Protein concentration gradients are a common strategy to compartmentalize activities within cells and tissues. Gradients position the division plane of bacterial cells, regulate the size of yeast cells, and pattern embryos (1–3). Among the most studied gradients is the Bicoid gradient of Drosophila. Bicoid protein is synthesized from a localized source of bcd mRNA at the anterior-most pole of the embryo (4, 5). In one model, newly synthesized Bicoid is proposed to diffuse away from this point source and to turn over at a constant rate throughout the cytoplasm, thus generating an anterior-rich protein concentration gradient (6) (Fig. 1A). Drosophila embryos, however, are unusually large (>400 μm) syncytial cells. Smaller cells are unlikely to support Bicoid-like gradients, since the rate of protein diffusion in the cytoplasm is typically too fast (10 μm2⋅s−1) to prevent proteins from sampling the entire cytoplasm within seconds (2, 7). If so, how do most cells generate protein gradients? In 2008, Lipkow and Odde (8) offered a simple solution to this dilemma. Using theoretical modeling, they demonstrated that protein gradients can be sustained in cells of any size by coupling regulation of protein diffusivity to a spatially segregated protein modification system. The reversible protein modification (e.g., phosphorylation) toggles the protein between two states with different diffusion coefficients (one fast and one slow). Imposing a spatial bias in the distribution of one of the protein modification enzymes (e.g., the kinase) locally increases the concentration of one diffusive state, thus generating a protein concentration gradient whose steepness is proportional to the amplitude of the difference in protein diffusion coefficients (8). In PNAS, Wu et al. (9) provide remarkable experimental evidence that such a mechanism drives the formation of cytoplasmic gradients in Caenorhabditis elegans zygotes.
Fig. 1.
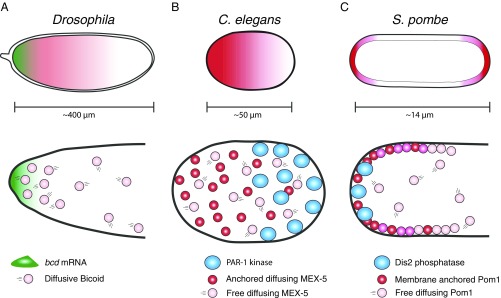
Role of protein diffusion in protein gradients. (A) In Drosophila, the anterior/posterior axis of the oocyte is patterned, in part, by a gradient of Bicoid protein. Bicoid protein is synthesized from a localized source of bicoid mRNA (green) at the anterior-most pole of the embryo. Newly synthesized Bicoid protein (pink circles) diffuses away from this point source and is turned over at a constant rate throughout the cytoplasm. (B) In the C. elegans zygote, somatic and germline cell fates are specified, in part, by a gradient of the protein MEX-5. MEX-5 diffuses throughout the cytoplasm in two states: an FD state (pink circles) and an SD state (red circles). The FD molecules are uniformly distributed. The FD-to-SD switch occurs at a higher frequency in the anterior, causing an enrichment of SD molecules in the anterior cytoplasm. In the posterior cytoplasm, the posterior-localized PAR-1 kinase (blue circles) antagonizes the FD-to-SD switch. (C) In S. pombe, entry into mitosis is regulated by a gradient of Pom1. Pom1 is a membrane-associated kinase that autoregulates its affinity for the plasma membrane by autophosphorylation. As a result, Pom1 exists in two states: a low-phosphorylation/slow diffusion/membrane-bound state and a high-phosphorylation/fast diffusion/cytoplasmic state. Dephosphorylation by the tip-localized phosphatase Dis2 recruits Pom1 to cell tips. Pom1 diffuses laterally from the tips until sufficiently phosphorylated to detach from membrane.
The C. elegans zygote is a 30 × 50-μm oblong cell that becomes polarized along its long axis shortly after fertilization. The sperm centrosome initiates a cascade of cytoskeletal movements and protein interactions that eventually enriches the polarity kinase PAR-1 in the posterior half of the zygote (10). Posterior PAR-1 phosphorylates the RNA-binding protein MEX-5, causing MEX-5 to accumulate in an anterior-rich gradient (11, 12). By an unknown mechanism, MEX-5, in turn, drives the RNA-binding PIE-1 in an opposite polarity, posterior-rich gradient (13, 14).
