Significance
SmgGDS plays a dual role in the cell and acts as not only a GEF specific for the Rho family but also a chaperone protein for small GTPases possessing a C-terminal polybasic region accompanied by the CaaX motif. SmgGDS folds into armadillo-repeat motifs, structurally distinct from the other GEFs and prenyl group-binding proteins. SmgGDS binding induces conformational changes in both switch regions, resulting in exposure of the nucleotide binding site. The prenyl group at the CaaX motif is inserted into the cryptic pocket of SmgGDS, which is newly created upon binding, shielding the lipid-modified C-terminal tail from the environment. Our structural characterization provides a detailed picture of how SmgGDS works as a GEF and implication for a chaperone mechanism.
Keywords: SmgGDS, guanine nucleotide exchange factor, prenyl group, crystal structure
Abstract
SmgGDS has dual functions in cells and regulates small GTPases as both a guanine nucleotide exchange factor (GEF) for the Rho family and a molecular chaperone for small GTPases possessing a C-terminal polybasic region followed by four C-terminal residues called the CaaX motif, which is posttranslationally prenylated at its cysteine residue. Our recent structural work revealed that SmgGDS folds into tandem copies of armadillo-repeat motifs (ARMs) that are not present in other GEFs. However, the precise mechanism of GEF activity and recognition mechanism for the prenylated CaaX motif remain unknown because SmgGDS does not have a typical GEF catalytic domain and lacks a pocket to accommodate a prenyl group. Here, we aimed to determine the crystal structure of the SmgGDS/farnesylated RhoA complex. We found that SmgGDS induces a significant conformational change in the switch I and II regions that opens up the nucleotide-binding site, with the prenyl group fitting into the cryptic pocket in the N-terminal ARMs. Taken together, our findings could advance the understanding of the role of SmgGDS and enable drug design strategies for targeting SmgGDS and small GTPases.
Small GTPases, including Rho, are key regulators in cellular events, and their dysfunction causes many types of cancer (1, 2). The core domain of small GTPases, the G domain, is composed of three conserved motifs: the phosphate-binding loop (P loop), switch I, and switch II. The P loop binds the alpha and beta phosphates of a guanine nucleotide, whereas the two switch motifs recognize the gamma phosphate (Fig. 1A). These three motifs cooperatively recognize guanine nucleotides and are responsible for GTPase activity. The intrinsic GTP hydrolysis activities of small GTPases are generally low, and they are accelerated by GTPase-activating proteins (GAPs). Guanine-nucleotide exchange factors (GEFs) promote GDP dissociation from small GTPases so that they can bind GTP. When GDP is released from small GTPases and GTP binds to them, a conformational rearrangement of switch I and switch II regions that allows binding to the gamma phosphate occurs and enables small GTPases to interact with downstream effectors (3). The C terminus region of small GTPases is known as the hypervariable region (HVR), which contains four C-terminal residues called the CaaX motif (“a” represents an aliphatic amino acid and “X” represents any amino acid). Cysteine residues in this motif are posttranslationally farnesylated by farnesyltransferase (FTase) or geranylgeranylated by geranylgeranyltransferase (GGTase) in the cytosol (4). This modification allows trafficking of small GTPases to membranes, a necessity for physiological activity.
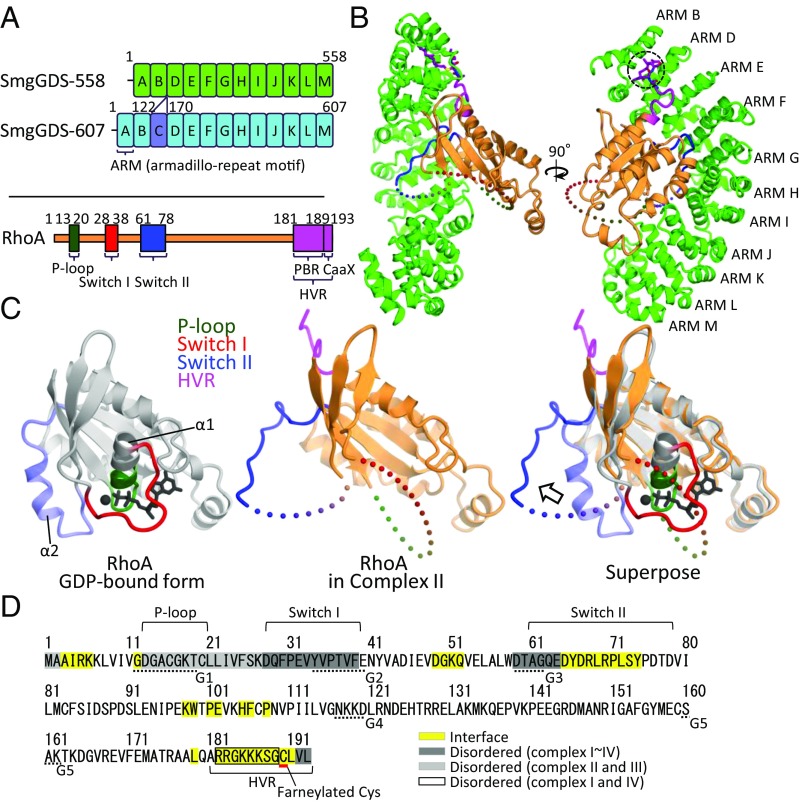
Fig. 1.
