Significance
We investigated the role of PKCε in driving lipid-induced hepatic insulin resistance beyond direct insulin receptor phosphorylation/inhibition using an in vivo model of acute hepatic insulin resistance and phosphoproteomic analysis. Many of the phosphoproteins we uncovered have not been previously associated with insulin signaling; to validate these connections, we developed a functional siRNA-based screen, which confirmed a direct role in regulating insulin signaling. We validated direct PKCε–substrate interactions using a recently developed peptide substrate library, which confirmed the cross talk between PKCε and p70S6K that our proteomic analysis suggested and which may result in aberrant negative feedback upon lipid-induced PKCε activation. Taken together, we expand the potential landscape of therapeutic targets for the treatment of insulin resistance and diabetes.
Keywords: insulin resistance, phosphoproteomics, cross talk, systems biology, PKCε
Abstract
Insulin resistance drives the development of type 2 diabetes (T2D). In liver, diacylglycerol (DAG) is a key mediator of lipid-induced insulin resistance. DAG activates protein kinase C ε (PKCε), which phosphorylates and inhibits the insulin receptor. In rats, a 3-day high-fat diet produces hepatic insulin resistance through this mechanism, and knockdown of hepatic PKCε protects against high-fat diet-induced hepatic insulin resistance. Here, we employed a systems-level approach to uncover additional signaling pathways involved in high-fat diet-induced hepatic insulin resistance. We used quantitative phosphoproteomics to map global in vivo changes in hepatic protein phosphorylation in chow-fed, high-fat–fed, and high-fat–fed with PKCε knockdown rats to distinguish the impact of lipid- and PKCε-induced protein phosphorylation. This was followed by a functional siRNA-based screen to determine which dynamically regulated phosphoproteins may be involved in canonical insulin signaling. Direct PKCε substrates were identified by motif analysis of phosphoproteomics data and validated using a large-scale in vitro kinase assay. These substrates included the p70S6K substrates RPS6 and IRS1, which suggested cross talk between PKCε and p70S6K in high-fat diet-induced hepatic insulin resistance. These results identify an expanded set of proteins through which PKCε may drive high-fat diet-induced hepatic insulin resistance that may direct new therapeutic approaches for T2D.
Insulin resistance, or a diminished tissue response to physiologic insulin concentrations, is the primary driving force behind type 2 diabetes (T2D). In liver, insulin resistance impairs insulin-mediated activation of hepatic glycogen synthesis and suppression of hepatic glucose production leading to increased fasting blood glucose (1, 2). Diacylglycerol (DAG) is a key mediator of lipid-induced hepatic insulin resistance; increased DAG content in human liver is strongly correlated with hepatic insulin resistance (3–5). In rats, a 3-d high-fat diet (HFD) is sufficient to increase hepatic DAG, which activates protein kinase C ε (PKCε) (6). PKCε in turn impairs INSR tyrosine kinase activity, inducing hepatic insulin resistance (7). Rats treated with 2′-O-methoxyethyl antisense oligonucleotides (ASOs) to knock down PKCε expression were protected from HFD-induced hepatic insulin resistance (7). Recently, we demonstrated that PKCε inhibits INSR tyrosine kinase activity directly through phosphorylation of INSR T1160 (1150 in mouse, 1161 in rat); a T1150A mutation protected mice from HFD-induced hepatic insulin resistance (8).
Although phosphorylation of INSR appears to be a key mechanism for PKCε-induced hepatic insulin resistance, DAG-activated PKCε might also act on additional substrates in an extended network of proteins that may contribute to the insulin-resistant phenotype. Furthermore, identifying PKCε targets in the HFD-fed liver could shed light not only on the molecular basis of insulin resistance but also on PKCε signaling in general. For these reasons, we sought to describe the PKCε network in HFD-induced hepatic insulin resistance.
To date, PKCε substrates have been investigated in a limited number of settings, including the tumor-promoting effects of phorbol esters (9–13), in protection against cardiac ischemia–reperfusion injury (14–17), in regulating cell–cell junctions (18, 19), in immune cell activation (20, 21), and in channel regulation (22, 23). However, most of these studies have investigated individual phosphosites or protein targets and have not evaluated the PKCε signaling network as a whole. In the two studies where PKCε was investigated on a proteome level, PKCε was implicated in glucose metabolism through modulation of the glycolytic enzymes pyruvate kinase, enolase, and lactate dehydrogenase (16) as well as in lipid metabolism through modulation of HTATIP2 (24) and the epoxyeicosatrienoic acid pathway (25). Another study investigated the PKCε phosphoproteome using 2D gel electrophoresis and Pro-Q Diamond phosphospecific staining with a peptide activator of PKCε translocation in cardiomyocytes (17). While this study did identify several additional PKCε targets involved in glucose and lipid metabolism and oxidative phosphorylation that may be relevant to insulin resistance, the difference in cell type (cardiac myocytes versus liver) and model (peptide activator versus DAG) may have important implications for the relevant PKCε network and targets. Furthermore, phosphosite localization information was not obtained for most of these substrates, and candidate site-specific kinase–substrate relationships were not directly tested.
