Significance
We attempt to quantify animal “bodyplans” and their variation within Metazoa. Our results challenge the view that maximum variation was achieved early in animal evolutionary history by nonuniformitarian mechanisms. Rather, they are compatible with the view that the capacity for fundamental innovation is not limited to the early evolutionary history of clades. We perform quantitative tests of the principal hypotheses of the molecular mechanisms underpinning the establishment of animal bodyplans and corroborate the hypothesis that animal evolution has been permitted or driven by gene regulatory evolution.
Keywords: Metazoa, disparity, evolution, morphology, Cambrian explosion
Abstract
The animal kingdom exhibits a great diversity of organismal form (i.e., disparity). Whether the extremes of disparity were achieved early in animal evolutionary history or clades continually explore the limits of possible morphospace is subject to continuing debate. Here we show, through analysis of the disparity of the animal kingdom, that, even though many clades exhibit maximal initial disparity, arthropods, chordates, annelids, echinoderms, and mollusks have continued to explore and expand the limits of morphospace throughout the Phanerozoic, expanding dramatically the envelope of disparity occupied in the Cambrian. The “clumpiness” of morphospace occupation by living clades is a consequence of the extinction of phylogenetic intermediates, indicating that the original distribution of morphologies was more homogeneous. The morphological distances between phyla mirror differences in complexity, body size, and species-level diversity across the animal kingdom. Causal hypotheses of morphologic expansion include time since origination, increases in genome size, protein repertoire, gene family expansion, and gene regulation. We find a strong correlation between increasing morphological disparity, genome size, and microRNA repertoire, but no correlation to protein domain diversity. Our results are compatible with the view that the evolution of gene regulation has been influential in shaping metazoan disparity whereas the invasion of terrestrial ecospace appears to represent an additional gestalt, underpinning the post-Cambrian expansion of metazoan disparity.
The diversity of animal organismal form (i.e., disparity) is decidedly nonrandom; members of one phylum share “bodyplan” characteristics distinct from those of other phyla, suggesting that only a very small subset of the universe of possible bodyplans has been realized. Paleontological analyses have suggested that the limits on organismal disparity were realized early in animal evolutionary history (1–3), inspiring the view that fundamental innovation has been precluded subsequently by gene regulatory developmental constraints (4–6), and that the evolutionary processes underlying the emergence of animals are nonuniformitarian (7, 8). However, this perspective is based largely on the timing of appearance of Linnean rank taxa in the fossil record (8, 9), assuming they provide an effective proxy for organismal disparity. Attempts to capture disparity by using morphometry have borne out the hypothesis of maximal initial disparity (3, 10), but this approach is limited practically to analysis at low taxonomic levels and it is not clear that the results can be generalized to higher taxa, including the phylum and kingdom levels. Here we attempt to map metazoan disparity within an empirical morphospace based on a large sampling of discrete characters from across the breadth of extant metazoan diversity. We use this to explore the impact of extinction on morphospace occupation and the relationship between organismal disparity and other phenomena such as complexity, body size, diversity, and Linnean rank. We then undertake quantitative tests of hypotheses of causality, including random variation, genome size, protein diversity, and gene regulatory complexity.
Mapping Metazoan Morphospace
The construction of a morphospace is dependent on methodology and the selection of relevant features. However, with a significantly large data source, the distances among taxa within morphospace will begin to approximate the evolutionary scale of the differences. Spatial landmark analysis is precluded at high taxonomic rank such as phylum because, by definition, phyla share few morphological homologies. Discrete characters provide a suitable alternative given that there are no practical limits to their scalability (2, 3), and comparative analyses have shown that continuous and discrete character datasets can capture the same phenomenon (11–14). The use of discrete characters produces results that have nonmetric properties (15–17), but this approach can and has been used to elucidate broad patterns of similarities and clustering within multidimensional space (18), particularly in formulating the hyoptheses we seek to test. To test between competing hypotheses for the evolution of disparity—whether the limits of disparity were established early or have continued to expand throughout evolutionary history—we compiled a cladistic character matrix derived from Peter Ax’s Phylogenetic System (19–21). This constitutes a single, densely sampled synthetic overview of character distribution among metazoans, including all phyla, by an individual who was not a taxonomic specialist in any of the groups that could perhaps be considered overrepresented in the dataset. Ax’s taxonomic sampling is not uniform across metazoans, but the number of characters and taxa within a phylum is representative of its intraphylum diversity (ref. 22; Spearman’s correlation, ρ = 0.821, P < 0.001). Therefore, we do not consider that any clades are significantly underrepresented or overrepresented in the dataset. We coded 1,767 characters for 212 extant, terminal taxa, including 34 animal phyla, most commonly down to the Linnean rank of order (SI Appendix, Table S1). The characters encompass all aspects of morphology (cellular, developmental, sexual, and skeletal and soft-tissue anatomy), including those minimally defining each clade, comprising 915 characters that are shared among the operational taxa (homologies and homoplasies) and 852 unique (i.e., autapomorphic) characters (SI Appendix, Table S2 and Dataset S1).
We mapped the relationships between features to identify characters that are nonapplicable rather than absent, which can be differentiated analytically by using Gower’s similarity metric (23–25). We subjected these data to a nonmetric multidimensional scaling (NMDS) analysis, a noneigenvector-based multivariate method that attempts to optimize the fit between the data and a preselected number of axes (Fig. 1). The use of a nonmetric ordination technique has several advantages (e.g., the ability to handle large amounts of absent data), but the distances between organisms may not be directly Euclidean, which may alter measurements of disparity. To control for the choice of ordination technique, we repeated all analyses by using principal coordinate analysis. The choice of ordination had no impact on the morphospace or any of the presented results (SI Appendix, Fig. S1). The distances between taxa using linear (i.e., principal coordinate analysis) and nonlinear (i.e., NMDS) methods are strongly and linearly correlated (Mantel test, R2 = 0.9764, P = 0.001), indicating that, even though the NMDS ordination is built by using a nonlinear method, it has linear properties. Absolute distances within the space can still be subject to nonmetric artifacts such that the distances between taxa should be taken as a qualitative metric of the overall distribution of metazoan morphologic diversity. An analysis of the stress (representing the goodness of fit) indicates that the majority of variance in the data is captured by the first two axes (SI Appendix, Fig. S2).
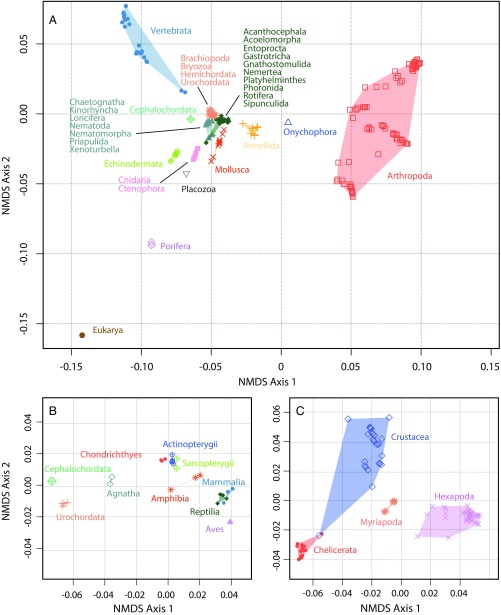
Fig. 1.