Structure of the SmgGDS-558/farnesylated RhoA complex. (A) Domain architectures of two SmgGDS isoforms and RhoA. ARM repeats of SmgGDS-558 and -607 except ARM C are shown in green and sky-blue boxes labeled ARM A–ARM M. ARM C is colored in violet. RhoA is shown in orange, and the three nucleotide recognition domains (P loop, switch I, and switch II) are shown in dark green, red, and blue, respectively. C-terminal HVR is shown colored in purple. The numbering schemes used are also shown. (B) Crystal structure of SmgGDS-558/farnesylated RhoA. SmgGDS-558 and RhoA are shown as a ribbon and surface model. C190 (S-farneylated) and L191 of RhoA are shown as a stick model and enclosed by a dotted circle. (C) Comparison of RhoA structure. Core domain of GDP-bound RhoA (PDB ID code: 1FTN) and RhoA located in complex II are colored in gray and orange, respectively (Left, Middle). Three nucleotide recognition domains (P loop, switch I, and switch II) and HVR are colored in green, red, blue, and purple, respectively. Superposition of these two structures is shown on the Right, and the arrows represent the movement of switch II. α1 and α2 helices of RhoA are labeled. (D) Interface and disordered region of RhoA in complex with SmgGDS-558. The protein–protein interface is highlighted in yellow. The interface was calculated by PISA. The amino acid residues were found to be disordered; complexes II and III, and complexes I and IV are highlighted in a light gray and black square, respectively. The amino acids that compose G boxes are underlined with a dotted line.
Chaperones are a group of proteins that assist noncovalent folding and assembly of macromolecules and/or prevent aggregation of newly synthesized polypeptides. Moreover, the functions of chaperones include the transport of proteins across the membranes of the subcellular organelles and escorting and shuttling of proteins to and from the membrane. SmgGDS also acts as a chaperone protein for small GTPases having a CaaX motif, in addition to a GEF for Rho family proteins (5–10). SmgGDS interacts with various small GTPases possessing a C-terminal polybasic region (PBR) upstream of the CaaX motif in a hypervariable region (HVR) (5, 6, 11–13). SmgGDS is overexpressed in nonsmall cell lung carcinoma (14, 15), prostate cancer (16), breast cancer (15, 17), and pancreatic cancer (15). SmgGDS is a unique architectural GEF that is entirely composed of armadillo-repeat motifs (ARMs) (18). Two SmgGDS splice variants, SmgGDS-558 and SmgGDS-607, which are only different in one insertion ARM (Fig. 1A), are reported to have different preferences in posttranslational modification of RhoA (8, 9). SmgGDS-558 prefers prenylated RhoA, whereas SmgGDS-607 prefers nonprenylated RhoA. Recently, we reported that these splice variants differ in GEF activity and binding affinity for RhoA, depending on their prenylation state (19). We also determined the crystal structure of SmgGDS-558 and revealed that the positively charged concave surface of SmgGDS-558 is a key region for GEF activity (19). However, the precise mechanism underlying how SmgGDS stimulates GDP/GTP exchange of RhoA and how SmgGDS isoforms can distinguish RhoA prenylation remain unclear.
Here, we determined the crystal structure of the SmgGDS-558/farnesylated RhoA complex. A large conformational change in the switch I and II regions opens the nucleotide binding region, facilitating GDP/GTP exchange. The prenyl group of RhoA fits into the hydrophobic cryptic pocket that is only formed by SmgGDS-558 when it binds RhoA. This is a structural study of GEF in complex with prenylated PBR–CaaX, containing small GTPases. Our findings should help advance the understanding of the role of each SmgGDS isoform and enable drug discovery strategies for SmgGDS and small GTPases.
Results
Overall Structure of the SmgGDS-558/Farnesylated RhoA Complex.
We determined the crystal structure of the SmgGDS-558/farnesylated RhoA complex at 3.5-Å resolution. RhoA is generally modified by a geranylgeranyl group at the CaaX motif, but geranylgeranylated RhoA is easily precipitated (20). A previous study reported that isoprenylation of RhoA is required for its biological activity and that the nature of the lipid modification is not critical (21). Therefore, we employed farnesylated RhoA in this work. The final structural model contained four complexes of SmgGDS-558/farnesylated RhoA (complex I–IV) in an asymmetric unit (SI Appendix, Fig. S1A and Table S1). SmgGDS/RhoA forms a 1:1 complex in solution revealed by small angle X-ray scattering, multiangle light scattering, and gel-filtration analyses (19). The N-terminal 10 residues (amino acids 77–86) and C-terminal 3 residues (amino acids 556–558) of SmgGDS-558 were disordered. The side chains for the 63 C-terminal amino acid residues of SmgGDS-558 were not assigned, owing to the poor electron density. In the RhoA structure, the N terminus (amino acids 1–2), the switch I region (amino acids 28–39), part of the switch II region (amino acids 59–64 each), and the C terminus (amino acids 192–193) were disordered in all complexes (Fig. 1 B–D and SI Appendix, Fig. S1B). In complexes II and III, the electron density of RhoA PBR (amino acids 181–189) was observed, but the density was so poor that only main-chain atoms were modeled. In addition, the P loop and its flanking region (amino acids 13–27) were disordered in complexes II and III (Fig. 1 B–D and SI Appendix, Fig. S1B). No guanine nucleotide and Mg2+ were observed, suggesting that these cofactors were released by SmgGDS-558 binding. The previous structural work suggested that the positively/negatively charged regions of SmgGDS-558 are important for GEF activity and recognition of the PBR (7, 19). Our current structure clearly shows that both regions play an important role for RhoA binding. The PBR lies on the negatively charged region formed by concentrated acidic resides in the N terminus, strongly suggesting that these regions are electrostatically attracted to each other, although interaction at an atomic level was unable to be described due to the poor electron density. The switch II region of RhoA interacted with the middle of SmgGDS, consisting of the positively charged region and its flanking region. Hereafter, this region is referred to as the switch II binding region (SI Appendix, Fig. S2).
