Significance
The conserved, membrane-proximal external region (MPER) of the HIV-1 envelope glycoprotein (Env) is a potential vaccine target. To visualize its structure in the context of a lipid-bilayer membrane, we have reconstituted a polypeptide containing the HIV-1 MPER and the contiguous transmembrane domain into a bilayer-like environment and determined its atomic structure by NMR. The MPER folds into a trimeric cluster, well exposed on the bilayer surface, even in the absence of the structural constraints from the rest of the Env ectodomain. Our analyses suggest that this structure probably represents a prefusion conformation of the MPER. The findings imply that presenting a well-defined structure will be important for MPER-based immunogen design.
Keywords: HIV-1 Env, membrane proximal region, NMR structure, transmembrane region
Abstract
The membrane-proximal external region (MPER) of the HIV-1 envelope glycoprotein (Env) bears epitopes of broadly neutralizing antibodies (bnAbs) from infected individuals; it is thus a potential vaccine target. We report an NMR structure of the MPER and its adjacent transmembrane domain in bicelles that mimic a lipid-bilayer membrane. The MPER lies largely outside the lipid bilayer. It folds into a threefold cluster, stabilized mainly by conserved hydrophobic residues and potentially by interaction with phospholipid headgroups. Antigenic analysis and comparison with published images from electron cryotomography of HIV-1 Env on the virion surface suggest that the structure may represent a prefusion conformation of the MPER, distinct from the fusion-intermediate state targeted by several well-studied bnAbs. Very slow bnAb binding indicates that infrequent fluctuations of the MPER structure give these antibodies occasional access to alternative conformations of MPER epitopes. Mutations in the MPER not only impede membrane fusion but also influence presentation of bnAb epitopes in other regions. These results suggest strategies for developing MPER-based vaccine candidates.
HIV-1 Env [trimeric (gp160)3, cleaved to (gp120/gp41)3], the sole antigen on the virion surface, induces strong antibody responses in infected individuals (1, 2). Env directs fusion of viral and host-cell membranes to initiate infection of a susceptible cell (3). Conformational changes accompany binding of the native Env trimer to receptor (CD4) and coreceptor (e.g., CCR5 or CXCR4), leading to a cascade of refolding events in gp41 (SI Appendix, Fig. S1A). The N-terminal fusion peptide of gp41 inserts into the target cell membrane, forming an extended conformation known as a “prehairpin intermediate” (4). Subsequent folding back of the C-terminal region of gp41 into a hairpin conformation creates the postfusion, six-helix bundle (5, 6), bringing together viral and cellular membranes to induce fusion and viral entry.
A ∼24-residue hydrophobic region (residues 660–683), immediately preceding the transmembrane domain (TMD) and known as the membrane-proximal external region (MPER), is one of the most conserved regions in gp41 (SI Appendix, Fig. S1B). The MPER, required for infectivity (7–9), is the epitope for several well-characterized broadly neutralizing antibodies (bnAbs) (10–13). MPER structures have been studied extensively by both NMR and X-ray crystallography under a wide range of conditions. It tends to adopt an α-helical conformation with or without a break in the middle. For example, a monomeric MPER peptide examined by NMR in detergent micelles folded into a kinked helix with many hydrophobic residues embedded in the micelles (14, 15). This result has led to a widely held belief that the MPER should be buried in viral membrane. Conversely, in the postfusion conformation of soluble gp41 trimer, the MPER forms a continuous helix (16–18). It is disordered in a cryo-EM structure of a detergent-solubilized clade B JR-FL EnvΔCT construct containing both the MPER and TMD (19), perhaps because of effects of the detergent micelle.
The MPER is the target of several anti-gp41 bnAbs, including 2F5, 4E10, and 10E8 (10–12). When liganded by bnAbs, the C-terminal half of the MPER retains its helical conformation, but its N-terminal half can adopt extended, nonhelical structures (13, 20–23), consistent with its conformational plasticity during membrane fusion. The MPER-directed bnAbs often contain a long heavy-chain third complementarity-determining region (HCDR3), with a hydrophobic surface, essential for neutralizing activity, that does not make direct contacts with gp41 but interacts instead with the membrane (24, 25). From the structure of an MPER peptide embedded in a detergent micelle, it has been proposed that these antibodies extract their epitopes out of the viral membrane (14, 15, 26). Other evidence suggests instead that these antibodies block HIV-1 infection by recognizing the prehairpin intermediate conformation of gp41 with the help of their lipid binding activity (22–24, 27, 28). Recent structural studies have shown that phospholipids may indeed be an integral component of the epitopes, at least, for 4E10 and 10E8 (22, 23). In addition to membrane binding, many MPER-targeted bnAbs also show polyreactivity to environmental antigens and/or autoreactivity to self antigens (29–31), raising concerns about how to induce them safely by vaccination.
To define the structure of the HIV-1 MPER under more physiologically relevant conditions than used for previous studies and to guide immunogen design, we have determined by NMR its structure when linked to the Env TMD in the context of a lipid bilayer.
Results
Structure Determination.
We previously determined the structure of the TMD reconstituted in bicelles using a fragment of gp41 (residues 677–716) derived from a clade D HIV-1 isolate 92UG024.2 [designated gp41HIV1D(677–716)] (32). The TMD forms a tightly assembled trimer stabilized by an N-terminal coiled-coil and a C-terminal hydrophilic core. (We discuss a subsequent challenge to this model at the end of this section.) To define the MPER–TMD structure in the context of a lipid bilayer, we used bicelle reconstitution as developed for the TMD and extended the protein sequence to include the MPER. We identified a gp41 fragment, residues 660–710 (SI Appendix, Fig. S1B), that had excellent solution NMR properties in dimyristoylphophatidylcholine/dihexanoylphosphatiylcholine (DMPC/DHPC) bicelles with 0.5 ≤ q ≤ 0.6 (SI Appendix, Fig. S1C). This construct, designated gp41HIV1D(660–710) or MPER-TMD, contains the entire MPER (residues 660–683) and TMD (residues 684–705) (33, 34). We purified MPER-TMD and reconstituted it into bicelles with procedures similar to those used for gp41HIV1D(677–716). The mobility of the reconstituted MPER-TMD in SDS/PAGE was close to that expected for a trimer (theoretical molecular weight 18.5 kDa) (SI Appendix, Fig. S1D), consistent with our earlier observation that the trimer of gp41HIV1D(677–716) resists SDS denaturation (32). We confirmed trimerization of MPER-TMD by cross-linking and urea-PAGE analysis (SI Appendix, Fig. S1E).