(A) Morphospace encompassing Ax’s 212 operational taxa representing 34 phyla. The character matrix was analyzed by using NMDS. (B) Morphospace of the Chordata, which includes 26 vertebrata taxa, 2 urochordate taxa, and a single cephalochordate taxon grouped by class. (C) Morphospace of the 94 arthropod taxa included in the study grouped by subphylum.
Concerned that different treatments of nonapplicable data (25) could significantly alter the structure of the ordination, we structured our data to reflect different coding strategies (Fig. 2). Treating nonapplicable characters as absent (Fig. 2A) or missing (Fig. 2B) produced statistically similar ordinations to the main analysis (Fig. 1) in which inapplicable characters are assigned a distinct state conferring distance (Mantel test, R = 0.907, P = 0.001). However, these differ in the (relative) displacement of the nonmetazoan eukaryotic outgroup, Porifera and Placozoa, into the central area of morphospace (Fig. 2A). Treating nonapplicable characters as absent or missing also increased intraphylum disparity at the cost of interphylum disparity (Fig. 2 A and B). However, the similarity in results from the different treatments of the data indicate the strength of the underlying structure of the data. These results indicate that the structure of disparity is robust to ordination and coding strategies.
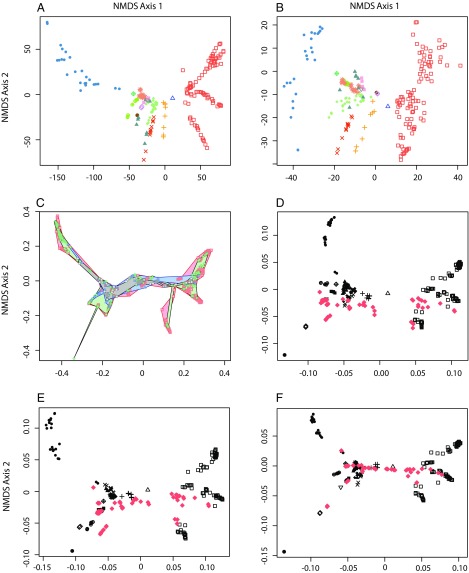
Fig. 2.
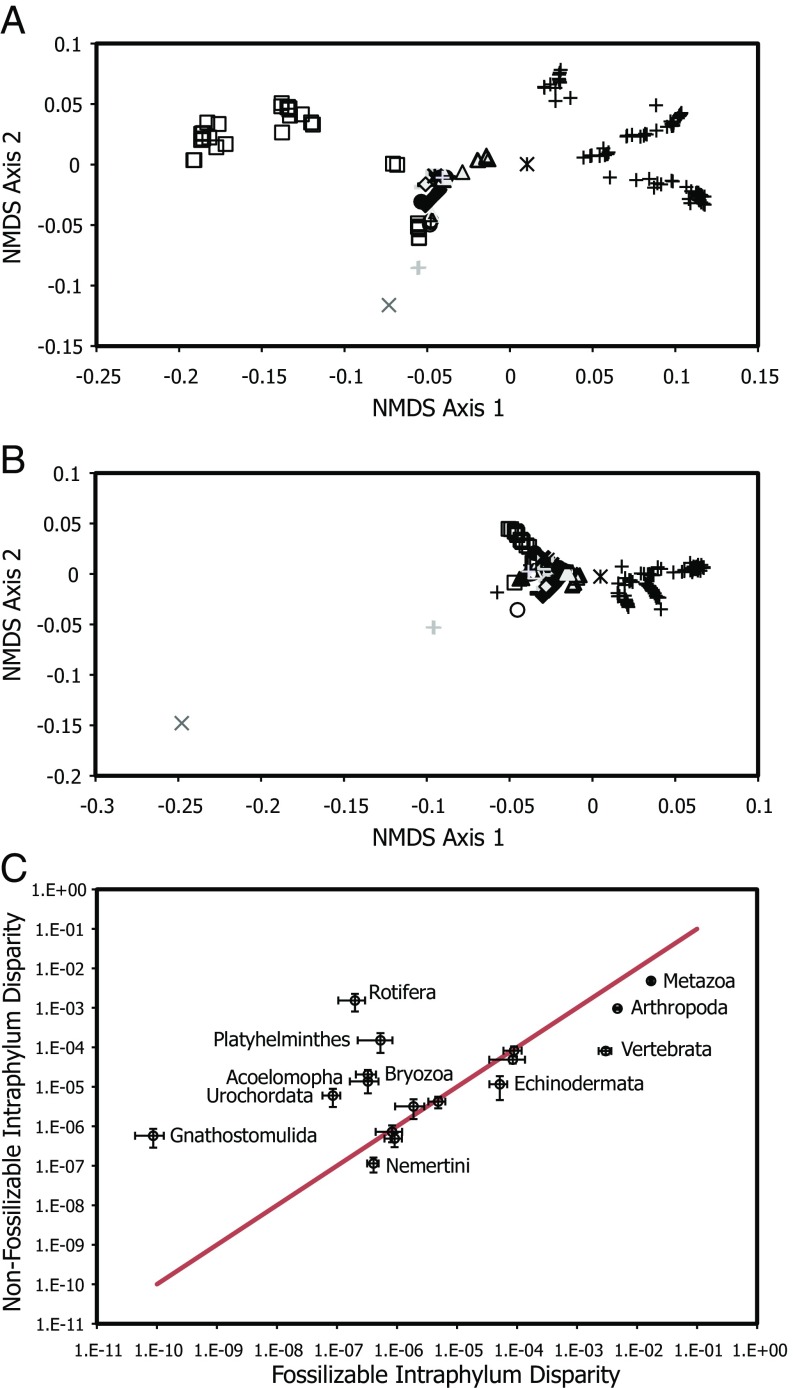
Exploration of the impact of ordination method, coding strategy, character fossilization potential, inclusion of fossil taxa, and controlling for missing data. (A and B) The effect on the morphospace with different treatment of nonapplicable data. (A) Morphospace constructed by coding nonapplicable characters as absent. (B) Morphospace constructed by coding nonapplicable characters as missing. (C) Ordination based on a Q-mode analysis of the character matrix, considering character variance based on their taxonomic distribution; characters are color-coded according to the taxonomic rank at which they exhibit greatest variance (black, greater than phylum; blue, phylum; green, subphylum to class; red, lower than class). (D–F) Incorporation of fossil taxa (fossils represented as red diamonds; extant taxa black following symbol scheme in Fig. 1). (D) Addition of fossil taxa with unknown character states treated as missing data. (E) Impact of the loss of nonpreservable characters on morphospace structure built by using 1,000 characters that were identified as preservable based on known fossils examples or theoretical preservabilty of the structures being characterized. (F) Addition of fossil taxa onto the morphospace in which missing data has been modeled based on their phylogenetic position. All morphospaces were constructed similarly to Fig. 1 by using NMDS and Gower’s similarity metric, with the exception of A and B, which used Manhattan distance.
