Significance
The efficiency of genome editing with CRISPR-Cas9 can vary widely at different targets and in different cells. Some of this variability may be due to the inherent quality of different guide RNAs, but it may also depend on the cellular context of the genomic target DNA. In this report, we demonstrate that targets bound by nucleosomes are cut much less efficiently than targets from which nucleosomes are absent or have been depleted. This information can inform target selection, particularly in cases where cells are quiescent or nucleosome mobility is limited.
Keywords: genome editing, CRISPR, zinc-finger nuclease, chromatin, nucleosomes
Abstract
Genome editing with CRISPR-Cas nucleases has been applied successfully to a wide range of cells and organisms. There is, however, considerable variation in the efficiency of cleavage and outcomes at different genomic targets, even within the same cell type. Some of this variability is likely due to the inherent quality of the interaction between the guide RNA and the target sequence, but some may also reflect the relative accessibility of the target. We investigated the influence of chromatin structure, particularly the presence or absence of nucleosomes, on cleavage by the Streptococcus pyogenes Cas9 protein. At multiple target sequences in two promoters in the yeast genome, we find that Cas9 cleavage is strongly inhibited when the DNA target is within a nucleosome. This inhibition is relieved when nucleosomes are depleted. Remarkably, the same is not true of zinc-finger nucleases (ZFNs), which cleave equally well at nucleosome-occupied and nucleosome-depleted sites. These results have implications for the choice of specific targets for genome editing, both in research and in clinical and other practical applications.
Genome editing describes the ability to introduce DNA sequence alterations at specific targets in complex genomes. Much of this work is based on site-specific cleavage by nucleases that can be targeted to essentially any chromosomal locus (1). Following feasibility studies with homing endonucleases, the first truly targetable editing enzymes were the zinc-finger nucleases (ZFNs). Work with these reagents laid much of the groundwork for subsequent platforms, including transcription activator-like effector nucleases and CRISPR-Cas9.
It is often said that the advent of the CRISPR-Cas9 platform has revolutionized or democratized the practice of genome editing (2–4). Instead of relying on protein–DNA recognition, Cas9 cleavage is based on Watson–Crick base pairing between a guide RNA, supplied by the experimenter, and the target DNA. The simplicity of guide RNA design for new targets; the need for only a single, constant protein; and the modest cost and complexity of this platform have made targeted genome modifications readily accessible to researchers around the world.
Typically, the approach to a new target is to design and test several guide RNAs corresponding to sequences close to the site of the desired genomic alteration. One or more of these will usually induce cleavage and sequence changes at frequencies that are sufficient for the ultimate application.
Several on-line informatics tools are available to help with target selection [e.g., Doench et al. (5)]. They use features of the candidate guide–target sequence and available experimental data to predict the efficiency of on-target cleavage and the potential for undesirable off-target cleavage. The underlying assumption is that the quality of any guide RNA is determined largely by inherent sequence features; however, to date, the predictions do not seem to reflect any known thermodynamic aspects of RNA–DNA duplex stability. In addition, different prediction tools may disagree with each other and with experimental observation (SI Appendix, Fig. S1).
In eukaryotic cells, genomic DNA is assembled into chromatin, and specific DNA sequences may exist in quite different chromatin contexts. Biochemical experiments with purified components have demonstrated that Streptococcus pyogenes Cas9, the most commonly used CRISPR nuclease, is strongly inhibited from binding and cleaving targets that have been assembled into nucleosomes (6–9). Experiments in cells have also made correlations between Cas9 cleavage efficiency and either general chromatin accessibility or transcription of target sequences (8, 10–13).
In this study, we examine Cas9 cleavage ability in real time at specific DNA sequences in two yeast (Saccharomyces cerevisiae) promoters, HO (14) and PHO5 (15), where nucleosome occupancy has been extensively characterized. Rather than relying on mutagenesis at the target sequences, we assess cleavage directly using Southern blots. We report that Cas9 cleavage is quite weak when nucleosomes are present and substantially enhanced when nucleosomes are depleted, either naturally or experimentally. Perhaps surprisingly, cleavage by a specific pair of ZFNs is affected very little by nucleosome occupancy of its target in these same promoters.
Results
Expression of Cas9 and sgRNA.
The protocol for expressing Cas9 protein and single-guide RNA (sgRNA) was critical for monitoring target cleavage in time course experiments. In initial trials, we used an integrated Cas9 gene controlled by a galactose-inducible promoter coupled with constitutive sgRNA expression from a low-copy plasmid. This led to low-level constitutive cleavage, indicating that there was significant unintended expression from the GAL promoter in cells grown in raffinose. For the experiments reported here, we retained the integrated, GAL-regulated Cas9 gene, but coupled it with tetracycline-regulated expression of the sgRNA gene on a low-copy plasmid (16). Cas9 expression was induced about 1.5 h before addition of doxycycline to initiate sgRNA production (detailed protocols are discussed in Materials and Methods). We occasionally saw a small amount of cleavage before addition of doxycycline, indicating that there is a low level of read-through from the sgRNA promoter as well, and we have verified this by RNA analysis (SI Appendix, Fig. S2).
We found the slow onset of Cas9 cleavage after sgRNA induction (discussed below) rather surprising. The sgRNAs are ∼100 nt long, so their synthesis should take seconds. Cas9 protein expression was induced well in advance of sgRNA induction, and we demonstrated that Cas9 protein was not limiting for cleavage. We did this by using longer periods of Cas9 induction, comparing time courses using our standard protocol with one based on a constitutive low level of Cas9, and on much higher levels of expression from a plasmid vector. None of these increased the extent or kinetics of cleavage (SI Appendix, Figs. S3 and S4). It seems unlikely that the search for the appropriate target is responsible, so we imagine that some aspect of Cas9-sgRNA assembly and/or localization may limit the kinetics.
Cas9 Cleavage in the HO Promoter.
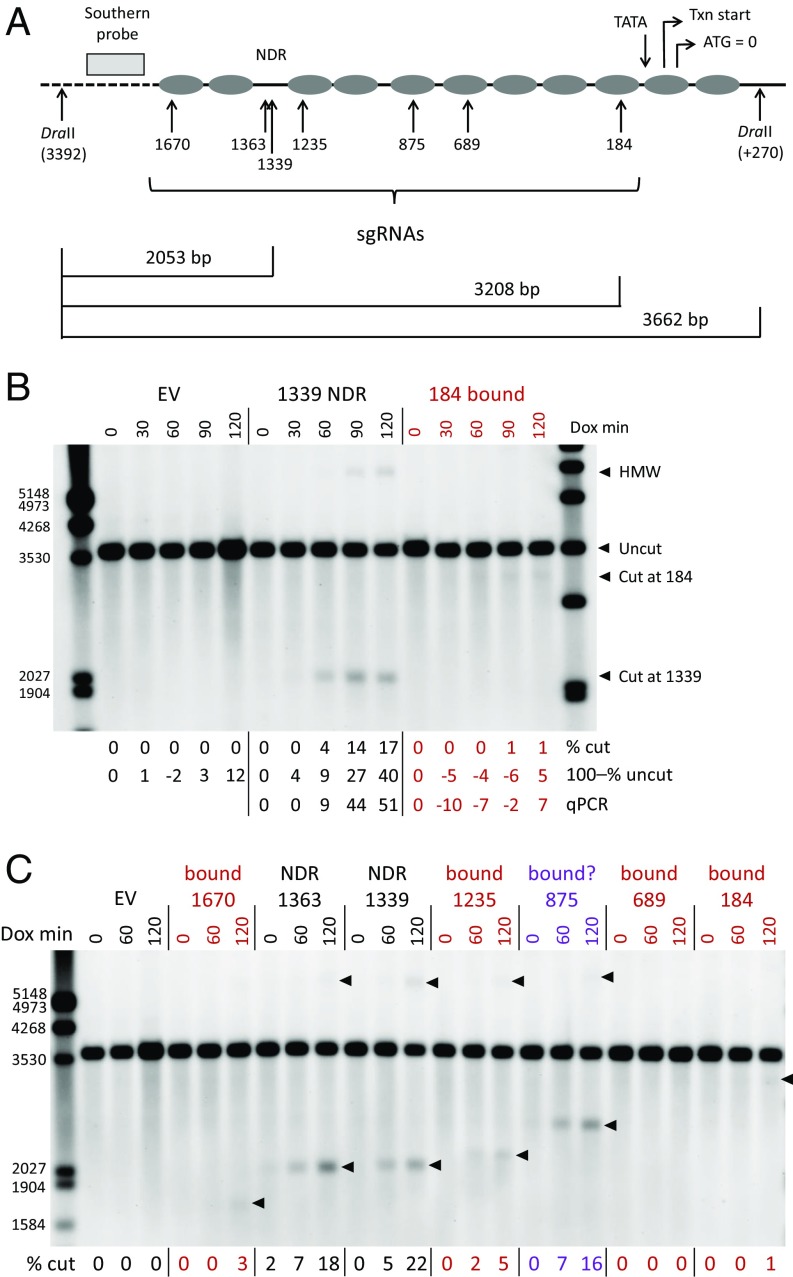
Nucleosome locations at the yeast HO promoter and their dynamics have been very well characterized (14, 17). To explore the effect of chromatin on Cas9 cleavage, we designed sgRNAs to DNA sequences in the constitutive nucleosome-depleted region (NDR) about 1,300 bp upstream of the translation start and to multiple nucleosome-bound sites (Fig. 1A). Cleavage was assessed after various times of sgRNA induction by Southern blot hybridization to DraII-digested genomic DNA. Probe location and some expected fragment sizes are shown in Fig. 1A.
