bound RAB5 persists on inclusions. Lysosomal fusion with the endosome requires hydrolysis of GTP bound to RAB5, with subsequent dissociation of RAB5-GDP from the membrane; its replacement with RAB7 is an essential step in late-endosome formation and subsequent fusion with lysosomes for degradation of the content of the endosomes (25). Thus, the persistence of RAB5-GTP on the inclusion membrane is the key for preventing Ehrlichia inclusions from maturing into late endosomes and subsequently fusing with lysosomes. In addition, RAB5-GTP and its effector EEA1 on Ehrlichia inclusions facilitate the expansion of the inclusion compartment via fusion with early endosomes, as demonstrated with GFP-RAB5–labeled endosomes and GFP-Rabankyrin-5 trafficking to established Ehrlichia inclusions (19).
Because RABs have a low rate of intrinsic nucleotide exchange and GTP hydrolysis, additional proteins called “GDP–GTP exchange factors” and “GTPase-activating proteins” (GAPs) regulate the RAB GTPase cycle (26). RAB5-GTP is indeed critical for Ehrlichia infection, as demonstrated in our previous findings: Ehrlichia infection is inhibited by ∼85% via overexpression of the RAB5-specific GAP, RABGAP5, but not by the catalytic site mutant RABGAP5R165A (19, 27) and by ∼50% via overexpression of dominant-negative RAB5S34N (RAB5-DN) that sequesters RAB5-GEF (19, 28). However, how Ehrlichia locks RAB5 in the GTP-bound state on Ehrlichia inclusions is unknown.
The bacterial type IV secretion system (T4SS) has been shown to translocate proteins and nucleoprotein complexes from bacteria to eukaryotic target cells across the eukaryotic cell membrane (29). There are several ancestral lineages for the T4SS including the archetype virB/virD system of Agrobacterium tumefaciens and the dot/icm system of Legionella pneumophila, sometimes referred as “T4aSS” and “T4bSS,” respectively (30). E. chaffeensis has T4aSS. T4SS functions through its effectors. T4bSS of L. pneumophila secretes ∼300 effectors with redundant functions; hence, each effector can be knocked out, but the mutant lacks a phenotype (31). In contrast, the total number of T4aSS effectors is much lower [for example, there are fewer than six effectors in A. tumefaciens (30)], but each effector has a crucial role in infection/disease. For obligatory intracellular bacteria, so far only a handful of T4SS effectors have been identified, and even fewer have been functionally characterized (32). By a bacterial two-hybrid screen using E. chaffeensis VirD4, a well-established coupling protein involved in escorting translocated DNA and proteins in A. tumefaciens (30), as the bait, we previously identified three Ehrlichia proteins that directly bind to Ehrlichia VirD4: ECH0825 (ehrlichial translocated factor-1, Etf-1), ECH0261 (Etf-2), and ECH0767 (Etf-3) (33). All three were formerly annotated as hypothetical proteins, as they lack homology to previously known proteins or protein domains or motifs. Etf-1 was recently found to have key roles in Ehrlichia infection of human cells (19, 33, 34), but the functions of the latter two Etfs remain unknown. Here we report that Etf-2 binds directly to RAB5-GTP and hence localizes to Ehrlichia inclusions and early endosomes. Although Etf-2 lacks homology to known prokaryotic and eukaryotic GAPs, manual alignment of amino acid sequences revealed that Etf-2 contains an Arg finger and a Gln finger motif of the Tre2-Bub2-Cdc16 (TBC) domain, the conserved RAB-GTP–interacting domain that has been identified in almost all RABGAP proteins (35). By taking advantage of latex beads that can be engulfed by biological membranes, we investigated the functions of Etf-2 during endosome maturation. Our data indicate that Etf-2 is a unique RAB5-GTP–binding factor that impedes early-endosome maturation by intervening in RABGAP5 localization to endosomes to the benefit of bacteria. Our data suggest that the TBC-like motif in Etf-2 is critical for this function.
Results
Native Etf-2 and FLAG-Etf-2C Are Secreted from Ehrlichia and Localize to the Ehrlichia Inclusion Membrane.
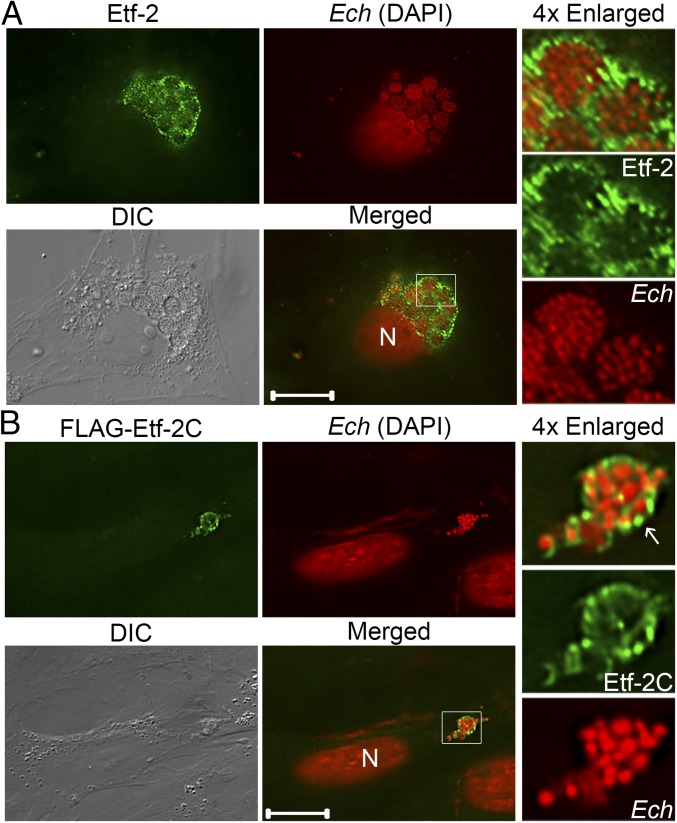
Etf-2 (264 amino acid residues, 28,560 Da) is highly conserved (99–100% amino acid sequence identity) among eight E. chaffeensis strains, which were isolated at Centers of Disease Control and Prevention via culture of blood samples from patients with monocytic ehrlichiosis in five different states in the United States during the period 1991–1998 and for which whole-genome sequences have been determined (SI Appendix, Fig. S1). Etf-2 mRNA/protein is expressed by Ehrlichia in the human monocyte cell line THP-1 (33), the human promyelocytic leukemia cell line HL-60 (36), the canine histiocytic leukemia cell line DH82, and the tick cell line ISE6 [Gene Expression Omnibus (GEO) accession no. GSE56339]. Secretion of native Etf-2 by Ehrlichia and its subcellular localization were determined by double fluorescence labeling using antigen-affinity–purified anti–Etf-2 IgG. Monkey RF/6A endothelial cells were used for subcellular localization studies, as this cell line manifests as thinly spread adherent host cells compatible with colocalization image analysis and can be readily infected with E. chaffeensis (33). As shown in Fig. 1A, the majority of Etf-2 was secreted and localized to the Ehrlichia inclusion membrane.
Fig. 1.
Native and FLAG-Etf-2 cloned into the Ehrlichia genome are secreted into host-cell cytoplasm and localize to the ehrlichial inclusion membrane. (A) Etf-2 was secreted and subsequently was trafficked to Ehrlichia-containing inclusion membranes. E. chaffeensis (Ech)-infected RF/6A cells at 2 dpi were fixed and labeled with rabbit anti–Etf-2C IgG and AF488 anti-rabbit IgG. (Scale bar: 10 μm.) (B) FLAG-Etf-2C cloned and expressed in Ehrlichia was secreted, and localized on inclusion membranes (white arrow). Transformed E. chaffeensis expressing FLAG-Etf-2C were selected with antibiotics in DH82 cells and used to infect RF/6A cells. Cells at 2 dpi were fixed and labeled with AF488 rat anti-FLAG mAb. (Scale bar: 5 μm.) Bacteria and the host nucleus (N) were labeled with DAPI (pseudocolored red). DIC, differential interference contrast; Merged, the fluorescence image is merged with the DIC image. Each boxed area is enlarged 4× on the right.
Currently, effector secretion analysis cannot be carried out using the autologous T4SS of Ehrlichia spp., because the bacterial genetics system is not readily applicable to these bacteria (37). Therefore, we employed the Himar 1 random mutagenesis system (38) to insert a gene encoding FLAG-Etf-2C (the C terminus of Etf-2 containing the T4SS effector signal, amino acids 152–264) into the E. chaffeensis chromosome and assess FLAG-Etf-2C secretion. As shown in Fig. 1B, FLAG-Etf-2C was produced by E. chaffeensis Himar 1 transposon mutants (inserted at E. chaffeensis genomic locus 25,974) and was distinctly secreted into the host-cell cytoplasm across the inclusion membrane, after which it localized to the cytoplasmic surface of the inclusion.
Ectopically Expressed Full-Length and C-Terminal Etf-2, but Not N-Terminal Etf-2, Localize to Ehrlichia Inclusions.