The position of taxa based on the first two axes is presented in Fig. 1A. Most of this variation is based on shared characters, as analysis of a dataset excluding autapomorphies has no significant impact on the structure of the morphospace (R2 > 0.92, P = 0.001; SI Appendix, Fig. S3). Phyla differ dramatically in the position and areal extent of their envelope of disparity. Although most taxa are clustered along both axes, the nonmetazoan eukaryote outgroup and Porifera plot separately from eumetazoans, principally for their lack of shared eumetazoan characters, even though neither of these groups occupy a large area of morphospace. Chordata (Fig. 1B), Arthropoda (Fig. 1C), and, to a lesser extent, Annelida, Echinodermata, and Mollusca, are much more disparate, each occupying larger ranges of morphospace than all other phyla combined, and defining the extremities of morphospace on the two principal axes. A Q-mode analysis of the distribution of characters (Fig. 2C) shows that the characters that describe intraphylum features load at the extremities of both axes. Superphylum- and phylum-level characters occupy approximately one fourth of the area of the lower-level characters and vary primarily along the first axis.
From Modern to Historical Disparity
Central to the thesis of maximal initial disparity in animal evolution was the discovery of distinct bodyplans among Cambrian Konservat–Lagerstätten (1), which were assigned historically to extinct phyla, classes, or orders. Thus, by comparing only living taxa, it could be argued that we have captured only net historical disparity. Therefore, we coded a phylogenetically diverse and representative sample of Cambrian taxa, principally the earliest representatives of ordinal level clades (26). This entailed coding 70 fossil taxa for the existing character set and adding 111 mostly autapomorphic characters. Coding these fossil taxa was potentially problematic in that most of the characters (54.1%) are not preserved, and therefore unknown. On average, only 8.6% of the characters were coded as applicable, resulting in the fossil taxa appearing more constrained and skewed toward lower values on the second axis, making the Cambrian taxa appear less complex (Fig. 2D); we interpret this result as an artifact of the great volume of data missing for the fossils. There are two possible solutions to accommodating fossil species. One approach is to subsample our dataset for fossilizable characters based on known examples of fossilized features or the anatomical nature of the character (1,000 characters). NMDS analysis of this subsampled dataset results in a plot of morphospace occupation with the same broad structure (Mantel test, R = 0.974, P = 0.001; Fig. 2E) as that recovered from analysis of the entire dataset (Fig. 1A). However, the resulting morphospace accentuates the relative disparity of vertebrates and arthropods while diminishing the relative disparity of all other phyla (nonbilaterians especially), individually and in combination, exaggerating the significance of skeletal and gross anatomical characters that are fossilized in instances of routine and exceptional preservation. Few of these characters are representative of bodyplans more generally, which are defined on the basis of soft-tissue, cellular, tissue, organ, and developmental characters that are not usually fossilized. Thus, restricting the analysis to only fossilizable characters cannot be considered to capture organismal disparity within metazoans in any meaningful way. These results are of concern because they suggest that the majority of disparity analyses, which have been applied principally to fossil groups, may have limited inferential power. This is because they are constrained to characterization of fossilizable characters that may be otherwise unrepresentative of phenotypic evolution.
An alternative approach to including fossil species exploits their known phylogenetic position among living and fossil relatives to infer character states that are lost during fossilization. There are obviously assumptions inherent in inferring missing data, including missing secondary reversals in soft tissues, the potential of differential evolutionary rates between preservable and nonpreservable characters, or limiting the coded fossil autapomorphies to preservable characteristics. However, given the rarity of reversals of superphylum-level nonfossilizable characters in extant taxa and the observation that autapomorphies contribute little to the construction of the morphospace (SI Appendix, Fig. S3), these assumptions are likely to have a minor impact on the projection of fossil taxa into the morphospace defined by the living species. The approach of inferring missing data likely strengthens the phylogenetic signal in the morphospace. However, a comparison with the taphonomically culled dataset (Fig. 2E) indicates a similar and robust placement of the fossil taxa within morphospace.
To implement this approach, we derived a consensus, time-scaled phylogenetic tree for the operational taxa, which differed from Ax’s original phylogenetic hypothesis (SI Appendix, Figs. S4 and S5). We completed the coding for the fossils through stochastic character mapping (27, 28), a probabilistic approach that accommodates the uncertainty in ancestral and tip states, based on current hypotheses of their phylogenetic position (SI Appendix, Fig. S5 and Dataset S2). On average, we were able to code 45.4% of the characters based on fossil material, and, of the remaining 54.6% of modeled characters, 98.8% were modeled as absent or nonapplicable. The majority of the traits that were inferred to be present from the character modeling are traits that are shared by all bilaterians. Fossil taxa were then included in the ordination among their extant relatives (Fig. 2F); comparison with the ordination of extant taxa alone (Fig. 1A) shows that their inclusion does not have a significant or qualitative impact on the universe of empirical morphospace defined by the living taxa (P = 0.001). We also reconstructed ancestral character states for all of the internal nodes in the tree, which represent hypothetical ancestors (Dataset S2), by using the same method of stochastic character state mapping (27, 28), and plotted the phylogeny into morphospace (Fig. 3A). We also subdivided the nodes (internal and terminal) into Cambrian and post-Cambrian origination (based on the fossil records of lineages and divergence time estimates for some ancestral nodes from ref. 29) to assess the scale of pre-Ordovician vs. post-Cambrian innovation (Fig. 3B).
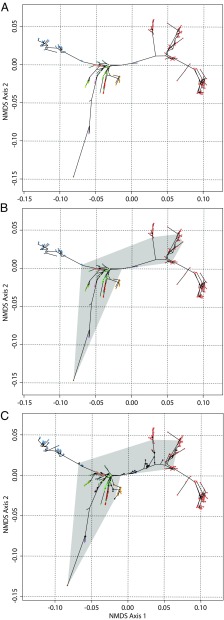
Fig. 3.
Phylomorphospace and circumscription of Cambrian vs. recent animal disparity. (A) Phylomorphospace derived by using a consensus phylogenetic tree for the included extant taxa (SI Appendix, Fig. S2) and character states inferred for all of the internal nodes and tips by using stochastic character state mapping. (B) Convex hull (gray) circumscribing clades established before the end of the Cambrian based on fossil and molecular clock data (29). (C) Phylomorphospace incorporating the earliest (Cambrian) representatives of animal orders, as identified in ref. 26, with a convex hull (gray) circumscribing clades established before the end of the Cambrian based on fossil ages and molecular clock data (29); the fossil taxa were included in the ordination, but this has little qualitative impact on the distances exhibited by extant taxa. Cambrian organisms are represented by black nodes, whereas the color scheme for extant taxa follows Fig. 1A.
