Significance
In eukaryotes, about one-third of all proteins are folded in the endoplasmic reticulum (ER). When protein folding demand exceeds capacity of the ER, the unfolded protein response (UPR) is activated to counteract ER stress. However, the mechanisms of adaptation to ER stress are not restricted to the UPR. Here, we report that gain of chromosome II in yeast allows adaptation of cells to protein misfolding. We narrowed down the effect of chromosome II duplication to three genes, which promote ER stress resistance independently of the UPR. Overexpression of these genes was sufficient to protect cells from ER stress and led to the elimination of aneuploidy. Thus, aneuploidy can serve as an important genome adaptation protecting cells against ER stress.
Keywords: aneuploidy, ER stress resistance, Saccharomyces cerevisiae
Abstract
The yeast genome becomes unstable during stress, which often results in adaptive aneuploidy, allowing rapid activation of protective mechanisms that restore cellular homeostasis. In this study, we performed a genetic screen in Saccharomyces cerevisiae to identify genome adaptations that confer resistance to tunicamycin-induced endoplasmic reticulum (ER) stress. Whole-genome sequencing of tunicamycin-resistant mutants revealed that ER stress resistance correlated significantly with gains of chromosomes II and XIII. We found that chromosome duplications allow adaptation of yeast cells to ER stress independently of the unfolded protein response, and that the gain of an extra copy of chromosome II alone is sufficient to induce protection from tunicamycin. Moreover, the protective effect of disomic chromosomes can be recapitulated by overexpression of several genes located on chromosome II. Among these genes, overexpression of UDP-N-acetylglucosamine-1-P transferase (ALG7), a subunit of the 20S proteasome (PRE7), and YBR085C-A induced tunicamycin resistance in wild-type cells, whereas deletion of all three genes completely reversed the tunicamycin-resistance phenotype. Together, our data demonstrate that aneuploidy plays a critical role in adaptation to ER stress by increasing the copy number of ER stress protective genes. While aneuploidy itself leads to proteotoxic stress, the gene-specific effects of chromosome II aneuploidy counteract the negative effect resulting in improved protein folding.
In eukaryotic cells, membrane and secreted proteins are folded in the endoplasmic reticulum (ER) lumen, before being exported to their target organelles (1). If the protein folding needs exceed the ER folding capacity, cells accumulate misfolded proteins and experience ER stress. The accumulation of misfolded proteins in the ER leads to activation of several intracellular signal transduction pathways, collectively named the unfolded protein response (UPR), to restore the ER homeostasis. Upon UPR activation, different mechanisms alleviate ER stress through increased expression of the ER-resident chaperones and enzymes involved in protein folding, the ER-associated degradation (ERAD) of the misfolded proteins, and the reduction of protein synthesis to limit the number of nascent peptides entering the ER lumen (2).
In yeast, the UPR is initiated by activation of the ER transmembrane sensor Ire1. Ire1 senses the accumulation of misfolded or unfolded proteins in the ER and then autophosphorylates and oligomerizes to activate its C-terminal RNase domain (3–6). Subsequently, the Ire1 RNase domain excises the translation-inhibitory intron in HAC1 mRNA, allowing translation of the Hac1 transcription factor (7–9). Hac1 then induces UPR target gene expression, including ER resident chaperones and enzymes involved in protein folding. However, the mechanisms of adaptation to ER stress are not restricted to the UPR. In addition, several Hac1-independent mechanisms that are activated in response to accumulation of misfolded proteins and are required for cell fitness during ER stress have been identified in yeast (10–12).
Gross genomic rearrangements such as aneuploidy (an alteration in chromosome number) are often associated with slow cell growth, imbalance in protein composition, and proteotoxicity, but may also serve as a mechanism to regulate gene dosage (13, 14). Recent studies have shown that adaptive aneuploidy can be beneficial under selective pressure and is often used by cells as a mechanism to up-regulate protective genes by changing chromosomal copy number (15). For example, in response to thiol-peroxidase deficiency, cells increase expression of genes that counteract oxidative stress through the duplication of chromosome (Chr) XI (16). In addition, aneuploidy has been shown to facilitate adaptation to telomerase deficiency induced by growth at elevated temperatures (17, 18).
Here, we report that aneuploidy also plays a key role in survival and adaptation of yeast during exposure to ER stress. In this study, we performed a genetic screen in Saccharomyces cerevisiae to identify mutants that are resistant to tunicamycin (TM)-induced ER stress. Whole-genome sequencing of TM-resistant mutants revealed that they acquired extra copies of multiple chromosomes. We find that the ER stress resistance in these mutants is specifically due to Chr II aneuploidy and is independent of the Hac1 transcription factor. Our analyses reveal that the protective effect of a Chr II disome against ER stress requires a combined function of at least three genes (ALG7, PRE7, and YBR085C-A) located on this chromosome. Among them, overexpression of ALG7, which encodes a key enzyme in the dolichol pathway of protein N-linked glycosylation, allowed cells to lose the extra copy of Chr II. Interestingly, we find that the N-glycan synthesis pathway is also activated in the UPR-deficient cells, which allows cells to compensate for the lack of the UPR by increasing production of UDP-N-acetylglucosamine (UDP-GlcNAc), a precursor for N-linked glycosylation. Taken together, our data demonstrate that adaptive aneuploidy is a critical mediator of ER stress resistance.
Results
Aneuploidy Allows Rapid Evolution of Yeast Cells Resistant to ER Stress.