Fig. 1.
Cas9 cleavage at nucleosome-bound (bound) and nucleosome-depleted (NDR) sites in the yeast HO promoter. (A) Diagram of nucleosome locations in the HO promoter and sites targeted by individual sgRNAs. The translation start (ATG) is designated as position 0, and cut site locations for each sgRNA are indicated by their distance upstream (in base pairs). Locations are also noted for the transcription (Txn) start and TATA box. Sites for the DraII restriction enzyme and the probe used for Southern blot analysis are shown; the dashed line indicates distances not shown to scale. The expected sizes for fragments generated by DraII and by DraII plus Cas9 cuts at the 184 and 1339 sites are shown. (B) Southern blot analysis of Cas9 cleavage in the NDR 1339 site and in the nucleosome-bound 184 site. Time courses, in minutes after sgRNA induction by doxycycline addition (Dox min), are shown for cells carrying an empty vector (EV) and each of the specific sgRNAs. Expected fragment sizes are shown at the right. HMW indicates the band in the 1339 lanes that we attribute to single-strand resection after Cas9 cleavage (Cas9 Cleavage in the HO Promoter). The percent cleavage is shown below each lane assessed in three ways. The % cut is the sum of the intensities of cut and HMW fragments as a proportion of all band intensities. The 100–% uncut reflects the loss of the uncut fragment relative to hybridization with a CLN2 probe in the same sample and normalized to the 0-min time point (SI Appendix, Fig. S5). qPCR reflects the loss of amplifiable (i.e., uncut) target DNA normalized to RPR1 in each sample and to the 0-min time point; the EV samples were not quantitated by qPCR. (C) Cleavage time courses for all sgRNAs indicated in A. EV and NDR lanes are labeled in black, lanes for nucleosome-bound sites are labeled in red, and the questionable nucleosome-bound site at 875 is labeled in purple. Locations of bands included in the quantitations for each target are indicated with arrowheads, and the % cut for each sample is shown below. In B and C, some marker fragment sizes are indicated on the left, in base pairs.
An initial time course showed that a target in the NDR (1,339 bp upstream of the translation start) was cut much more efficiently than one in the nucleosome-bound site 184 bp upstream of ATG. At the 1339 target, very little cleavage was seen at 30 min of induction, but cut bands were clearly visible at 60 min and continued to increase up to 120 min, the limit of these experiments (Fig. 1B). Cut bands were barely detectable for the 184 target.
In addition to the expected band for the 1339 target, at about 2,050 bp, a slow-migrating high-molecular-weight (HMW) band also correlated with the degree of cleavage. We interpret this as representing cleavage products that have experienced 5′-3′ nuclease resection on one strand beyond the left DraII site; that site cannot be cut, and larger, partly single-stranded, fragments result. This interpretation was offered by Sugawara and Haber (18) for very similar observations after HO nuclease cleavage. The migration of these slow bands correlates with the locations of various Cas9 cut sites (Figs. 1C and 2C), the location of the next upstream DraII site (4.2 kb upstream of the left one shown in Fig. 1A), and the estimated rate of resection (100 nt⋅min−1) (19). At this rate, HMW products should start appearing 20–30 min after the initial cleavage.
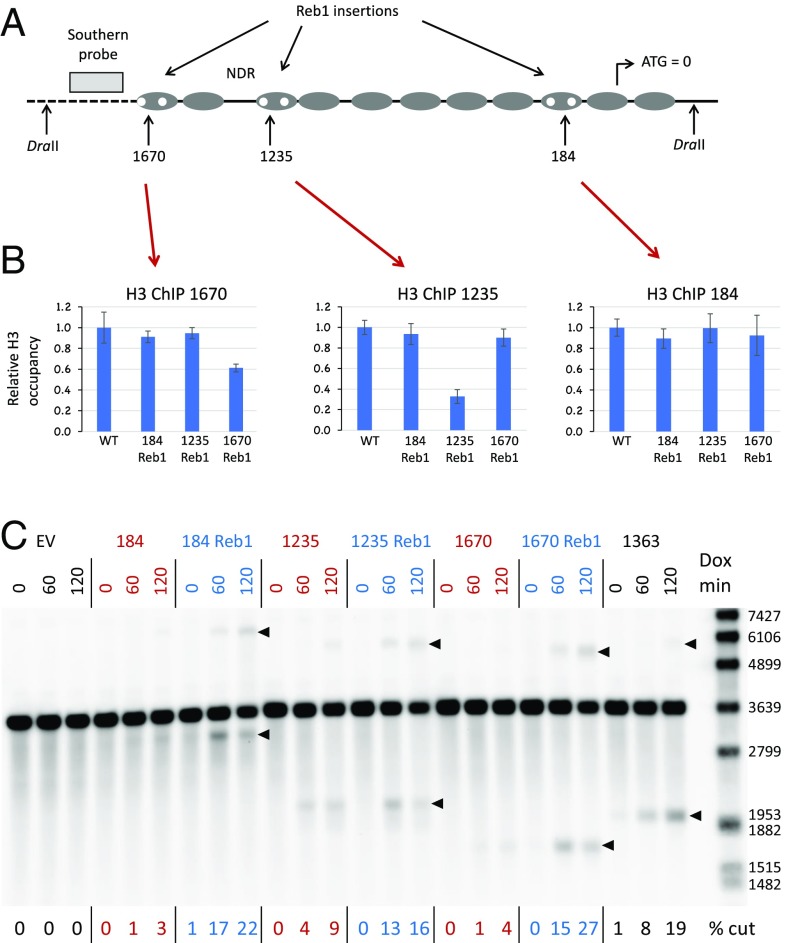
Fig. 2.
Cas9 cleavage after nucleosome depletion by Reb1 site insertions in the HO promoter. (A) Pairs of 8-bp Reb1 sites (TTTACCCG) were inserted in the nucleosome-bound sequences near the 1670, 1235, and 184 targets as replacements for sequences of the same length; thus, spacing in the region was not altered. Importantly, the Reb1 sites did not overlap or disrupt the sgRNA recognition sequences. Each pair of sites was present in independent strains. (B) ChIP experiments on the Reb1 insertion strains. H3 occupancy was determined for each of the Reb1 strains relative to wild type (WT). (Left) ChIP data using primers for sequences flanking the 1670 target in all four strains. (Center) ChIP data for primers flanking the 1235 target. (Right) ChIP data for primers flanking the 184 target. (C) Southern blot showing Cas9 cleavage at the 184, 1235, and 1670 targets in strains without (red) and with (blue) Reb1 insertions. Bands used for quantitation for each target are indicated with arrowheads. Some marker fragment sizes are indicated on the right, in base pairs. Dox min, minutes after sgRNA induction by doxycycline addition; EV, empty vector.