Superimposition of the tree topology (Fig. 3A) reveals that living clades do not deviate significantly from the paths that animal phylogeny is inferred to have coursed through morphospace, and extinct taxa (fossils and internal nodes) plot intermediate of their living relatives. These results support the view that the apparent distinctiveness of phylum-level crown groups and, more generally, the “clumpiness” of animal disparity, are consequences of the extinction of phylogenetic intermediates. By implication, the aspect of morphological disparity recognized by the Linnean ranks (e.g., ref. 9) is largely an artifact of later Phanerozoic extinction, not of late Neoproterozoic–Cambrian innovation. For example, the addition of the Cambrian stem-arthropods, Anomalocaris, Aysheaia, and Opabinia, does expand the envelope of morphospace occupied by the arthropods, but does so by bridging the gap to onychophorans (Fig. 3C). Hence, the distinctiveness of panarthropod phyla has increased over time with the extinction of these now-“stem” arthropods. There is no evidence, however, that the overall envelope of metazoan morphospace occupation has diminished significantly as a consequence of extinction since the Cambrian, nor that maximal disparity was achieved early in animal evolutionary history (compare Fig. 3B vs. Fig. 3C). Quite to the contrary, the inclusion of these Cambrian arthropods only expanded the region and density of morphospace occupied by arthropods, and only then in diminishing the distance between arthropods and their nearest living relatives, the onychophorans. With the exclusion of Cambrian vertebrates, the inclusion of Cambrian taxa does not in itself increase the envelope of net metazoan morphospace, which remains defined by living clades. The envelope of metazoan disparity expanded post-Cambrian, and numerous reversals are represented by crossing evolutionary pathways in Fig. 3. Reversals, and the obvious overlap in morphospace occupation by the majority of phyla, reflect the role of convergence and constraint in metazoan diversification (30). Evidently, there is no general trend in the tempo of clade disparity: the majority of phylum-level clades exhibit maximal disparity achieved by the Cambrian (Fig. 3, gray), compatible with previous studies at low taxonomic rank (18, 31, 32), whereas others exhibit a progressive exploration of morphospace: principally arthropods and chordates (corroborating refs. 33–35), but also annelids, echinoderms, and mollusks. Thus, the envelope of disparity explored by Kingdom Metazoa has increased through geological time.
Relationship Between Disparity and Complexity, Body Size, and Diversity.
Having codified metazoan disparity, we next attempted to understand the relationship between morphology and other primary biologic metrics. To achieve this, taxonomic rank had to be normalized to the phylum level. The morphologic position of phyla was determined by using two methods: (i) including the crown ancestor of each phylum in the preexisting morphospace and (ii) independently analyzing the modeled characters for the crown ancestor of each phylum (SI Appendix, Fig. S6). The first method is influenced by differences in diversity or disparity between phyla, whereas the second disregards those differences between phyla in an effort to control for potential sampling biases. As expected, the two morphospaces differ in the uniqueness of arthropods and chordates, which alter the strength of correlation between morphology and some other datasets. However, these two methods maintain the structure of the morphospace (Mantel test, R = 0.946 P = 0.001; SI Appendix, Fig. S6). Thus, differences in sampling are not the controlling factor in correlations between morphology and other datasets. In all of the following tests of correlation, we undertook parallel analyses by using both methods, the results of which were similar; we present only those results based on the independent ordination of the character sets inferred for the phylum crown-ancestors. The distances between the morphological position of the phyla are then considered as a qualitative measure of the overall similarity of the phyla and can be used as a guide to compare morphology to other primary biological metrics.
The concept of disparity has been linked to, and sometimes even conflated with, the concept of organismal complexity (36). To explore their relationship, we compiled a new dataset of metazoan cell type diversity, the only widely accepted proxy for morphological complexity (e.g., ref. 11). We questioned whether the two phenomena were correlated by using a nonlinear (Spearman) Mantel test to compare pairwise distances derived from the morphology and complexity datasets (Table 1). The possibility that the two datasets are uncorrelated can be rejected at a high level of significance (Table 1). This relationship can be rationalized because only simple body plans are possible with few cell types, compatible with the view that expansion in cell type diversity has underpinned the expansion of metazoan disparity (37).
Table 1.
Statistical comparison of morphology against other biologic features
| Feature | Mantel r | P value | No. of phyla |
| No. of cell types | 0.3387 | 0.01 | 29 |
| Minimum body size | 0.09537 | 0.248 | 28 |
| Maximum body size | 0.4875 | 0.004 | 28 |
| Range in body size | 0.4862 | 0.007 | 28 |
| Species level diversity | 0.3678 | 0.007 | 32 |
| Genome length | 0.278 | 0.028 | 22 |
| Protein (superfamily) | 0.2512 | 0.141 | 12 |
| Protein (family) | 0.1701 | 0.218 | 12 |
| Protein (architecture) | −0.288 | 0.886 | 12 |
| microRNA | 0.39 | 0.005 | 24 |
The positions of phyla were calculated in two manners. First, the modeled character suites for the ancestral nodes of the individual phyla were projected onto the morphospace constructed with Ax’s 212 operational taxa. This method includes the disparity contained within phyla to some degree and therefore could be potentially biased by differential sampling. Second, the modeled character suites for the ancestral nodes were independently ordinated by using NMDS. This method treats each phylum equally and disregards the synapomorphies contained within each phylum such that the structure of the data are not controlled by differential sampling. The results were the same, so only the later is presented. Correlations between matrices were analyzed by using Mantel tests (Spearman), which compare the rank-order distance between phyla within the two matrices. The number of phyla included in the test varies with the availability of data (lists of phyla included in the different comparisons are presented in SI Appendix, Table S3). Multivariate datasets (miRNA and proteins) were analyzed in a similar manner to the morphological dataset (NMDS). Body size data are from McClain and Boyer (38). Genome length data were accessed from the animal genome size database
Morphologic distances between phyla were also compared with compilations of minimum, maximum, and range in body size within each phylum (38). Body size is correlated with many ecological and evolutionary traits and has increased by 16 orders of magnitude during the history of life (39). Morphologic distance correlates significantly with maximum body size and range of body size, but not with minimum body size (Table 1). This correlation reflects the greater physical demands and adaptive solutions to body form required by larger body size. It also suggests that there is a threshold in body size below which broad phenotypic disparity may not be possible, perhaps linked to the greater diversity of cell types that characterize organisms that achieve large body size (40, 41).