NMR spectra and carbon secondary chemical shifts of the core region of the TMD (residues 685–702) in MPER-TMD are almost identical to those of the same segment in gp41HIV1D(677–716) (SI Appendix, Fig. S2 A–E), indicating that addition of the MPER did not alter the TMD conformation. This result justified the use during the MPER-TMD structure calculation of NMR-derived structural restraints for residues 685–702, assigned previously for the TMD. These restraints were valuable, because the same region of the MPER-TMD did not generate sufficient NOE data, owing to fast signal relaxation resulting from the larger size of the construct. The regions of MPER-TMD outside the TM core nevertheless showed excellent spectroscopic properties, because of their greater local dynamics, allowing a comprehensive analysis of NOE data. We completed the structure of the MPER-TMD trimer by a strategy similar to the one we had used previously for the TMD alone (32, 35). We first determined the local MPER structures and then measured interprotomer NOEs between structurally equivalent but isotopically differently labeled protomers (SI Appendix, Fig. S3A). A control analysis of homotrimeric (15N, 2H)-labeled MPER-TMD, prepared from the same labeling batch used for mixing, ensured that NOEs were not due to low levels of 1H in the samples (SI Appendix, Fig. S3 B and C). We also confirmed assignment of intermolecular NOE cross-peaks by selectively detecting NOEs between 1H(15N) of one strand and 1H(13C) of a neighboring strand (SI Appendix, Fig. S3D). Finally, we showed that those NOE cross-peaks we could detect from the TM core of the MPER-TMD corresponded to strong peaks from the published TM NOE analysis (SI Appendix, Fig. S4A). The final ensemble of structures converged to rmsd of 1.19 Å and 1.71 Å for backbone and all heavy atoms, respectively (SI Appendix, Fig. S4 B and C and Table S1).
In addition to NOE-based structure determination, we performed paramagnetic relaxation enhancement (PRE) analysis of a mixed sample, in which half of the MPER-TMDs were spin-labeled at either the N terminus (L660) or the C terminus (Q710) and the other half 15N-labeled. If the MPER-TMD forms oligomers, the NMR resonances of the relevant region will show strong PRE. With the N-terminal spin label, our measurements showed strong PRE in the N-terminal helix (residues 660–672), with an average signal loss of ∼50% (SI Appendix, Fig. S5A), as expected from 1:1 random mixing of spin-labeled and unlabeled protomers. In contrast, the TMD, which is far from the spin label, was unaffected. Furthermore, L663 and A667, the two residues closest to the N terminus of the neighboring protomer in our NMR structure, showed the strongest PRE (SI Appendix, Fig. S5A), offering further validation of the calculated model. We obtained the converse result—strong PRE in the TM region and none in the MPER—with the C-terminal spin label (SI Appendix, Fig. S5B). In that case, L704 showed the strongest PRE. C-terminal spin labeling of the TMD alone (a construct that extended to residue 716) resulted in strong PRE for residues near the C terminus of the 15N-labeled chain (SI Appendix, Fig. S5C), including L704. These data show that the TMD C termini lie relatively close together in both constructs.
An NMR study of a bicelle-embedded, TMD construct identical to ours (36) has suggested that this peptide is a monomer tilted with respect to the membrane normal, rather than the trimer we described (32). We have shown here by cross-linking that our MPER-TMD construct is trimeric and by PRE experiments with spin label at either end that there are close trimer interactions at both termini of the fragment. Moreover, the mixed NOE experiment definitively identifies intersubunit NOEs in the MPER-TMD spectra. We have also now carried out the C-terminal spin-label PRE experiment on the TMD fragment, showing that it too is an oligomer. We describe in SI Appendix possible sources of the discrepant interpretations, but we believe that the data presented here confirm our previously published interpretation of the TMD structure.
MPER-TMD Structure and Its Transmembrane Partition.
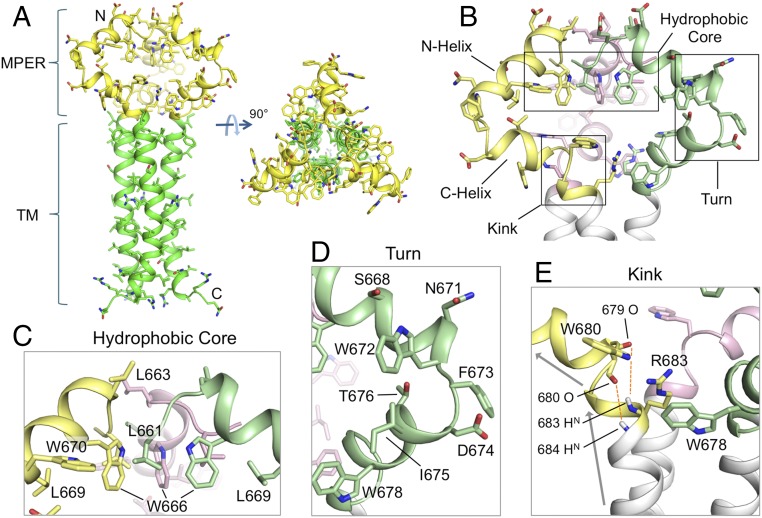
The MPER in this trimeric assembly is well-ordered (Fig. 1A), showing substantial interchain contacts not expected from any of the previously reported structures (14–16). The MPER folds into two α-helices connected by a sharp turn (Fig. 1B). The C-terminal helix (C-helix) connects into the coiled-coil region of the TMD through a kink at K/R683 (Fig. 1C). The broken main-chain hydrogen bond(s) at this kink (O679–HN683, and possibly O680–HN684) expose donors and acceptors that could be stabilized by bound water or lipid headgroups. Moreover, the guanidinium group of R683, although not precisely defined by NMR data, could form a hydrogen bond with the backbone oxygen of either L679 or W680 (Fig. 1C). The N-terminal helix (N-helix) connects to the C-helix by a ∼90° turn at conserved residues W672 and F673 (Fig. 1D). The N-helices from the three protomers converge around the threefold axis and create a hydrophobic core sequestering L661 and W666 (Fig. 1E); L669 and W670 appear to extend the hydrophobic core at the periphery. An L669S substitution leads to prolonged exposure of the MPER neutralizing epitopes (37), suggesting that the hydrophobic core is important for maintaining the antigenic structure of prefusion Env. Overall, some of the most conserved residues in this region determine the MPER trimer conformation. The structure can thus explain the strict conservation of these residues, rather than just retention of the hydrophobicity that would be needed for the membrane insertion suggested by others (14, 15).