To characterize the mechanisms by which cells adapt to ER stress, we performed a genetic screen in S. cerevisiae. To this end, wild-type haploid BY4741 cells were grown in the presence of a lethal dose of tunicamycin (TM), an inhibitor of N-linked glycosylation in the ER (19). We identified several tunicamycin-resistant (TM-R) clones that spontaneously evolved stress resistance and then selected five of them for further analysis (Fig. 1 A and B). We found that TM resistance in these strains was associated with a decrease in growth rate compared with wild-type cells (Fig. 1C and SI Appendix, Fig. S1A). We also observed that TM-R strains had decreased replicative lifespan and were not able to grow on glycerol, a nonfermentable carbon source, suggesting that TM-R cells lack respiratory capacity (SI Appendix, Figs. S1B and S2A). Prior work has indicated that a reduction in global mRNA translation through depletion of ribosomal proteins can enhance resistance to TM in yeast (20). Although TM-R strains demonstrate slow growth and decreased lifespan, we found that there were no significant changes in the rate of protein translation in these mutants as evident from polysome profile analysis (SI Appendix, Fig. S2B).
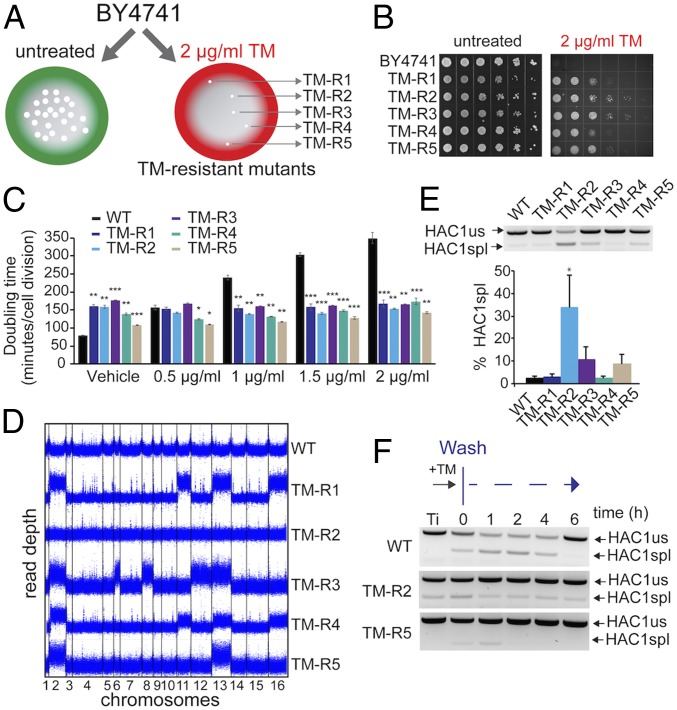
Fig. 1.
Genetic screen identifies gains of Chr II and XIII to be associated with resistance to ER stress. (A) Experimental design of the genetic screen used to isolate mutants that acquired resistance to TM-induced ER stress. (B) Representative images of wild-type (BY4741) cells and TM-resistant (TM-R) mutants grown in the presence of indicated concentrations of TM. Serial dilutions (10×) of logarithmically growing cells were spotted on control agar plates or plates with TM and incubated for 48 h at 30 °C. (C) Doubling time for TM-R strains in the presence of indicated concentrations of TM. Doubling time was calculated using the Yeast Outgrowth Data Analyzer software. Error bars represent SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, Welch’s t test. (D) Duplication of chromosomes in TM-R strains was determined by whole-genome sequencing. Read depth was calculated in 100-bp windows. (E) Analysis of HAC1 mRNA splicing in TM-R mutants. Levels of spliced (spl) and unspliced (us) HAC1 mRNA were detected by RT-PCR. Error bars represent SEM (n = 3). *P < 0.05, Student’s t test. (F) Analysis of spliced HAC1 mRNA after TM washout in the TM-R2 and TM-R5 mutants. Logarithmically growing cells were treated with 1 μg/mL TM for 30 min; cells were washed and incubated in fresh media for indicated time. Levels of spliced (spl) and unspliced (us) HAC1 mRNA were detected by RT-PCR.
To identify genome adaptations that may explain resistance to ER stress, we performed whole-genome sequencing of the TM-resistant mutants. Surprisingly, we found that many TM-R mutants acquired extra copies of multiple chromosomes (Fig. 1D). Aneuploidy, or an abnormal number of chromosomes, is associated with multiple human disorders including cancer, Down syndrome, and aging. Although aneuploidy in general leads to genome instability and proteotoxic stress, it can also be adaptive (16, 21). Previous studies have shown that aneuploidy can induce cell survival and rapid adaptation in response to stress (22, 23). Interestingly, Chr II and Chr XIII were duplicated in four different mutants, whereas the TM-R2 strain did not show any aneuploidy.
To establish whether ER stress-resistant mutants activate the UPR signaling, we analyzed splicing of the HAC1 mRNA. We found that the fraction of spliced HAC1 mRNA was significantly (P < 0.05) increased in the nonaneuploid TM-R2 strain compared with wild-type cells (34% of HAC1 mRNA was spliced for TM-R2) (Fig. 1E). However, in the aneuploid TM-R strains, the fraction of the spliced HAC1 mRNA was not significantly increased. We also monitored recovery kinetics of the UPR in the TM-R strains by monitoring levels of the spliced HAC1 mRNA over time following removal of TM (Fig. 1F and SI Appendix, Fig. S3). Interestingly, the UPR activity deviated more significantly in wild-type cells compared with TM-R2 and TM-R5. However, unlike wild-type and TM-R5 cells, the TM-R2 strain continued to display the spliced form of HAC1 even several hours after TM washout. Together, these data suggest that aneuploidy allows adaptation of yeast cells to ER stress and promotes resistance to TM independently of the UPR.
Chr II Duplication Confers Resistance to TM-Induced ER Stress.