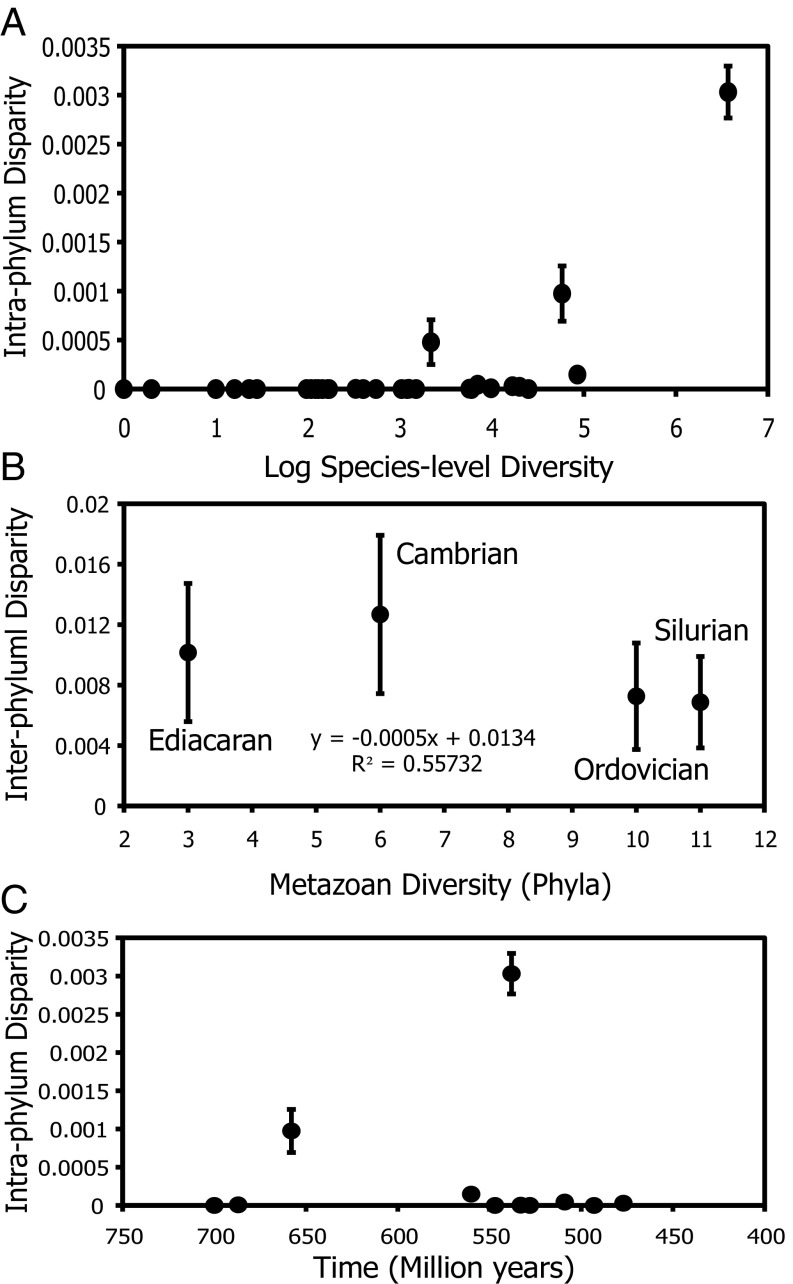
The relationship between diversity and disparity has been an area of intense study in deciphering the meaning of the two metrics (42) as well as the use of higher-level diversity as a proxy for disparity (9). Many metrics have been used to calculate disparity from constructed morphospaces, but the average squared distance between taxa within morphospace shows the greatest stability with smaller sample sizes (43). Differences in modern species-level diversity within phyla (22) correlate to morphologic distances between phyla (Table 2), and the number of species within a phylum correlates strongly to the disparity contained within (Fig. 4A and Table 2). This indicates that, at a higher taxonomic level, these metrics are closely related. However, a comparison of phylum-level diversity and disparity through time (based on origination data from ref. 29) indicates that there is no correlation between these two aspects of variance. Indeed, the relationship is static through time (Fig. 4B), indicating that the number of phyla provides a poor measure of metazoan disparity. However, our results cannot reject equivalence between our measure of disparity and diversity measured by counts of Linnean ranks below the phylum level. In sum, Linnean rank taxonomic measures of disparity have overestimated the scale of early metazoan disparity and therefore the phenomenon to be explained by intrinsic and extrinsic causal factors.
Table 2.
Statistical comparison of morphology against diversity and origination
| Diversity/origination | Spearman’s ρ | P value | No. of phyla |
| Species level diversity | 0.698 | <0.001 | 32 |
| Species level diversity (excl. 0s) | 0.865 | <0.001 | 16 |
| Origination | 0.041 | 0.905 | 11 |
| Origination (excl. 0s) | 0.047 | 0.903 | 9 |
The positions of phyla were calculated as described in Table 1. The number of phyla included in the test varies with the availability of data (lists of phyla included in the different comparisons are presented in SI Appendix, Table S3). Morphological disparity was calculated as the average squared distance between taxon within a phylum. Diversity was compiled by Chapman (22), and origination data are from Erwin et al. (29)
Fig. 4.
Comparisons between taxonomic diversity and morphologic disparity. Disparity is calculated as the average squared distance between taxa within the morphospace. Intraphylum disparity is calculated by using the distance between Ax’s operational taxa within the morphospace for each phylum, and interphylum disparity is calculated by using the distance between phyla (SI Appendix, Fig. S5). (A) Intraphyla disparity compared with estimates of modern diversity. If there was a single operational taxon within a phylum, the disparity is given a value of 0. (B) Comparison between metazoan diversity and interphylum disparity for 4 time intervals for the 11 phyla with well-resolved origination dates (SI Appendix, Table S5). For each time bin, the number of phyla that were present during the beginning of the interval were tallied (i.e., metazoan diversity) so, for example, 3 of the 11 phyla originated before the beginning of the Ediacaran, whereas an additional 3 phylum-level crown groups had appeared by the beginning of the Cambrian. Interphylum disparity was then calculated for the phyla present during that interval. (C) Intraphyla disparity compared with the age of the phylum. Origination data are from Erwin et al. (29) and diversity data are from Chapman (22). Error bars are calculated as the SE of 1,000 bootstrap replicates.
Testing Hypotheses of Causality.
Distilling the phenomenon of animal disparity is one thing; establishing its causality is another. Explanations encompass intrinsic causes, including expansions in genome size (44), the diversification of protein domains and domain architectures (45), the origin of a “developmental toolkit” of transcription factors and cell signaling molecules (46), and the evolutionary assembly of gene regulatory networks (GRNs) (5), although it has also been argued that the exploration of metazoan morphospace is largely a time-dependent random walk (36). To test among these hypotheses, we compiled datasets of phylum origination dates (29), protein domains and their architectures (47), average genome sizes (48), and microRNAs (49) to serve in proxy for the diversity of GRNs. The multivariate protein and microRNA data were also analyzed by using NMDS. Our tests are limited to correlation with the use of a Mantel test. The position of a phylum within morphospace was taken as the position of its crown ancestor (SI Appendix, Fig. S6).
To explore whether the observed patterns in phylum-level morphologic differences could be the result of accumulated random evolutionary processes over time (36), we compared origination time to within-phylum disparity (Fig. 4C). In the absence of morphological constraint or stabilizing selection, clades that originated earlier would have more time to accumulate random changes and therefore to accumulate a higher level of disparity. No significant trend is seen between disparity and origination time (Table 2), indicating that, even though random processes may be contributing to disparity, there must be other, dominant factors that cause the vast difference in morphologic expansion between phyla regardless of origination time.
To test among the intrinsic causes of disparity (SI Appendix, Tables S3 and S4), we first compared differences in genome size (c-value) to the distances between phyla within morphospace. Genome size has been linked to several biological features such as organismal complexity, metabolic rates, and developmental rate (50), and we can reject the hypothesis that genome size and morphological complexity are uncorrelated at a high level of significance (Table 1). Genome size is not correlated to the number of genes (50), suggesting that the link between genome size and morphology might instead be effected largely through gene regulation, rather than the number of genes per se. Although the expansions of some protein-domain superfamilies have been correlated with the evolution of complexity (51), our data show no overall correlation between disparity and the repertoire of protein domains or architectures (Table 1 and SI Appendix, Fig. S7). In contrast, the number of miRNA families correlates significantly with morphological disparity (Table 1 and SI Appendix, Fig. S8), lending correlative support to the hypothesis that expansions in gene regulatory complexity underlie the evolution of metazoan morphological complexity (5, 7, 52).