Fig. 1.
Structure of the MPER-TMD of HIV-1 Env. (A) Ribbon representation of the average structure of the calculated ensemble. The MPER (residues 660–683) and the TMD (residues 684–710) are shown in yellow and green, respectively. (B) A close-up view of the MPER trimer showing the three protomers in different colors, as well as the characteristic features including the N- and C-helices, the hydrophobic core, the turn connecting the N- and C-helices, and the kink between the C-helix and TMD. (C) The “hydrophobic core” consisting of N-helix hydrophobic residues (W666, W670, L661, and L669). (D) The “turn” region containing residues 671–676. (E) The “kink” at residues 680–683 resulting in a ∼45° change in helix orientation (indicated by the arrows). The O(i)−HN(i + 4) distances of 679–683 and 680–684, indicated by red dashed lines, are >5 Å, much greater than a standard hydrogen bond distance (∼2.5 Å).
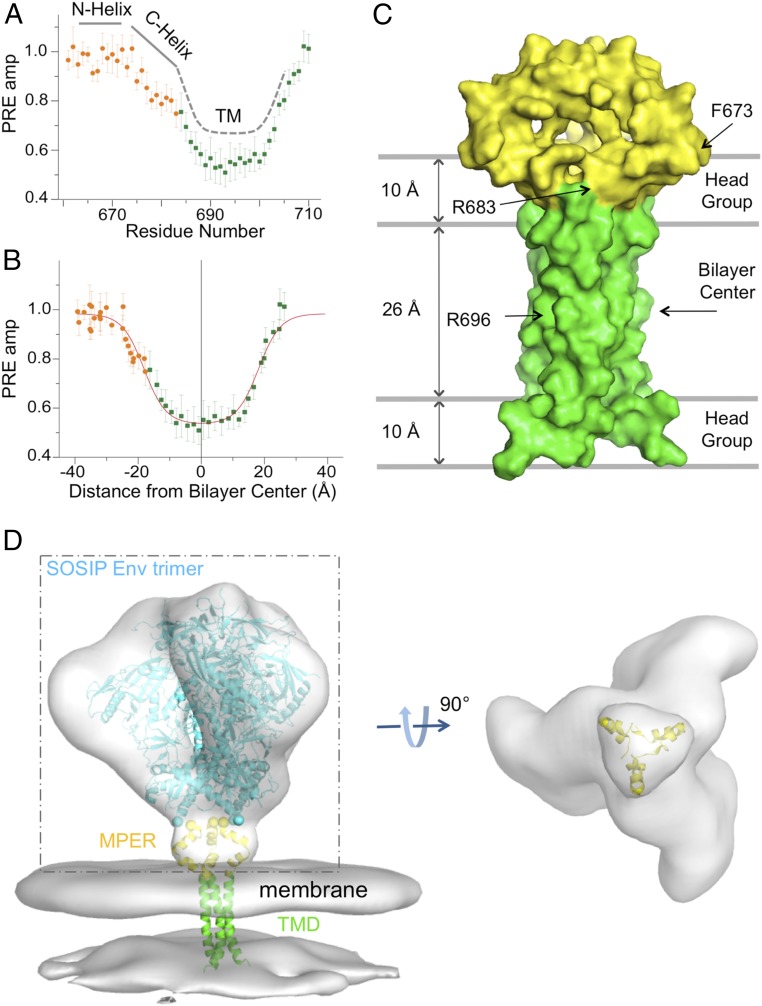
To position the MPER with respect to the lipid bilayer, we analyzed the transmembrane partition of the MPER-TMD trimer using our previously developed paramagnetic probe titration (PPT) method (38). We reconstituted the MPER-TMD protein in bicelles with q = 0.6 and titrated the water-soluble paramagnetic probe Gd-DOTA into the aqueous phase surrounding the bicelles, allowing us to measure residue-specific PRE amplitudes (PREamp) (Fig. 2A and SI Appendix, Fig. S6). A plot of PREamp vs. (residue number) changes slope by ∼30° at R683, between the C-helix of MPER and the N-terminal portion of the TM helix (Fig. 2A); this change provides independent evidence for the kink in the calculated NMR structure (Fig. 1C). The PREamp is flat at value ∼1.0 for the N-helix of MPER, indicating that this helical segment is completely solvent-exposed. To position the MPER-TMD relative to the bilayer, we calculated, for each residue, the distance along the protein symmetry axis (), assumed normal to the bilayer, from the amide proton to an arbitrary reference point. This calculation converted PREamp vs. (residue number) to PREamp vs. , which we then analyzed by sigmoidal fitting (Materials and Methods) to position the structure relative to the bilayer center (Fig. 2B). The results indicate that part of the C-helix lies in the lipid headgroup region, while the rest of the MPER lies outside the membrane (Fig. 2C). In particular, none of the conserved hydrophobic residues in the MPER is submerged in the hydrophobic part of the lipid bilayer.
Fig. 2.
Transmembrane partition of the MPER-TMD in bicelles. (A) Solvent PRE amplitude is plotted against residue number for the MPER-TMD with the MPER shown in orange and the TMD in green. (B) PRE amplitude is plotted against distance from the bilayer center along the trimer axis, fitted to the sigmoidal function. The fit is shown in red. (C) Position of the MPER-TMD trimer in surface representation relative to the lipid bilayer and the bilayer center with the MPER in yellow and the TMD in green. (D) Fit of the MPER-TMD into EM density of the HIV-1 Env trimer on the surface of virion. (Left) Density of the Env trimer [Electron Microscopy Data Bank (EMDB) ID: EMD-5019] and viral membrane (EMDB ID: EMD-5020), derived from cryo-ET (39), is shown in gray. The backbone trace of a natively glycosylated HIV-1 BG505 SOSIP.664 Env trimer [Protein Data Bank (PDB) ID code 5T3Z] (43), fitted to the cryo-ET density, is in light blue, the MPER in yellow, and the TMD in green. (Right) A view from below of the cryo-ET density within the dashed box, with the membrane density and the SOSIP backbone omitted for clarity.