Structural Changes in the Switch Regions: Implications of the GEF Mechanism.
The most remarkable feature of the structure is the drastic conformational changes that occur in both switch I and II regions. The switch I region was completely disordered and was not able to participate in nucleotide recognition (Fig. 1C). SmgGDS-558 pulled out a part of switch II into the switch II binding region at the middle of its own concave surface, resulting in a drastic conformational change accompanied by the disruption of the α2 helix (Fig. 1C). This conformational change in the switch I and II regions could induce disruption of important interactions among RhoA, Mg2+, and the guanine nucleotide, facilitating guanine nucleotide dissociation. This strongly suggests the involvement of a GDP/GTP exchange mechanism mediated by SmgGDS. The binding interface was analyzed by PISA (22) and showed a wide range of interactions (a buried surface area of 1816.8 Å2), including the N-terminal and C-terminal regions of RhoA in addition to the switch II region. This suggested that SmgGDS forms a broad interface (Fig. 1D). Comparison of the SmgGDS-558/farnesylated RhoA structure to other complex structures of GEFs and small GTPases showed that only SmgGDS-558 does not recognize switch I, whereas, in general, other GEFs recognize switch I and II cooperatively (SI Appendix, Fig. S3).
To date, over 35 structures of RhoA were deposited in the Protein Data Bank, and 7 of these structures (PDB ID codes: 1LB1, 1X86, 1XCG, 3T06, 4XH9, 5JHG, and 5JHH) were guanine nucleotide and magnesium-free forms of RhoA. Notably, none of these were similar to our crystal structure in that both switch regions were disordered, demonstrating that SmgGDS has a unique guanine nucleotide exchange mechanism.
Mutational Analysis for the Interaction Interface Between SmgGDS and RhoA.
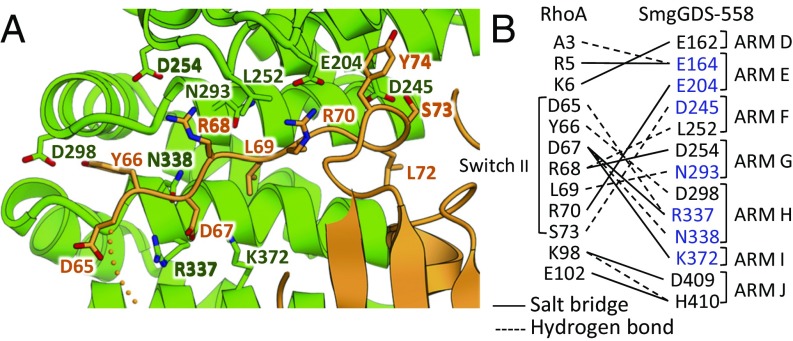
The structure provides an atomic view of the interaction interface, which is highly consistent with the reported mutational effects. The N338 residue of SmgGDS-558, which was reported to be essential for interaction and GEF activity toward RhoA, made a hydrogen bond to the D67 residue of RhoA (Fig. 2) (7, 12, 19). The switch II region and some other residues, including the N-terminal (A3, R5, and K6) and C-terminal flanking region (K98 and E102) of RhoA, formed interaction networks toward SmgGDS-558 (Fig. 2B). Our previous mutational analysis of SmgGDS-558 showed that four residues (H330, R337, N338, and K372) are important for GEF activity and interaction with RhoA. Three of the four residues (R337, N338, and K372) were observed to interact with the switch II region of RhoA in our crystal structure.
Fig. 2.
Interaction of SmgGDS-558/farnesylated RhoA complex. (A) Enlarged view of switch II region of RhoA in complex with SmgGDS-558. Side chains of RhoA (amino acids 65–74) are shown as a stick model. Side chains of SmgGDS-558, which interact with switch II, are also shown as a stick model. (B) Interaction between RhoA and SmgGDS-558. Interactions observed in multiple heterodimers of the asymmetric unit are shown. Dashed and solid lines represent the presence of a hydrogen bond and salt bridge, respectively. SmgGDS residues that are reported to be important for interaction are shown in blue (7, 19).