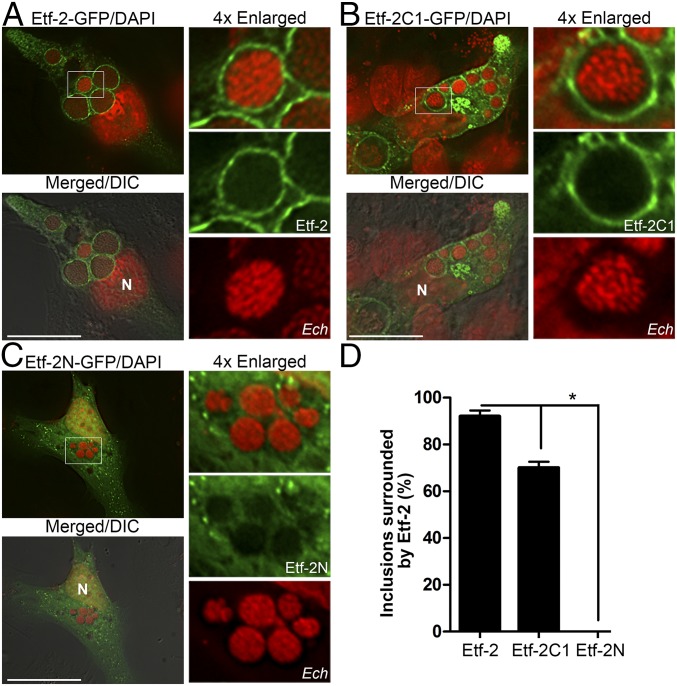
Given that both native Etf-2 and FLAG-Etf-2C were secreted and localized to the ehrlichial inclusion membrane, we examined whether Etf-2-GFP expressed in mammalian cells also localized to inclusions. Indeed, Etf-2-GFP completely lined the membrane of individual inclusions (Fig. 2 A and D), indicating that Eft-2 and its fusion proteins consistently traffic to inclusions. To determine which segments of Etf-2 are critical for Etf-2 localization to ehrlichial inclusions, we assessed the subcellular localization of the GFP-tagged N terminus of Etf-2 (Etf-2N; amino acids 1–114) and C terminus of Etf-2 (Etf-2C1; amino acids 135–264). Etf-2C1-GFP, but not Etf-2N-GFP, localized to the inclusion membrane (Fig. 2 B–D); although Etf-2C1-GFP localization was slightly less efficient than that of full-length Etf-2-GFP, this result demonstrated that C-terminal Etf-2 is sufficient for localization to inclusions.
Fig. 2.
Ectopically expressed full-length Etf-2 and its C-terminal fragment, but not the N-terminal fragment, traffic to Ehrlichia inclusions. (A–C) Ehrlichia-infected RF/6A cells at 1 dpi were transfected with Etf-2-GFP (A), Etf-2C1-GFP (B), or Etf-2N-GFP (C). At 15 hpt [39 h postinfection (hpi)], cells were fixed and stained with DAPI for DNA (pseudocolored red). Merged/DIC, the fluorescence image is merged with the DIC image; N, nucleus. Each boxed area is enlarged 4× on the right. (Scale bars: 10 µm.) (D) Quantification of the localization of Etf-2, Etf-2C1, or Etf-2N-GFP to 120 Ehrlichia inclusions in 20–30 transfected cells from three independent experiments. Data are presented as the mean ± SD. *P < 0.05, ANOVA.
C-terminal Etf-2 Is Crucial for Etf-2 Localization to the Early-Endosome Membrane and Interaction with RAB5-GTP.
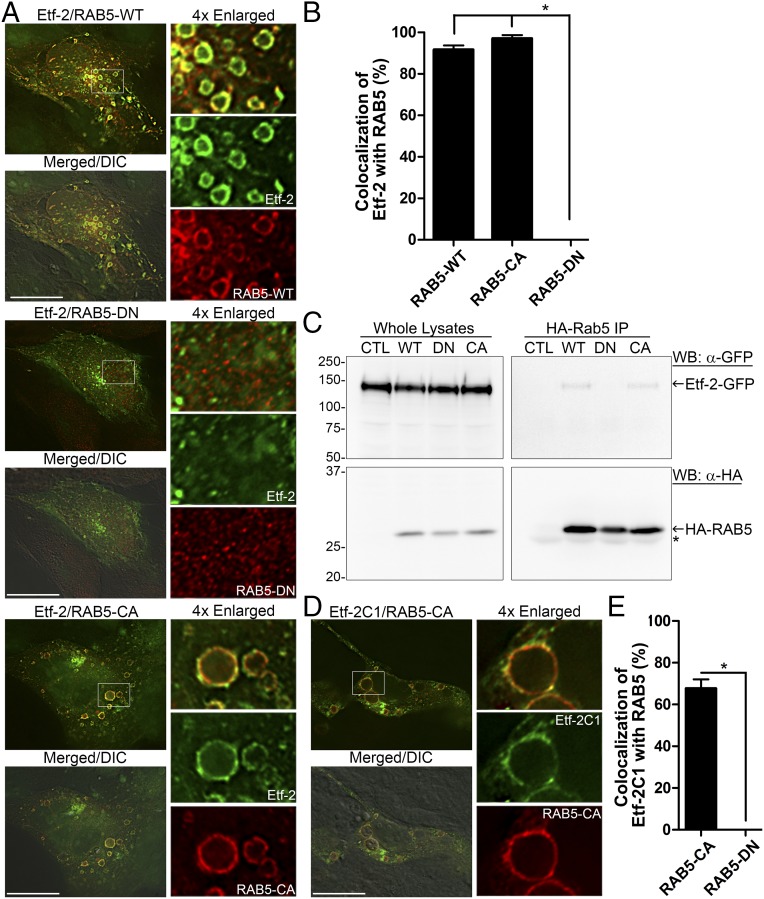
Both endogenous and ectopically expressed RAB5 (RAB5A) localize to ehrlichial inclusions (18, 19). Because we found that the majority of Etf-2 also localized to the inclusion membrane, we examined whether Etf-2 would localize to early endosomes (marked with RAB5) in uninfected cells. As shown in Fig. 3 A and B, Etf-2-GFP colocalized almost completely with both RAB5-WT and RAB5-CA (constitutively active RAB5Q79L) on the endosomal membrane; note that expression of RAB5-CA resulted in a preponderance of larger endosomes, as previously reported (39). However, Etf-2-GFP did not colocalize with RAB5-DN. Moreover, coimmunoprecipitation revealed that Etf-2-GFP interacted physically with RAB5-WT and RAB5-CA but not with RAB5-DN in mammalian cells (Fig. 3C). Coimmunoprecipitation also showed Etf-2-GFP interacts specifically with RAB5 but not with RAB7 (SI Appendix, Fig. S2A). This finding indicates that in the absence of any other ehrlichial molecules Etf-2 targets early endosomes, and this targeting is dependent on active RAB5.
Fig. 3.
Etf-2 localizes to the early-endosome membrane and interacts with RAB5-GTP. (A) RF/6A cells cotransfected with Etf-2-GFP and HA-RAB5 (WT, DN, or CA). (B) Quantification of 150 vesicles in 20–30 cells cotransfected with Etf-2 and RAB5 (WT, CA, or DN) from three independent experiments. *P < 0.05, ANOVA. (C) HEK-293 cells were cotransfected with plasmids expressing Etf-2-GFP and HA-RAB5 (WT, DN, or CA) or none (CTL), lysed at 2 dpt, immunoprecipitated with mouse anti-HA–linked protein G-Sepharose beads, and analyzed by Western blotting with anti-GFP and anti-HA. The asterisk indicates the IgG light chain. (D) RF/6A cells were cotransfected with Etf-2C1-GFP and HA-RAB5-CA. In A and D cells were subjected to immunofluorescence labeling with mouse anti-HA (red; AF555) at 2 dpt. Merged/DIC, the fluorescence image is merged with the DIC image. Each boxed area is enlarged 4× on the right. (Scale bars: 10 µm.) (E) Quantification of Etf-2C1-GFP colocalization to 100 vesicles or dots of HA-RAB5-CA or HA-RAB5-DN in 30 cells. *P < 0.05, two-tailed t test.
Because Etf-2C1-GFP localized to the inclusion membrane (Fig. 2 B and D), we examined whether Etf-2C1-GFP, like Etf-2-GFP, also localized to early endosomes. As shown in Fig. 3 D and E, Etf-2C1-GFP localized to early endosomes marked with RAB5-CA but not with RAB5-DN, demonstrating that C-terminal Etf-2 is sufficient for localization to early endosomes. A single hydrophobic (HY) domain is predicted in the central region (amino acids 132–151) of Etf-2 by the program Protean in the DNASTAR Lasergene suite or TMpred server (https://embnet.vital-it.ch/software/TMPRED_form.html) (Fig. 4A). To determine whether this hydrophobic segment is required for endosome membrane localization, we assessed the localization of Etf-2ΔHY (Etf-2Δamino acids 132–151)-GFP and Etf-2C (amino acids 152–264)-GFP, each of which lacks the hydrophobic domain, and obtained the same result shown in Fig. 3 (SI Appendix, Fig. S2 B–D), indicating that the central HY domain of Etf-2 is not required for Etf-2 localization to the early-endosome membrane.
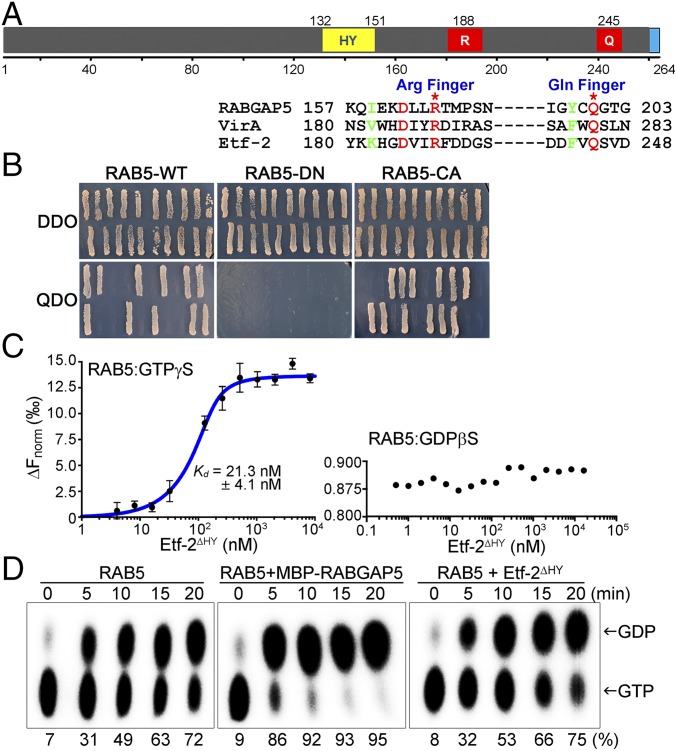
Fig. 4.