To overcome these limitations, we investigated the PKCε signaling network using quantitative phosphoproteomics in a physiologically relevant model of HFD-induced hepatic insulin resistance with knockdown of PKCε, evaluated the role of our PKCε network in insulin signaling using an siRNA-based functional screen, and evaluated direct site-specific kinase–substrate relationships using a recently developed substrate peptide display library, which were further validated in cells. Our phosphoproteomics and kinase–substrate validation suggest that PKCε-mediated HFD-induced hepatic insulin resistance may, in part, be the result of cross talk between PKCε and p70S6K in phosphorylating IRS1 and RPS6. Additionally, our functional siRNA-based screen identified 16 phosphoproteins (two inhibitors and 14 activators beyond IRS1 and RPS6) whose role in regulating the insulin signaling pathway has not been previously reported, one inhibitor and eight activators of which may play additional roles in promoting the rescue of insulin sensitivity upon PKCε knockdown. These results provide insights into the signaling dynamics that occur during acute hepatic HFD-induced insulin resistance, which may be some of the earliest signaling events in the pathogenesis of insulin resistance and T2D.
Results
Investigation of HFD-Induced Insulin Resistance by Phosphoproteomics.
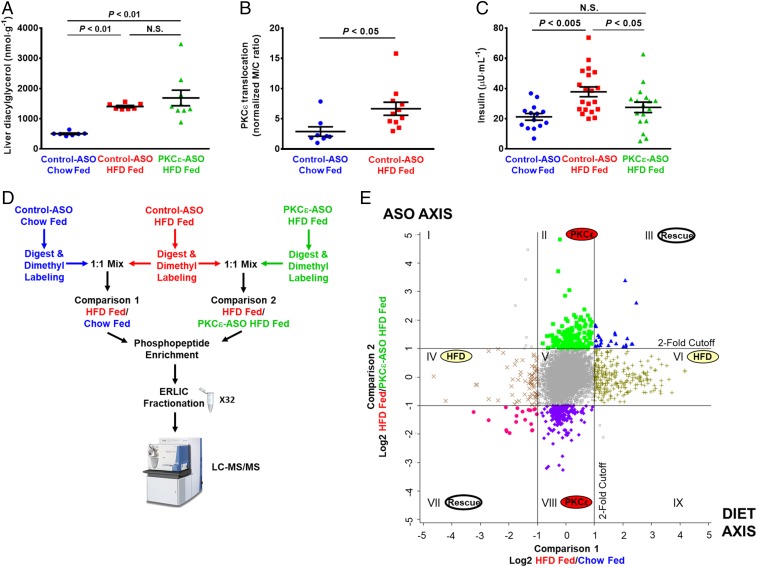
To study the signaling events and pathways that drive insulin resistance through DAG-activated PKCε, we examined protein phosphorylation in rat liver in the basal, overnight fasted state in regular chow-fed rats treated with control nontargeted ASO and in 3-d HFD and sucrose water-fed rats treated with control or PKCε-targeted ASO. ASO was delivered by i.p. injection for 4 wk before experiments, with the animals divided into normal chow and HFD for the last 3 d. This approach was chosen to isolate the effects of PKCε activation, without confounding signal from insulin stimulation, muscle or adipose insulin resistance, obesity, or chronic inflammation (26, 27). ASOs are used clinically to target hepatic gene expression; PKCε ASO safely and effectively decreases hepatic but not muscle PKCε protein expression in rats (7). Study animals showed no difference in weight gain over the course of ASO treatment (SI Appendix, Fig. S1A). Also, consistent with previous work, rats fed a HFD displayed increased liver triglyceride and DAG content (Fig. 1A and SI Appendix, Fig. S1 B–D) and increased PKCε activation as measured by membrane translocation (Fig. 1B and SI Appendix, Fig. S1 E and F). HFD-fed rats also exhibited higher basal plasma insulin concentrations compared with chow-fed rats (Fig. 1C and SI Appendix, Fig. S1G) despite similar plasma glucose (SI Appendix, Fig. S1 H and I), reflecting mild hepatic insulin resistance. The PKCε-directed ASO was >90% effective at reducing PKCε protein levels in liver after 4 wk of treatment (SI Appendix, Fig. S1J) and, importantly, was able to protect rats from HFD-induced hepatic insulin resistance (Fig. 1C and SI Appendix, Fig. S1G). These results are consistent with previous studies using this model (6, 7). Three animals from each experimental group were selected as representative samples for phosphoproteomic analysis; animals selected were chosen to best represent the relevant phenotypes based on several parameters including quality of study, weight gain, weight gain on HFD, liver lipid levels, basal plasma glucose, and basal plasma insulin (phenotyping values and selection criteria are in Dataset S1). In study animals, there were no significant differences in serum transaminase activity, alkaline phosphatase activity, or albumin, indicating no effect of the diet or ASO on liver toxicity (SI Appendix, Fig. S1K and Dataset S1).