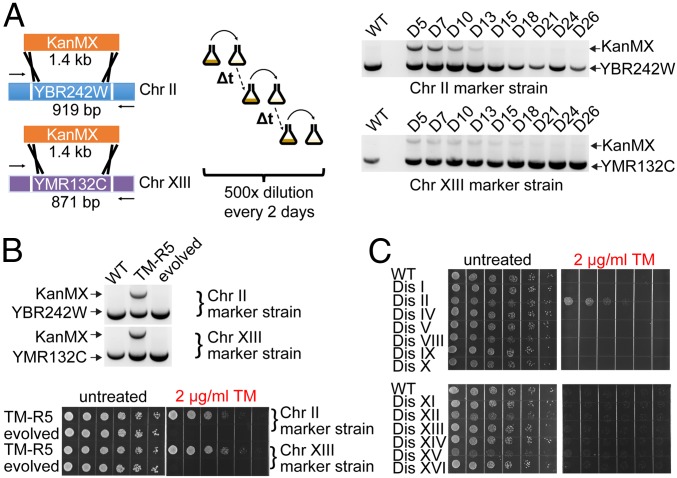
We next tested whether the extra chromosomes in the TM-R strains would be lost if the TM stress is removed. We generated two TM-R5 marker strains in which one of two copies of either Chr II or Chr XIII were labeled with the KanMX selectable marker conferring resistance to G418 antibiotic. We found that the extra copies of both Chr II and Chr XIII were lost when cells were grown in the absence of TM (Fig. 2A), and their loss reversed the TM resistance phenotype in the evolved TM-R5 populations (Fig. 2B). To narrow down which of the chromosome duplications are responsible for the ER stress resistance in our aneuploid mutants, we used a set of disomes, haploid yeast strains harboring an extra copy of a single chromosome (14). We observed that a haploid yeast strain possessing an extra Chr II, but not other disomic strains, is resistant to TM-induced ER stress (Fig. 2C and SI Appendix, Fig. S4). It is worth noting that these disomic strains were derived in a different genetic background (W303) from our standard wild-type strain (BY4741). Together, these data indicate that the duplication of Chr II is necessary and sufficient to confer ER stress resistance to cells in at least two different laboratory yeast strains.
Fig. 2.
Chr II duplication is necessary and sufficient to confer ER stress resistance to cells. (A) The extra copies of Chr II or Chr XIII were labeled with the KanMX cassette. The laboratory evolution experiment was carried out by serial dilution in YPD medium. The loss of the extra Chr II or XIII was detected by PCR at the indicated days. (B) The loss of extra copies of the Chr II and XIII in the absence of TM reverted the TM resistance phenotype in the evolved TM-R5 populations. (C) Chr II duplication is sufficient to confer ER stress resistance to cells.
Protective Effect of Disomic Chromosomes Can Be Recapitulated by Overexpression of Several Genes Located on Chr II.
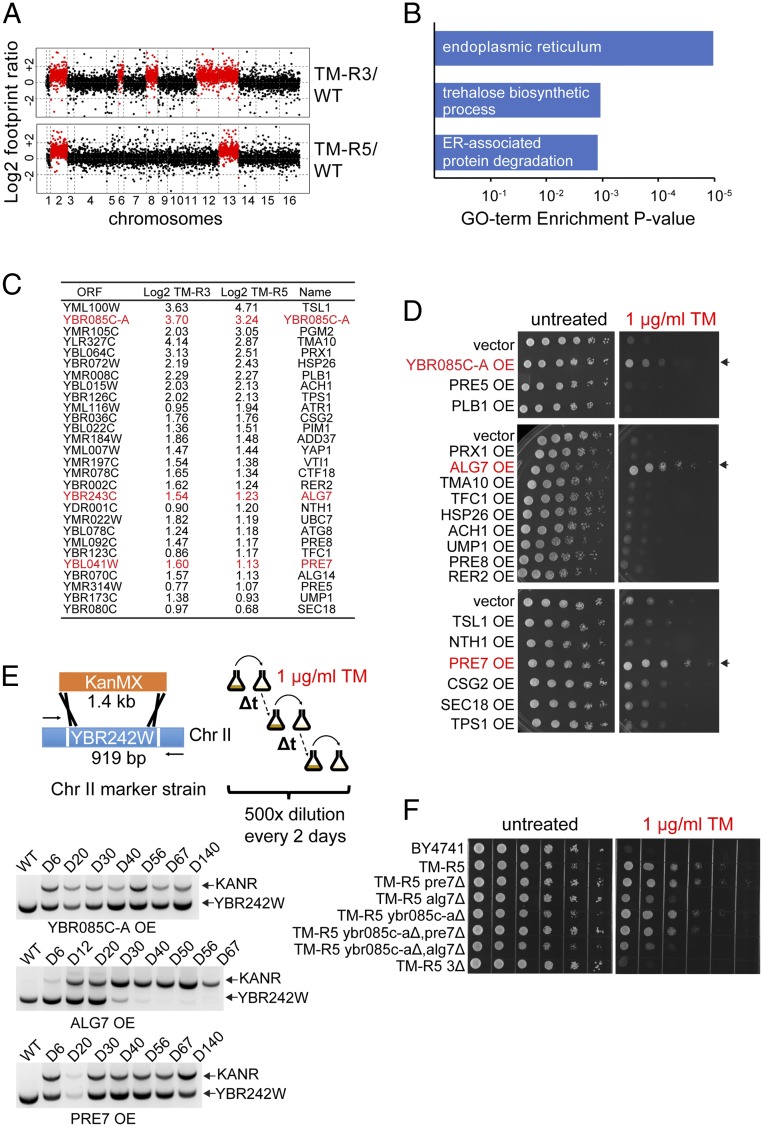
ER stress resistance could be explained by increased copy number of protective genes on duplicated chromosomes (24, 25) or duplication of genes encoding regulatory factors that affect expression of genes throughout the genome (26). To find which pathways are activated in aneuploid mutants, we analyzed protein translation in the TM-R3 and TM-R5 mutants using Ribo-Seq. Consistent with our expectation, translation of most genes on the duplicated chromosomes was doubled due to increased copy number (Fig. 3A and Dataset S1). Gene ontology analysis of genes activated in the TM-R mutants showed enrichment of genes encoding proteins involved in ER function, trehalose biosynthesis, and ERAD (Fig. 3B). Among genes that were up-regulated in the TM-R mutants we also identified several genes involved in chitin biosynthesis and its cell wall deposition (CHS2, CHS3, CHS7, RCR1). Chitin is a component of the cell wall that may confer stress resistance to yeast cells by increasing stability of the cell wall. However, aneuploid TM-R mutants did not exhibit increased resistance to cell wall stress induced by calcofluor white and Congo red, whereas the TM-R3 strain had a defect in cell wall integrity as was evident by its inability to grow on medium containing cell wall-damaging agents (SI Appendix, Fig. S5).