Etf-2 has a GAP TBC-like dual catalytic finger motif that directly binds to RAB5-GTP but lacks RAB5 GAP activity. (A, Upper) Putative domain structures of Etf-2. Numbers indicate amino acid positions. Q, glutamine finger; R, arginine finger; the blue boxed domain is the putative T4SS signal. (Lower) Amino acid sequence alignment of Etf-2 with TBC domains of RABGAP5 and Shigella VirA that have RAB1 GAP activity. Etf-2 has the GAP TBC-like dual (Arg and Glu) catalytic finger motif (red) near its C terminus. (B) Direct interaction of full-length Etf-2 with RAB5-WT, -DN, and -CA was tested using the yeast two-hybrid assay. Twenty independent colonies were tested on SD/−Leu/−Trp double-dropout (DDO) and SD/−Leu/−Trp/−His/−Ade quadruple-dropout (QDO) agar plates. Growth on QDO plates indicated a specific interaction between Etf-2 and RAB5-WT or -CA. (C) MST assay with purified proteins demonstrating binding of RAB5A to Etf-2∆HY in a GTP-dependent manner. RAB5A was fluorescently labeled with Tris-nitrilotriacetic acid at fixed concentrations ranging from 5 to 200 nM and was preincubated with GTPγS or GDPβS; binding was assessed with unlabeled Etf-2∆HY from the low picomolar range to 100 µM. RAB5A binding to Etf-2∆HY was observed only in RAB5A-GTPγS samples. The determined Kd value is shown. Data are shown as mean ± SD (n = 4). (D) GAP activity of Etf-2∆HY. GST-RAB5 that had been affinity-purified on GST resin was preloaded with [α-32P]GTP followed by a GTP hydrolysis reaction in the absence or presence of RABGAP5 or Etf-2∆HY. Samples were taken at the indicated times and subjected to TLC followed by autoradiography and PhosphorImager analysis to visualize and quantify the radiolabeled hydrolysis product (32P-GDP) from substrate (32P-GTP) as indicated. The percentage values indicate the amount of GDP produced from GTP.
Etf-2 Has a GAP TBC-Like Dual Catalytic Finger Motif and Directly Binds RAB5-GTP but Lacks RAB5-GAP Activity.
Etf-2 has a T4SS effector motif in its C-terminal region that directly binds E. chaffeensis VirD4 (Fig. 4A) (33). Based on a bioinformatics analysis of Etf-2, however, no conserved domains could be identified. Although Etf-2 lacks homology to known prokaryotic and eukaryotic GAPs, manual alignment of the Etf-2 amino acid sequence with that of several RABGAPs, including RABGAP5 (27) and Shigella flexneri VirA (40), revealed that Etf-2 contains an Arg finger and a Gln finger motif of the TBC domain, the conserved RAB-GTP–interacting domain that has been identified in almost all RABGAP proteins (Fig. 4A) (41). Such a motif is absolutely conserved among the eight strains of E. chaffeensis (SI Appendix, Fig. S1), suggesting that this is functionally critical. The RAB5-specific GAP RABGAP5 directly binds to RAB5-CA but not to RAB5-DN (27), and Etf-2 selectively interacts with RAB5-WT or RAB5-CA but not with RAB5-DN (Fig. 3). We therefore examined whether Etf-2 could bind directly to RAB5-CA in a yeast two-hybrid analysis, revealing that full-length Etf-2 bound directly to RAB5-WT and RAB5-CA but not to RAB5-DN (Fig. 4B).
To determine the affinity of Etf-2 for RAB5-GTP, we used microscale thermophoresis (MST), a highly sensitive technology for measuring the relative strength of interactions between biomolecules (42). As full-length Etf-2 is poorly expressed in Escherichia coli, Etf-2∆HY was cloned, expressed in E. coli, and purified to assess the ability of Etf-2 to bind RAB5-GTP and promote GTP hydrolysis. Etf-2∆HY bound specifically and with high affinity to RAB5-GTPγS (GTPγS is a hydrolysis-resistant GTP analog, Kd = 21.3 ± 4.1 nM, n = 4) but not to RAB5-GDPβS (Fig. 4C). This result also demonstrated that the hydrophobic domain is not required for the direct binding of Etf-2 to RAB5-GTPγS. We also determined whether Etf-2 has GAP activity toward RAB5-GTP with an in vitro GTP hydrolysis assay using α-32P–labeled GTP. This GTP hydrolysis assay showed that RAB5 has relatively strong intrinsic GTPase activity (Fig. 4D). In the presence of RABGAP5 near-full conversion of [α-32P]GTP to [α-32P]GDP occurred in 10 min (Fig. 4D). Compared with RABGAP5, the addition of Etf-2∆HY in the assay did not affect the extent of [α-32P]GTP hydrolysis, indicating that Etf-2 has negligible GAP activity toward RAB5 (Fig. 4D).
Mutation of the Arg or Gln Finger of the TBC-Like Motif in Etf-2 Impairs Etf-2 Localization to RAB5-CA Endosomes.
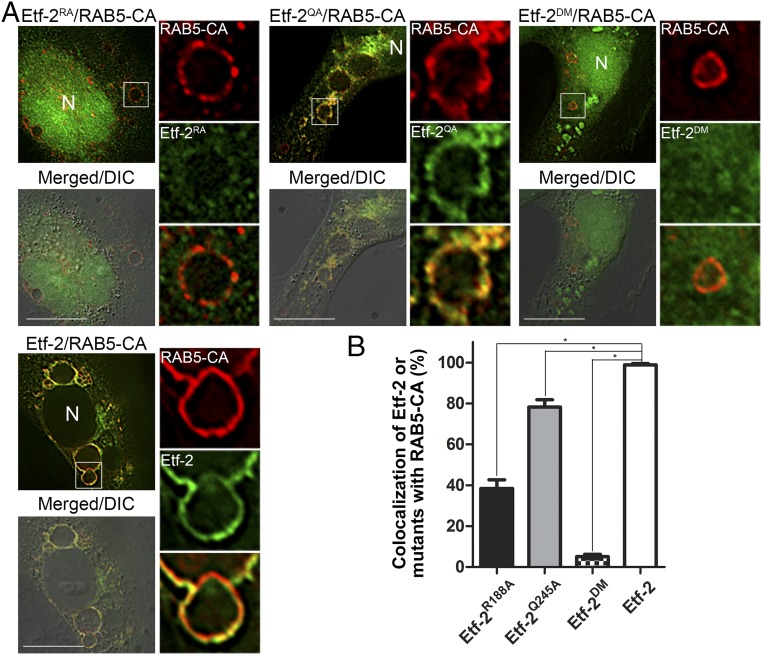
To analyze whether the TBC motif is required for Etf-2 colocalization with RAB5 on endosomes, Arg188 and Gln245 within the Arg and Gln fingers of the TBC motif were mutated to Ala by site-directed mutagenesis. The colocalization of mutants Etf-2R188A-GFP and Etf-2Q245A-GFP with RAB5-CA (Fig. 5) and RAB5-WT (SI Appendix, Fig. S3) on endosomal membranes was significantly reduced compared with Etf-2, and the mutation of both Arg188 and Gln245 to Ala [Etf-2R188A/Q245A (Etf-2DM)-GFP] essentially abolished the colocalization on early endosomes (Fig. 5 and SI Appendix, Fig. S3), indicating a crucial role for Arg188 and Gln245 in the endosomal localization of Etf-2.
Fig. 5.
Mutation of the Arg and Gln fingers of the TBC-like motif of Etf-2 impairs Etf-2 localization to RAB5-CA endosomes. (A) RF/6A cells were cotransfected with HA-RAB5-CA and with Etf-2R188A (Etf-2RA), Etf-2Q245A (Etf-2QA), Etf-2R188A/Q245A (Etf-2DM), or Etf-2-GFP. At 2 dpt, cells were subjected to immunofluorescence labeling with mouse anti-HA (red; AF555). Merged/DIC, the fluorescence image is merged with the DIC image. Boxed areas are enlarged 4× on the right. (Scale bars: 10 μm.) (B) Quantification of 100–120 RAB5-CA endosomes in 20–30 cotransfected RF/6A cells from three independent experiments. *P < 0.05, ANOVA.
Etf-2 Localizes to Phagosomes Loaded with Latex Beads and Delays the Maturation of Phagosomes to Phagolysosomes.
Because Etf-2 is targeted to endosomes, we investigated the effects of Etf-2 on phagocytosis and phagosome maturation. To rule out the possible involvement of other ehrlichial factors, we monitored phagocytosis via the uptake of latex beads (43) in the DH82 cell line that has been used to culture and isolate all E. chaffeensis strains from the blood of patients afflicted with monocytic ehrlichiosis. The advantages of using DH82 cells in our study included good transfection efficiency and the ability to assess phagocytosis readily.