Fig. 1.
Quantitative phosphoproteomic analysis of acute hepatic insulin resistance and rescue by PKCε knockdown. Rats were fed either a control, chow diet or a HFD and treated with ASO that was either scrambled (control ASO) or targeted to PKCε (PKCε ASO). (A) HFD resulted in increased liver diacylglycerol independent of PKCε ASO. (B) PKCε translocation was quantified as a marker of PKCε activity [membrane-to-cytosol (m/c) ratio]; from immunoblot in SI Appendix, Fig. S1E. (C) HFD feeding increased basal plasma insulin, which was rescued by PKCε ASO. (D) Phosphoproteomics workflow, indicating three experimental groups and two pairwise comparisons. Phospho-enriched samples were separated into 32 fractions by ERLIC to reduce sample complexity and increase proteomic coverage; three technical replicates of each fraction were analyzed by LC-MS/MS using a 120 method (three replicates of 32 fractions = 96 120-min runs). (E) Effect of HFD and PKCε knockdown on the phosphoproteome. Phosphopeptide ratios obtained from two technical replicates of each comparison were averaged and log2 transformed, and peak intensity ratios were plotted against each other. The y axis represents the effect of the PKCε ASO on the phosphoproteome, whereas the x axis represents the effect of the HFD on the phosphoproteome; the lines represent twofold change in the phosphopeptide ratio. The twofold cutoff lines designate 2D enrichment categories of phosphopeptides that are designated by roman numerals. Error bars represent mean ± SEM (A–C).
Quantitative phosphoproteomic analysis was carried out with an established dimethyl labeling strategy to enable direct pairwise comparisons between experimental groups (28). We compared control ASO-treated, chow-fed rats to control ASO-treated, HFD-fed rats to determine which phosphorylation events or pathways are altered in the setting of acute hepatic HFD-induced insulin resistance (Fig. 1D, comparison 1). We also compared control ASO-treated, HFD-fed rats to PKCε ASO-treated, HFD-fed rats to determine which phosphorylation events and signaling networks are affected by PKCε knockdown (Fig. 1D, comparison 2). Following labeling, phosphopeptides were enriched with TiO2 and fractionated using electrostatic repulsion liquid chromatography (ERLIC) into 32 fractions to improve phosphoproteome coverage (Fig. 1D). In total, 11,224 sites of protein phosphorylation (phosphosites) were reproducibly quantified across all experimental conditions. In total, 779 phosphosites from 560 distinct proteins showed a greater than twofold change (Fig. 1E).
Mapping Phosphorylation Dynamics to Physiological Response.
Phosphosites showing a greater than twofold change were sorted into biologically meaningful categories by 2D enrichment analysis (Fig. 1E and Dataset S3). This analysis enabled us to relate how changes in the phosphoproteome connect to the physiology of HFD feeding and the effect of knocking down PKCε, as well as how PKCε knockdown may protect against HFD-induced insulin resistance. Categories II and VIII represent PKCε-dependent but diet-independent changes in the phosphoproteome, with category II containing phosphosites reduced by PKCε knockdown and category VIII containing phosphosites increased by PKCε knockdown. As expected, three phosphosites on PKCε were observed in category II, which is in accordance with PKCε knockdown at the protein level. Category II is predicted to be enriched for direct substrates of PKCε and phosphosites that are downstream of PKCε signaling. Category VIII contains phosphosites that were increased by PKCε knockdown; these are expected to be sites normally inhibited by PKCε activity or related to compensatory mechanisms responding to PKCε knockdown. Categories IV and VI represent diet-dependent but PKCε-independent changes in the phosphoproteome. Finally, categories III and VII represent changes in the phosphoproteome that were both diet and PKCε dependent; because these phosphosites were altered by the HFD but “rescued” or returned to control-like levels by PKCε knockdown, they may be directly linked to the mechanism by which PKCε drives HFD-induced insulin resistance and are therefore of particular interest.