We docked the MPER-TMD structure into low-resolution density, derived from cryo-electron tomography (cryo-ET), of unliganded, prefusion Env trimer on the surface of HIV-1 virions (39). When placed according to the PRE-derived TMD position in the viral membrane (Fig. 2D), the MPER matches a triangle-shaped density that bridges the gap between the docked model of an SOSIP.664 trimer (40–43), which terminates just at the N-terminal end of the MPER, and the viral membrane. Thus, the MPER conformation we see by NMR is consistent with the structure of prefusion Env on the virion. The published gp140 SOSIP structures extend, as a C-terminal helix, to residue 664 and hence overlap very slightly the sequence included in our MPER construct. A cryo-EM structure that includes the MPER and TM (5FUU) (19) becomes disordered at residue 656, however, suggesting that residues 656–660 may depart from the helical structure seen in the truncated constructs. We therefore cannot make any conclusions about how the major part of the ectodomain would connect to the MPER in the structure we have determined.
Antigenic Properties of the MPER-TMD.
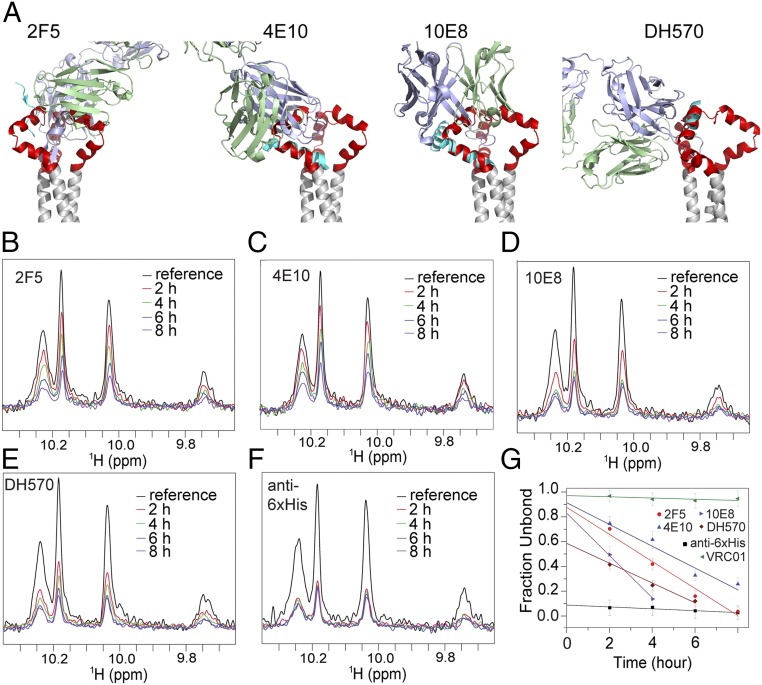
Many known MPER-specific bnAbs, such as 2F5, 4E10, and 10E8, do not bind the prefusion Env trimer; they neutralize primarily by targeting the receptor-bound conformation (24, 27, 28). The 2F5 epitope has an α-helical conformation in the MPER-TMD trimer (Fig. 3A) incompatible with 2F5 binding (20). Likewise, when the structures of 4E10 and 10E8 in complex with their epitope peptides are superposed on the MPER-TMD structure, both antibodies overlap with other parts of the trimer (Fig. 3A). A weakly neutralizing antibody, DH570 (44), elicited by vaccination of rhesus macaques, could bind the MPER-TMD without major hindrance, although optimal binding might require slight rotation of the epitope helix (Fig. 3A).
Fig. 3.
Antibody accessibility of the MPER-TMD in bicelles. (A) The MPER-TMD structure, superposed on crystal structures of the MPER epitope peptides in the complex with their corresponding antibodies, 2F5 [PDB ID code 1TJH (20)], 4E10 [PDB ID code 2FX7 (71)], 10E8 [PDB ID code 4G6F (12)], and DH570 [PDB ID code 5DD0 (44)]. Antibody heavy and light chains are in blue and green, respectively; the epitope peptides are in cyan, the MPER trimer in red, and the TMD in gray. (B) Binding of the MPER-TMD to 2F5 Fab was monitored by loss of NMR signal (due to rapid signal relaxation upon Fab binding). The 1D 1H-15N TROSY-HSQC spectrum of the tryptophan indole amines was recorded for the MPER-TMD in bicelles (q = 0.55) at various time points, shown in different colors, after addition of 2F5. The reference spectrum in black was recorded without 2F5. The MPER-TMD:antibody molar ratio was 1.0:0.7. (C–E) Same as in B performed for the 4E10, 10E8, and DH570 Fabs, respectively. (F) Same as in B performed for the anti-6xHis Fab (prepared from antibody MA1-21315) using the MPER-TMD with an N-terminal 6xHis tag. (G) Fraction of Fab not bound to the MPER-TMD at various time points, calculated as (I − 0.3)/(I0 − 0.3), where I0 is the reference peak intensity normalized to 1, I is the fraction peak intensity at a particular time relative to I0, and subtraction of 0.3 corrected for the 30% molar excess of MPER-TMD in the mixture. The y axis intercepts indicate the fraction of the MPER-TMD in a conformation that is incompatible with antibody binding. The essentially flat line for VRC01 shows little or no binding to MPER, as expected.
To test the structural modeling experimentally, we determined antibody binding of the bicelle-reconstituted MPER-TMD in solution by monitoring loss of its NMR signals when bound to an antibody. Addition of three antigen-binding fragments (Fabs), a total of ∼150 kDa, should make the MPER-TMD essentially NMR-invisible, due to the increased molecular mass of the antibody complex. When we added the Fab of 2F5, 4E10, or 10E8 to the MPER-TMD at a molar ratio of ∼1.0:0.7, the NMR peak intensity diminished very slowly and steadily over a course of 4–10 h (Fig. 3 B–E). In contrast, addition of an anti–His-tag Fab (prepared from MA1-21315) to the MPER-TMD bearing an N-terminal 6His-tag led to instant loss of NMR signals (Fig. 3F). No obvious changes were detected after addition of an anti-gp120 antibody VRC01 (45), as expected (Fig. 3G). These results indicate that the MPER conformation is incompatible with binding of the three MPER bnAbs, as predicted, but that a slow conformational change allows the Fabs to associate. The peak intensities at each time point in Fig. 3 B–F represent the amount of MPER-TMD in the NMR samples not bound to the Fabs. The plots of (fraction unbound) vs. time are linear (Fig. 3G); extrapolation to time 0 determines the fraction of MPER-TMD that cannot readily bind the Fabs. For the 2F5, 4E10, and 10E8 Fabs, although their time dependences show different slopes, the plots extrapolate to ∼90% at time 0, indicating that less than 10% of the MPER-TMD conformation in the NMR sample exposed the relevant epitopes for Fab binding. As expected, over 90% of the His-tagged MPER-TMD is in a conformation compatible with binding to the anti–His-tag Fab. DH570 is an intermediate case; slightly over 40% of the MPER-TMD binds this antibody at time 0. We conclude that the MPER-TMD in bicelles transiently samples conformations that allow antibody binding; the MPER appears to be accessible up to ∼10% of the time to the 2F5, 4E10, and 10E8 Fabs but ∼40% of time to the DH570 Fab. Constraints from the ectodomain would further restrict these fluctuations in the context of an intact Env trimer.