The phagocytosis of latex beads by DH82 cells was significantly reduced in Etf-2-GFP–transfected DH82 cells compared with GFP-transfected control cells at 60–80 min postincubation (SI Appendix, Fig. S4 A, B, and D). At 20 min postincubation nearly 100% of Etf-2-GFP was found on phagosomes containing 1-µm Flash Red (near-infrared) latex beads (SI Appendix, Fig. S4 A and C). Even after 80-min coincubation, >90% of the bead-containing phagosomes retained Etf-2-GFP (SI Appendix, Fig. S4C). To examine the effects of Etf-2 during phagolysosome maturation following the uptake of latex beads, colocalization studies were carried out using tricolor fluorescence microscopy. In control GFP-transfected DH82 cells, RAB5 localized to 70% of the bead-containing phagosomes at 20 min after beads were added to DH82 cells, whereas at 80 min the bead-containing phagosomes did not retain RAB5 (SI Appendix, Fig. S5 A and C). In Etf-2-GFP–transfected cells, however, RAB5 localized to almost 100% of the bead-containing phagosomes at 20 min after beads were added, and at 80 min 60% of the bead-containing phagosomes still retained RAB5 (SI Appendix, Fig. S5 B and C).
GTP-bound RAB5 and RAB7 localize to early and late endosomes, respectively, where they regulate trafficking of endocytosed cargoes from the plasma membrane to lysosomes (28). In control GFP-transfected DH82 cells, RAB7 localized to ∼25% of the bead-containing phagosomes at 20 min after beads were mixed with cells, and at 80 min RAB7 localized to 100% of the bead-containing phagosomes (SI Appendix, Fig. S5 D and F). In Etf-2-GFP–transfected cells, however, RAB7 localization occurred in less than 5% of the bead-containing phagosomes at 20 min after beads were mixed with cells, and at 80 min RAB7 localized to ∼60% of the bead-containing phagosomes (SI Appendix, Fig. S5 E and F).
LysoTracker Red labels and tracks acidic organelles such as late endosomes and phagolysosomes in live cells (44). In GFP-transfected DH82 cells, LysoTracker Red localized to ∼40% of the bead-containing phagosomes at 20 min after beads were mixed with live cells, and at 80 min this percentage rose to 100% (SI Appendix, Fig. S5 G and I). In Etf-2-GFP–transfected cells, however, no LysoTracker Red localized in bead-containing phagosomes after 20 min; after 80 min this percentage rose to 70% (SI Appendix, Fig. S5 H and I). These results indicated that Etf-2 quickly colocalized with RAB5 on bead-containing phagosomes, prolonged RAB5 localization on the phagosome membrane, and delayed phagosomal maturation and acidification.
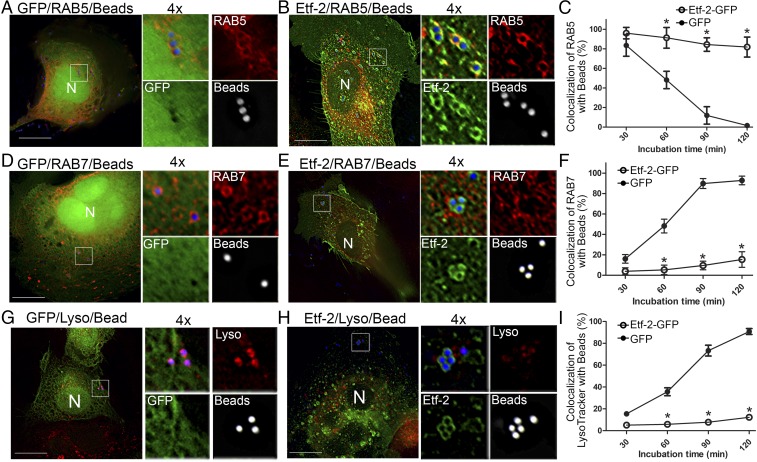
E. chaffeensis enters host cells via receptor-mediated endocytosis rather than phagocytosis (45); thus, phagosomes containing latex beads may not represent true E. chaffeensis inclusions. Therefore, we assessed the uptake of latex beads coated with EtpE-C (the C terminus of the E. chaffeensis invasin EtpE, the entry-triggering protein of Ehrlichia); this uptake mimics the E. chaffeensis entry pathway into early endosomes in relevant E. chaffeensis host cells (both nonphagocytes and phagocytes) (45). Beads lacking EtpE-C cannot be taken up by nonphagocytes (45). In GFP-transfected RF/6A cells (nonphagocytes), RAB5 localized to ∼85% of the endosomes containing EtpE-C–coated beads at 30 min after the beads were mixed with cells, but at 120 min RAB5 did not localize to any of the bead-containing endosomes (Fig. 6 A and C). In Etf-2-GFP–transfected cells, however, RAB5 localized to 100% of the endosomes containing EtpE-C–coated beads at 30 min after beads were mixed with cells, and at 120 min RAB5 remained localized to ∼90% of the bead-containing endosomes (Fig. 6 B and C). In GFP-transfected RF/6A cells, RAB7 localized to ∼20% of the endosomes containing EtpE-C–coated beads at 30 min after the beads were mixed with cells, and at 120 min RAB7 localized to almost all the bead-containing endosomes (Fig. 6 D and F). In Etf-2-GFP–transfected cells, however, RAB7 did not localize to any of the endosomes containing EtpE-C–coated beads at 30 min after beads were mixed with cells, and at 120 min RAB7 localization remained at less than 20% of the bead-containing endosomes (Fig. 6 E and F). Similarly, Etf-2-GFP–transfected RF/6A cells also exhibited delayed localization of LysoTracker Red to phagosomes containing EtpE-C–coated beads, in which LysoTracker Red localized to less than 20% of the bead-containing endosomes compared with nearly 100% of the bead-containing endosomes in GFP-transfected cells (Fig. 6 G–I).
Fig. 6.
Etf-2 localizes to endosomes containing EtpE-C–coated latex beads and delays the maturation of endosomes in nonphagocytic RF/6A cells. RF/6A cells were cotransfected with GFP (A, D, and G) or Etf-2-GFP (B, E, and H) and HA-RAB5 (A and B) or HA-RAB7 (D and E) for 2 d and then were incubated for 30–120 min with EtpE-C–coated Flash Red latex beads (1 µm, pseudocolored blue for merged images or white for enlarged single-channel panels). At 30–120 min after the addition of latex beads, cells were treated with LysoTracker Red for 10 min before fixation and were immediately observed with a DeltaVision microscope (G and H). In A, B, D, E, G, and H, representative images at 60 min after bead uptake are shown, and each boxed area is enlarged 4× on the right. (Scale bars: 10 μm.) (C, F, and I) Quantification of the localization of RAB5 (C), RAB7 (F), and LysoTracker Red in 100 endosomes that had taken up latex beads (I) in Etf-2-GFP–transfected or GFP-transfected RF/6A cells. Data are presented as the mean ± SD from three independent experiments. *P < 0.05, two-tailed t test.
These results indicated that, similar to observations with phagosomes containing noncoated beads in DH82 cells, Etf-2 quickly localized with RAB5 on endosomes containing EtpE-C–coated beads in RF/6A cells, prolonged RAB5 localization on the endosomal membrane, and delayed endosomal maturation. In the GFP control, the maturation of endosomes containing EtpE-C–coated beads in RF/6A cells was slower than the maturation of phagosomes containing noncoated beads in DH82 cells. In the presence of Etf-2, RAB5 retention and blockade of RAB7 localization were more efficient on endosomes containing EtpE-C–coated beads in RF/6A cells than on phagosomes containing noncoated beads in DH82 cells (SI Appendix, Fig. S5).
RABGAP5 Does Not Localize to Ehrlichial Inclusions, and Etf-2 Prevents RABGAP5 Localization to Endosomes Containing EtpE-C–Coated Latex Beads.
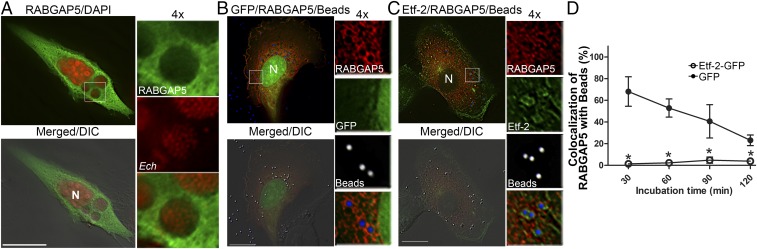
Expression of any of the three GFP-RAB5 isoforms (A, B, or C) results in the production of puncta (vesicles), and each RAB has been found to accumulate continuously on established Ehrlichia inclusions (19). RABGAP5 can interact with each of the three RAB5 isoforms (27) to stimulate GTP hydrolysis in vitro and disrupt endocytosis when overexpressed in cultured cells (27). Furthermore, RNAi of RABGAP5 induces a large early-endosome phenotype as seen when cells accumulate RAB5-GTP (27). Thus, we assessed RABGAP5 localization to ehrlichial inclusions. As shown in Fig. 7A, RABGAP5 did not localize to inclusions. Because RABGAP5 binds RAB5-GTP but not RAB5-GDP (27), and Etf-2 prolongs the localization of RAB5 to phagosomes and endosomes (SI Appendix, Figs. S5 and S6), we determined whether RABGAP5 could localize to endosomes containing EtpE-C–coated beads upon cotransfection of RF/6A cells with Etf-2-GFP or GFP (control). In GFP-transfected cells, RABGAP5 localized to ∼70% of the endosomes containing EtpE-C–coated beads at 30 min after the beads were added to cells, and at 120 min RABGAP5 localized to <20% of bead-containing endosomes (Fig. 7 B and D). In Etf-2-GFP–transfected cells, however, RABGAP5 did not localize to any of the endosomes containing EtpE-C–coated beads from 30–120 min (Fig. 7 C and D), indicating that Etf-2 prevents RABGAP5 localization to endosomes.