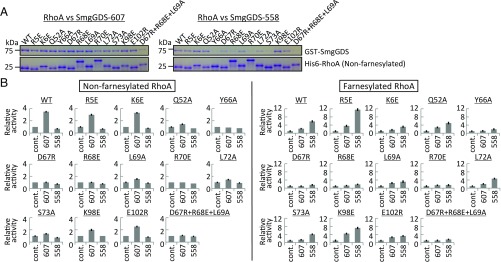
To further confirm the validity of the crystal structure, we performed structure-based pulldown (Fig. 3A) and GEF assays (Fig. 3B and SI Appendix, Fig. S4) to examine the role of the residues in the interaction between SmgGDS-558 and RhoA. We also performed the same experiment using SmgGDS-607. Wild type, 12 single mutants (R5E, K6E, Q52A, Y66A, D67R, R68E, L69A, R70E, L72A, S73A, K98E, and E102R), and one triple mutant (D67R+R68E+L69A) of RhoA were expressed in Escherichia coli and purified. A pulldown assay was performed for nonfarnesylated RhoA and a GEF assay was performed for both nonfarnesylated and farnesylated RhoA.
Fig. 3.
Mutational analysis. (A) Pulldown assay of SmgGDS-558 and SmgGDS-607. Pulldown results in which His-RhoA was used as bait are shown. Nonfarnesylated RhoA was used for this assay. Different mobility of RhoA observed on the SDS-PAGE was in the nature of each mutant. (B) GEF assay of SmgGDS-558 and SmgGDS-607. Guanine nucleotide dissociation was measured by monitoring fluorescence from BODYPY-GDP bound farnesylated or nonfarnesylated RhoA. The same assay was performed three times. The relative GEF activity and its SE are shown in bar chart.
Y66A, R70E, S73A, and triple (D67R+R68E+L69A) mutants significantly reduced binding to SmgGDS-558 relative to that found in wild type. Moreover, R68E and L72A mutants showed weaker binding, suggesting that switch II of RhoA is the key region for SmgGDS-558 binding (Fig. 3A). The other single mutations in the N-terminal and C-terminal flanking regions at the interaction interface tended to be tolerated by the broad contact surface areas. The reduced binding found for the S73A mutant in interacting with D245 of SmgGDS-558 suggests that D245 is important for RhoA recognition. In sharp contrast, SmgGDS-607 did not show any changes as a result of RhoA mutation except for the triple mutant. This clearly demonstrates that SmgGDS-607 has a strong interaction with the RhoA HVR, as we previously reported (19). The result of the GEF assay coincided with that of the pulldown assay. GEF activities of the lower affinity mutants are significantly decreased both with and without farnesylation (Fig. 3 and SI Appendix, Fig. S4).
In previous studies, D239K+E242K+E246K mutations in SmgGDS-607 (corresponding residues: D190, E193, and E197 in SmgGDS-558) reduced the GEF activity toward RhoA (7). These residues constitute the negatively charged region, where K187 of RhoA PBR is located nearby, thus the mutation of these residues would reduce the interaction (SI Appendix, Fig. S5). M307A mutant was also reported to reduce the activity (7), but no interaction was observed between M307 and RhoA. M307 of SmgGDS-558 forms a hydrophobic core, influencing its interaction with RhoA. Our structural work can explain almost all previous mutational studies in addition to those in the present study.
SmgGDS-558 Forms a Cryptic Pocket for Accommodating the Prenyl Group of RhoA.
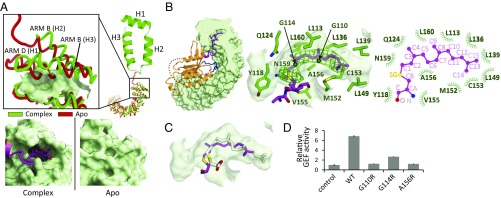
Of particular interest, a farnesyl group at the CaaX motif was inserted into the cryptic pocket, a newly created pocket between the ARM B and D regions in SmgGDS-558. This pocket is not formed in the apo structure of SmgGDS-558; however, upon farnesylated RhoA binding, the second and third helix of ARM B (H2 and H3) and the first helix of ARM D (H1) moved outward to enable SmgGDS-558 to make a new pocket and accommodate the farnesyl group of RhoA (Fig. 4A). Notably, most of the structure, excluding ARM B and ARM D in the apo form, superposed well with the complex structure, with a root-mean-square deviation (rmsd) value of 1.2 Å.
Fig. 4.
Farnesyl group of RhoA C190 inserted into the cryptic pocket of SmgGDS-558. (A, Left) Typical ARM and movement of α-helices of SmgGDS-558, as determined by making a complex with farnesylated RhoA. A typical ARM (H1–3) is shown as a ribbon model. Apo and RhoA-bound form of SmgGDS-558 are shown in red and green, respectively. Enlarged view of the N-terminal ARMs is shown (Inset). The surface of the cryptic pocket is shown. (A, Right) Enlarged view of surface models of SmgGDS-558 apo and complex forms. (B) Cross-section (Left) and enlarged views (Middle) of the insertion of farnesyl group of RhoA C190 into SmgGDS-558. RhoA is shown in orange using the ribbon model. C190 (S-farnesylated) and L191 of RhoA are shown as stick models. Molecular surface of SmgGDS-558 is shown in gray and side chains of SmgGDS-558, which form the pocket, are shown as a green stick model. The electron density map (mFo–DFc) of the side chain of S-farnesylated C190 contoured at 2.5 σ is shown in green mesh. Schematic representation of interactions between C190 and SmgGDS-558 (Right). (C) Docking model of S-geranylgeranylated cysteine to the crystal structure. Docking model of S-geranylgeranylated cysteine is shown as a white stick model. (D) GEF assay of SmgGDS-558 and its mutants interfering with the lipid binding for farnesylated RhoA. Guanine nucleotide dissociation was measured by monitoring fluorescence from BODYPY-GDP bound RhoA. The same assay was performed three times. The relative GEF activity and its SE are shown in bar chart.