Phosphoproteomics Identifies Regulators of Insulin Signaling.
Our phosphoproteomic analysis revealed changes in phosphorylation on proteins with mostly no known connection to insulin signaling. In general, we did not observe many known insulin-stimulated phosphosites; however, because (i) we investigated the basal (rather than insulin stimulated) state and (ii) we did not perform enrichment for tyrosine-phosphorylated peptides [which comprise only 1–2% of phosphorylation events (28)], this observation is not too surprising. We were also disappointed, but not surprised, that INSR T1161 phosphorylation was not observed in category III because of the low abundance of the INSR (observation of this site in our previous work required two-step isolation and immunoprecipitation of the INSR) (8). The INSR phosphopeptides that were observed in category V are autophosphorylation events and are not associated with any known function (29, 30).
Given the strong physiological basis of our experimental model and the changes in phosphorylation that were observed in categories III and VII, we hypothesized that some of these sites and proteins are likely to actively regulate insulin signaling and may be drivers of insulin resistance in our model. In addition to categories III and VII, changes in categories IV and VI may also be relevant to insulin signaling and the metabolic changes that occur on a HFD even if they are unlikely to drive insulin resistance through PKCε, while changes in categories II and VIII may represent how PKCε knockdown “rewires” signaling to promote insulin sensitivity beyond preventing phosphorylation of insulin resistance drivers. To elucidate whether a phosphoprotein we identified in any of these categories may regulate insulin signaling, we developed a siRNA-based functional screen in rat hepatoma cells for canonical hepatic insulin signaling. Although gene knockdown does not recapitulate the consequences of the altered protein phosphorylation that we observed in vivo, we reasoned that if knockdown of a gene in our siRNA screen does not affect insulin signaling, then a simple direct connection to insulin signaling cannot be made and further in vivo experimentation would be required to discern whether the phosphoprotein is involved in insulin resistance or is a collateral consequence of the altered signaling and physiology. However, a significant response in the siRNA screen would allow us to connect these genes and their phosphorylated protein products to the mechanism of insulin action in vivo.
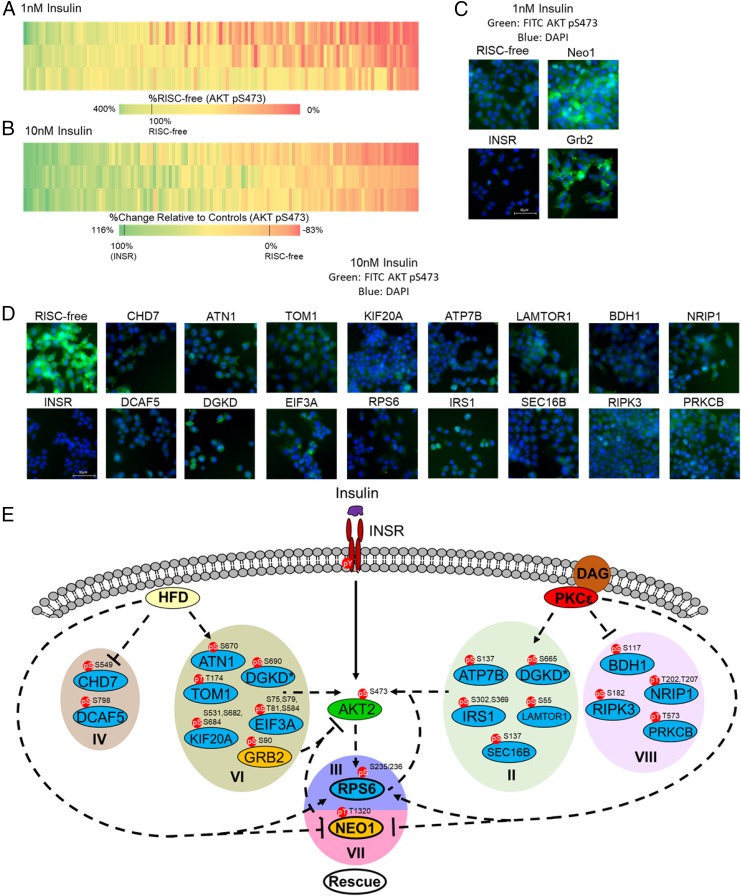
We used rat hepatoma cells (McArdle hepatoma cells, ATCC McA-RH7777) and AKT phosphorylation at S473 (31) to develop an image-based, single-cell quantitative assay for canonical insulin signaling. Using a commercially available phosphospecific antibody, we observed an insulin dose-dependent increase in AKT pS473 phosphorylation (SI Appendix, Fig. S2A). Single-cell quantitation was achieved by measuring the intensity of AKT pS473 staining, determining baseline intensity using the cells treated with 0 nM insulin (SI Appendix, Fig. S2B), and plotting the data as the percentage of cells with intensity greater than baseline (SI Appendix, Fig. S2C). We then validated siRNA knockdown in this system using a positive control siRNA targeting INSR and a negative control siRNA that does not engage the RNA-induced silencing complex (RISC-free) (SI Appendix, Fig. S3A). As expected, the RISC-free siRNA-transfected cells responded dose dependently to insulin in a similar manner to mock-transfected cells, while INSR siRNA-transfected cells were unresponsive to insulin (SI Appendix, Fig. S3 A and B). Because we did not know a priori whether a given phosphoprotein target might be an activator or inhibitor of insulin signaling, we screened with both 1 and 10 nM insulin stimulation. Knocking down an activator should result in a decrease in AKT pS473 signal that would be more easily observed at a 10 nM insulin dose, while knocking down an inhibitor should result in an increase in AKT pS473 signal that would be more easily observed at a 1 nM insulin dose.