The percentage cut (% cut) shown for each sample includes quantitation of both the slow- and fast-moving bands. These estimates of the extent of cleavage at longer times are very likely underestimates. Measuring cleavage as loss of the uncut band relative to an unaffected site (100–% uncut) was unreliable at low levels of cutting but consistently showed much higher values, approaching 50% at 120 min, when cleavage was extensive (Fig. 1B; more examples are shown in SI Appendix, Fig. S5). The same was true for quantitation by qPCR (Fig. 1B and SI Appendix, Fig. S5). The discrepancy among the methods at high levels of cleavage is likely due to loss of hybridizable cut DNA by continuing resection, degradation by other nuclease activities, cell death, and double-strand break (DSB) repair, none of which we can assess accurately. Hybridization to an unaffected site was not always performed. In most succeeding figures, only the “% cut” measurements are reported, despite underestimating high levels of cleavage.
Repair of Cas9-induced DSBs may occur by homologous recombination with the sister chromatid or by accurate or inaccurate nonhomologous end joining (NHEJ). It does not appear that target mutations accumulate significantly in the short time courses in these experiments, since long-term expression of Cas9 and sgRNA leads to substantial inviability (SI Appendix, Fig. S6). Accurate NHEJ would restore the target sequence and appear as uncut in the Southern blots, thereby reducing the apparent % cut.
Multiple Targets in the HO Promoter.
Cleavage induced by multiple sgRNAs across the HO promoter, including those from Fig. 1B, are shown in Fig. 1C. Two targets in the NDR (1363 and 1339) are effectively cleaved, while four nucleosome-bound sites (1670, 1235, 689, and 184) show weak or no cleavage. All of the sgRNAs induced robust cleavage in biochemical experiments (SI Appendix, Fig. S7). This supports the suggestion that nucleosomes inhibit Cas9 cleavage in vivo.
In Fig. 1C, strong cleavage at the 875 site is an anomaly. Cleavage at the 1235 site is also somewhat elevated compared with other nucleosome-bound targets. We believe this is due to the fact that the nucleosomes at these locations are absent during a significant portion of the cell cycle (14). An indication of this is seen in experiments monitoring histone occupancy across the promoter in unsynchronized cells, where occupancy at the 875 and 1235 sites is clearly lower than at other nucleosome-bound sites (SI Appendix, Fig. S8). This issue is addressed experimentally below.
Nucleosome Depletion at HO.
The foregoing results are based on cleavage mediated by different sgRNAs at the various sites, so it is possible that those sgRNAs have different inherent activities, despite their apparently comparable in vitro efficacies. To address this more directly, we took advantage of the observation that the Reb1 protein (20, 21) opposes nucleosome binding (22) and is classified as a strong nucleosome-displacing factor (23). To evict nucleosomes, we introduced pairs of Reb1 binding sites at several of the nucleosome-bound locations (Fig. 2A). The 8-bp Reb1 insertions replaced sequences of the same length, avoiding the sgRNA targets. We then showed reduced histone occupancy at those sites by chromatin immunoprecipitation (ChIP) with a histone 3 (H3) antibody (Fig. 2B). H3 depletion in the 184 region was not significant, but the signal may reflect overlap with neighboring nucleosomes or nucleosome shifting, as discussed further below.
In all three cases (184, 1235, and 1670 sites), cleavage was enhanced at the Reb1 targets (Fig. 2C and SI Appendix, Fig. S5B), rising to levels comparable to those seen at the NDR targets (e.g., 1363 target in Fig. 2C). The same sgRNAs that cut poorly at the wild-type locus cut better when nucleosomes were displaced by Reb1, ruling out simple differences in sgRNA quality.
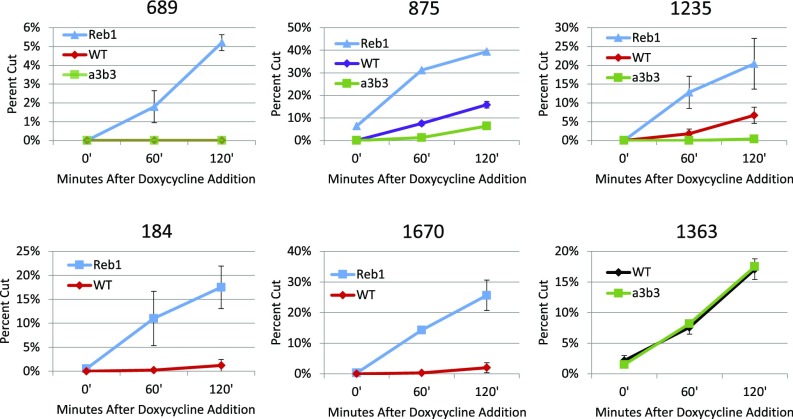
Stimulation of Cas9 cleavage by Reb1 insertions was also observed for the 689 and 875 targets (SI Appendix, Fig. S9). The increased cleavage of the 875 and 1235 Reb1 targets above what was seen in the wild-type context demonstrates that there is some nucleosome inhibition of cleavage in wild type that can be relieved. As suggested above, we hypothesized that the cleavage observed at the 875 and 1235 targets in wild-type cells was due to nucleosome depletion, initiated by the Swi5 transcription factor, which occurs during part of the cell cycle in these unsynchronized cells (14). Therefore, we also examined cleavage of these targets in a strain in which two critical Swi5 binding sites had been mutated. In the mutant (a3b3) (24), Cas9 cleavage at both sites was sharply reduced, while cleavage at the 1363 NDR was unaffected, as expected (SI Appendix, Fig. S10). There was still noticeable cutting at the 875 target, which may be due to an especially effective sgRNA or to incomplete nucleosome occupancy. Graphical comparisons showing the Reb1 and a3b3 effects on various targets are shown in Fig. 3. Overall, we found that high nucleosome occupancy inhibited Cas9 cleavage at all sites tested, while decreased nucleosome occupancy enhanced cleavage.
Fig. 3.
Graphical comparisons of Cas9 cleavage at sites in the HO promoter in various conditions. The sgRNAs for the targets were tested in strains with the WT promoter (red, purple, or black), in strains with the a3b3 mutations in the Swi5 transcription factor binding sites (green), and in strains in which Reb1 site insertions were present at the indicated sites (blue). Note that the y axes are all different, while the trends are consistent. All data reflect the average and SD based on multiple independent experiments. WT, wild type.
It seems likely that inhibition of cleavage is due to inhibition of Cas9 binding, but it is also possible that binding occurs within nucleosomes and cleavage activity, per se, is blocked. To test this, we examined the binding of catalytically inactive Cas9 (dCas9) to two nucleosome-bound sites (184 and 1670) in wild-type and Reb1 insertion strains (SI Appendix, Fig. S11). In both cases, binding to the target was approximately fivefold higher in the Reb1 strain. This also supports the suggestion that the ChIP result for the 184 Reb1 site (Fig. 2B) was influenced by nearby nucleosomes, and not a failure of nucleosome depletion at the target.
Cas9 Cleavage at the PHO5 Promoter.
The yeast PHO5 promoter also has very well-characterized, strongly positioned nucleosomes (15) (Fig. 4A). This is a smaller and less complex promoter than that of HO, and it is subject to rather simple regulation. The Pho5 protein, a secreted acid phosphatase, is required only when available phosphate is limiting. Under conditions of plentiful phosphate, PHO5 transcription is tightly repressed. The Pho85 kinase phosphorylates the Pho4 activator protein, which excludes the latter from the cell nucleus and leads to its degradation. In low-phosphate conditions, Pho85 is inhibited and unphosphorylated Pho4 rapidly enters the nucleus and activates a battery of genes, including PHO5. Pho4 binding at sites in the PHO5 NDR leads to eviction of the first, second, and possibly third nucleosomes upstream of the translation start (15, 25, 26).