The cryptic pocket was mainly formed by the hydrophobic residues (G110, L113, G114, Y118, Q124, L136, L139, L149, M152, C153, V155, A156, N159, and L160) of SmgGDS-558 and was sufficient to accommodate a farnesyl group consisting of 15 carbon atoms (Fig. 4B). RhoA is naturally modified with a geranylgeranyl group, which has 5 additional carbon atoms (20 carbon atoms). We were able to generate a docking model of geranylgeranylated C190 that showed no unfavorable steric clash, suggesting that the pocket was large enough to accommodate a geranylgeranyl group in size and depth (Fig. 4C).
To confirm the functional importance of the cryptic pocket, we performed the GEF assay of the mutant interfering with the lipid binding (Fig. 4D and SI Appendix, Fig. S6). We chose G110, G114, and A156, which form a cryptic pocket (Fig. 4B). As expected, the mutation of these residues resulted in a great reduction of the activity, demonstrating that the prenyl group at the CaaX motif inserted into the pocket is obviously the prerequisite for GEF activity of SmgGDS-558. Although no direct interaction between the GTPase and lipid binding sites, binding of the prenyl group in RhoA to the pocket significantly accelerates GEF activity by bringing GTPase in proximity to the concave surface of SmgGDS.
Homology Modeling and Mutational Analysis of SmgGDS-607.
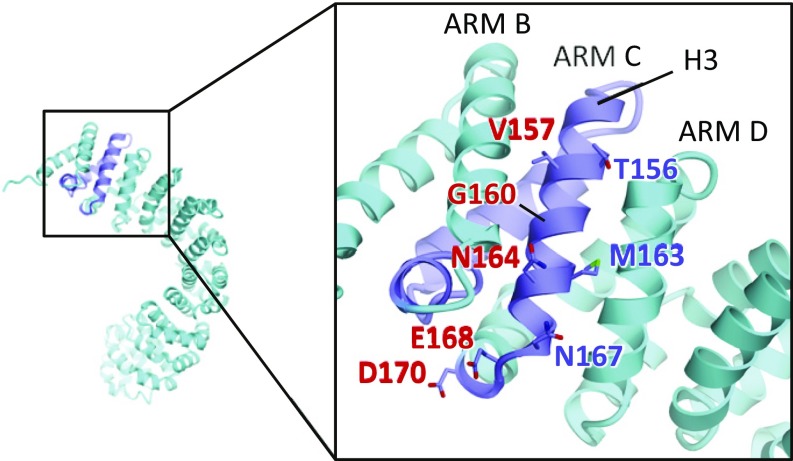
The splicing variant SmgGDS-607 exhibits different biochemical features in RhoA binding and GEF activity. To consider the differences from an atomic viewpoint, we created a homology model of SmgGDS-607 with the structures of SmgGDS-558 and β-catenin as templates (Fig. 5). According to this model, two acidic residues E168 and D170 located at the loop between ARM C and D are likely to widen the negatively charged region. Moreover, H3 of ARM C is wedged between ARM B and D, which form a hydrophobic pocket for accommodating a farnesyl group in SmgGDS-558, preventing the generation of the cryptic pocket in SmgGDS-607. To experimentally determine the binding interface between SmgGDS-607 and the RhoA PBR, we carried out mutational studies. We prepared eight single mutants (T156A, V157A, G160R, M163A, N164A, N167A, E168R, and D170R) and two double mutants (G160R+N164A and E168R+D170R), and then conducted an isothermal titration calorimetry (ITC) experiment to directly determine the binding affinity (SI Appendix, Fig. S7 and Table S2). Compared with the wild-type protein with a dissociation constant (Kd) of 0.2 µM, mutation at G160 and N164 showed a significant decrease in binding, with Kd values of 5.5 µM and 12.8 µM, respectively. Double mutation (G160R+N164R) further decreased the affinity (Kd 18.9 µM). Presence of either the E168R and D170R mutations resulted in a slight decrease in binding affinity (Kd 0.83 µM and 0.48 µM, respectively). These findings suggest that PBR–CaaX binds to the surface of H3 in the newly inserted ARM C, but electrostatic interaction with the PBR slightly contributes to the binding.
Fig. 5.
Homology model of SmgGDS-607. SmgGDS-607 is shown in sky-blue and the newly created ARM C is shown in violet. Side chains of SmgGDS-607 that were mutated in this study are shown as a stick model. Residues, showing reduced binding affinity to the PBR–CaaX peptide by mutation, are colored in red.
Discussion
In this study, we report the SmgGDS-558/farnesylated RhoA complex structure, which reveals drastic conformational changes in RhoA and the formation of a pocket in SmgGDS to accommodate the prenyl group. These findings offer a mechanism for both GEF activity and prenyl group-binding proteins.