Fig. 7.
RABGAP5 does not localize to ehrlichial inclusions, and Etf-2 prevents RABGAP5 localization to endosomes containing EtpE-C–coated beads. (A) Ehrlichia-infected RF/6A cells were transfected with Myc-RABGAP5 at 1 dpi. Cells were fixed at 2 dpt (3 dpi) and were immunofluorescently labeled with anti-Myc (green) and DAPI (pseudocolored red). (B and C) RF/6A cells were cotransfected with Myc-RabGAP5 and GFP (B) or Etf-2-GFP (C) for 2 d and then were incubated for 30–120 min with EtpE-C–coated beads (pseudocolored blue for merged images or white for enlarged single-channel panels). Cells were fixed and immunostained with anti-Myc (red). Representative images at 60 min after bead uptake are shown. Boxed areas are enlarged 4× on the right. Merged/DIC, the fluorescence image is merged with the DIC image. (Scale bars: 10 μm.) (D) Quantification of the localization of Myc-RabGAP5 to 100 endosomes that had taken EtpE-C–coated beads in GFP- or Etf-2-GFP–transfected RF/6A cells. Data are presented as the mean ± SD from three independent experiments. *P < 0.05, two-tailed t test.
Etf-2 Enhances Ehrlichia Infection, and an Etf-2–Specific Antisense Peptide Nucleic Acid Inhibits Infection.
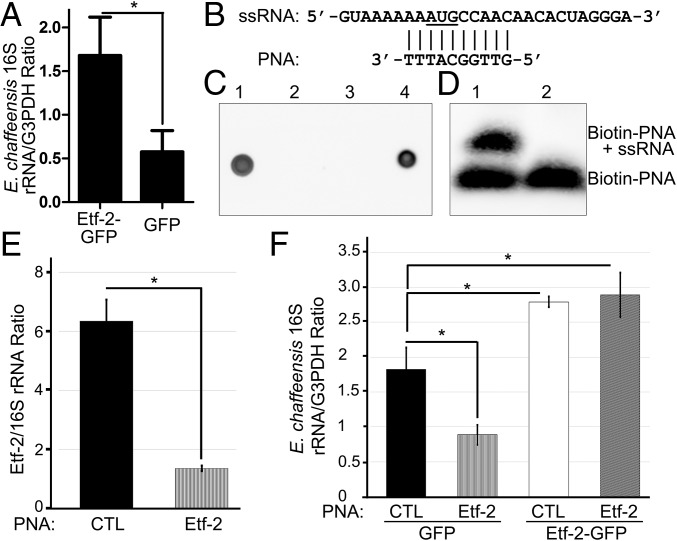
Because Etf-2 blocks endosomal maturation to late endosomes and RABGAP5 localization to endosomes containing EtpE-C–coated beads, we used qPCR to examine whether Etf-2 overexpression affects Ehrlichia infection. Compared with GFP-overexpressing control cells, Etf-2 overexpression significantly enhanced Ehrlichia infection (Fig. 8A). No Etf-2 mutants are currently available, because classical bacteriology techniques such as targeted mutagenesis are not readily applicable to obligatory intracellular bacteria, which include all members of the order Rickettsiales (E. chaffeensis belongs to this order), and, unlike facultative intracellular bacteria, knockout mutants for genes essential for intracellular infection (e.g., Etf-2) cannot be recovered (37). Therefore, we designed an antisense peptide nucleic acid (PNA) specific for Etf-2 (Fig. 8 B–D). Electroporation of Ehrlichia with this PNA significantly reduced Etf-2 mRNA expression (Fig. 8E) as well as its ability to infect host cells (Fig. 8F). Etf-2 PNA-mediated inhibition of Etf-2 expression and infection could be complemented by ectopic expression of Etf-2-GFP in host cells (Fig. 8F). These results indicated the Etf-2 is critical for Ehrlichia infection of human cells.
Fig. 8.
Overexpression of Etf-2 promotes ehrlichial infection, and Etf-2–specific antisense PNA inhibits ehrlichial infection. (A) HEK293 cells were transfected with Etf-1-GFP or GFP. At 8 hpt, freshly isolated E. chaffeensis was added, and cells were harvested at 2 dpi. Bacterial numbers in each sample were determined by qPCR using specific primers for Ehrlichia 16S rDNA and were normalized against the level of human G3PDH (glyceraldehyde 3-phosphate dehydrogenase). Data are presented as the mean ± SD from three independent experiments. *P < 0.05, two-tailed t test. (B–D) Etf-2 PNA binds the target ssRNA. (B) Etf-2 PNA targets the putative translation start site (underlined) of the mRNA. The upper line shows the ssRNA sequence designed for the hybridization assay. (C) Dot blot analysis of biotinylation of PNAs using HRP-conjugated streptavidin. Dots 1–4: 1, biotin-Etf-2 PNA; 2, unlabeled Etf-2 PNA; 3, unlabeled control (CTL) PNA; 4, biotin-CTL PNA. (D) EMSA of biotinylated Etf-2 PNA bound to ssRNA. Lane 1, labeled Etf-2 PNA incubated with ssRNA; lane 2, labeled Etf-2 PNA only. (E) Etf-2 PNA significantly reduces ehrlichial etf-2 mRNA expression. HEK293 cells infected with Etf-2–transfected or CTL PNA-transfected E. chaffeensis were harvested at 48 hpi and subjected to qRT-PCR analysis. The values reflect bacterial etf-2 mRNA normalized against 16S rRNA. (F) HEK293 cells expressing Etf-2 complemented the Etf-2 PNA knockdown of Ehrlichia infection. RNA samples prepared from HEK293 cells expressing Etf-2-GFP or GFP incubated with Etf-2–transfected or CTL PNA-transfected Ehrlichia were harvested at 48 hpi and subjected to qRT-PCR analysis. The values reflect bacterial 16S rRNA normalized against human G3PDH mRNA. Data indicate the mean ± SD from three independent experiments that had three replicates per sample. *P < 0.05, two-tailed t test or ANOVA.
Discussion
RAB GTPases are the central regulators of membrane trafficking and organelle identity in eukaryotic cells (28). RAB5 is found in nascent phagosomes and early endosomes that lack the microbicidal capacity required to kill invading pathogens; this requires subsequent maturation into late endosomes via the RAB5-to-RAB7 transition and ensuring fusion with lysosomes. In the present study, we established that E. chaffeensis has evolved a unique molecule, Etf-2, and a strategy to prevent the maturation of inclusions and thereby prevent their fusion with lysosomes. (A model of Etf-2 function in E. chaffeensis-infected cells is shown in SI Appendix, Fig. S6.) Proteins containing a TBC-like motif are commonly found among eukaryotes and function as GAPs for multiple families of small GTPases including RAB GTPases (46). Several bacterial proteins are also known to have GAP activity toward small GTPases, Cdc42, Rac, and/or Rho (47) Recently, TBC-domain GAPs specific to RAB1 were discovered among bacteria (Shigella, enteropathogenic E. coli, and Legionella), which are secreted into mammalian host cells by a bacterial secretion system to subvert ER membrane traffic (40, 48). For example, S. flexneri VirA, a type III secretion effector, contains TBC-like dual finger motifs and exhibits potent RABGAP activity toward RAB1, and specific inactivation of RAB1 by VirA disrupts ER-to-Golgi transport and suppresses ER-derived autophagy, facilitating bacterial intracellular survival (40).
Etf-2 harbors an unexpected TBC-like RABGAP motif and binds RAB5-GTP in vivo and in vitro with high affinity, but, remarkably, Etf-2 lacks RABGAP5 activity. Thus, in this sense Etf-2 behaves like a RAB5 effector. In a classic sense, the binding of RAB effectors to their cognate RABs is a dynamic process that is necessary for cyclic RAB activation/inactivation via GTP hydrolysis upon association with a GAP. Because effectors and GAPs share overlapping binding sites on RABs, dynamic binding ensures access of the GAP to the RAB upon dissociation of the effector. Because a GAP cannot act directly on a RAB–effector complex, dissociation must occur to allow access of the GAP to the active site of the GTPase to promote GTP hydrolysis. The dissociation rate of the effector protein can greatly influence the accessibility of the RAB to binding by its cognate GAP and therefore can indirectly regulate the speed of deactivation. GAPs have low affinity for GTP-bound RABs (Kd 20–200 μM) (41, 49). RAB5 interacts weakly with its effector molecules (Kd >0.9 µM) (50–52), and the affinity of Etf-2 for RAB5-GTP is much higher (Kd 21.3 nM). In this respect, the spontaneous dissociation of Etf-2–RAB5 complexes appears to be orders of magnitude too slow. Thus, possible roles for Etf-2 include blocking RAB5 inactivation and/or sequestering RAB5 on the surface of Ehrlichia inclusions. For Ehrlichia, which thrives in RAB5-decorated vacuoles in the host cytoplasm, preventing RAB5 GTPase activation by Etf-2 contributes to bacterial proliferation by blocking endosome maturation to late endosomes and subsequent fusion with lysosomes. Orthologous proteins of Etf-2 were identified in all sequenced Ehrlichia spp., including Ehrlichia canis, Ehrlichia ruminantium, Ehrlichia muris, and Ehrlichia sp. HF, suggesting that Etf-2 plays a critical role in the establishment of Ehrlichia infection. Among the family Anaplasmataceae, orthologs of Etf-2 were also identified in Neorickettsia and Wolbachia species, although the similarities are low (E-value >e−5). However, no homologs were found in Anaplasma spp. including Anaplasma phagocytophilum, which replicates in membrane-bound inclusions that are decorated with markers of autophagosomes but not those of endosomes (18, 53). Nonetheless, TBC-like motifs lacking RABGAP activity might be used by other unidentified prokaryotic or eukaryotic proteins to regulate RAB activity.