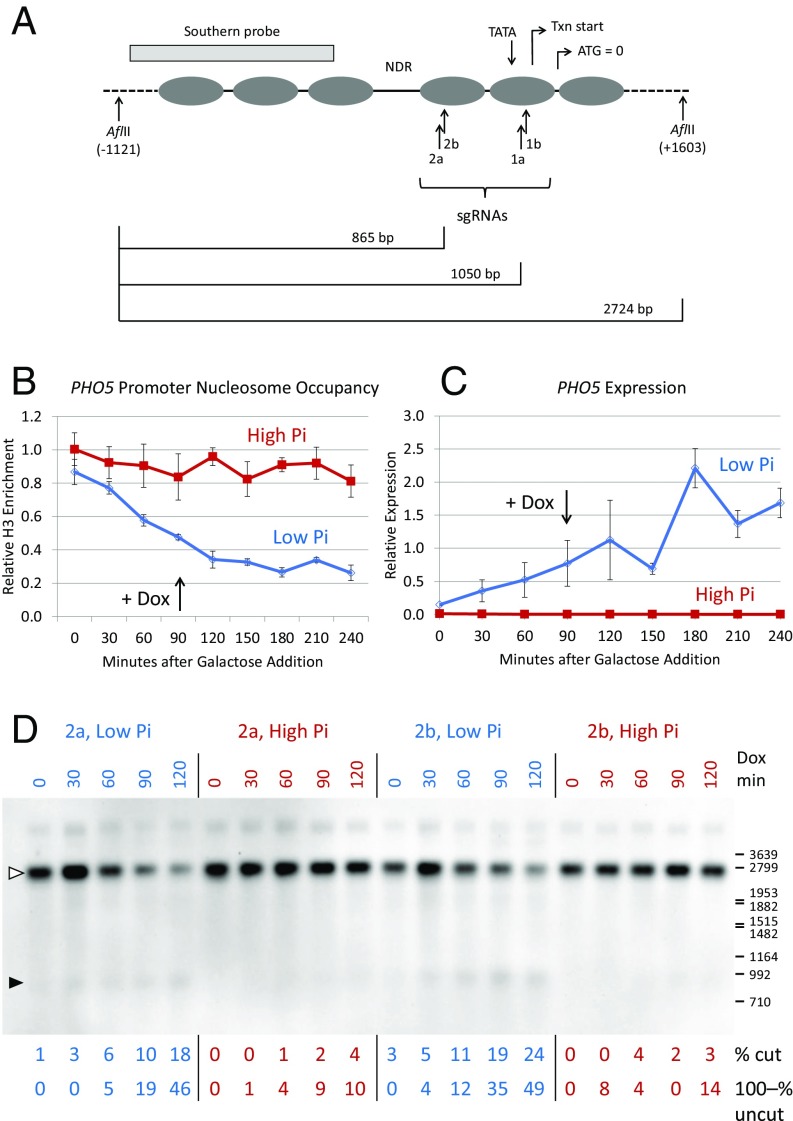
Fig. 4.
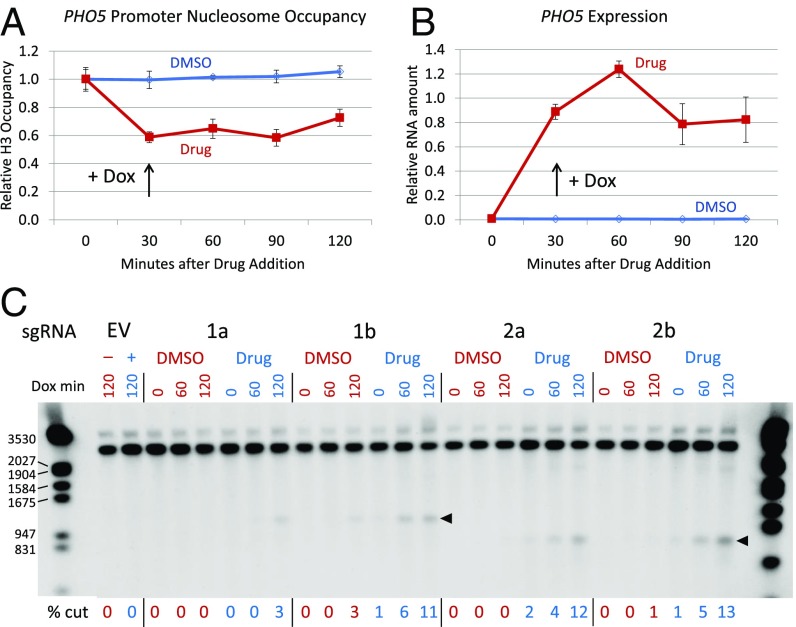
Cas9 cleavage at the PHO5 promoter in low- and high-phosphate conditions. (A) Diagram of nucleosome locations in the PHO5 promoter and sites targeted in the −1 (1a, 1b) and −2 (2a, 2b) nucleosomes. Locations of the translation start (ATG), transcription (Txn) start, and TATA box are indicated. The restriction enzyme sites and probe used for Southern blots are also shown, as are expected fragment sizes for cleavage at the −1 and −2 targets; the dashed lines indicate distances not shown to scale. (B) ChIP data showing depletion of nucleosomes −1 and −2 in low-phosphate medium, but not in high-phosphate medium. The sgRNA was induced with doxycycline 90 min after galactose induction of Cas9 expression. The primers for qPCR analysis of the ChIP samples included parts of both nucleosomes. (C) PHO5 RNA levels measured by quantitative RT-PCR in the same time course as in B. (D) Southern blot showing Cas9 cleavage at sites 2a and 2b in the −2 nucleosome, in low-phosphate (blue) and high-phosphate (red) media. The time course reflects minutes after sgRNA induction by doxycycline addition (Dox min), starting 90 min after galactose addition, as indicated in B and C. Quantitation by both % cut and 100–% uncut is shown for each lane, as in Fig. 1B. The open arrowhead shows the location of the uncut target, and the solid arrowhead shows the expected cleavage products. The locations of some size markers from a nearby lane are shown on the right, in base pairs. Pi, inorganic phosphate.
We designed four sgRNAs to target sequences within the evictable −1 and −2 nucleosomes (Fig. 4A) and tested their ability to direct cleavage in low- and high-phosphate media. To ensure good control of phosphate concentrations, cells from overnight cultures were washed and resuspended in media containing raffinose and 0.04 mM (low) or 6.8 mM (high) buffered potassium phosphate. After growth for several hours, Cas9 expression was induced with galactose. About 1.5 h later, sgRNA expression was induced with doxycycline.
H3 ChIP experiments showed that nucleosomes were depleted from the −1 and −2 sites (Fig. 4B), and quantitative RT-PCR (Fig. 4C) showed that PHO5 RNA levels rose sharply in low phosphate. These effects took several hours to develop, due to the need to deplete vacuolar phosphate stores. The time course of PHO5 expression is consistent with earlier studies (27) and is independent of doxycycline addition.
Southern blot analysis showed much stronger cleavage in low phosphate by the sgRNAs directed to the −2 nucleosome in a time course after sgRNA induction with doxycycline (Fig. 4D). Cleavage was estimated by quantitation of the cut bands and by loss of the uncut band. Very similar results were obtained with the sgRNAs for the −1 nucleosome (SI Appendix, Fig. S13). As at the HO promoter, the same sgRNAs mediate rather ineffective cleavage when their targets are nucleosome-bound but induce much more efficient cleavage when nucleosomes are depleted.
Chemical Inhibition of Pho85.
Nucleosome eviction at the PHO5 promoter can also be achieved by direct inhibition of Pho85 in normal phosphate conditions. Carroll et al. (28) introduced an amino acid substitution in the ATP binding site of this kinase, making it uniquely sensitive to the inhibitor, 1Na-PP1. We generated this pho85F82G mutation in our strains and tested Cas9 cleavage at the −1 and −2 nucleosomes in the absence and presence of the drug, which was added from a DMSO solution 30 min before sgRNA induction.
H3 ChIP (Fig. 5A) and quantitative RT-PCR (Fig. 5B) showed rapid depletion of nucleosomes and induction of PHO5 transcripts following drug addition. Southern blots showed little or no cleavage in samples from DMSO-treated cells, but much more efficient cleavage in the drug-induced samples with all four sgRNAs in a time course after sgRNA induction with doxycycline (Fig. 5C). The 1a sgRNA consistently produced lower levels of cleavage than the others, perhaps due to lower inherent binding efficiency at its target or to its location within the nucleosome.
Fig. 5.
Cas9 cleavage at the PHO5 promoter after Pho85 inhibition. (A) H3 ChIP data in a time course after addition of 1Na-PP1 (Drug) to a culture of the pho85F82G mutant strain. The drug was added 60 min after Cas9 induction with galactose, and doxycycline was added to induce sgRNA expression 30 min after addition of the drug. (B) PHO5 RNA expression in the same time course. (C) Southern blot showing Cas9 cleavage at all four sgRNA targets without (DMSO, red) and with (Drug, blue) 1Na-PP1. The time course reflects minutes after sgRNA induction (Dox min), starting 30 min after drug addition. The cleavage time course extended 30 min longer than those shown in A and B. The locations of cut bands are shown with arrowheads. EV indicates an empty sgRNA vector; in these lanes, a minus symbol indicates no drug (DMSO only) and a plus symbol indicates with drug added. The sizes of some marker fragments are indicated on the left, in base pairs.