Roles of the MPER in Membrane Fusion.
To assess possible functional roles for the MPER in membrane fusion, we generated 17 Env mutants using the sequence of a clade A isolate, 92UG037.8, guided by the new structure. We mutated each of the three structural elements: hydrophobic core, turn, and kink. When introduced into pseudoviruses, all mutants gave wild-type-like Env incorporation and processing, except the one bearing W666A in the hydrophobic core, which gave increased Env incorporation (SI Appendix, Fig. S7A). Infectivity of all mutant pseudoviruses, except the one bearing L663A near the hydrophobic core, decreased substantially (SI Appendix, Figs. S7 B–D and S8F). In particular, W666 in the hydrophobic core and K683 at the kink appeared to be the two most critical residues, as all of the mutants with changes at these positions lost infectivity almost completely. For the rest of the mutants, infectivity ranged from 4.3 to 50.8% of that of the wild type. Likewise, when transiently transfected in 293T cells in the absence of Gag protein, these MPER mutants expressed comparable levels of Env, with similar extents of cleavage between gp120 and gp41, as well as similar cell-surface levels (SI Appendix, Fig. S8 A–C). At a low Env expression level that mimics the low surface density on HIV-1 virions (46), most MPER mutants induced cell–cell fusion at a significantly lower level than did the wild-type Env (SI Appendix, Fig. S8 D and F), largely in agreement with the observations on pseudoviruses. Interactions between Env and Gag may contribute to the small discrepancies between viral infectivity and cell–cell fusion. A high Env expression level appeared to compensate for defects in membrane fusion of all less-active mutants, including those containing mutations at W666 and K683 (SI Appendix, Fig. S8E). Overall, these data indicate that the key residues important for stabilizing the MPER structure determined by NMR are also critical for Env-induced membrane fusion activity, especially in the context of viral infection.
Effect of the MPER on Antibody Sensitivity of the Env Ectodomain.
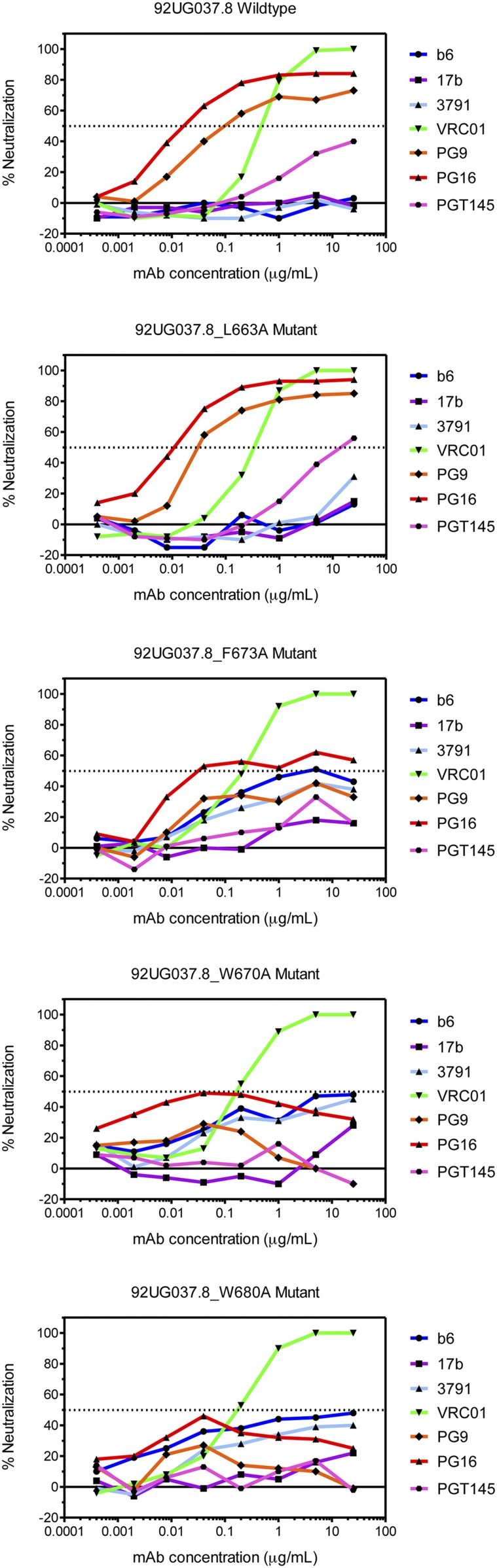
We reported previously that the TMD and the cytoplasmic tail (CT) both influence the antigenic structure of the Env ectodomain (32, 47). The MPER—the only direct link connecting the TMD and CT to the ectodomain—could in principle also modulate the Env antigenic properties, and indeed increased antibody-neutralization sensitivity has been attributed to single residue changes in the MPER of certain naturally selected escape viruses (48). To determine whether or not mutations in the MPER affect epitope accessibility in other regions of the Env ectodomain, we tested several of the most active MPER mutants in a pseudovirus-based neutralization assay using bnAbs PG9, PG16, and PGT145 (trimer-specific) (49, 50) and VRC01 (CD4 binding site) (45), as well as nonneutralizing or strain-specific neutralizing antibodies, including b6 (CD4 binding site) (45), 3791 (V3) (51), and 17b (CD4-induced) (52). Pseudoviruses bearing the wild-type Env were sensitive to VRC01, PG9, PG16, and PGT145 but resistant to b6, 3791, and 17b, as expected (Fig. 4). The mutant L663A, which is near the hydrophobic core but not critical for stabilizing the observed MPER conformation, had a wild-type level of viral infectivity and an antibody sensitivity profile almost identical to that of wild-type Env. In contrast, the mutants W670A (hydrophobic core), F673A (turn), and W680A (kink), while still sensitive to VRC01, became much more resistant to the trimer-specific bnAbs and also gained sensitivity to b6, 3791, and 17b. Thus, the MPER mutations can destabilize the Env ectodomain and shift it toward an open conformation (39, 53, 54). These findings support the notion that the MPER is a control relay that modulates open and closed states of the Env trimer and exposure of other epitopes.