The mechanism of action of Etf-2 is also unique in that it binds RAB5-GTP at high affinity, which does not alter the membrane localization of certain downstream RAB5 effectors but seems to block RABGAP5 localization. Etf-2 localization to endosomes is dependent on the TBC-like motif, suggesting that this motif is critical for Etf-2 binding to RAB5-GTP and consequently blocking RABGAP5 binding to RAB5. Thus, Etf-2 binding may keep RAB5 in the active GTP-bound state, which in turn recruits and activates VPS34 (the endosomal PIK3C3), increases phosphatidylinositol 3-phosphate (PtdIns3P) prevalence on the membrane, and enhances recruitment of RAB5 effectors containing a FYVE domain, such as EEA1 and Rabankyrin-5, independently of RAB5 binding. These effectors normally bind to both RAB5-GTP and PtdIns3P, but a substantial increase in PtdIns3P in ehrlichial inclusions (19) is sufficient to recruit these effectors to the membrane (54). Indeed, a double FYVE-containing protein that lacks an RAB5-binding domain can be recruited to E. chaffeensis inclusions (19). Unlike L. pneumophila LidA, which lacks a TBC motif and binds at very high affinity to GTP- and GDP-bound RAB1b, RAB8a, and RAB6a (55, 56), Etf-2 binds specifically to GTP-bound RAB5. Thus, Etf-2 introduces a unique virulence mechanism for hijacking host vesicle trafficking. Whether the observed high-affinity binding of Etf-2 is specific to RAB5 remains to be analyzed.
Several intracellular pathogens prevent or delay the RAB5-to-RAB7 transition on endosomes. For example, Mycobacterium tuberculosis localizes in the RAB5-positive endocytic compartment by reducing the prevalence of PtdIns3P in the phagosomal membrane, thereby delaying phagosome maturation (57, 58). The mannose-capped lipoarabinomannan in the bacterial membrane is released into the phagosomal membrane and inactivates VPS34 that regenerates PtdIns3P (57). Concurrently, an M. tuberculosis lipid phosphatase, SapM, consumes PtdIns3P and arrests phagosome maturation (58, 59). PtdIns3P is specifically enriched in early endosome/phagosome membranes and stabilizes RAB5 and all its effectors. Its absence in M. tuberculosis infection interferes with the recruitment of the RAB5 effectors, and therefore M. tuberculosis-containing phagosomes mature to the RAB7-predominant state (60). The nucleoside diphosphate kinase of M. tuberculosis is a GAP specific for RAB5 and RAB7, interrupts VPS34 recruitment to phagosomes via hydrolysis of RAB5-GTP, and blocks the interaction of RAB7 with its effector RILP, ultimately leading to reduced phagolysosome fusion (61). Listeria monocytogenes GAPDH binds and carries out ADP ribosylation of RAB5, thereby impairing GDP/GTP exchange and blocking phagosome maturation (62). Tropheryma whipplei, which has a homolog of L. monocytogenes GAPDH, resides and replicates within RAB5- and RAB7-positive phagosomes that do not mature into phagolysosomes (63). L. pneumophila VipD is an RAB5-activated phospholipase A1 that catalyzes the removal of PtdIns3P and EEA1 from the vacuolar membrane to prevent Legionella-containing vacuoles from acquiring RAB5 endosomal components (64). Arrest of the maturation of RAB5-positive Ehrlichia inclusions is either the opposite of or distinct from these examples because (i) PtdIns3P is highly enriched in the inclusion membrane; (ii) GFP-RAB5 endosomes are continuously acquired by Ehrlichia inclusions; and (iii) RAB5 and the RAB5 effectors EEA1, VPS34, and Rabankyrin-5 are stably present on the inclusion membrane (19).
Our data suggest that Etf-2 can inhibit bead phagocytosis. Overexpression of RAB5CA was reported to increase phagocytosis of serum protein-opsonized latex beads but not C3- or IgG-opsonized beads (65). We suspect Etf-2, by keeping endogenous RAB5 at the active state, interferes with the recycling of endogenous RAB5 that is required for new bead uptake. This would be also beneficial for E. chaffeensis as a population, because Etf-2 synthesized and secreted by strong intracellular bacteria will interfere with new bacterial uptake, consequently reducing competition for scarce host cell energy, nutrients, and resources for efficient bacterial proliferation. Compared with persistent blockade of lysosomal fusion with Ehrlichia inclusions (18), we found that ectopic expression of Etf-2 led to only a transient blockade of lysosomal fusion with latex bead-containing phagosomes. Because Ehrlichia cannot replicate or even survive outside a eukaryotic cell, continuous biosynthesis and T4SS-mediated secretion of Etf-2 by Ehrlichia likely ensure sustained blockade of lysosomal fusion with ehrlichial inclusions. The blockade persists much longer with endosomes containing EtpE-C–coated beads than with phagosomes containing noncoated beads. Whether EtpE-C has a synergistic effect on blocking endosome maturation or this effect is due to different host cell types, i.e., phagocytes vs. nonphagocytes, remains to be studied. It is also possible that an additional ehrlichial factor or mechanism exists that prolongs the inhibition of RAB5 GTPase.
Our study reveals that Etf-2 promotes infection of eukaryotic cells by E. chaffeensis and that PNA-mediated inhibition of Etf-2 counters infection. Etf-1, another ehrlichial T4SS effector, recruits the master regulator of autophagy, VPS34-Beclin 1, to endosomes in the presence of RAB5-GTP to induce cellular autophagy (19). Through stabilization of GTP-bound RAB5, Etf-2 potentially serves another function—ensuring that RAB5-GTP is available for Etf-1–induced autophagy (19). Furthermore, the blockade of lysosomal fusion by Etf-2 on Ehrlichia inclusions prevents the maturation of Etf-1–induced autophagosomes to autolysosomes. Thus, Etf-2, via its high-affinity binding to GTP-bound RAB5 and maintenance of RAB5 in the activated state, may have at least three functions that promote ehrlichial infection, namely blocking endosomal maturation and subsequent fusion with lysosomes (summarized in SI Appendix, Fig. S6), expansion of the inclusion compartment by homotypic fusion of endosomes, and aiding RAB5-regulated autophagy.
The Etf-2 knockin approach used in this study would facilitate the identification of T4SS effectors and other bacterial factors and their functions not only in E. chaffeensis but also in other bacteria in which classic genetic manipulation is difficult.
Materials and Methods
Additional experimental details are provided in SI Appendix, Supplemental Materials and Methods. Supplementary figures and tables are presented in SI Appendix.
Bacteria, Cell Culture, and Antibodies.
E. chaffeensis Arkansas strain (66) was cultured in THP-1 cells (ATCC) (67). The detailed culture method and a list of antibodies used are provided in SI Appendix.
Transformation of E. chaffeensis with FLAG-Etf-2C Himar Plasmid.
The pCis-FLAG-Etf-2-SS-Himar A7 plasmid expressing FLAG-tagged Etf-2C (the C terminus of Etf-2 containing a T4SS signal, amino acids 152–264) and the spectinomycin/streptomycin antibiotic resistance gene (aad) was created from the pCis-mCherry-SS-Himar A7 construct (38). The detailed method is described in SI Appendix.
Cloning.
A gene encoding Etf-2∆HY was PCR amplified from Ehrlichia genomic DNA using two-step PCR with overlapping primers (SI Appendix, Table S1) and subsequently was cloned into pET33b(+) (Novagen) to create a plasmid expressing 6× His-tagged Etf-2∆HY. For expression in mammalian cells, full-length Etf-2 was codon optimized, custom synthesized (GenScript) (SI Appendix, Table S1), and recloned into pEGFP-N1 (Clontech) to create plasmids encoding full-length Etf-2 (amino acids 1–264)-GFP, Etf-2N (amino acids 1–114)-GFP, Etf-2C1 (amino acids 135–264)-GFP, Etf-2C (amino acids 152–264)-GFP, and Etf-2 (amino acids Δ132–151)-GFP. Etf-2R188A-GFP, Etf-2Q245A-GFP, and Etf-2DM-GFP were made using the QuikChange Site-Directed Mutagenesis kit (Stratagene). HA-tagged RAB5 (RAB5A), RAB5-DN, and RAB5-CA were constructed previously (19).
Yeast Two-Hybrid Assay.
Codon-optimized full-length Etf-2 was cloned into vector pGBKT7 (Clontech) and transformed into yeast strain Y187 using the Quick and Easy yeast transformation kit (Clontech). RAB5-WT, RAB5-CA, and RAB5-DN were cloned into vector pGADT7 and were transformed into yeast strain AH109. Yeast two-hybrid assays were performed according to the user manual for the Matchmaker Gold Yeast Two-Hybrid System (Clontech).
MST and RAB5 GAP Assay.
Full-length human RAB5 (RAB5A) was expressed and purified as described (68). GTPγS or GDPβS was added to apoRAB5 with incubation for 30 min on ice. RAB5 complexed with GTPγS (or GDPβS) was then labeled using an Alexa Fluor 647 Protein Labeling Kit (Molecular Probes). The MST concentration screen and experiments were also performed with Monolith NT.115, and data were collected and analyzed as described (69). For RAB5 GAP assay, affinity-purified GST-RAB5 on glutathione-Sepharose resin was preloaded with [α-32P]GTP (0.1 μM) for 15 min at room temperature followed by a GTP hydrolysis reaction at 30 °C in 20 mM Tris⋅HCl (pH 8.0), 10 mM MgCl2, and 1 mM DTT in the absence or presence of RABGAP5 or Etf-2∆HY (10 nM). Samples were taken at the indicated time and were subjected to TLC to separate the hydrolysis product [α-32P]GDP from the substrate [α-32P]GTP, followed by autoradiography and quantification using a PhosphorImager (GE Healthcare Life Sciences).