ZFN Cleavage at Nucleosome-Bound Sites.
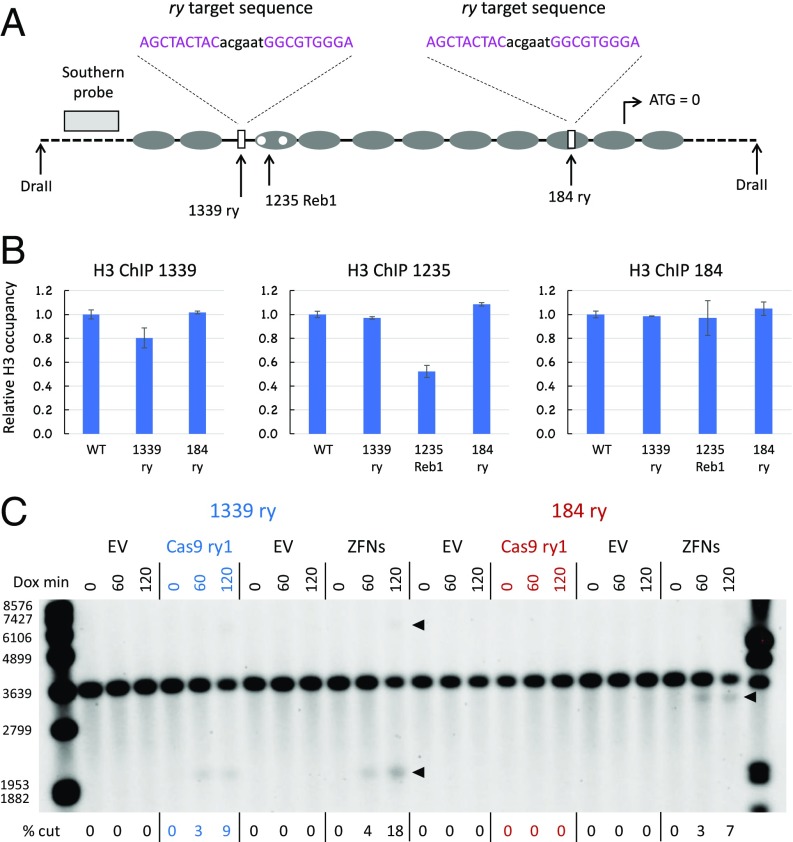
We wondered whether nucleosomes also limit target access by other genome editing nucleases, particularly ZFNs. As it is difficult to design new ZFNs to arbitrary target sites, we instead inserted target sequences for a very specific and efficient pair of three-finger ZFNs identified in previous work targeting the Drosophila rosy (ry) gene (29). The 24-bp recognition sequence (two recognized 9-mers with a 6-bp spacer) was introduced as a replacement for Cas9 targets at two sites in the HO promoter (Fig. 6A) and at site 1a in the PHO5 promoter. In addition, we designed an sgRNA to target this sequence.
Fig. 6.
Lack of nucleosome inhibition of ZFN cleavage at the HO promoter. (A) Diagram showing locations of ZFN target (ry) replacements at the 1339 and 184 sites in the HO promoter. The 1235 Reb1 insertions are also diagrammed. The 9-bp sequences bound by each of the three-finger ZFNs are shown in pink, and the 6-bp spacer is shown in black. (B) ChIP analysis of H3 occupancy relative to wild type (WT) in the ry replacement and 1235 Reb1 strains. (C) Southern blot showing cleavage in the 1339 ry and 184 ry replacement strains by Cas9 with an sgRNA targeting the ry sequence (ry1) and by the ry ZFNs. The locations of cut bands are shown with arrowheads. The sizes of some marker fragments are indicated on the left, in base pairs. Dox min, minutes after sgRNA induction by doxycycline addition; EV, empty vector.
We determined that these sequence substitutions did not disrupt nucleosome occupancy. At HO, H3 ChIP analysis showed no depletion by the substitution at position 184 and very modest depletion at position 1339, which is a natural NDR and normally has little histone occupancy (Fig. 6B). The positive control in the 1235 Reb1 strain showed 50% H3 depletion at that site.
The ry ZFNs produced substantial cleavage at both the nucleosome-bound 184 target and the 1339 NDR (Fig. 6C). In contrast, and like the sgRNAs directed to the natural sequences, the ry-targeted sgRNA (ry1) induced cleavage only at the 1339 NDR, but not at the 184 target, where the nucleosome remained in place (Fig. 6C). The induction of cleavage by the ZFNs was faster than typically seen for Cas9, with good cutting evident by 30 min (SI Appendix, Fig. S12). The levels of cleavage obtained were also higher in this experiment, and cutting at the late time points is underestimated with the % cut approach, as shown with the 100–% uncut assessments. Cleavage at the 184 target was somewhat lower than at the 1339 NDR, but was still strong.
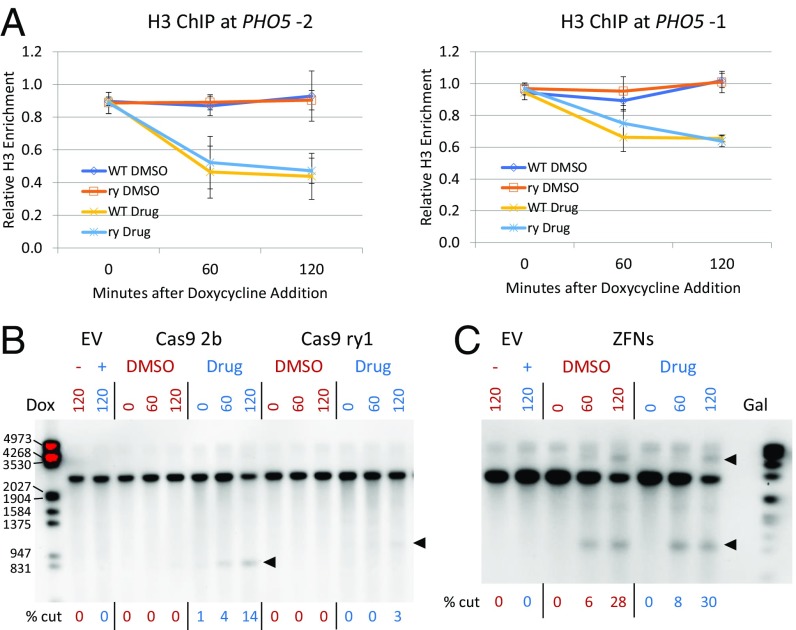
At the PHO5 promoter, a ry replacement was made in the −1 nucleosome in the pho85F82G strain. H3 ChIP analysis showed that the −1 and −2 nucleosomes remained in place and were evicted upon drug treatment (Fig. 7A), similar to strains without the substitution. Cleavage mediated by both the ry ZFNs and the ry sgRNA was tested with and without the Pho85 inhibitor (Fig. 7 B and C). We found that the ZFNs efficiently cut the ry targets both in the presence and absence of drug, apparently undeterred by the chromatin environment, as at HO. The ry sgRNA and the control 2b sgRNA mediated Cas9 cleavage only in the presence of the drug. The ZFNs also cleaved efficiently at the −1 nucleosome in both low- and high-phosphate media (SI Appendix, Fig. S13), again indicating that ZFNs are largely impervious to nucleosomes on their target.
Fig. 7.
Lack of nucleosome inhibition of ZFN cleavage at the PHO5 promoter. (A) ChIP analysis of H3 occupancy relative to wild type (WT) for the ry replacement at the −1 nucleosome in the pho85F82G strain in the absence (DMSO) and presence (Drug) of 1Na-PP1. Data are shown for both the −1 and −2 nucleosomes in the −1 ry replacement strain. (B) Southern blot showing Cas9 cleavage mediated by the 2b and ry1 sgRNAs in the −1 ry substitution pho85F82G strain in the absence and presence of 1Na-PP1. EV is empty sgRNA vector; in these lanes, a minus symbol indicates no drug (DMSO only) and a plus symbol indicates with drug added. Times shown above each lane are minutes after doxycycline (Dox) addition to induce sgRNA expression. Arrowheads show the positions of cut bands. (C) Southern blot showing ZFN cleavage in the −1 ry substitution pho85F82G strain at the −1 nucleosome in the absence and presence of 1Na-PP1. Times shown above each lane are minutes after galactose (Gal) addition to induce expression of the two ZFNs. Cleavage was strong enough at 120 min that both the lower and HMW cut bands were seen. The locations of cut bands are shown with arrowheads. The sizes of some marker fragments are indicated on the left, in base pairs.