Fig. 4.
Effect of mutations in the MPER on Env antibody sensitivity. Antibody neutralization of pseudoviruses containing either the wild-type 92UG037.8 Env or one of the MPER mutants shown was determined for the ordinarily nonneutralizing antibodies, b6 (CD4 binding site; blue), 3791 (V3; cyan), and 17b (CD4-induced; purple) and for the trimer-specific bnAbs, PG9 (orange), PG16 (red), and PGT145 (magenta). The CD4 binding site bnAb VRC01 (green) was a control. The experiment was performed in duplicate.
Discussion
We have shown that the HIV-1 gp41 MPER when connected to its TMD forms a trimeric cluster that is largely solvent-exposed on the membrane surface. The width and depth of the MPER trimer match membrane-proximal density from a low-resolution, cryo-ET–derived structure of the prefusion Env trimer on virions (Fig. 2D). The TMD in this construct has an NMR spectrum identical to that of the TMD alone (32). The antigenic properties of the bicelle-associated MPER-TMD are also consistent with previous studies showing that most of the known MPER-directed bnAbs recognize the prehairpin conformation of this region, rather than its prefusion structure. From the effects on ectodomain antigenicity of mutations that destabilize the observed MPER structure, from its resemblance to the cryo-ET density, and from the likelihood that the MPER postfusion conformation will involve interaction with residues close to the fusion peptide, we propose that the conformation described here corresponds to the MPER-TMD in prefusion Env. Structures of membrane-anchored gp160 with better resolution in this region than those currently available will be needed to validate this proposal.
Prevalence of anti-MPER neutralizing antibodies in HIV-1–infected patients varies among different cohorts (12, 55–58). When present, MPER-directed neutralizing antibodies appear to correlate with greater breadth and potency of anti-HIV-1 activity (59). Early attempts to elicit 2F5- or 4E10-like antibodies primarily by MPER peptides were unsuccessful (60). Immune tolerance mechanisms have been proposed to restrict induction of these antibodies (29–31, 44, 61–63). Appreciation that phospholipids may form part of the bnAb epitopes and isolation of 10E8-like antibodies with great breadth and potency but low polyreactivity and autoreactivity have prompted renewed enthusiasm for using the MPER as a vaccine target (64, 65). Our MPER structure raises the possibility that an appropriately configured immunogen might elicit bnAbs that react with an epitope exposed on the membrane surface without accompanying polyreactivity. Indeed, a weakly neutralizing antibody (DH570) induced by vaccination in nonhuman primates shows detectable binding to the MPER in the conformation described here (Fig. 3). These results suggest that the MPER-TMD in a lipid bilayer might be a more effective immunogen than the membrane-linked peptides used in published studies. Any neutralizing antibody targeting the MPER would probably have considerable breadth, because of the high degree of sequence conservation. Whether this structure can serve as a useful vaccine template for inducing a new class of bnAbs will require further investigation.
Potent bnAbs recognize epitopes in the CD4 binding site, the V1V2-glycan region, the V3-glycan area, and the gp120–gp41 interface (66, 67). Optimal presentation of these epitopes depends on the organization of the Env trimer. We do not yet know which conformation(s) of each epitope should be presented at different stages of bnAb development, such as B cell activation and affinity maturation. Our data indicate that the MPER is critical for trimer stability and antigenicity, suggesting that we can use it as a structural modulator to alter exposure of other bnAb epitopes. The atomic details of the MPER trimer structure should enable rational protein engineering to manipulate presentation of the antigenic surfaces of Env to induce effective antibody responses.
Materials and Methods
For complete methods see SI Appendix.
Sample Preparation.
A fragment (MPER-TMD) spanning residues 660–710 of HIV-1 gp41 (clade D, isolate 92UG024.2) was expressed in Escherichia coli strain BL21 (DE3) cells as a trpLE fusion and isolated by Ni-NTA affinity purification, cyanogen bromide cleavage, and reverse-phase HPLC as previously described (32). Pure MPER-TMD was reconstituted in DMPC/DHPC bicelles with [DMPC]/[DHPC] (q) between 0.5 and 0.6. A typical NMR sample contained ∼0.8 mM MPER-TMD (monomer), ∼50 mM DMPC, ∼100 mM DHPC, 40 mM MOPS (pH 6.7), 0.02% NaN3, and 5% D2O. For perdeuterated proteins, cells were adapted in 100 mL M9 minimal media (∼97% D2O) and then grown at large scale (4 L) in 99.8% D2O with deuterated glucose. For NOE experiments, the protein was reconstituted using DMPC and DHPC with perdeuterated acyl chains.
Oligomeric State.
The bicelle-reconstituted MPER-TMD was mixed with a SDS sample buffer without boiling, followed by SDS/PAGE (SI Appendix, Fig. S1C). The MPER-TMD protein migrated at ∼17 kDa, the size of a trimer (∼18.5 kDa). We confirmed this interpretation by chemical cross-linking and urea-PAGE (SI Appendix, Fig. S1D). We made the double mutant K665R/D674K, designed retrospectively based on the MPER-TMD structure, to facilitate cross-linking with dithiobis-succinimidyl propionate.
NMR Resonance Assignment.
All NMR data were collected at 35 °C. Sequence-specific assignment of backbone chemical shifts used transverse relaxation optimized spectroscopy (TROSY)-enhanced triple resonance experiments (68), confirmed with a 3D 15N-edited NOESY-TROSY-HSQC (heteronuclear single-quantum correlation) spectrum. Protein sidechain resonances were assigned by a combination of 3D 15N-edited NOESY-TROSY and 13C-edited NOESY-HSQC spectra, recorded using a (15N, 13C)-labeled protein sample in deuterated bicelles. For residues 685–702, the methyl group assignments were taken from those of the TMD in our previous study, as their chemical shifts were essentially identical.
NMR Structure Determination.