Coimmunoprecipitation.
HEK293 cells (2 × 106) were cotransfected with 5 μg each of plasmids Etf-2-GFP and HA-RAB5 (WT, DN, or CA) or HA-RAB7 by electroporation (Bio-Rad Gene Pulser Xcell electroporation system) at 100 V and 1,000 μF in a 2-mm cuvette and were cultured for 2 d in a 25-cm2 flask. Cells were lysed for 15 min in lysis buffer [20 mM Hepes (pH 7.5), 100 mM NaCl, 5 mM MgCl2, 1% (wt/vol) Triton X-100] and then were incubated for 2 h with mouse monoclonal anti-HA cross-linked to protein G-Sepharose beads (BioLegend). After extensive washing with lysis buffer, bound proteins were dissociated from the beads with 2× SDS/PAGE sample buffer, and Western blotting was subsequently performed with anti-GFP or anti-HA.
Transfection and Fluorescence Labeling.
RF/6A cells cultured on coverslips in the wells of a 24-well plate were cotransfected with plasmids encoding Etf-2- or Etf-2 mutant-GFP, and HA-RAB5 (WT, DN, or CA) using Lipofectamine 2000 (Invitrogen). Subcellular localization was determined at 40–48 h posttransfection (hpt). Cells were fixed in 4% paraformaldehyde (PFA) and incubated with mouse monoclonal anti-HA in PGS [PBS supplemented with 0.5% BSA (Sigma), 0.1% gelatin (Sigma), and 0.1% saponin (Sigma)] followed by AF555-conjugated goat anti-mouse IgG in PGS.
Ehrlichia-infected RF/6A cells on coverslips in the wells of a 24-well plate at 1 d postinfection (dpi) were transfected with a plasmid encoding Etf-2-GFP, Etf-2N-GFP, Etf-2C-GFP, or Myc-RABGAP5. Cells were fixed in 4% PFA at 15–17 hpt (for Etf-2 localization) or 40–48 hpt (for RABGAP5). RABGAP5 localization was detected by labeling with mouse anti-Myc and AF488-conjugated anti-mouse IgG in PGS. DAPI was used to label DNA in the host-cell nucleus and Ehrlichia.
To detect native Etf-2 localization, E. chaffeensis-infected RF/6A cells at 2 dpi were fixed in 4% PFA and then were labeled with antigen-affinity–purified rabbit anti–Etf-2C IgG preadsorbed with uninfected RF/6A cells in PGS.
Internalization of EtpE-C–Coated Latex Beads in RF/6A Cells.
Flash Red latex beads at 5–6 × 106 beads in 180 μL of 25-mM MES buffer (pH 6.0) were coated with 200 ng of recombinant EtpE-C proteins (45). RF/6A cells cultured on coverslips in a 24-well plate were cotransfected with plasmids encoding Etf-2-GFP or GFP and HA-RAB5 (WT), MYC-RABGAP5, or HA-RAB7 with Lipofectamine 3000 (Invitrogen) for 2 d. Freshly prepared EtpE-C–coated beads were added to the wells of a 24-well plate (∼5 × 106 beads per well). After incubation at 37 °C with 5% CO2 in a humidified atmosphere for 30–120 min, uninternalized beads were removed by washing with PBS. Cells were then fixed in 4% PFA and incubated with mouse monoclonal anti-Myc or monoclonal anti-HA followed by AF555-conjugated goat anti-mouse IgG in PGS.
For labeling with LysoTracker Red, RF/6A cells cultured on coverslips in a 24-well plate were cotransfected with a plasmid encoding Etf-2-GFP or GFP for 2 d. Freshly prepared EtpE-C–coated beads were added to the wells of a 24-well plate (∼5 ×106 beads per well). After incubation at 37 °C with 5% CO2 in a humidified atmosphere for 30–120 min, cells were incubated with LysoTracker Red for 10 min. Uninternalized beads were removed by washing with PBS, and cells were fixed in 3.2% PFA and then were immediately observed using a DeltaVision Deconvolution microscope. Detailed methods, image analysis, and uptake of latex beads in DH82 cells are described in SI Appendix.
Effects of Etf-2-GFP Overexpression, etf-2 Knockdown with PNA, and Complementation Analysis of Ehrlichia Infection.
Details of the effects of Etf-2-GFP overexpression, etf-2 knockdown with PNA, and complementation analysis of Ehrlichia infection are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. John Brumell at the University of Toronto for providing the plasmid encoding GFP-RAB5A, Dr. Francis Barr at the University of Oxford for providing plasmid pcDNA3.1-MYC-RABGAP5, Dr. Ulrike Munderloh at the University of Minnesota for providing plasmid pCis-mCherry-SS-Himar A7, and Dr. Yongcong Tan for his pilot studies on Etf-2 localization. This work was supported in part by NIH Grants R01AI054476 (to Y.R.) and R01GM074692 (to G.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806904115/-/DCSupplemental.
References
- 1.Pizarro-Cerdá J, et al. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect Immun. 1998;66:5711–5724. doi: 10.1128/iai.66.12.5711-5724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Celli J, Salcedo SP, Gorvel JP. Brucella coopts the small GTPase Sar1 for intracellular replication. Proc Natl Acad Sci USA. 2005;102:1673–1678. doi: 10.1073/pnas.0406873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 2002;295:679–682. doi: 10.1126/science.1067025. [DOI] [PubMed] [Google Scholar]
- 4.Hackstadt T, Rockey DD, Heinzen RA, Scidmore MA. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 1996;15:964–977. [PMC free article] [PubMed] [Google Scholar]
- 5.Sturgill-Koszycki S, et al. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science. 1994;263:678–681. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- 6.D’Costa VM, et al. Salmonella disrupts host endocytic trafficking by SopD2-mediated inhibition of Rab7. Cell Rep. 2015;12:1508–1518. doi: 10.1016/j.celrep.2015.07.063. [DOI] [PubMed] [Google Scholar]
- 7.Spanò S, Gao X, Hannemann S, Lara-Tejero M, Galán JE. A bacterial pathogen targets a host Rab-family GTPase defense pathway with a GAP. Cell Host Microbe. 2016;19:216–226. doi: 10.1016/j.chom.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinzen RA, Hackstadt T, Samuel JE. Developmental biology of Coxiella burnettii. Trends Microbiol. 1999;7:149–154. doi: 10.1016/s0966-842x(99)01475-4. [DOI] [PubMed] [Google Scholar]
- 9.Straley SCHP, Harmon PA. Yersinia pestis grows within phagolysosomes in mouse peritoneal macrophages. Infect Immun. 1984;45:655–659. doi: 10.1128/iai.45.3.655-659.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marquis H, Doshi V, Portnoy DA. The broad-range phospholipase C and a metalloprotease mediate listeriolysin O-independent escape of Listeria monocytogenes from a primary vacuole in human epithelial cells. Infect Immun. 1995;63:4531–4534. doi: 10.1128/iai.63.11.4531-4534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernardini ML, Mounier J, d’Hauteville H, Coquis-Rondon M, Sansonetti PJ. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc Natl Acad Sci USA. 1989;86:3867–3871. doi: 10.1073/pnas.86.10.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rikihisa Y, Ito S. Intracellular localization of Rickettsia tsutsugamushi in polymorphonuclear leukocytes. J Exp Med. 1979;150:703–708. doi: 10.1084/jem.150.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitworth T, Popov VL, Yu XJ, Walker DH, Bouyer DH. Expression of the Rickettsia prowazekii pld or tlyC gene in Salmonella enterica serovar typhimurium mediates phagosomal escape. Infect Immun. 2005;73:6668–6673. doi: 10.1128/IAI.73.10.6668-6673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paddock CD, Childs JE. Ehrlichia chaffeensis: A prototypical emerging pathogen. Clin Microbiol Rev. 2003;16:37–64. doi: 10.1128/CMR.16.1.37-64.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker DH, et al. Ehrlichia chaffeensis: A prevalent, life-threatening, emerging pathogen. Trans Am Clin Climatol Assoc. 2004;115:375–382, discussion 382–384. [PMC free article] [PubMed] [Google Scholar]
- 16.Adams DA, et al. Nationally Notifiable Infectious Conditions Group Summary of notifiable infectious diseases and conditions - United States, 2015. MMWR Morb Mortal Wkly Rep. 2017;64:1–143. doi: 10.15585/mmwr.mm6453a1. [DOI] [PubMed] [Google Scholar]
- 17.Barnewall RE, Rikihisa Y, Lee EH. Ehrlichia chaffeensis inclusions are early endosomes which selectively accumulate transferrin receptor. Infect Immun. 1997;65:1455–1461. doi: 10.1128/iai.65.4.1455-1461.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mott J, Barnewall RE, Rikihisa Y. Human granulocytic ehrlichiosis agent and Ehrlichia chaffeensis reside in different cytoplasmic compartments in HL-60 cells. Infect Immun. 1999;67:1368–1378. doi: 10.1128/iai.67.3.1368-1378.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin M, et al. Ehrlichia secretes Etf-1 to induce autophagy and capture nutrients for its growth through RAB5 and class III phosphatidylinositol 3-kinase. Autophagy. 2016;12:2145–2166. doi: 10.1080/15548627.2016.1217369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li G, Marlin MC. Rab family of GTPases. Methods Mol Biol. 2015;1298:1–15. doi: 10.1007/978-1-4939-2569-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfeffer SR. Rab GTPase regulation of membrane identity. Curr Opin Cell Biol. 2013;25:414–419. doi: 10.1016/j.ceb.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ullrich O, et al. Rab GDP dissociation inhibitor as a general regulator for the membrane association of rab proteins. J Biol Chem. 1993;268:18143–18150. [PubMed] [Google Scholar]
- 23.Christoforidis S, McBride HM, Burgoyne RD, Zerial M. The Rab5 effector EEA1 is a core component of endosome docking. Nature. 1999;397:621–625. doi: 10.1038/17618. [DOI] [PubMed] [Google Scholar]