Discussion
Previous work has shown that assembly of an sgRNA-targeted DNA substrate into a nucleosome protects it from Cas9 cleavage in vitro (6–9). Experiments in cells have suggested that the same may be true in a nuclear context (8, 10–13). In this study, we confirmed this inhibition by examining a number of individual targets when nucleosome-bound and largely nucleosome-depleted. We measured cleavage efficiency directly using Southern blots, rather than relying on surrogate measures like target mutagenesis via NHEJ. The interference by nucleosomes with Cas9 cleavage that we document is quite striking.
At the HO promoter, naturally nucleosome-depleted targets were efficiently cut by Cas9, while most nucleosome-bound targets were not. Cleavage improved at the latter targets when nucleosomes were depleted by the insertion of binding sites for the Reb1 protein. When average nucleosome occupancy was increased by mutation of binding sites for the Swi5 transcription factor, cleavage was reduced. At the PHO5 promoter, strongly positioned nucleosomes upstream of the inactive gene prevented Cas9 cleavage. Activation of the gene and eviction of those nucleosomes, either by limiting phosphate availability or by inhibiting the activity of the Pho85 kinase, led to efficient cleavage. Overall, every manipulation of occupancy had the effect expected for nucleosome inhibition of Cas9 cleavage.
What impact will our findings have on experimental genome editing? Chromatin structure will be more important in some contexts than others, and there will certainly be differences in the inherent effectiveness of different guide RNAs. In many situations, finding a good Cas9 target and producing an effective guide RNA is simply a matter of testing a modest number of candidates, just as illustrated in SI Appendix, Fig. S1. Gene knockout applications frequently do not demand that mutations be created in a narrow sequence window. In addition, the effect of chromatin structure may be small when the experimental time frame is long enough. Chromatin dynamics could expose most regions of the genome long enough to allow hit-and-run access by the Cas9–sgRNA complex, for example, during DNA replication or active transcription.
Knowledge of nucleosome locations could, however, be quite important for target choice in some situations. In nondividing cells, nucleosome positioning may be more static. Making animal models of specific human genetic diseases requires precise placement of the offending mutation. When introducing sequence changes from a donor DNA in mammalian cells, the induced break must be quite close (within about 100 bp) to the site of the desired change, due to the limited length of conversion tracts. In applications to human somatic therapies, not only will the target cells likely not be dividing but inefficiencies in delivering genome-editing reagents to those cells make it imperative that the efficiency of editing be high in cells that are successfully accessed. It should be noted that mapping nucleosome locations in target cells will not always be easy, and information derived from established cell lines may not be relevant.
The recently developed base-editors constructed from Cas9 (30–32) should be even more affected by the nucleosome inhibition we describe, since editing occurs only within a window of a few bases in the strand displaced by sgRNA binding. Finding the desired target between nucleosomes could be quite challenging. The impact of nucleosomes on transcription factors based on dCas9 has been reported. In a broad study, sgRNAs that target sequences in NDRs close to the transcription start site were more effective in directing transcription repression than ones targeting sites in the upstream nucleosomes (8). In fact, the impact on transcription factors may be more substantial, since their effect requires continuous residence at the promoter, while the nucleases and base-editors can execute their permanent modifications in a narrow time window of access. Some Cas9 variants may be more or less susceptible than the S. pyogenes enzyme to nucleosome inhibition, and it seems likely that there will be effects on more distant relatives like Cas12a and Cas12b.
While the CRISPR platform was substantially inhibited by nucleosomes in our experiments, ZFNs were not. In its natural bacterial setting, Cas9 would not encounter nucleosomes, so there would be no pressure to acquire that capability. Obviously, this is not a significant impediment to general use of Cas9 in eukaryotic cells, perhaps due to the factors outlined above. Zinc fingers, in contrast, are found in large families of natural eukaryotic transcription factors, and they have evolved to function in a chromatin context. Some, like GATA-4 (33), are able to bind to a target located within a nucleosome.
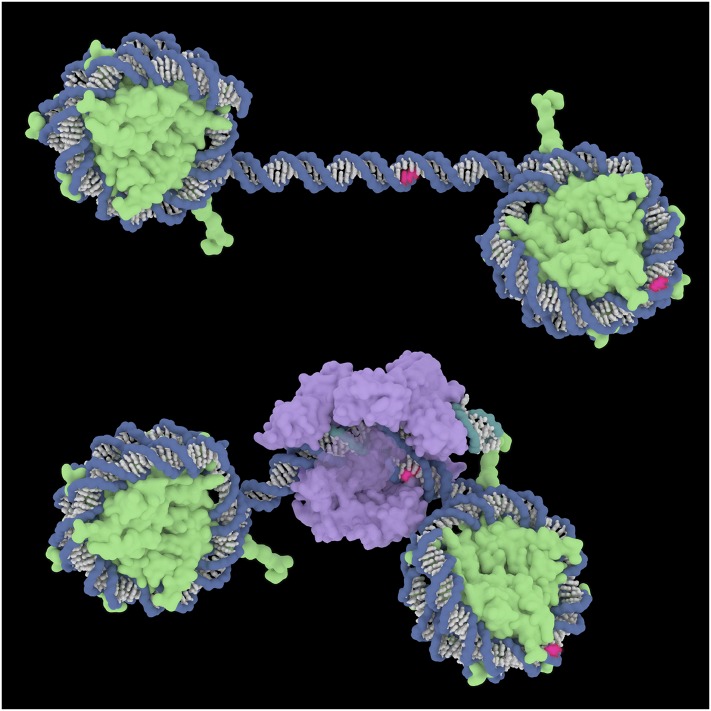
Examination of the structures of a nucleosome and of Cas9-sgRNA on DNA suggests an explanation for the failure of Cas9 to cleave at nucleosome-bound targets (Fig. 8). Cas9 searches DNA initially for protospacer adjacent motif (PAM) sequences, which are recognized by binding within a deep cleft in the protein. While this occurs readily on free DNA, there does not seem to be any way for it to get started on the nucleosome surface. In contrast, zinc fingers recognize DNA sequences by inserting the tip of each finger into the major groove and interacting with base-specific functional groups displayed there. While we do not know how ZFNs cleave nucleosome-bound targets, short stretches of the major groove, about six consecutive base pairs, are exposed on the surface of the nucleosome. This may be enough for a set of fingers to initiate binding and perhaps to begin displacing adjacent base pairs from the histone core.
Fig. 8.
Illustration of Cas9 binding to DNA. (Top) Two nucleosomes surrounding a nucleosome-free stretch of DNA. Hypothetical PAM sites for Cas9 targets in the central region and the right-hand nucleosome are shown in red. (Bottom) Cas9 (with sgRNA) bound to the central target, with the PAM and flanking DNA held deep in the protein cleft. The PAM in the nucleosome could not be accessed without dissociating the DNA from the histone core. DNA backbones are blue; the RNA backbone is teal; DNA and RNA bases are white, except for the PAM; histones are green; and Cas9 is purple. Protein Data Bank ID codes are 1aoi for the nucleosome (49) and 4un3 for Cas9-sgRNA bound to DNA (50). Image courtesy of Janet Iwasa (University of Utah, Salt Lake City).
Knowing that nucleosomes interfere directly with Cas9 cleavage, but not with ZFN cleavage, is useful information. It may explain some cases of unanticipated sgRNA failure, and it should guide specific applications where precisely located, high-efficiency cleavage is necessary.
Materials and Methods
Yeast Strains.