We used the following general approach: (i) determine local structure of an individual chain in the oligomer, (ii) apply intermonomer distance restraints to derive the oligomer structure, (iii) cross-validate the NOE-derived structure using PRE from spin labels, and (iv) determine the transmembrane partition in bicelles by PPT (38). We determined secondary structures of the subunits in the oligomeric complex using local NOE restraints and backbone dihedral restraints derived from chemical shifts and TALOS+ (69). We then identified intermonomer contacts using a mixed sample with differentially labeled monomers to record NOEs between the 15N-attached protons of one monomer and nonexchangeable protons of the neighboring monomers. We used a negative control sample to confirm the intermonomer NOEs. The MPER-TMD trimer structure was calculated using X-PLOR-NIH (70) and refined against all dihedral and NOE restraints. As independent validation, we performed PRE analysis on a mixed sample containing ∼1:1 ratio of (15N, ∼70% 2H)-labeled MPER-TMD and 14N MPER-TMD tagged with MTSL at position 660 or 710. We determined the transmembrane partition of the MPER-TMD trimer in bicelles by PPT (38).
Fab Binding to the MPER-TMD in Bicelles.
We tested Fab binding to (15N, 80% 2H)-labeled MPER-TMD in bicelles (q = 0.5); association with Fab will completely attenuate NMR signals in 1D, 15N-edited TROSY-HSQC spectra from the MPER-TMD. For binding to 2F5 or 4E10 Fab, we used the wild-type MPER-TMD. For binding to the anti-His Fab, we used the MPER-TMD with N-terminal 6His-tag.
Supplementary Material
Acknowledgments
We thank J. Chen for technical assistance, D. Barouch and B. Haynes for critical reading of the manuscript, and the NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases for reagents. This work was supported by NIH Grants AI127193 (to B.C. and J.J.C.), AI106488 (to B.C.), and AI129721 (to B.C.), the Center for HIV/AIDS Vaccine Immunology–Immunogen Design Grant AI-100645 (to S.C.H.), a Merck Fellowship (Q.F.), and Collaboration for AIDS Vaccine Discovery Grant OPP1169339 (to B.C. from the Bill and Melinda Gates Foundation). The NMR data were collected at the NMR facility of National Center for Protein Science Shanghai (supported by Chinese Academy of Sciences Grant XDB08030301) and MIT-Harvard CMR (supported by NIH Grant P41 EB-002026). S.C.H. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic structure coordinate and structural constraints have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6E8W). The chemical shift values have been deposited in the Biological Magnetic Resonance Data Bank, www.bmrb.wisc.edu (accession no. 30503).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807259115/-/DCSupplemental.
References
- 1.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 2.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison SC. Mechanism of membrane fusion by viral envelope proteins. Adv Virus Res. 2005;64:231–261. doi: 10.1016/S0065-3527(05)64007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan DC, Kim PS. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 5.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 6.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 7.Dimitrov AS, Rawat SS, Jiang S, Blumenthal R. Role of the fusion peptide and membrane-proximal domain in HIV-1 envelope glycoprotein-mediated membrane fusion. Biochemistry. 2003;42:14150–14158. doi: 10.1021/bi035154g. [DOI] [PubMed] [Google Scholar]
- 8.Muñoz-Barroso I, Salzwedel K, Hunter E, Blumenthal R. Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J Virol. 1999;73:6089–6092. doi: 10.1128/jvi.73.7.6089-6092.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salzwedel K, West JT, Hunter E. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J Virol. 1999;73:2469–2480. doi: 10.1128/jvi.73.3.2469-2480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muster T, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stiegler G, et al. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2001;17:1757–1765. doi: 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- 12.Huang J, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams LD, et al. Potent and broad HIV-neutralizing antibodies in memory B cells and plasma. Sci Immunol. 2017;2:eaal2200. doi: 10.1126/sciimmunol.aal2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun ZY, et al. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity. 2008;28:52–63. doi: 10.1016/j.immuni.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 15.Kim M, et al. Antibody mechanics on a membrane-bound HIV segment essential for GP41-targeted viral neutralization. Nat Struct Mol Biol. 2011;18:1235–1243. doi: 10.1038/nsmb.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Deng Y, Dey AK, Moore JP, Lu M. Structure of the HIV-1 gp41 membrane-proximal ectodomain region in a putative prefusion conformation. Biochemistry. 2009;48:2915–2923. doi: 10.1021/bi802303b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buzon V, et al. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog. 2010;6:e1000880. doi: 10.1371/journal.ppat.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi W, et al. Structural characterization of HIV gp41 with the membrane-proximal external region. J Biol Chem. 2010;285:24290–24298. doi: 10.1074/jbc.M110.111351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JH, Ozorowski G, Ward AB. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ofek G, et al. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol. 2004;78:10724–10737. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pejchal R, et al. A conformational switch in human immunodeficiency virus gp41 revealed by the structures of overlapping epitopes recognized by neutralizing antibodies. J Virol. 2009;83:8451–8462. doi: 10.1128/JVI.00685-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irimia A, Sarkar A, Stanfield RL, Wilson IA. Crystallographic identification of lipid as an integral component of the epitope of HIV broadly neutralizing antibody 4E10. Immunity. 2016;44:21–31. doi: 10.1016/j.immuni.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irimia A, et al. Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: Insights for vaccine and therapeutic design. PLoS Pathog. 2017;13:e1006212. doi: 10.1371/journal.ppat.1006212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alam SM, et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci USA. 2009;106:20234–20239. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scherer EM, Leaman DP, Zwick MB, McMichael AJ, Burton DR. Aromatic residues at the edge of the antibody combining site facilitate viral glycoprotein recognition through membrane interactions. Proc Natl Acad Sci USA. 2010;107:1529–1534. doi: 10.1073/pnas.0909680107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song L, et al. Broadly neutralizing anti-HIV-1 antibodies disrupt a hinge-related function of gp41 at the membrane interface. Proc Natl Acad Sci USA. 2009;106:9057–9062. doi: 10.1073/pnas.0901474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frey G, et al. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc Natl Acad Sci USA. 2008;105:3739–3744. doi: 10.1073/pnas.0800255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, et al. Mechanism of HIV-1 neutralization by antibodies targeting a membrane-proximal region of gp41. J Virol. 2014;88:1249–1258. doi: 10.1128/JVI.02664-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haynes BF, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 30.Yang G, et al. Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies. J Exp Med. 2013;210:241–256. doi: 10.1084/jem.20121977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu M, et al. Polyreactivity and autoreactivity among HIV-1 antibodies. J Virol. 2015;89:784–798. doi: 10.1128/JVI.02378-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dev J, et al. Structural basis for membrane anchoring of HIV-1 envelope spike. Science. 2016;353:172–175. doi: 10.1126/science.aaf7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rowell JF, Stanhope PE, Siliciano RF. Endocytosis of endogenously synthesized HIV-1 envelope protein. Mechanism and role in processing for association with class II MHC. J Immunol. 1995;155:473–488. [PubMed] [Google Scholar]