Fig. 3.
Overexpression of YBR085C-A, ALG7, and PRE7 in wild-type cells induces resistance to TM. (A) Translation levels of genes in TM-R3 and TM-R5 cells compared with a wild-type strain identified by Ribo-Seq. (B) Gene ontology enrichment analysis. Common genes up-regulated more than twofold in TM-R3 and TM-R5 mutants were analyzed using DAVID. (C) Common genes that were up-regulated more than 1.5-fold (0.6 in log2 scale) in both TM-R3 and TM-R5 mutants. Genes whose overexpression induces resistance to TM are shown in red. (D) Overexpression (OE) of YBR085C-A, ALG7, and PRE7 in wild-type cells induces resistance to TM. Resistance of strains to ER stress was determined using spot assays on SD agar plates containing 1 μg/mL TM. (E) ALG7 overexpression induces the loss of the extra Chr II in TM-R5 cells treated with TM. For the laboratory evolution experiment, the extra copy of Chr II in TM-R5 was labeled with the G418 resistance marker by replacing YBR242W with the KanMX cassette. TM-R5 marker strains overexpressing (OE) YBR085C-A, ALG7, or PRE7 were cultured in SD medium lacking leucine in the presence of 1 μg/mL TM. The laboratory evolution experiments were carried out by serial dilution with fresh medium every 2 d, and the loss of the extra copy of Chr II was detected by PCR at the indicated days. (F) Simultaneous deletion of extra copies of YBR085C-A, ALG7, and PRE7 (3Δ) prevents TM resistance in TM-R5 cells.
To identify specific genes that contribute to the ER stress resistance phenotype in the TM-R mutants, we selected common genes that were up-regulated in both TM-R3 and TM-R5 aneuploid mutants at the footprint level and tested the impact of overexpression of these genes on TM resistance in a wild-type BY4741 strain (Fig. 3C). We found that the protective effect of disomic chromosomes against ER stress can be recapitulated by overexpression of several genes located on Chr II. Among these genes, overexpression of UDP-N-acetylglucosamine-1-P transferase (ALG7), a subunit of the 20S proteasome (PRE7), and YBR085C-A induced TM resistance in wild-type cells (Fig. 3D and SI Appendix, Fig. S6A). We also found that ALG7 overexpression induces the loss of the extra Chr II in TM-R5 cells treated with TM (Fig. 3E). Although deletion of the extra copy of these genes alone in aneuploid strains did not reverse the TM resistance phenotype, simultaneous deletion of the extra copy of all three genes completely prevented TM resistance, suggesting that it results from a combined effect of multiple genes (Fig. 3F and SI Appendix, Fig. S6B).
Chromosome Duplications Are Not Gained in Response to UPR Deficiency.
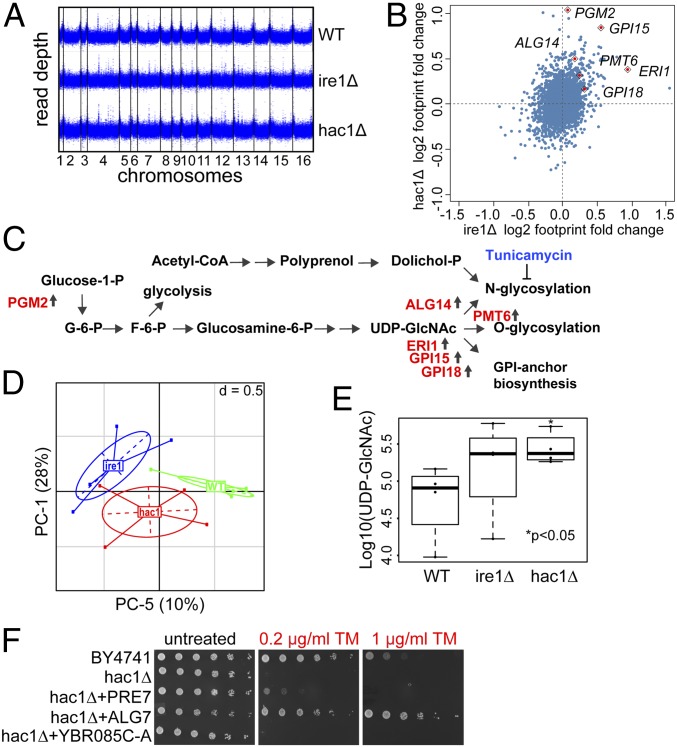
Previous studies in yeast revealed that aneuploidy is often acquired as a “crude” solution to stress, which is then eliminated and replaced by gene-specific solutions (15). To test if chromosome duplications are gained in response to low-dose ER stress, we exposed wild-type yeast cells to sublethal concentrations of TM, and then selected individual colonies and analyzed their growth rates in the media without TM (SI Appendix, Fig. S7A). Similar to aneuploid TM-R mutants, a large fraction of the analyzed clones displayed increased population doubling times, a hallmark of aneuploidy, when cells were pretreated with high doses of TM (0.5–2 μg/mL). But at low concentrations (0.1 and 0.2 μg/mL), TM did not induce the growth-inhibitory effects. To further test whether prolonged ER stress leads to aneuploidy, we performed whole-genome sequencing of mutants lacking IRE1 and HAC1 genes that have been previously characterized by ER stress and widespread protein aggregation (27). Our analysis did not reveal any gross genomic rearrangements in the ire1Δ and hac1Δ mutants (Fig. 4A). Together, these data and the fact that ire1Δ and hac1Δ have lifespans and growth rates similar to wild-type cells (SI Appendix, Fig. S7 B and C) suggest that these mutants may have evolved alternative mechanisms to maintain protein homeostasis.