Generally, GEFs catch the switch I region which is regarded to be intrinsically structurally flexible, displacing this region away from the nucleotide binding site. Simultaneously, GEFs engage extensive contacts with switch II, which adopts an ordered conformation in the complex. Consequently, GEF stabilizes nucleotide-free small GTPase. In addition to binding to the switch regions, binding of the GEF in many cases involves projection of residues toward steric hindrance of Mg2+ binding as well as electrostatic repulsion between acidic residues of GEFs to expel Mg2+ and GDP. In contrast to the switch regions, the conformational change of the G1 motif (P loop) is usually less dramatic.
In humans, there are 69 typical RhoGEFs in the Dbl family and 11 atypical RhoGEFs known as DOCK family proteins (SI Appendix, Fig. S3A) (23–25). GEF mechanisms for proteins in both families have been well characterized by biochemical and structural studies (23, 24). Dbl family proteins contain a Dbl homology (DH) domain associated with a PH domain and DOCK homology region 2 (DHR2) as a catalytic domain. These GDP/GTP exchange reactions are fundamentally achieved by steric hindrance and electrostatic repulsion for GDP.
One intriguing example is the Rab8/Mss4 complex (26). Mss4 binds to the switch I and interswitch regions, forming a complex with Rab8 (SI Appendix, Fig. S8A). The complex formation disorders the G1 motif and adjacent residues, switch II, and additional loops of the Rab8 nucleotide-binding pocket. This Mss4-induced GTPase unfolding is proposed as a key element in the nucleotide exchange mechanism. SmgGDS also employs the similar strategy for exerting GEF activity in that SmgGDS disorders G1 motif and both switch regions. Our close inspection reveals that the GEF mechanism is distinctly different. While Mss4 binds the switch I region, forming a complex, SmgGDS binds a part of the switch II region. SmgGDS-558 draws the switch II close to its own concave surface with the disruption of the α-helix and makes the whole switch I region disordered. Accordingly, the nucleotide binding region becomes open, facilitating GDP/GTP exchange. Of interest, the G1 motif is disordered (complexes II and III) or shows a large conformational change (complexes I and IV), moving toward the phosphate binding site (Fig. 1 B and C and SI Appendix, Figs. S1B and S8 B and C). This disorder or conformation change would facilitate expelling GDP from RhoA (SI Appendix, Fig. S8 B and C). More importantly, SmgGDS-558 harbors the cryptic pocket accommodating the prenyl group of RhoA, which is required for exerting GEF activity. SmgGDS exhibits a unique GEF mechanism through the cooperation of the lipidation and the GEF activity accompanied by the conformational changes in RhoA that have not been reported to date.
Two structures of prenyl group-accommodating proteins for small GTPases have been reported. One is the Rho guanine nucleotide dissociation inhibitor (RhoGDI), and the other is phosphodiesterase-δ (PDEδ). Both proteins shield the lipid modification added to small GTPases, sequestering the small GTPases in the cytosol to protect them from aggregation and degradation or enhancing their diffusion in the cell (20, 27, 28). SmgGDS-558 also allows the prenyl group of RhoA to bind in the hydrophobic pocket in a similar manner as these two proteins, strongly suggesting that SmgGDS-558 acts as a chaperone. However, SmgGDS-558 shows significant differences in its structure and recognition mechanism. RhoGDI and PDEδ fold into a similar Ig-like β-sandwich comprising two antiparallel β-sheets. The prenyl group plunges into a hydrophobic pocket of the Ig-like fold (20, 27, 28). This pocket is open in the apo form of SmgGDS, which is an entirely α-helical protein composed of ARMs. SmgGDS-558 moves the α-helix of its ARM B and D outward to make cryptic a pocket between ARM B and D and thus accommodating the prenyl group of RhoA. SmgGDS-607 cannot make this pocket, because the space between ARM B and D is occupied by ARM C. It was reported that the knockdown of SmgGDS-558, but not SmgGDS-607, decreases proliferation of tumors in vivo as well as RhoA activity in breast cancer cells (17). The fact that only SmgGDS-558 can recognize the prenyl group of RhoA may suggest that only SmgGDS-558 can act as a chaperone of matured RhoA, and consequently SmgGDS-558 retains its activity (17). A previous study showed that SmgGDS-607 uniquely recognizes the CaaX motif (9). Consistently, our homology modeling and mutational study of SmgGDS-607 indicated that SmgGDS-607 harbors a binding interface distinct from SmgGDS-558 toward PBR–CaaX with nonprenylated cysteine. Wilson et al. (29) reported that phosophorylation of two Ser residues of Rap1B in the PBR reduces the binding to SmgGDS-607, indicating that electrostatic interaction would be responsible for binding between SmGDS-607 and Rap1B. Our model coincides with this, because positively charged PBR–CaaX in RhoA interacts with the negatively charged surface of H3 formed by the inserted ARM C. An ITC experiment showed that nonpolar residues, such as G160 and N164, in ARM C were important for PBR–CaaX recognition, suggesting that nonpolar interaction with CaaX is also important in addition to electrostatic interaction with PBR.