Earlier studies have shown that formation of the MEX-5 and PIE-1 gradients coincides with changes in their diffusive mobility along the anterior/posterior axis (11, 12, 14–16). MEX-5 mobility, on average, increases in the posterior, and PIE-1 mobility, on average, increases in the anterior. Fluorescence correlation spectroscopy measurements suggested that each protein exists in at least two diffusive states whose relative ratio changes along the anterior/posterior axis during gradient formation (12, 15, 16). Because those studies lacked single-molecule resolution, the diffusive behavior of each species and their interconversion dynamics were not known. In particular, weak directional transport of the slow species could not be excluded as a potential contributor to gradient formation. Using total internal reflection fluorescence microscopy, Wu et al. (9) tracked individual MEX-5 and PIE-1 molecules labeled with GFP. As expected, they detected two species for each protein: a fast-diffusing (FD) species that samples the entire cytoplasm evenly and a slow-diffusing (SD) species that accumulates in the high-concentration region of each gradient (anterior for MEX-5 and posterior for PIE-1) (Fig. 1B). Molecules in the SD state appear throughout the cytoplasm and undergo short-range motions with no directional bias, consistent with being tethered to a distributed, subdiffusive anchor. For both MEX-5 and PIE-1, the rate at which SD-state molecules appear varies along the anterior/posterior axis, with the highest frequency in the high-concentration areas of the gradient. Remarkably, the authors find that the SD MEX-5 and PIE-1 species persist only for a few seconds before returning to the FD state. This rapid switching implies that all molecules in the gradient sample the entire cytoplasm and only rest intermittently on the anchor. Together, these observations suggest that local differences in the rate of FD-to-SD switching is what drives gradient formation.
Using mathematical modeling, the authors asked which parameters were critical for gradient generation. First, they found that rates of SD-to-FD and FD-to-SD switching estimated from the experimental data were sufficient to produce gradients of the correct amplitude within minutes in silico, as is true in vivo. The authors also found that coordinately changing the diffusion coefficients of the SD and FD states had minimal effects on gradient shape as long as the difference between the two values remained high (>10-fold). These findings suggest that kinetic switching could be used to generate stable gradients over a wide range of diffusion constants. The amplitude of the gradient was maintained over a range of cell sizes from 16 to 50 μm but decreased below 16 μm, as the mobility of the SD species became sufficient to dissipate the gradient. Gradient amplitude was also sensitive to the relative rates of interconversion between FD and SD states. Faster FD-to-SD rates increase gradient steepness but only within a certain range beyond which the gradient no longer forms (9). These studies highlight the importance of balancing the on/off rates of the protein modifications that mediate the kinetic switch.
The modification that regulates MEX-5 and PIE-1 kinetic switching is likely to be phosphorylation. MEX-5 has been shown to be phosphorylated by PAR-1 on a residue required for gradient formation in vivo (11, 12). The kinase that phosphorylates PIE-1 is not yet known but is likely to be PLK-1, a kinase that binds to MEX-5 and accumulates with it in an anterior-rich gradient (9, 17). The prediction is that phosphorylation antagonizes the transition from FD to SD by reducing the affinity with which MEX-5 and PIE-1 molecules interact with the anchor. The uniformly distributed PP2A phosphatase has been proposed to function as the opposing phosphatase that dephosphorylates MEX-5, returning it to the SD state (12). The identity of the anchor is not yet known. MEX-5 and PIE-1 both contain RNA-binding domains, which are required for gradient formation (12, 18, 19). mRNAs associated with the endoplasmic reticulum, which fills the entire cytoplasm in the zygote, could, in principle, function as subdiffusive anchors (20). Interestingly, regulation of membrane binding by a spatially segregated phosphorylation/dephosphorylation cycle has already been suggested to promote the formation of another intracellular gradient. Pom1 is a kinase in Schizosaccharomyces pombe that regulates mitotic entry. Pom1 forms a membrane gradient, with the highest concentration at cell tips (21, 22). Pom1 association with the membrane is antagonized by phosphorylation (23). The tip-enriched Dis2 phosphatase promotes loading of unphosphorylated Pom1 at cell tips (23, 24). Pom1 diffuses laterally and autophosphorylates, gradually causing its own detachment from the membrane (23) (Fig. 1C). In principle, variations in the rate of kinetic switching and in the distributions of the kinase, phosphatase, and anchor could generate gradients with different steepness, shape, and location. Connecting the formation of one gradient to that of another, as in the case of MEX-5 and PIE-1, expands gradient possibilities even further. The versatility and rapidity of gradient formation by kinetic switching suggest that this mechanism may be a common strategy to pattern cells in a wide range of developmental contexts.
Footnotes
The authors declare no conflict of interest.
See companion article on page E8440 in issue 36 of volume 115.