Fig. 4.
IRE1 and HAC1 deletion mutants activate enzymes involved in protein glycosylation and GPI anchor synthesis. (A) Genome coverage of the ire1Δ and hac1Δ mutants. Read depth was calculated in 100-bp windows. (B) Comparison of protein translation changes in the ire1Δ and hac1Δ mutants using Ribo-seq. Log2 footprint changes in the ire1Δ mutant compared with wild-type cells are plotted in x axis, and log2 footprint changes in the hac1Δ mutant are plotted in y axis. (C) Deletion of IRE1 and HAC1 genes activates protein glycosylation and GPI anchor synthesis. (D) Principal component analysis plot showing clustering of metabolites analyzed in the ire1Δ and hac1Δ mutants. (E) Analysis of UDP-GlcNAc levels. (F) Overexpression of ALG7 and PRE7, but not YBR085C-A, can rescue the growth of the hac1Δ mutant under ER stress.
To identify mechanisms that compensate for the lack of the UPR, we analyzed gene expression changes in the ire1Δ and hac1Δ mutants by Ribo-Seq (Fig. 4B and Dataset S2). Our analysis revealed that lack of IRE1 and HAC1 in these strains is associated with increased expression of genes involved in the ER secretory pathway processes including glycophosphatidylinositol (GPI) anchor (ERI1, GPI15, GPI18) synthesis and protein glycosylation (ALG14, PGM2, PMT6) (Fig. 4C). We hypothesized that increased N- and O-glycan synthesis and GPI anchor production may allow cells to counteract ER stress by increasing export of glycosylated and GPI-anchored proteins. We further used targeted metabolite analysis to characterize metabolic profiles in wild-type cells and the UPR-deficient mutants and identify which metabolic pathways are activated in ire1Δ and hac1Δ cells. Clustering analysis revealed a significant difference in metabolome profiles between the three genotypes (Fig. 4D). Strikingly, we observed an approximately three- and fivefold increase in intracellular levels of UDP-GlcNAc in cells lacking IRE1 and HAC1, respectively, which was among the most up-regulated metabolites (Fig. 4E and Dataset S3). UDP-GlcNAc is utilized in protein glycosylation and GPI anchor synthesis reactions, which further supports a possibility that cells lacking IRE1 and HAC1 could restore protein homeostasis through activation of O- and N-linked glycosylation.
Finally, we tested whether the overexpression of genes involved in ER stress resistance due to Chr II aneuploidy can confer TM resistance to the UPR-deficient cells. Our analysis revealed that overexpression of ALG7 and PRE7, but not YBR085C-A, can rescue the growth of the hac1Δ mutant under ER stress (Fig. 4F). Together, these data suggest that ALG7 and PRE7 can confer ER stress resistance independently of the UPR.
Discussion
In this study, we performed a genetic screen in S. cerevisiae to understand the mechanisms by which cells can resist ER stress. Surprisingly, we found that ER stress resistance was associated with gains of multiple chromosomes. The number of duplicated chromosomes varied from two in the TM-R5 (aneuploid for Chr II and XIII) to five extra chromosomes in the TM-R3 (aneuploid for Chr II, VI, VIII, XII, and XIII) clones. We narrowed down the effect of disomic chromosomes to several specific genes located on Chr II. We show that Chr II duplication promotes expression of genes that support the ER stress resistance through increased gene dosage. However, the TM-resistant cells were short-lived and had slow growth compared with wild-type cells. These observations are consistent with previous studies showing that acquisition of extra chromosomes leads to proteotoxicity (14, 28, 29). Moreover, disomic yeast strains containing an extra copy of a single chromosome have been shown to generally have a decreased replicative lifespan (30). Together, these data indicate that protective genes located on duplicated chromosomes are beneficial upon ER stress, but the proteotoxic effect of aneuploidy leads to reduced fitness, as evidenced by shortened lifespan and slow growth.
Previous studies have shown that the expression of a majority of genes encoded on aneuploid chromosomes correlates with the relative chromosome copy number (29, 31, 32). We hypothesized that ER stress resistance in the aneuploid mutants can be explained by increased dosage of specific genes located on duplicated chromosomes. We have found that the protective effect of duplicated chromosomes can be recapitulated by overexpression of three genes located on Chr II that are involved in regulation of protein homeostasis in the ER, ALG7, PRE7, and YBR085C-A. ALG7 encodes a UDP-GlcNAc:dolichol phosphate N-acetylglucosamine-1-phosphate transferase that mediates the first step in the protein N-glycosylation pathway (33, 34). N-glycosylation plays an important role in protein folding and glycan-dependent quality control in the ER. In turn, PRE7 encodes a subunit of the 20S proteasome involved in proteasome-mediated protein degradation (35). The physiological role of YBR085C-A, which may represent a UPR signaling factor, is unknown. Interestingly, overexpression of ALG7 and PRE7, but not YBR085C-A, can partially rescue the growth of the hac1Δ mutant in the presence of TM, suggesting that YBR085C-A may act upstream of the UPR.
Although Chr II and Chr XIII were both aneuploid in four different mutants, we found that only Chr II disome had a protective effect, but not Chr XIII. Because disomic strains used for these experiments were derived in a different genetic background from our standard wild-type strain (BY4741), we cannot exclude the possibility that Chr XIII has a protective role only in this specific background. An alternative explanation could be that the Chr XIII duplication helps with other stresses such as aneuploidy itself and, as a consequence, may not be implicated directly in the ER stress resistance, but could still be needed for the cell survival.