SmgGDS acts as a GEF only for RhoA and RhoC (7), although Rho family proteins possess almost the same amino residues in the switch II region. The N terminus of RhoA (R5 and K6) was found to make a hydrogen bond or salt bridge with SmgGDS-558. However, other Rho family proteins, such as Cdc42 and Rac1, lack these residues, suggesting that SmgGDS might show GEF activity only for RhoA and RhoC. RhoB is not activated by SmgGDS although it has R5 and K6, because RhoB has no PBR (7). SmgGDS can bind to not only Rho family members but also multiple Ras family proteins such as K-Ras4B and Rap1A. These proteins exhibit diverse sequences; the switch II region, in particular, is not well conserved. The PBR–CaaX motif may provide sufficient affinity to SmgGDS.
There are many RhoA-specific GEFs and GDI. Jaiswal et al. (25) reported that catalytic efficiency and substrate selectively of GEFs are significantly different from each other. Among GEFs, SmgGDS shows a unique Rho activation mechanism. Two splice variants of SmgGDS use different mechanisms depending on a lipidation state of GTPases. On the other hand, SmgGDS-558 harbors a lipid binding pocket reminiscent of GDI. While GDI negatively regulates RhoA and SmgGDS can act as a GEF for RhoA, both proteins are similarly involved in the regulation of intracellular transport. SmgGDS is also involved in Rac1 translocation into the nucleus (30). This functional redundancy and diversity would be required for maintaining the physiological cellular processes.
The CaaX motif is considered to be posttranslationally modified in three steps, and these modifications facilitate plasma membrane localization. In the first step, the cysteine residue in the CaaX motif is prenylated by FTase or GGTase in the cytosol. The other two steps are performed at the ER membrane. Three terminal residues of CaaX are removed by the Ras converting enzyme 1 (RCE1) following methylation for the prenylated-cysteine residue by isoprenylcysteine methyl transferase (ICMT) (31, 32). These modifications are essential for small GTPases to recruit them to their correct localization and to exert their function. Further studies are required to clarify the influence of the other two steps on SmgGDS/RhoA interaction. The side chain atoms of L191 in the CaaX motif make no specific interaction with SmgGDS, while main-chain atoms make contact, suggesting the second amino acid in the CaaX motif is not directly recognized by SmgGDS.
Several papers reported that SmgGDS works as a chaperone (for examples, see refs. 8, 9). For example, Berg et al. (8) reported that the prenylation and trafficking of PBR-containing small GTPases (K-Ras, Rap1, RhoA, and Rac1) are regulated by GDP/GTP exchange and by interactions with two splice variants of SmgGDS (SmgGDS-558 and SmgGDS-607). And they demonstrated that SmgGDS-607 specifically interacts with nonprenylated small GTPases and regulates their entry into the prenylation pathway, whereas SmgGDS-558 specifically associates with prenylated small GTPases and regulates trafficking to the plasma membrane. We revealed that SmgGDS-558 has a pocket accommodating the prenyl group, implicating that SmgGDS-558 could protect newly prenylated RhoA from aggregation or degradation. This structural feature suggested that SmgGDS-558 possibly works as a chaperone in the prenylation pathway. Further biochemical and structural studies should be performed to clarify the chaperone mechanism of SmgGDS.
Our findings reveal a cryptic pocket by which SmgGDS-558 accommodates RhoA prenylation, loosening the structure of Rho when it interacts with SmgGDS, which may enable drug development strategies for targeting SmgGDS and small GTPases.
Materials and Methods
All experimental procedures can be found in SI Appendix, including sample preparation, crystallization, data collection, structural determination, pulldown assay, GEF assay, docking simulation, homology modeling, protein-peptide modeling, and ITC.
Supplementary Material
Acknowledgments
This work was supported by a grant-in-aid from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (to S.T.-F., K.K., T.K., and T.S.), and by Core Research for Evolutional Science & Technology (CREST), Japan Science & Technology Agency (JST) (T.S.), the Takeda Science Foundation (T.S.), the Uehara Memorial Foundation (T.S.), and the Naito Foundation (T.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.S.G. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5ZHX).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804740115/-/DCSupplemental.