Discussion
Our mapping of metazoan morphospace has shown that, even though some animal phyla demonstrate maximal initial disparity, others, notably chordates, arthropods, annelids, and mollusks, have progressively expanded on the limits of phylum and kingdom level morphospace post-Cambrian. This contradicts the generally held view of maximal initial disparity for the animal kingdom, which has been based largely on two assumptions: (i) that Linnean ranks serve as an effective proxy for organismal disparity and (ii) that patterns of morphospace occupation captured by continuous or discrete character observations at low taxonomic rank reflect the same phenomenon at phylum and kingdom levels. These are assumptions we refute. Not all studies have returned a pattern of maximal initial disparity. Indeed, disparity analyses based on continuous (53) and discrete characters (18) have identified instances in which maximal disparity is achieved in the middle period or late in the evolutionary history of lineages. Nevertheless, recent compilations have shown that, more often than not, maximal disparity is achieved early in the evolutionary history clades (10, 18). However, it does not follow logically that, at the comparatively low taxonomic level at which these studies have been conducted, a pattern of maximal initial disparity will scale to a self-similar pattern at the highest taxonomic levels. Nevertheless, disparity at low and high taxonomic levels is linked hierarchically such that, regardless of whether its zenith is achieved early or late, increasing disparity at low taxonomic levels must scale to gross disparity increasing progressively at the highest taxonomic levels.
The clumpiness of metazoan morphospace occupation has been alternately argued to be a consequence of (i) gene regulatory and/or other constraints such as integration or modularity, (ii) extinction, and (iii) incomplete exploration of morphospace. Our analyses demonstrate that, even though many (but not all) modern clades occupy discrete regions of morphospace, the inclusion of Cambrian taxa indicates that phylogenetic intermediates of living clades occupy concomitantly intermediate regions of morphospace. Combined with our inferences of the course of metazoan phylogeny through morphospace, these results indicate that the morphological discreteness of modern clades is largely a consequence of the extinction of phylogenetic intermediates. Nevertheless, the results of our analyses including fossils and phylogenetic ancestors indicate that the majority of the empirical morphospace circumscribed by our dataset has not been explored in metazoan evolutionary history. This may be because insufficient time has elapsed for all of these theoretical phenotypes to have been realized during metazoan phylogeny. However, there is a high level of phylogenetic linkage among many of the phenotypic characters in the dataset (Dataset S1): approximately half of the characters in the dataset are contingent directly on just 20 characters, and almost all of the characters are contingent ultimately on just a few characters (epithelia; ontogenesis; somatic differentiation). This reflects a high level of phylogenetically rooted developmental constraint underpinning the distribution of characters and their possible combinations. Thus, it is likely that the majority of the theoretical morphologies represented by unoccupied volumes of morphospace are unrealizable because the implied character combinations are not possible (e.g., ref. 54).
Evidently, the heterogeneity in the phylogenetic linkage of characters reflects the fact that some have contributed more than others to the realization of metazoan bodyplans. These characters can be identified based simply on their contingent linkage, and also through ordination of the phenotypic characters based on their distribution among species rather than through ordination of the species based on their distribution among characters (Fig. 2C). This reveals that the large distance of morphospace that separates nonmetazoans from metazoans is influenced strongly by those characters with a heavy contingent burden that evolved early within the metazoan stem-lineage. Crown metazoan characters, such as a defined head, stomochord, protocoel, ecdysis, segmentation, and features of nephridial cells, all load strongly along NMDS axis one, indicating that these characters are important in the establishment of metazoan body plans.
Undoubtedly, it would be useful to explore the impact of including a greater number of fossil taxa in future analyses of evolutionary disparity that build upon the dataset we present. However, fossil species will always be limited in the amount and class of data they contribute to disparity analyses. Reducing the dataset to features that can be preserved in the fossil record fails to encompass the richness of characters that diagnose the bodyplans of high-rank taxa, and the ordination of such a dataset results in a perspective on metazoan diversification that is reduced dramatically, demonstrating that many of the key superphylum characteristics defining axis one are lost taphonomically. Even though the structure of the morphospace is retained by using preserved or nonpreserved subsets of the characters, there are major differences between the resulting ordinations (Fig. 5). This is a consequence of an increase in apparent disparity within highly skeletonized phyla (Arthropoda, Vertebrata, and Echinodermata) at the expense of those phyla that are extensively soft-bodied (e.g., Rotifera, Platyhelminthes, and Gnathostomulida). The absence of soft-tissue characteristics exaggerates the disparity within the independently derived skeletal features, masking the differences between the majority of phyla, which are based largely on soft-tissue characters, and diminishing the scale of the morphological radiation within metazoans as a whole. Therefore, it is likely that, because the anatomical features that underlie the deep connections within metazoans are not preserved, trends in metazoan disparity cannot be addressed with fossil taxa alone.
Fig. 5.
Distribution of taxa within morphospace based on fossilizable and nonfossilizable characters. (A) Morphospace based on 912 fossilizable characters. (B) Morphospace based on 878 characters unlikely to be observable in fossil organisms. (C) Intrametazoan and intraphylum disparity based on ordinations of only fossilizable characters and only nonfossilizable characters. Phyla above red line have higher disparity within nonpreserved soft-tissue characters, whereas those below have higher disparity within preserved gross anatomical or skeletal characters.
The results of this analysis also suggest that the perception of metazoan diversification represented in the fossil record serves to exaggerate the increase in disparity associated with the origin of fossilizable characters. Thus, the “Cambrian Explosion” phenomenon may more represent an explosion in fossilizable characters, and therefore fossils (55), rather than the dramatic increase in phenotypic disparity it has long been interpreted to represent. In demonstrating a dramatic post-Cambrian expansion in the maximum variance in phenotypic disparity, our results indicate that the scale of the Cambrian Explosion has been grossly overestimated. This effectively mirrors the extensive pre-Cambrian evolutionary history of metazoans estimated by molecular clock methodology (56), in providing more time for the accrual of metazoan disparity achieved before the end of the Cambrian and thereby making the challenge of identifying causality far more tractable.