Inhibition of protein synthesis is an essential mechanism for cell adaptation in response to accumulation of misfolded proteins in the ER (2). While aneuploid mutants were characterized by slow growth, increased resistance to ER stress in these strains was not due to defects in protein translation. These data are in line with previous studies in disomic S. cerevisiae strains showing that aneuploidy causes a defect in cell growth by delaying the G1 phase of the cell cycle as a result of slower accumulation of G1 cyclins rather than due to defects in protein translation (25, 36). Alternatively, the TM-resistant mutants may have acquired extra gene dosages to decrease TM import into the cell (37). Accumulation of chitin, a structural component of the cell wall, has been associated with increased resistance to multiple stresses (38, 39) and may be able to explain decreased permeability of the cell wall to TM. However, we found that aneuploid mutants did not show increased resistance to cell wall stress, suggesting that ER stress resistance in aneuploid mutants is unlikely due to decreased permeability of the cells to TM.
Notably, increased protection against ER stress in the TM-R2 mutant is not caused by aneuploidy, but instead can be explained by increased UPR signaling. We have found that activation of the UPR in this mutant is caused by inhibited attenuation of HAC1 splicing, which resembles previously described Ire1 kinase domain mutants (4, 40). The level of HAC1 mRNA splicing and production of the Hac1 protein is tightly controlled by dephosphorylation of Ire1, which is essential for attenuation of the UPR, suggesting potential involvement of posttranslational modifications in the UPR activation in the TM-R2 strain.
Furthermore, our analysis of the mutants lacking IRE1 and HAC1 genes demonstrates that aneuploidy is not gained in response to prolonged ER stress, and that these strains evolved aneuploidy-independent mechanisms to compensate for the lack of the UPR. To identify specific pathways that cells use to compensate for UPR deficiency, we analyzed translation changes in ire1Δ and hac1Δ by Ribo-Seq. We found that lack of IRE1 and HAC1 genes in these mutants is associated with increased expression of enzymes involved in ER secretory pathways processes. Specifically, we found up-regulated translation of genes involved in GPI anchor synthesis (ERI1, GPI15, GPI18) as well as N- and O-linked glycosylation (ALG14, PGM2, PMT6). Consistent with the activation of the glycosylation pathway, the ire1Δ and hac1Δ mutants had increased levels of UDP-GlcNAc. UDP-GlcNAc is an important substrate for N-linked glycosylation and GPI anchor synthesis, which may allow cells to counteract ER stress by increasing export of N-glycosylated and GPI-anchored glycoproteins. Indeed, in a recent study, Denzel et al. (41) observed that the activation of the UDP-GlcNAc synthesis pathway or the supplementation with GlcNAc can alleviate ER stress and even increase lifespan in worms.
Together, our data demonstrate that aneuploidy plays a key role in adaptation to ER stress, which is mediated by a synergistic effect of at least three genes located on Chr II. Although aneuploidy is known to cause proteotoxicity, the beneficial effect of Chr II duplication results from a balance between gene-specific and global consequences of aneuploidy.
Materials and Methods
Yeast Strains and Plasmids.
The yeast strains used in this study and their genotypes are listed in SI Appendix, Table S1. Cells were grown at 30 °C in standard YPD medium (1.0% yeast extract, 2.0% peptone, and 2.0% glucose) unless otherwise stated. The TM-resistant strains were generated by plating BY4741 parental cells on YPD plates containing 2 μg/mL TM. Spontaneously evolved TM-resistant mutants, TM-R1 to TM-R5, isolated from single colonies, were then selected for further analysis. One-step PCR-mediated gene disruption was performed using standard techniques. For overexpression of genes of interest, the pATP425 plasmid containing three yeast expression cassettes driven by constitutive ADH1, TDH3, and PGK1 promoters was used (42). Oligonucleotides used for gene disruption and cloning are listed in SI Appendix, Table S2.
Spot Assays.
Resistance of strains to TM, calcofluor white, and Congo red was determined using spot assays. Cells were initially grown in liquid culture without the drugs until OD600 = 0.6, and 10× serial dilutions for each strain were spotted on YPD or synthetic defined (SD) agar plates containing indicated concentrations of the drugs. The plates were incubated at 30 °C, and images were taken 48 h after plating.
Replicative Lifespan and Growth Rate Analyses.
Lifespan assays were carried out as described (43, 44). For the replicative lifespan assay, cells were grown on freshly prepared YPD plates for 2 d at 30 °C. For each strain, founder cells were plated on agar plates by selecting the newborn daughter cells using micromanipulator. Cells were monitored for cell divisions, and subsequent budded daughter cells were separated and removed as they formed. The process continued until cells stopped dividing. Replicative lifespan was calculated as the number of times each mother cell divided before it underwent senescence. Yeast growth rates were analyzed using the Bioscreen C automated microbiology growth curve analysis system (Growth Curves USA) as described (45, 46).
Genome Sequencing.
Whole-genome sequencing of yeast strains was performed as described (16). To calculate the depth of coverage for each chromosome, we used the Genome Analysis Toolkit (47). The “DepthOfCoverage” tool was applied to sorted *.bam files to generate a genome-wide table of sequencing depth. Chromosome coverage was then normalized by the total number of reads per sample. The graph was plotted with the ggplot2 package within the R statistical environment (https://www.r-project.org). Each dot is an average of the sequencing depths of 100 nt. There are 100,000 dots per graph.
Ribosome Profiling.