All yeast strains used in this study are listed in SI Appendix, Table S1 and are isogenic in the W303 background (34). Standard genetic methods were used for strain construction (35, 36). A yeast codon-optimized Cas9 gene (yCas9) was generously provided by Giacomo Bastianelli and Alain Nicolas, Meiogenix, Paris. This gene was integrated at the URA3 locus by selection for the linked KanMX marker with G418. The marker was subsequently converted to HphMX (hygromycin resistance). Other gene manipulations in this study were constructed using pCORE-UK and the delitto perfetto method (37), and by transformation with the appropriate PCR products. Reb1 binding sites (TTTACCCG) were inserted by replacement of existing 8-bp sequences at various sites in the HO promoter. Recognition sites for the ry ZFNs (AGCTACTACACGAATGGCGTGGGA) were inserted by replacement of existing 24-bp sequences that included sgRNA targets in the HO (184 and 1339 sites) and PHO5 (1a site) promoters. Plasmids introduced into the various strains are listed in SI Appendix, Table S2.
sgRNAs.
Cas9 target sites (SI Appendix, Table S3) were chosen with guidance from the Broad (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design) and ChopChop (chopchop.cbu.uib.no/) web tools. Guide sequences were inserted into the pRS416gT plasmid vector (16), where expression of the sgRNA is controlled by a tetracycline-inducible promoter. Cloning via Gibson assembly was as described by the Smith et al. (16). Individual guide sequences are shown in SI Appendix, Table S3.
ZFNs.
Coding sequences for the ZFNs (ryA and ryB) designed for the Drosophila ry gene (29) were inserted into yeast YCp plasmids under the control of a GAL1 promoter and with the auxotrophic markers URA3 (ryA) and LEU2 (ryB). Source plasmids were the Gateway entry vectors described previously (38).
Culture Protocols.
For cleavage experiments at the HO promoter, cells were grown overnight in synthetic complete medium (SC) minus Ura (to select for the sgRNA plasmid) with 2% raffinose as a carbon source. Secondary cultures were inoculated at an OD660 of 0.1 in the same medium. When they reached an OD660 of 0.2, galactose was added to a final concentration of 2% to induce Cas9 expression. After about 1.5 h (OD660 = 0.3), the cultures were split and doxycycline hyclate was added to a final concentration of 10 μg/mL at 30- or 60-min intervals to generate time points that had experienced sgRNA induction for 0–120 min. Doxycycline was not added to zero-time samples. Cells were harvested together at the same time and approximately the same cell concentration, pelleted, and flash-frozen in liquid nitrogen.
For experiments involving the PHO5 promoter, cells were grown overnight as above, and then pelleted and washed twice with water. When testing the effects of phosphate concentration, cells were resuspended in SC minus Ura without phosphate, and buffered potassium phosphate was then added to a final concentration of 0.04 mM (low) or 6.8 mM (high). Growth and induction of Cas9 and sgRNA were as above. For experiments with the pho85F82G mutant, cells were washed and resuspended in SC minus Ura with 2% raffinose and induced as above. The Pho85 inhibitor, 1Na-PP1 (39), was added from a 10 mM stock in DMSO to a final concentration of 10 μM, 30 min before the addition of doxycycline.
ZFN-carrying strains were grown overnight in SC minus Leu and Ura with 2% raffinose. Secondary cultures were inoculated at an OD660 of 0.1 in the same medium. When they reached an OD660 of 0.3, galactose was added to a final concentration of 2% to induce ZFN expression.
Southern Blots.
Genomic DNA was prepared from individual samples by established methods (40) with slight modifications. Restriction enzymes were purchased from New England Biolabs, Inc., and used according to the manufacturer’s instructions. Samples were subjected to electrophoresis in 1% or 1.2% agarose gels. Denaturation, transfer to nylon membranes, digoxygenin-labeled probe preparation, and hybridization were performed as described in the DIG Application Manual for Filter Hybridization published by Roche Diagnostics. Templates for probe labeling were prepared using the PCR DIG Probe Synthesis Kit (Roche Diagnostics). PCR products for probe preparation were amplified from genomic DNA or from a plasmid carrying the entire HO promoter, using primers shown in SI Appendix, Table S4.
Southern Blot Quantitation.
Chemiluminescence from each Southern blot was collected on a Bio-Rad ChemiDoc MP Imaging System. The data were analyzed with both Image Lab (Bio-Rad) and Fiji (https://fiji.sc/) software. All quantitation was performed on unsaturated images. Contrast has been enhanced in many of the presented images to make faint bands more visible.
Bands representing uncut and cut targets in each lane were quantitated, and the amount of cleavage was calculated as the proportion of cut over total. In many cases, particularly for HO promoter samples, “cut” bands included both the expected faster moving band and the slower moving HMW band behind “uncut” that we attribute to extensive resection of one strand after cleavage and loss of the proximal restriction site (18). Because of this interpretation, the intensity of the HMW bands was doubled in the calculation reflecting the loss of half the homology to the double-stranded probe. The values obtained with Image Lab and Fiji were generally in good agreement. The reported values are from Fiji, where the method used for correction for background intensity was more transparent.
We also quantitated cleavage by measuring loss of the uncut band relative to hybridization to a nontarget band in each sample (100–% uncut), but this proved quite unreliable at low levels of cleavage (Figs. 1B and 4 and SI Appendix, Figs. S5 and S12). The same was true for analysis of loss of uncut target by qPCR (41), using primers specific for each target site (SI Appendix, Table S4). The signal at each time point was normalized to RPR1 as an internal reference and to the zero-time control (Fig. 1B and SI Appendix, Fig. S5). At high levels of cleavage at longer times, we believe these methods may more accurately reflect actual cutting, because the signal from cut bands is lost due to continuing degradation.
ChIP.
Immunoprecipitations were performed as described earlier (42, 43) using anti-H3 antibodies (07-690; Upstate) and antibody-coated magnetic beads (rabbit IgG beads; Life Technologies). Samples were cross-linked in 1% formaldehyde overnight on ice. ChIP assays were analyzed by real-time qPCR as described (41), with primers listed in SI Appendix, Table S4. Experimental samples were first normalized to the ChIP signal at the IGR-I gene-free reference region on chromosome I (44) and then normalized to their respective input DNA samples. HO ChIP data are graphed relative to wild type, and PHO5 ChIPs are graphed relative to zero-time control conditions (normal phosphate levels or DMSO). Error bars in the H3 ChIP assays reflect the SD of at least four measurements from two to four biological replicates.
dCas9 Immunoprecipitation.
The M5771 plasmid expressing dCas9 (catalytically dead D10A H840A Cas9) fused to protein A was generously provided by Alan Tackett, University of Arkansas for Medical Sciences, Little Rock, AR. Overnight cultures were grown in SC minus Trp and Ura (to select for the dCas9 and sgRNA expression plasmids) with 2% glucose as the carbon source. Secondary cultures were inoculated at an OD660 of 0.1 in the same medium, and doxycycline was added 2 h later. Cells were harvested at an OD660 of ∼0.7–0.8. Cross-linking and immunoprecipitations were performed as above using IgG beads (IgG Sepharose 6 Fast Flow; GE Healthcare). No antibody was added as binding was dependent on interaction between the Fc region of IgG and protein A. ChIP assays were analyzed as above, with primers listed in SI Appendix, Table S4. Experimental samples were first normalized to the ChIP signal at the IGR-V gene-free reference region on chromosome V (45) and then normalized to their respective input DNA samples.
RNA Analysis.
To assess the induction of transcription at PHO5, RNA was isolated from logarithmically growing cells at the indicated times, and PHO5 RNA levels were measured by quantitative RT-PCR as described (43). RNA levels were normalized to RPR1 levels and graphed relative to time after addition of galactose or 1Na-PP1. Error bars reflect the SD of at least four measurements from two biological samples. In the case of sgRNA induction, primers specific for the 5′ and 3′ ends of the HO 1363 sgRNA were used. Quantitative RT-PCR primers are listed in SI Appendix, Table S4.
Western Blots.
Cultures were grown, induced, and harvested as described for cleavage experiments. Proteins were extracted from 2.5 OD660 of cells in mild alkali (46) and fractionated by SDS/PAGE electrophoresis. Western blots were probed with mouse anti-Cas9 mAb (1:500; Epigentek) and secondary goat anti-mouse IgG-HRP conjugate (Millipore). Blots were developed with ECL Western blotting substrate (Pierce). Antibody to yeast PGK1 (Abcam) was used as a loading control.
In Vitro Assays.