References
- 1.Wartlick O, Kicheva A, González-Gaitán M. Morphogen gradient formation. Cold Spring Harb Perspect Biol. 2009;1:a001255. doi: 10.1101/cshperspect.a001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kiekebusch D, Thanbichler M. Spatiotemporal organization of microbial cells by protein concentration gradients. Trends Microbiol. 2014;22:65–73. doi: 10.1016/j.tim.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Griffin EE. Cytoplasmic localization and asymmetric division in the early embryo of Caenorhabditis elegans. Wiley Interdiscip Rev Dev Biol. 2015;4:267–282. doi: 10.1002/wdev.177. [DOI] [PubMed] [Google Scholar]
- 4.Berleth T, et al. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Driever W, Nüsslein-Volhard C. A gradient of bicoid protein in Drosophila embryos. Cell. 1988;54:83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- 6.Ephrussi A, St Johnston D. Seeing is believing: The bicoid morphogen gradient matures. Cell. 2004;116:143–152. doi: 10.1016/s0092-8674(04)00037-6. [DOI] [PubMed] [Google Scholar]
- 7.Howard M. How to build a robust intracellular concentration gradient. Trends Cell Biol. 2012;22:311–317. doi: 10.1016/j.tcb.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Lipkow K, Odde DJ. Model for protein concentration gradients in the cytoplasm. Cell Mol Bioeng. 2008;1:84–92. doi: 10.1007/s12195-008-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Y, et al. Rapid diffusion-state switching underlies stable cytoplasmic gradients in the Caenorhabditis elegans zygote. Proc Natl Acad Sci USA. 2018;115:E8440–E8449. doi: 10.1073/pnas.1722162115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Motegi F, Seydoux G. The PAR network: Redundancy and robustness in a symmetry-breaking system. Philos Trans R Soc Lond B Biol Sci. 2013;368:20130010. doi: 10.1098/rstb.2013.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tenlen JR, Molk JN, London N, Page BD, Priess JR. MEX-5 asymmetry in one-cell C. elegans embryos requires PAR-4- and PAR-1-dependent phosphorylation. Development. 2008;135:3665–3675. doi: 10.1242/dev.027060. [DOI] [PubMed] [Google Scholar]
- 12.Griffin EE, Odde DJ, Seydoux G. Regulation of the MEX-5 gradient by a spatially segregated kinase/phosphatase cycle. Cell. 2011;146:955–968. doi: 10.1016/j.cell.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schubert CM, Lin R, de Vries CJ, Plasterk RH, Priess JR. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol Cell. 2000;5:671–682. doi: 10.1016/s1097-2765(00)80246-4. [DOI] [PubMed] [Google Scholar]
- 14.Wu Y, Zhang H, Griffin EE. Coupling between cytoplasmic concentration gradients through local control of protein mobility in the Caenorhabditis elegans zygote. Mol Biol Cell. 2015;26:2963–2970. doi: 10.1091/mbc.E15-05-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniels BR, Perkins EM, Dobrowsky TM, Sun SX, Wirtz D. Asymmetric enrichment of PIE-1 in the Caenorhabditis elegans zygote mediated by binary counterdiffusion. J Cell Biol. 2009;184:473–479. doi: 10.1083/jcb.200809077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniels BR, Dobrowsky TM, Perkins EM, Sun SX, Wirtz D. MEX-5 enrichment in the C. elegans early embryo mediated by differential diffusion. Development. 2010;137:2579–2585. doi: 10.1242/dev.051326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han B, et al. Polo-like kinase couples cytoplasmic protein gradients in the C. elegans zygote. Curr Biol. 2018;28:60–69.e8. doi: 10.1016/j.cub.2017.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reese KJ, Dunn MA, Waddle JA, Seydoux G. Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol Cell. 2000;6:445–455. doi: 10.1016/s1097-2765(00)00043-5. [DOI] [PubMed] [Google Scholar]
- 19.Smith J, et al. Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3. eLife. 2016;5:5. doi: 10.7554/eLife.21337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reid DW, Nicchitta CV. Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2015;16:221–231. doi: 10.1038/nrm3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bähler J, Pringle JR. Pom1p, a fission yeast protein kinase that provides positional information for both polarized growth and cytokinesis. Genes Dev. 1998;12:1356–1370. doi: 10.1101/gad.12.9.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Padte NN, Martin SG, Howard M, Chang F. The cell-end factor pom1p inhibits mid1p in specification of the cell division plane in fission yeast. Curr Biol. 2006;16:2480–2487. doi: 10.1016/j.cub.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 23.Hachet O, et al. A phosphorylation cycle shapes gradients of the DYRK family kinase Pom1 at the plasma membrane. Cell. 2011;145:1116–1128. doi: 10.1016/j.cell.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 24.Alvarez-Tabarés I, Grallert A, Ortiz JM, Hagan IM. Schizosaccharomyces pombe protein phosphatase 1 in mitosis, endocytosis and a partnership with Wsh3/Tea4 to control polarised growth. J Cell Sci. 2007;120:3589–3601. doi: 10.1242/jcs.007567. [DOI] [PubMed] [Google Scholar]