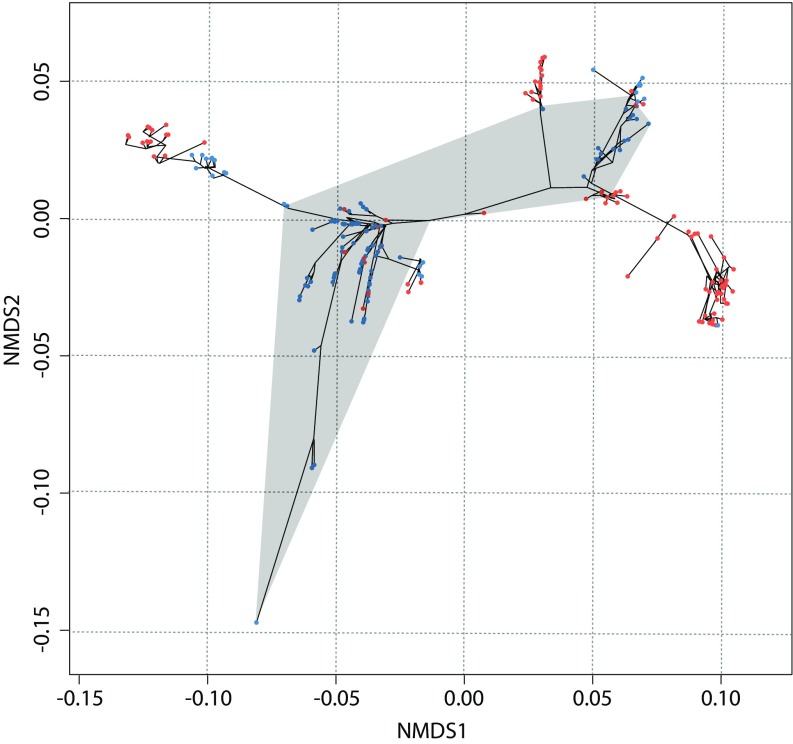
Our tests of hypotheses of causality cannot reject a role for random variation in effecting metazoan phenotypic disparity, but we find no correlative support for the role of expansions in the protein-coding repertoire of the genome. However, our results lend correlative support to the hypothesis that expansions in gene regulatory complexity underlie the evolution of metazoan morphological complexity. This does not preclude a role for extrinsic mechanisms such as the expansion of ecospace (57). Indeed, the phyla that deviate most significantly from the thesis of maximal initial disparity (arthropods, mollusks, annelids, chordates) did so in large part within the hitherto underexplored terrestrial environment (Fig. 6). In so doing, these lineages were released from the physical constraints of an aqueous marine environment to explore new realms of morphospace. Although the causal factors underlying metazoan organismal disparity have been considered nonuniformitarian (7, 8), our analysis shows that the capacity for novelty in metazoan evolution has not disappeared since the Cambrian. Of course, this may be because the intrinsic processes generating the underlying genetic and developmental variation are the same.
Fig. 6.
The role of transitioning to terrestrial environments in the post-Cambrian exploration of morphospace. The phylogenetic pathway within morphospace as well as the area occupied during the Cambrian is shown as in Fig. 3. Organisms that are exclusively aquatic (marine, brackish, freshwater, or parasitic) are colored in blue, whereas taxonomic groups that have some terrestrial representation are colored in red. Terrestrial groups are included within arthropods, chordates, onychophorans, tardigrades, gastropods, rotifers, annelids, nematodes, nemerteans, and Platyhelminthes.
Our results also suggest that debate on whether early animal evolution has been underpinned by uniformitarian or nonuniformitarian processes has been misplaced. Animal evolutionary history does not appear to have been characterized by a uniform rate and scale of change but rather by a high frequency of small changes and low frequency of changes of large magnitude within the context of intrinsic genetic and developmental variation and extrinsic environmental change. Such patterns are readily open to modeling in the same manner as nucleotide and amino acid substitution frequencies. Future research in this direction will inform understanding of the nature of phenotypic evolution, its relation to molecular evolution, underpinning the development of phylogenetic methods. However, it will also provide for a more precise characterization of the tempo of metazoan diversification and the processes that underpinned the establishment of animal bodyplans.
Supplementary Material
Acknowledgments
We thank Neil Shubin (Chicago), Peter Wagner (Lincoln, NE) and two anonymous referees for their patience in assisting in the development of the manuscript. This work was funded through grants from the Natural Environment Research Council (to P.C.J.D.), the National Science Foundation (to B.D. and K.J.P.), the NASA National Astrobiology Institute (to K.J.P.), and a Benjamin Meaker Visiting Professorship from the Institute of Advanced Studies, University of Bristol (to B.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810575115/-/DCSupplemental.
References
- 1.Gould SJ. Wonderful Life: The Burgess Shale and the Nature of History. Huchinson Radius; London: 1990. [Google Scholar]
- 2.Briggs DEG, Fortey RA, Wills MA. Morphological disparity in the cambrian. Science. 1992;256:1670–1673. doi: 10.1126/science.256.5064.1670. [DOI] [PubMed] [Google Scholar]
- 3.Foote M. The evolution of morphological diversity. Annu Rev Ecol Syst. 1997;28:129–152. [Google Scholar]
- 4.Hall BK. Baupläne, phylotypic stages, and constraint. Why there are so few types of animals. Evol Biol. 1996;29:215–261. [Google Scholar]
- 5.Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 6.Davidson EH, Erwin DH. Evolutionary innovation and stability in animal gene networks. J Exp Zoolog B Mol Dev Evol. 2010;314:182–186. doi: 10.1002/jez.b.21329. [DOI] [PubMed] [Google Scholar]
- 7.Erwin DH, Davidson EH. The evolution of hierarchical gene regulatory networks. Nat Rev Genet. 2009;10:141–148. doi: 10.1038/nrg2499. [DOI] [PubMed] [Google Scholar]
- 8.Erwin DH. Evolutionary uniformitarianism. Dev Biol. 2011;357:27–34. doi: 10.1016/j.ydbio.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 9.Erwin DH, Valentine JW, Sepkoski JJ. A comparative-study of diversification events–The early Paleozoic versus the Mesozoic. Evolution. 1987;41:1177–1186. [PubMed] [Google Scholar]
- 10.Erwin DH. Disparity: Morphologic pattern and developmental context. Palaeontology. 2007;50:57–73. [Google Scholar]
- 11.Villier L, Eble GJ. Assessing the robustness of disparity estimates: The impact of morphometric scheme, temporal scale, and taxonomic level in spatangoid echinoids. Paleobiology. 2004;30:652–665. [Google Scholar]
- 12.Foth C, Brusatte SL, Butler RJ. Do different disparity proxies converge on a common signal? Insights from the cranial morphometrics and evolutionary history of Pterosauria (Diapsida: Archosauria) J Evol Biol. 2012;25:904–915. doi: 10.1111/j.1420-9101.2012.02479.x. [DOI] [PubMed] [Google Scholar]
- 13.Hopkins MJ. How well does a part represent the whole? A comparison of cranidial shape evolution with exoskeletal character evolution in the trilobite family Pterocephaliidae. Palaeontology. 2017;60:309–318. [Google Scholar]
- 14.Hetherington AJ, et al. Do cladistic and morphometric data capture common patterns of morphological disparity? Palaeontology. 2015;58:393–399. [Google Scholar]
- 15.Huttegger SM, Mitteroecker P. Invariance and meaningfulness in phenotype spaces. Evol Biol. 2011;38:335–351. [Google Scholar]
- 16.Mitteroecker P, Huttegger SM. The concept of morphospaces in evolutionary and developmental biology: Mathematics and metaphors. Biol Theory. 2009;4:54–67. [Google Scholar]
- 17.Gerber S. The geometry of morphospaces: Lessons from the classic Raup shell coiling model. Biol Rev Camb Philos Soc. 2016;92:1142–1155. doi: 10.1111/brv.12276. [DOI] [PubMed] [Google Scholar]
- 18.Hughes M, Gerber S, Wills MA. Clades reach highest morphological disparity early in their evolution. Proc Natl Acad Sci USA. 2013;110:13875–13879. doi: 10.1073/pnas.1302642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ax P. Multicellular Animals: A New Approach to the Phylogenetic Order in Nature. Vol 1. Springer; Berlin: 1996. p. 225. [Google Scholar]