References
- 1.Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 2.Fernández-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011;2:344–358. doi: 10.1177/1947601911411084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dvorsky R, Ahmadian MR. Always look on the bright site of Rho: Structural implications for a conserved intermolecular interface. EMBO Rep. 2004;5:1130–1136. doi: 10.1038/sj.embor.7400293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casey PJ, Seabra MC. Protein prenyltransferases. J Biol Chem. 1996;271:5289–5292. doi: 10.1074/jbc.271.10.5289. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto T, et al. Purification and characterization from bovine brain cytosol of proteins that regulate the GDP/GTP exchange reaction of smg p21s, ras p21-like GTP-binding proteins. J Biol Chem. 1990;265:16626–16634. [PubMed] [Google Scholar]
- 6.Mizuno T, et al. A stimulatory GDP/GTP exchange protein for smg p21 is active on the post-translationally processed form of c-Ki-ras p21 and rhoA p21. Proc Natl Acad Sci USA. 1991;88:6442–6446. doi: 10.1073/pnas.88.15.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamel B, et al. SmgGDS is a guanine nucleotide exchange factor that specifically activates RhoA and RhoC. J Biol Chem. 2011;286:12141–12148. doi: 10.1074/jbc.M110.191122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berg TJ, et al. Splice variants of SmgGDS control small GTPase prenylation and membrane localization. J Biol Chem. 2010;285:35255–35266. doi: 10.1074/jbc.M110.129916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuld NJ, et al. The chaperone protein SmgGDS interacts with small GTPases entering the prenylation pathway by recognizing the last amino acid in the CAAX motif. J Biol Chem. 2014;289:6862–6876. doi: 10.1074/jbc.M113.527192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams C. A new signaling paradigm to control the prenylation and trafficking of small GTPases. Cell Cycle. 2013;12:2933–2934. doi: 10.4161/cc.26230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams CL. The polybasic region of Ras and Rho family small GTPases: A regulator of protein interactions and membrane association and a site of nuclear localization signal sequences. Cell Signal. 2003;15:1071–1080. doi: 10.1016/s0898-6568(03)00098-6. [DOI] [PubMed] [Google Scholar]
- 12.Ogita Y, et al. Di-Ras2 protein forms a complex with SmgGDS protein in brain cytosol in order to be in a low affinity state for guanine nucleotides. J Biol Chem. 2015;290:20245–20256. doi: 10.1074/jbc.M115.637769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergom C, et al. The tumor-suppressive small GTPase DiRas1 binds the noncanonical guanine nucleotide exchange factor SmgGDS and antagonizes SmgGDS interactions with oncogenic small GTPases. J Biol Chem. 2016;291:6534–6545. doi: 10.1074/jbc.M115.696831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tew GW, et al. SmgGDS regulates cell proliferation, migration, and NF-kappaB transcriptional activity in non-small cell lung carcinoma. J Biol Chem. 2008;283:963–976. doi: 10.1074/jbc.M707526200. [DOI] [PubMed] [Google Scholar]
- 15.Schuld NJ, et al. SmgGDS-558 regulates the cell cycle in pancreatic, non-small cell lung, and breast cancers. Cell Cycle. 2014;13:941–952. doi: 10.4161/cc.27804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhi H, et al. SmgGDS is up-regulated in prostate carcinoma and promotes tumour phenotypes in prostate cancer cells. J Pathol. 2009;217:389–397. doi: 10.1002/path.2456. [DOI] [PubMed] [Google Scholar]
- 17.Hauser AD, et al. The SmgGDS splice variant SmgGDS-558 is a key promoter of tumor growth and RhoA signaling in breast cancer. Mol Cancer Res. 2014;12:130–142. doi: 10.1158/1541-7786.MCR-13-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peifer M, Berg S, Reynolds AB. A repeating amino acid motif shared by proteins with diverse cellular roles. Cell. 1994;76:789–791. doi: 10.1016/0092-8674(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 19.Shimizu H, et al. Structure-based analysis of the guanine nucleotide exchange factor SmgGDS reveals armadillo-repeat motifs and key regions for activity and GTPase binding. J Biol Chem. 2017;292:13441–13448. doi: 10.1074/jbc.M117.792556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhlmann N, et al. Structural and mechanistic insights into the regulation of the fundamental Rho regulator RhoGDIα by lysine acetylation. J Biol Chem. 2016;291:5484–5499. doi: 10.1074/jbc.M115.707091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allal C, et al. RhoA prenylation is required for promotion of cell growth and transformation and cytoskeleton organization but not for induction of serum response element transcription. J Biol Chem. 2000;275:31001–31008. doi: 10.1074/jbc.M005264200. [DOI] [PubMed] [Google Scholar]
- 22.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 23.Rossman KL, Der CJ, Sondek J. GEF means go: Turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 24.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: Critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 25.Jaiswal M, Dvorsky R, Ahmadian MR. Deciphering the molecular and functional basis of Dbl family proteins: A novel systematic approach toward classification of selective activation of the Rho family proteins. J Biol Chem. 2013;288:4486–4500. doi: 10.1074/jbc.M112.429746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Itzen A, Pylypenko O, Goody RS, Alexandrov K, Rak A. Nucleotide exchange via local protein unfolding–Structure of Rab8 in complex with MSS4. EMBO J. 2006;25:1445–1455. doi: 10.1038/sj.emboj.7601044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Mata R, Boulter E, Burridge K. The ‘invisible hand’: Regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol. 2011;12:493–504. doi: 10.1038/nrm3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dharmaiah S, et al. Structural basis of recognition of farnesylated and methylated KRAS4b by PDEδ. Proc Natl Acad Sci USA. 2016;113:E6766–E6775. doi: 10.1073/pnas.1615316113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson JM, Prokop JW, Lorimer E, Ntantie E, Williams CL. Differences in the phosphorylation-dependent regulation of prenylation of Rap1A and Rap1B. J Mol Biol. 2016;428:4929–4945. doi: 10.1016/j.jmb.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanning CC, Daddona JL, Ruiz-Velasco R, Shafer SH, Williams CL. The Rac1 C-terminal polybasic region regulates the nuclear localization and protein degradation of Rac1. J Biol Chem. 2004;279:44197–44210. doi: 10.1074/jbc.M404977200. [DOI] [PubMed] [Google Scholar]
- 31.Wang M, Casey PJ. Protein prenylation: Unique fats make their mark on biology. Nat Rev Mol Cell Biol. 2016;17:110–122. doi: 10.1038/nrm.2015.11. [DOI] [PubMed] [Google Scholar]
- 32.Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: Post-translational modification of RAS. Nat Rev Mol Cell Biol. 2011;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.