- 34.Boge M, Wyss S, Bonifacino JS, Thali M. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J Biol Chem. 1998;273:15773–15778. doi: 10.1074/jbc.273.25.15773. [DOI] [PubMed] [Google Scholar]
- 35.Fu Q, et al. Structural basis and functional role of intramembrane trimerization of the Fas/CD95 death receptor. Mol Cell. 2016;61:602–613. doi: 10.1016/j.molcel.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiliveri SC, Louis JM, Ghirlando R, Baber JL, Bax A. Tilted, uninterrupted, monomeric HIV-1 gp41 transmembrane helix from residual dipolar couplings. J Am Chem Soc. 2018;140:34–37. doi: 10.1021/jacs.7b10245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen X, et al. Prolonged exposure of the HIV-1 gp41 membrane proximal region with L669S substitution. Proc Natl Acad Sci USA. 2010;107:5972–5977. doi: 10.1073/pnas.0912381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piai A, Fu Q, Dev J, Chou JJ. Optimal bicelle size q for solution NMR studies of the protein transmembrane partition. Chemistry. 2017;23:1361–1367. doi: 10.1002/chem.201604206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Julien JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lyumkis D, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pancera M, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gristick HB, et al. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat Struct Mol Biol. 2016;23:906–915. doi: 10.1038/nsmb.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang R, et al. Initiation of immune tolerance-controlled HIV gp41 neutralizing B cell lineages. Sci Transl Med. 2016;8:336ra62. doi: 10.1126/scitranslmed.aaf0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu P, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 47.Chen J, et al. HIV-1 ENVELOPE. Effect of the cytoplasmic domain on antigenic characteristics of HIV-1 envelope glycoprotein. Science. 2015;349:191–195. doi: 10.1126/science.aaa9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bradley T, et al. Amino acid changes in the HIV-1 gp41 membrane proximal region control virus neutralization sensitivity. EBioMedicine. 2016;12:196–207. doi: 10.1016/j.ebiom.2016.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker LM, et al. Protocol G Principal Investigators Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker LM, et al. Protocol G Principal Investigators Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swetnam J, Shmelkov E, Zolla-Pazner S, Cardozo T. Comparative magnitude of cross-strain conservation of HIV variable loop neutralization epitopes. PLoS One. 2010;5:e15994. doi: 10.1371/journal.pone.0015994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thali M, et al. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H, et al. Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop. Proc Natl Acad Sci USA. 2016;113:E7151–E7158. doi: 10.1073/pnas.1615939113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ozorowski G, et al. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature. 2017;547:360–363. doi: 10.1038/nature23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morris L, et al. Isolation of a human anti-HIV gp41 membrane proximal region neutralizing antibody by antigen-specific single B cell sorting. PLoS One. 2011;6:e23532. doi: 10.1371/journal.pone.0023532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Molinos-Albert LM, et al. Anti-MPER antibodies with heterogeneous neutralization capacity are detectable in most untreated HIV-1 infected individuals. Retrovirology. 2014;11:44. doi: 10.1186/1742-4690-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pietzsch J, et al. Anti-gp41 antibodies cloned from HIV-infected patients with broadly neutralizing serologic activity. J Virol. 2010;84:5032–5042. doi: 10.1128/JVI.00154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Landais E, et al. Broadly neutralizing antibody responses in a large longitudinal Sub-Saharan HIV primary infection cohort. PLoS Pathog. 2016;12:e1005369. doi: 10.1371/journal.ppat.1005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jacob RA, et al. Anti-V3/glycan and anti-MPER neutralizing antibodies, but not anti-V2/glycan site antibodies, are strongly associated with greater anti-HIV-1 neutralization breadth and potency. J Virol. 2015;89:5264–5275. doi: 10.1128/JVI.00129-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Montero M, van Houten NE, Wang X, Scott JK. The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: Dominant site of antibody neutralization and target for vaccine design. Microbiol Mol Biol Rev. 2008;72:54–84. doi: 10.1128/MMBR.00020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verkoczy L, et al. Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proc Natl Acad Sci USA. 2010;107:181–186. doi: 10.1073/pnas.0912914107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Y, et al. Common tolerance mechanisms, but distinct cross-reactivities associated with gp41 and lipids, limit production of HIV-1 broad neutralizing antibodies 2F5 and 4E10. J Immunol. 2013;191:1260–1275. doi: 10.4049/jimmunol.1300770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verkoczy L, et al. Induction of HIV-1 broad neutralizing antibodies in 2F5 knock-in mice: Selection against membrane proximal external region-associated autoreactivity limits T-dependent responses. J Immunol. 2013;191:2538–2550. doi: 10.4049/jimmunol.1300971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Donius LR, et al. Generation of long-lived bone marrow plasma cells secreting antibodies specific for the HIV-1 gp41 membrane-proximal external region in the absence of polyreactivity. J Virol. 2016;90:8875–8890. doi: 10.1128/JVI.01089-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lutje Hulsik D, et al. A gp41 MPER-specific llama VHH requires a hydrophobic CDR3 for neutralization but not for antigen recognition. PLoS Pathog. 2013;9:e1003202. doi: 10.1371/journal.ppat.1003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kwong PD, Mascola JR, Nabel GJ. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: The end of the beginning. Nat Rev Immunol. 2013;13:693–701. doi: 10.1038/nri3516. [DOI] [PubMed] [Google Scholar]
- 67.Klein F, et al. Antibodies in HIV-1 vaccine development and therapy. Science. 2013;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salzmann M, Wider G, Pervushin K, Wüthrich K. Improved sensitivity and coherence selection for [15N,1H]-TROSY elements in triple resonance experiments. J Biomol NMR. 1999;15:181–184. doi: 10.1023/a:1008358030477. [DOI] [PubMed] [Google Scholar]
- 69.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 71.Cardoso RM, et al. Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10. J Mol Biol. 2007;365:1533–1544. doi: 10.1016/j.jmb.2006.10.088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.