Yeast cultures were grown to OD600 = 0.5 in 500 mL of standard YPD medium, and cells were collected by filtering through a 0.45-μm filter (Millipore) with glass holder. Pellets were scraped with spatula, flash frozen in liquid nitrogen, and stored at −80 °C. Yeast extracts were prepared by cryogrinding the cell paste with BioSpec cryomill. Aliquots of cell lysates were used for footprint extraction. The Ribo-Seq libraries were prepared using ARTseq Ribosome Profiling kit (Illumina) as described (48, 49) and sequenced using the Illumina HiSeq platform. Ribosomal footprint reads were aligned to the S. cerevisiae genome from the Saccharomyces Genome Database (https://www.yeastgenome.org/, release number R64-2-1). Sequence alignment was performed using Bowtie 1.1.2 software (50) allowing two mismatches per read, and rpkm (reads per kilobase per million mapped reads) values were calculated using custom scripts. Gene ontology and pathways enrichment analysis were performed using DAVID database (51).
HAC1 mRNA Splicing.
An aliquot of cell lysates was used for RNA isolation. RNA was treated with DNaseI, and first-strand cDNA was synthesized using the SuperScript III reverse transcriptase (Life Technologies) with random hexamer primers. Analysis of the HAC1 mRNA splicing was performed by RT-PCR with primers flanking the HAC1 intron. PCR fragments were resolved on 2% agarose gels and quantified using ImageJ. Results are represented as means ± SEM of three independent experiments. Statistical significance of the data was determined by calculating P values using Student’s t test.
Laboratory Evolution Experiment.
TM-R5 marker strains, in which one of two copies of either Chr II or Chr XIII were labeled with the G418 resistance marker, were prepared by replacing YBR242W and YMR132C coding regions with the KanMX cassette, respectively. The laboratory evolution experiment was carried out by serial dilution. Cells were grown until reaching stationary phase at 30 °C with moderate shaking and then diluted by a factor of 1:500 with fresh medium. This procedure was repeated until the loss of the extra chromosome was detected by PCR. Genotyping was performed by isolating genomic DNA from an aliquot of the yeast culture using the Master Pure Yeast DNA purification Kit (Epicentre), which was then used as a template for PCR with primers that align outside of the coding regions of YBR242W and YMR132C.
Metabolite Profiling.
Yeast cells were inoculated overnight in YPD medium. The next morning, the cultures were diluted to OD600 < 0.1 and grown at 30 °C until OD600 = 0.5. Cells were washed in sterile H2O, and 20 million cells per sample were flash frozen in liquid nitrogen and stored at −80 °C. Metabolites were extracted from frozen samples with acetonitrile and were analyzed using a liquid chromatography-mass spectrometry (LC-MS) platform at the University of Washington Northwest Metabolomics Research Center as described (52). Stable isotope-labeled internal standards were also run as a control alongside the experimental samples to ensure reproducibility of the results. Four independent biological replicates were analyzed per each strain. Statistical significance of the metabolomics data was determined by calculating P values (not corrected for false discovery rate) using a Wilcoxon rank-sum test (53).
Supplementary Material
Acknowledgments
We thank Angelika Amon and Vadim Gladyshev for providing yeast strains. This work was supported by the National Institutes of Health Grants AG040191 and AG054566 (to V.M.L.), R01AG049494 (to D.E.L.P.), and P30AG013280 (to M.K.). This research was conducted while V.M.L. was an American Federation for Aging Research (AFAR) Research Grant recipient from AFAR. M.B.L. was supported by the Howard Hughes Medical Institute (HHMI) Gilliam Fellowship for Advanced Study. L.D. was supported by the Brazilian Scientific Mobility Program/CAPES, Coordination for the Improvement of Higher Education Personnel—Brazil.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804264115/-/DCSupplemental.
References
- 1.Parodi AJ. Role of N-oligosaccharide endoplasmic reticulum processing reactions in glycoprotein folding and degradation. Biochem J. 2000;348:1–13. [PMC free article] [PubMed] [Google Scholar]
- 2.Walter P, Ron D. The unfolded protein response: From stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 3.Gardner BM, Pincus D, Gotthardt K, Gallagher CM, Walter P. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol. 2013;5:a013169. doi: 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 5.Aragón T, et al. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2009;457:736–740. doi: 10.1038/nature07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee KP, et al. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell. 2008;132:89–100. doi: 10.1016/j.cell.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 8.Di Santo R, Aboulhouda S, Weinberg DE. The fail-safe mechanism of post-transcriptional silencing of unspliced HAC1 mRNA. eLife. 2016;5:e20069. doi: 10.7554/eLife.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 10.Pincus D, Aranda-Díaz A, Zuleta IA, Walter P, El-Samad H. Delayed Ras/PKA signaling augments the unfolded protein response. Proc Natl Acad Sci USA. 2014;111:14800–14805. doi: 10.1073/pnas.1409588111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leber JH, Bernales S, Walter P. IRE1-independent gain control of the unfolded protein response. PLoS Biol. 2004;2:E235. doi: 10.1371/journal.pbio.0020235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schröder M, Clark R, Kaufman RJ. IRE1- and HAC1-independent transcriptional regulation in the unfolded protein response of yeast. Mol Microbiol. 2003;49:591–606. doi: 10.1046/j.1365-2958.2003.03585.x. [DOI] [PubMed] [Google Scholar]