- 20.Ax P. Multicellular Animals: The Phylogenetic System of the Metazoa. Vol 2. Springer; Berlin: 2000. p. 369. [Google Scholar]
- 21.Ax P. Multicellular Animals: Order in Nature–System Made by Man. Vol 3. Springer; Berlin: 2003. p. 317. [Google Scholar]
- 22.Chapman A. Numbers of Living Species in Australia and the World. Australian Biological Resources Study; Canberra, Australia: 2005. p. 61. [Google Scholar]
- 23.Gower J. A general coefficient of similarity and some of its properties. Biometrics. 1971;27:857–871. [Google Scholar]
- 24.Deline B. The effects of rarity and abundance distributions on measurements of local morphological disparity. Paleobiology. 2009;35:175–189. [Google Scholar]
- 25.Deline B, Ausich W. Testing the plateau: A reexamination of disparity and morphologic constraints in early Paleozoic crinoids. Paleobiology. 2011;37:214–236. [Google Scholar]
- 26.Tweedt S. The Cambrian Explosion: The Construction of Animal Biodiversity. Roberts; Greenwood Village, CO: 2013. First appearences of major metazoan clades in the fossil record; pp. 343–354. [Google Scholar]
- 27.Huelsenbeck JP, Nielsen R, Bollback JP. Stochastic mapping of morphological characters. Syst Biol. 2003;52:131–158. doi: 10.1080/10635150390192780. [DOI] [PubMed] [Google Scholar]
- 28.Revell LJ. Phytools: An R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2012;3:217–223. [Google Scholar]
- 29.Erwin DH, et al. The Cambrian conundrum: Early divergence and later ecological success in the early history of animals. Science. 2011;334:1091–1097. doi: 10.1126/science.1206375. [DOI] [PubMed] [Google Scholar]
- 30.Conway Morris S. Life’s Solution: Inevitable Humans in a Lonely Universe. Cambridge Univ Press; Cambridge, UK: 2003. p. 650. [Google Scholar]
- 31.Wills M. Cambrian and recent disparity: The picture from priapulids. Paleobiology. 1998;24:177–199. [Google Scholar]
- 32.Foote M. Morphological diversity in the evolutionary radiation of Paleozoic and post-Paleozoic crinoids. Paleobiology. 1999;25:1–116. [Google Scholar]
- 33.Stockmeyer Lofgren A, Plotnick RE, Wagner PJ. Morphological diversity of Carboniferous arthorpods and insights on disparity patterns of the Phanerozoic. Palaeobiology. 2003;29:350–369. [Google Scholar]
- 34.Wagner PJ, Ruta M, Coates MI. Evolutionary patterns in early tetrapods. II. Differing constraints on available character space among clades. Proc Biol Sci. 2006;273:2113–2118. doi: 10.1098/rspb.2006.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sallan LC, Friedman M. Heads or tails: Staged diversification in vertebrate evolutionary radiations. Proc Biol Sci. 2012;279:2025–2032. doi: 10.1098/rspb.2011.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McShea DW, Brandon RN. Biology’s First Law: The Tendency for Diversity and Complexity to Increase in Evolutionary Systems. Univ Chicago Press; Chicago: 2010. p. 170. [Google Scholar]
- 37.Valentine JW, Collins AG, Meyer CP. Morphological complexity increase in metazoans. Paleobiology. 1994;20:131–142. [Google Scholar]
- 38.McClain CR, Boyer AG. Biodiversity and body size are linked across metazoans. Proc Biol Sci. 2009;276:2209–2215. doi: 10.1098/rspb.2009.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Payne JL, et al. Two-phase increase in the maximum size of life over 3.5 billion years reflects biological innovation and environmental opportunity. Proc Natl Acad Sci USA. 2009;106:24–27. doi: 10.1073/pnas.0806314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonner JT. Size and Cycle: An Essay on the Structure of Biology. Princeton Univ Press; Princeton, NJ: 1965. p. 260. [Google Scholar]
- 41.Bonner JT. The Evolution of Complexity. Princeton Univ Press; Princeton, NJ: 1988. [Google Scholar]
- 42.Foote M. Contributions of individual taxa to overall morphological disparity. Paleobiology. 1993;19:403–419. [Google Scholar]
- 43.Ciampaglio CN, Kemp M, McShea DW. Detecting changes in morphospace occupation patterns in the fossil record: Characterization and analysis of measures of disparity. Paleobiology. 2001;27:695–715. [Google Scholar]
- 44.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 45.Tordai H, Nagy A, Farkas K, Bányai L, Patthy L. Modules, multidomain proteins and organismic complexity. FEBS J. 2005;272:5064–5078. doi: 10.1111/j.1742-4658.2005.04917.x. [DOI] [PubMed] [Google Scholar]
- 46.Carroll SB. Homeotic genes and the evolution of arthropods and chordates. Nature. 1995;376:479–485. doi: 10.1038/376479a0. [DOI] [PubMed] [Google Scholar]
- 47.de Lima Morais DA, et al. SUPERFAMILY 1.75 including a domain-centric gene ontology method. Nucleic Acids Res. 2011;39:D427–D434. doi: 10.1093/nar/gkq1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gregory TR. 2012 Animal Genome Size Database. www.genomesize.com.
- 49.Tarver JE, et al. miRNAs: Small genes with big potential in metazoan phylogenetics. Mol Biol Evol. 2013;30:2369–2382. doi: 10.1093/molbev/mst133. [DOI] [PubMed] [Google Scholar]
- 50.Gregory TR. Genome size evolution in animals. In: Gregory TR, editor. The Evolution of the Genome. Elsevier; San Diego: 2005. pp. 3–87. [Google Scholar]
- 51.Vogel C, Chothia C. Protein family expansions and biological complexity. PLOS Comput Biol. 2006;2:e48. doi: 10.1371/journal.pcbi.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peterson KJ, Dietrich MR, McPeek MA. MicroRNAs and metazoan macroevolution: Insights into canalization, complexity, and the Cambrian explosion. BioEssays. 2009;31:736–747. doi: 10.1002/bies.200900033. [DOI] [PubMed] [Google Scholar]
- 53.Foote M. Paleozoic record of morphologica diversity in blastozoan echinoderms. Proc Natl Acad Sci USA. 1992;89:7325–7329. doi: 10.1073/pnas.89.16.7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Foote M. Morphological disparity in Ordovician-Devonian crinoids and the early saturation of morphological space. Palaeobiology. 1994;20:320–344. [Google Scholar]
- 55.Runnegar B. The Cambrian explosion–animals or fossils. J Geol Soc Aust. 1982;29:395–411. [Google Scholar]
- 56.dos Reis M, et al. Uncertainty in the timing of origin of animals and the limits of precision in molecular timescales. Curr Biol. 2015;25:2939–2950. doi: 10.1016/j.cub.2015.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Erwin DH. Early introduction of major morphological innovations. Acta Palaeontol Pol. 1994;38:281–294. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.