Samples of purified Cas9 protein were generously provided by Chris Richardson (Innovative Genomics Institute, University of California, Berkeley) and Daniel Reich (Department of Biochemistry, University of Utah School of Medicine). The sgRNAs were prepared by in vitro transcription of synthetic templates as described by DeWitt et al. (47, 48). Individual guide sequences are shown in SI Appendix, Table S3. Digestion reactions were performed essentially as described at the New England BioLabs website (https://www.neb.com/protocols/2018/01/30/in-vitro-digestion-of-dna-with-engen-cas9-nls-s-pyogenes-m0646), but with the following concentrations: 3 nM DNA substrate, 60 nM Cas9 protein, and 90 nM sgRNA. Reaction products were analyzed on a 1% agarose gel.
Supplementary Material
Acknowledgments
We thank David Stillman, in particular, and Tim Formosa, as well as members of their laboratories, for continuing advice during the course of this project and for comments on the manuscript. Emily Parnell showed the efficacy of nucleosome removal by inserting Reb1 sites at the 1235 nucleosome. We thank Giacomo Bastianelli and Alain Nicolas for the yeast codon-optimized Cas9 gene, Justin Smith for the pRS416gT plasmid, Alan Tackett for the M5771 plasmid, Kevan Shokat for 1Na-PP1, and Chris Richardson and Dan Reich for Cas9 protein. We thank Janet Iwasa for the image in Fig. 8. Kelly Beumer contributed to early work on this project, and Zachary Harmer participated in some of the experiments reported. The research was supported by NIH Grant R01 GM078571 and, in part, by a University of Utah Cancer Center support grant.
Footnotes
Conflict of interest statement: D.C. receives license royalties from Sangamo Therapeutics, Inc., a company that employs zinc-finger nucleases for genome editing.
See Profile on page 9331.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810062115/-/DCSupplemental.
References
- 1.Carroll D. Genome engineering with targetable nucleases. Annu Rev Biochem. 2014;83:409–439. doi: 10.1146/annurev-biochem-060713-035418. [DOI] [PubMed] [Google Scholar]
- 2.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 3.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komor AC, Badran AH, Liu DR. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinz JM, Laughery MF, Wyrick JJ. Nucleosomes inhibit Cas9 endonuclease activity in vitro. Biochemistry. 2015;54:7063–7066. doi: 10.1021/acs.biochem.5b01108. [DOI] [PubMed] [Google Scholar]
- 7.Hinz JM, Laughery MF, Wyrick JJ. Nucleosomes selectively inhibit Cas9 off-target activity at a site located at the nucleosome edge. J Biol Chem. 2016;291:24851–24856. doi: 10.1074/jbc.C116.758706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horlbeck MA, et al. Nucleosomes impede Cas9 access to DNA in vivo and in vitro. eLife. 2016;5:12677. doi: 10.7554/eLife.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Isaac RS, et al. Nucleosome breathing and remodeling constrain CRISPR-Cas9 function. eLife. 2016;5:13450. doi: 10.7554/eLife.13450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X, Liu J, Janssen JM, Gonçalves MAFV. The chromatin structure differentially impacts high-specificity CRISPR-Cas9 nuclease strategies. Mol Ther Nucleic Acids. 2017;8:558–563. doi: 10.1016/j.omtn.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, et al. Probing the impact of chromatin conformation on genome editing tools. Nucleic Acids Res. 2016;44:6482–6492. doi: 10.1093/nar/gkw524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daer RM, Cutts JP, Brafman DA, Haynes KA. The impact of chromatin dynamics on Cas9-mediated genome editing in human cells. ACS Synth Biol. 2017;6:428–438. doi: 10.1021/acssynbio.5b00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen KT, et al. Chromatin accessibility and guide sequence secondary structure affect CRISPR-Cas9 gene editing efficiency. FEBS Lett. 2017;591:1892–1901. doi: 10.1002/1873-3468.12707. [DOI] [PubMed] [Google Scholar]
- 14.Stillman DJ. Dancing the cell cycle two-step: Regulation of yeast G1-cell-cycle genes by chromatin structure. Trends Biochem Sci. 2013;38:467–475. doi: 10.1016/j.tibs.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korber P, Barbaric S. The yeast PHO5 promoter: From single locus to systems biology of a paradigm for gene regulation through chromatin. Nucleic Acids Res. 2014;42:10888–10902. doi: 10.1093/nar/gku784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith JD, et al. Quantitative CRISPR interference screens in yeast identify chemical-genetic interactions and new rules for guide RNA design. Genome Biol. 2016;17:45. doi: 10.1186/s13059-016-0900-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yarrington RM, Goodrum JM, Stillman DJ. Nucleosomes are essential for proper regulation of a multigated promoter in Saccharomyces cerevisiae. Genetics. 2016;202:551–563. doi: 10.1534/genetics.115.183715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugawara N, Haber JE. Characterization of double-strand break-induced recombination: Homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White CI, Haber JE. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartley PD, Madhani HD. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009;137:445–458. doi: 10.1016/j.cell.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubik S, et al. Nucleosome stability distinguishes two different promoter types at all protein-coding genes in yeast. Mol Cell. 2015;60:422–434. doi: 10.1016/j.molcel.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Bai L, Ondracka A, Cross FR. Multiple sequence-specific factors generate the nucleosome-depleted region on CLN2 promoter. Mol Cell. 2011;42:465–476. doi: 10.1016/j.molcel.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan C, Chen H, Bai L. Systematic study of nucleosome-displacing factors in budding yeast. Mol Cell. 2018;71:294–305. doi: 10.1016/j.molcel.2018.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McBride HJ, Brazas RM, Yu Y, Nasmyth K, Stillman DJ. Long-range interactions at the HO promoter. Mol Cell Biol. 1997;17:2669–2678. doi: 10.1128/mcb.17.5.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almer A, Hörz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986;5:2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Almer A, Rudolph H, Hinnen A, Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbaric S, Walker J, Schmid A, Svejstrup JQ, Hörz W. Increasing the rate of chromatin remodeling and gene activation–A novel role for the histone acetyltransferase Gcn5. EMBO J. 2001;20:4944–4951. doi: 10.1093/emboj/20.17.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carroll AS, Bishop AC, DeRisi JL, Shokat KM, O’Shea EK. Chemical inhibition of the Pho85 cyclin-dependent kinase reveals a role in the environmental stress response. Proc Natl Acad Sci USA. 2001;98:12578–12583. doi: 10.1073/pnas.211195798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D. Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics. 2006;172:2391–2403. doi: 10.1534/genetics.105.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaudelli NM, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishida K, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353:aaf8729. doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 33.Cirillo LA, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 34.Thomas BJ, Rothstein R. The genetic control of direct-repeat recombination in Saccharomyces: The effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics. 1989;123:725–738. doi: 10.1093/genetics/123.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rothstein R. Targeting, disruption, replacement, and allele rescue: Integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 36.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 37.Storici F, Lewis LK, Resnick MA. In vivo site-directed mutagenesis using oligonucleotides. Nat Biotechnol. 2001;19:773–776. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
- 38.Beumer KJ, et al. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105:19821–19826. doi: 10.1073/pnas.0810475105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bishop AC, et al. Generation of monospecific nanomolar tyrosine kinase inhibitors via a chemical genetic approach. J Am Chem Soc. 1999;121:627–631. [Google Scholar]
- 40.Rose MD, Winston F, Hieter P. Methods in Yeast Genetics, a Laboratory Course Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1990. [Google Scholar]
- 41.Eriksson P, Biswas D, Yu Y, Stewart JM, Stillman DJ. TATA-binding protein mutants that are lethal in the absence of the Nhp6 high-mobility-group protein. Mol Cell Biol. 2004;24:6419–6429. doi: 10.1128/MCB.24.14.6419-6429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhoite LT, Yu Y, Stillman DJ. The Swi5 activator recruits the mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 2001;15:2457–2469. doi: 10.1101/gad.921601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voth WP, et al. Forkhead proteins control the outcome of transcription factor binding by antiactivation. EMBO J. 2007;26:4324–4334. doi: 10.1038/sj.emboj.7601859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 45.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 47.DeWitt MA, Corn JE, Carroll D. Genome editing via delivery of Cas9 ribonucleoprotein. Methods. 2017;121–122:9–15. doi: 10.1016/j.ymeth.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeWitt MA, et al. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci Transl Med. 2016;8:360ra134. doi: 10.1126/scitranslmed.aaf9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 50.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.