- 13.Mulla W, Zhu J, Li R. Yeast: A simple model system to study complex phenomena of aneuploidy. FEMS Microbiol Rev. 2014;38:201–212. doi: 10.1111/1574-6976.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 15.Yona AH, et al. Chromosomal duplication is a transient evolutionary solution to stress. Proc Natl Acad Sci USA. 2012;109:21010–21015. doi: 10.1073/pnas.1211150109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaya A, et al. Adaptive aneuploidy protects against thiol peroxidase deficiency by increasing respiration via key mitochondrial proteins. Proc Natl Acad Sci USA. 2015;112:10685–10690. doi: 10.1073/pnas.1505315112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Millet C, Makovets S. Aneuploidy as a mechanism of adaptation to telomerase insufficiency. Curr Genet. 2016;62:557–564. doi: 10.1007/s00294-015-0559-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millet C, Ausiannikava D, Le Bihan T, Granneman S, Makovets S. Cell populations can use aneuploidy to survive telomerase insufficiency. Nat Commun. 2015;6:8664. doi: 10.1038/ncomms9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehrman MA. Oligosaccharide-based information in endoplasmic reticulum quality control and other biological systems. J Biol Chem. 2001;276:8623–8626. doi: 10.1074/jbc.R100002200. [DOI] [PubMed] [Google Scholar]
- 20.Steffen KK, et al. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics. 2012;191:107–118. doi: 10.1534/genetics.111.136549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan Z, et al. Aneuploidy underlies a multicellular phenotypic switch. Proc Natl Acad Sci USA. 2013;110:12367–12372. doi: 10.1073/pnas.1301047110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rancati G, et al. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135:879–893. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen G, Bradford WD, Seidel CW, Li R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature. 2012;482:246–250. doi: 10.1038/nature10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pavelka N, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Birchler JA. Reflections on studies of gene expression in aneuploids. Biochem J. 2010;426:119–123. doi: 10.1042/BJ20091617. [DOI] [PubMed] [Google Scholar]
- 27.Hamdan N, Kritsiligkou P, Grant CM. ER stress causes widespread protein aggregation and prion formation. J Cell Biol. 2017;216:2295–2304. doi: 10.1083/jcb.201612165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oromendia AB, Dodgson SE, Amon A. Aneuploidy causes proteotoxic stress in yeast. Genes Dev. 2012;26:2696–2708. doi: 10.1101/gad.207407.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dephoure N, et al. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. eLife. 2014;3:e03023. doi: 10.7554/eLife.03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sunshine AB, et al. Aneuploidy shortens replicative lifespan in Saccharomyces cerevisiae. Aging Cell. 2016;15:317–324. doi: 10.1111/acel.12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torres EM, et al. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheltzer JM, Torres EM, Dunham MJ, Amon A. Transcriptional consequences of aneuploidy. Proc Natl Acad Sci USA. 2012;109:12644–12649. doi: 10.1073/pnas.1209227109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 34.Bretthauer RK. Structure, expression, and regulation of UDP-GlcNAc: Dolichol phosphate GlcNAc-1-phosphate transferase (DPAGT1) Curr Drug Targets. 2009;10:477–482. doi: 10.2174/138945009788488369. [DOI] [PubMed] [Google Scholar]
- 35.Kleiger G, Mayor T. Perilous journey: A tour of the ubiquitin-proteasome system. Trends Cell Biol. 2014;24:352–359. doi: 10.1016/j.tcb.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorburn RR, et al. Aneuploid yeast strains exhibit defects in cell growth and passage through START. Mol Biol Cell. 2013;24:1274–1289. doi: 10.1091/mbc.E12-07-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiling JH, et al. A haploid genetic screen identifies the major facilitator domain containing 2A (MFSD2A) transporter as a key mediator in the response to tunicamycin. Proc Natl Acad Sci USA. 2011;108:11756–11765. doi: 10.1073/pnas.1018098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valdivieso MH, Ferrario L, Vai M, Duran A, Popolo L. Chitin synthesis in a gas1 mutant of Saccharomyces cerevisiae. J Bacteriol. 2000;182:4752–4757. doi: 10.1128/jb.182.17.4752-4757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levin DE. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics. 2011;189:1145–1175. doi: 10.1534/genetics.111.128264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chawla A, Chakrabarti S, Ghosh G, Niwa M. Attenuation of yeast UPR is essential for survival and is mediated by IRE1 kinase. J Cell Biol. 2011;193:41–50. doi: 10.1083/jcb.201008071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Denzel MS, et al. Hexosamine pathway metabolites enhance protein quality control and prolong life. Cell. 2014;156:1167–1178. doi: 10.1016/j.cell.2014.01.061. [DOI] [PubMed] [Google Scholar]
- 42.Ishii J, et al. Three gene expression vector sets for concurrently expressing multiple genes in Saccharomyces cerevisiae. FEMS Yeast Res. 2014;14:399–411. doi: 10.1111/1567-1364.12138. [DOI] [PubMed] [Google Scholar]
- 43.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steffen KK, Kennedy BK, Kaeberlein M. Measuring replicative life span in the budding yeast. J Vis Exp. 2009;28:e1209. doi: 10.3791/1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee MB, et al. A system to identify inhibitors of mTOR signaling using high-resolution growth analysis in Saccharomyces cerevisiae. Geroscience. 2017;39:419–428. doi: 10.1007/s11357-017-9988-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olsen B, Murakami CJ, Kaeberlein M. YODA: Software to facilitate high-throughput analysis of chronological life span, growth rate, and survival in budding yeast. BMC Bioinformatics. 2010;11:141. doi: 10.1186/1471-2105-11-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Labunskyy VM, et al. Lifespan extension conferred by endoplasmic reticulum secretory pathway deficiency requires induction of the unfolded protein response. PLoS Genet. 2014;10:e1004019. doi: 10.1371/journal.pgen.1004019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beaupere C, Chen RB, Pelosi W, Labunskyy VM. Genome-wide quantification of translation in budding yeast by ribosome profiling. J Vis Exp. 2017;130:e56820. doi: 10.3791/56820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 52.Hoffman JM, et al. Effects of age, sex, and genotype on high-sensitivity metabolomic profiles in the fruit fly, Drosophila melanogaster. Aging Cell. 2014;13:596–604. doi: 10.1111/acel.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilcoxon F. Individual comparisons of grouped data by ranking methods. J Econ Entomol. 1946;39:269. doi: 10.1093/jee/39.2.269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.