- 24.Rybin V, et al. GTPase activity of Rab5 acts as a timer for endocytic membrane fusion. Nature. 1996;383:266–269. doi: 10.1038/383266a0. [DOI] [PubMed] [Google Scholar]
- 25.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 26.Vitale G, et al. The GDP/GTP cycle of Rab5 in the regulation of endocytotic membrane traffic. Cold Spring Harb Symp Quant Biol. 1995;60:211–220. doi: 10.1101/sqb.1995.060.01.024. [DOI] [PubMed] [Google Scholar]
- 27.Haas AK, Fuchs E, Kopajtich R, Barr FA. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat Cell Biol. 2005;7:887–893. doi: 10.1038/ncb1290. [DOI] [PubMed] [Google Scholar]
- 28.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 29.Grohmann E, Christie PJ, Waksman G, Backert S. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol Microbiol. 2018;107:455–471. doi: 10.1111/mmi.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 32.Rikihisa Y. Role and function of the type IV secretion system in anaplasma and ehrlichia species. Curr Top Microbiol Immunol. 2017;413:297–321. doi: 10.1007/978-3-319-75241-9_12. [DOI] [PubMed] [Google Scholar]
- 33.Liu H, Bao W, Lin M, Niu H, Rikihisa Y. Ehrlichia type IV secretion effector ECH0825 is translocated to mitochondria and curbs ROS and apoptosis by upregulating host MnSOD. Cell Microbiol. 2012;14:1037–1050. doi: 10.1111/j.1462-5822.2012.01775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma P, Teymournejad O, Rikihisa Y. Peptide nucleic acid knockdown and intra-host cell complementation of Ehrlichia type IV secretion system effector. Front Cell Infect Microbiol. 2017;7:228. doi: 10.3389/fcimb.2017.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barr F, Lambright DG. Rab GEFs and GAPs. Curr Opin Cell Biol. 2010;22:461–470. doi: 10.1016/j.ceb.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin M, Kikuchi T, Brewer HM, Norbeck AD, Rikihisa Y. Global proteomic analysis of two tick-borne emerging zoonotic agents: Anaplasma phagocytophilum and Ehrlichia chaffeensis. Front Microbiol. 2011;2:24. doi: 10.3389/fmicb.2011.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McClure EE, et al. Engineering of obligate intracellular bacteria: Progress, challenges and paradigms. Nat Rev Microbiol. 2017;15:544–558. doi: 10.1038/nrmicro.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng C, et al. Targeted and random mutagenesis of Ehrlichia chaffeensis for the identification of genes required for in vivo infection. PLoS Pathog. 2013;9:e1003171. doi: 10.1371/journal.ppat.1003171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stenmark H, et al. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong N, et al. Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell. 2012;150:1029–1041. doi: 10.1016/j.cell.2012.06.050. [DOI] [PubMed] [Google Scholar]
- 41.Pan X, Eathiraj S, Munson M, Lambright DG. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 2006;442:303–306. doi: 10.1038/nature04847. [DOI] [PubMed] [Google Scholar]
- 42.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 43.Werb Z, Cohn ZA. Plasma membrane synthesis in the macrophage following phagocytosis of polystyrene latex particles. J Biol Chem. 1972;247:2439–2446. [PubMed] [Google Scholar]
- 44.Chazotte B. Labeling lysosomes in live cells with LysoTracker. Cold Spring Harb Protoc. 2011;2011:pdb.prot5571. doi: 10.1101/pdb.prot5571. [DOI] [PubMed] [Google Scholar]
- 45.Mohan Kumar D, et al. Ehrlichia chaffeensis uses its surface protein EtpE to bind GPI-anchored protein DNase X and trigger entry into mammalian cells. PLoS Pathog. 2013;9:e1003666. doi: 10.1371/journal.ppat.1003666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernards A. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim Biophys Acta. 2003;1603:47–82. doi: 10.1016/s0304-419x(02)00082-3. [DOI] [PubMed] [Google Scholar]
- 47.Stebbins CE, Galán JE. Structural mimicry in bacterial virulence. Nature. 2001;412:701–705. doi: 10.1038/35089000. [DOI] [PubMed] [Google Scholar]
- 48.Ingmundson A, Delprato A, Lambright DG, Roy CR. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature. 2007;450:365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- 49.Albert S, Will E, Gallwitz D. Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO J. 1999;18:5216–5225. doi: 10.1093/emboj/18.19.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eathiraj S, Pan X, Ritacco C, Lambright DG. Structural basis of family-wide Rab GTPase recognition by rabenosyn-5. Nature. 2005;436:415–419. doi: 10.1038/nature03798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mishra A, Eathiraj S, Corvera S, Lambright DG. Structural basis for Rab GTPase recognition and endosome tethering by the C2H2 zinc finger of early endosomal autoantigen 1 (EEA1) Proc Natl Acad Sci USA. 2010;107:10866–10871. doi: 10.1073/pnas.1000843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rai A, Goody RS, Müller MP. Multivalency in Rab effector interactions. Small GTPases. 2017:1–7. doi: 10.1080/21541248.2016.1265700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niu H, Xiong Q, Yamamoto A, Hayashi-Nishino M, Rikihisa Y. Autophagosomes induced by a bacterial Beclin 1 binding protein facilitate obligatory intracellular infection. Proc Natl Acad Sci USA. 2012;109:20800–20807. doi: 10.1073/pnas.1218674109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dumas JJ, et al. Multivalent endosome targeting by homodimeric EEA1. Mol Cell. 2001;8:947–958. doi: 10.1016/s1097-2765(01)00385-9. [DOI] [PubMed] [Google Scholar]
- 55.Schoebel S, Cichy AL, Goody RS, Itzen A. Protein LidA from Legionella is a Rab GTPase supereffector. Proc Natl Acad Sci USA. 2011;108:17945–17950. doi: 10.1073/pnas.1113133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng W, et al. Structural insights into a unique Legionella pneumophila effector LidA recognizing both GDP and GTP bound Rab1 in their active state. PLoS Pathog. 2012;8:e1002528. doi: 10.1371/journal.ppat.1002528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fratti RA, Chua J, Vergne I, Deretic V. Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. Proc Natl Acad Sci USA. 2003;100:5437–5442. doi: 10.1073/pnas.0737613100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Puri RV, Reddy PV, Tyagi AK. Secreted acid phosphatase (SapM) of Mycobacterium tuberculosis is indispensable for arresting phagosomal maturation and growth of the pathogen in guinea pig tissues. PLoS One. 2013;8:e70514. doi: 10.1371/journal.pone.0070514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saikolappan S, et al. The fbpA/sapM double knock out strain of Mycobacterium tuberculosis is highly attenuated and immunogenic in macrophages. PLoS One. 2012;7:e36198. doi: 10.1371/journal.pone.0036198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Purdy GE, Owens RM, Bennett L, Russell DG, Butcher BA. Kinetics of phosphatidylinositol-3-phosphate acquisition differ between IgG bead-containing phagosomes and Mycobacterium tuberculosis-containing phagosomes. Cell Microbiol. 2005;7:1627–1634. doi: 10.1111/j.1462-5822.2005.00580.x. [DOI] [PubMed] [Google Scholar]
- 61.Sun J, et al. Mycobacterial nucleoside diphosphate kinase blocks phagosome maturation in murine RAW 264.7 macrophages. PLoS One. 2010;5:e8769. doi: 10.1371/journal.pone.0008769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alvarez-Dominguez C, et al. Characterization of a Listeria monocytogenes protein interfering with Rab5a. Traffic. 2008;9:325–337. doi: 10.1111/j.1600-0854.2007.00683.x. [DOI] [PubMed] [Google Scholar]
- 63.Mottola G, et al. Tropheryma whipplei, the agent of Whipple’s disease, affects the early to late phagosome transition and survives in a Rab5- and Rab7-positive compartment. PLoS One. 2014;9:e89367. doi: 10.1371/journal.pone.0089367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gaspar AH, Machner MP. VipD is a Rab5-activated phospholipase A1 that protects Legionella pneumophila from endosomal fusion. Proc Natl Acad Sci USA. 2014;111:4560–4565. doi: 10.1073/pnas.1316376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duclos S, Corsini R, Desjardins M. Remodeling of endosomes during lysosome biogenesis involves ‘kiss and run’ fusion events regulated by rab5. J Cell Sci. 2003;116:907–918. doi: 10.1242/jcs.00259. [DOI] [PubMed] [Google Scholar]
- 66.Dawson JE, et al. Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J Clin Microbiol. 1991;29:2741–2745. doi: 10.1128/jcm.29.12.2741-2745.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barnewall RE, Rikihisa Y. Abrogation of gamma interferon-induced inhibition of Ehrlichia chaffeensis infection in human monocytes with iron-transferrin. Infect Immun. 1994;62:4804–4810. doi: 10.1128/iai.62.11.4804-4810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu G, et al. High resolution crystal structures of human Rab5a and five mutants with substitutions in the catalytically important phosphate-binding loop. J Biol Chem. 2003;278:2452–2460. doi: 10.1074/jbc.M211042200. [DOI] [PubMed] [Google Scholar]
- 69.Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol. 2011;9:342–353. doi: 10.1089/adt.2011.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.