We selected 125 candidate proteins to screen (Dataset S5), which included all phosphoproteins from categories III and VII because of their correlation with insulin resistance, phosphoproteins with a known or putative role in insulin signaling, and phosphoproteins with greater than fourfold change in at least one experimental condition (SI Appendix, Table S1, and Dataset S5). We then performed three screening replicates for both the 1 and 10 nM screen. We observed high reproducibility for the 10 nM screen (Pearson correlation coefficient R ≥ 0.85). Signal-to-background ratio (mean of the positive control over the mean of the negative control) and Z′ factor (screening statistic reflecting both assay window and assay variability, calculated from the positive and negative control mean signals and their SDs) were monitored for each screening plate to evaluate performance of the 10 nM screen. Cell-based assays with signal-to-background ratios greater than 3 and positive Z′ are amenable to screening. We observed acceptable signal-to-background (11.4, 15.4, and 6.2) ratios and Z′ (0.29, 0.73, and 0.30) factors, validating robustness of the 10 nM screen. In the 1 nM screen, signal-to-background ratios and Z′ factors were not calculated because of a lack of a positive control, and reproducibility was not as high, but still acceptable (Pearson correlation coefficients of 0.83, 0.57, and 0.63 between 1 and 2, 1 and 3, and 2 and 3, respectively). To cross-validate putative activators and inhibitors of insulin signaling, we repeated the siRNA transfection of the top hits and remeasured AKT S473 phosphorylation by Western blot. Candidate knockdowns that produced a fold change greater than or equal to IRS2, a known member of the insulin signaling pathway, were considered validated (SI Appendix, Fig. S4 A and B).
In the screen for inhibitors, we identified growth factor receptor-bound protein 2 (GRB2) and neogenin 1 (NEO1) as strong inhibitors of insulin signaling with 184% effect and 270% effect, respectively, compared with the RISC-free control set as 100% signal (Fig. 2 A and C and SI Appendix, Fig. S4 and Table S1). Although GRB2 is an adaptor protein that is known to connect insulin signaling to the RAS and MAP kinase pathways (32), GRB2 has not previously been described as an inhibitor of insulin signaling, nor is the mechanism by which knockdown of GRB2 increases AKT pS473 phosphorylation immediately clear. However, while we observed that phosphorylation on GRB2 S90 was increased by HFD in vivo, its phosphorylation was unchanged by PKCε knockdown (category VI), suggesting that while this site is dynamically regulated by the HFD and that GRB2 knockdown does increase AKT pS473 phosphorylation, it is unlikely that this site contributes to PKCε-mediated HFD-induced insulin resistance. Conversely, phosphorylation of NEO1 T1320 was significantly down-regulated by the HFD but restored to WT levels by PKCε knockdown (category VII). Given that NEO1 knockdown had a profound stimulatory effect on AKT S473 phosphorylation, and that NEO1 T1320 phosphorylation was significantly regulated by both the HFD and PKCε knockdown, it is likely that this phosphoprotein is involved in PKCε-mediated HFD-induced insulin resistance and that T1320 regulates NEO1 function with respect to insulin signaling; however, the connection between NEO1 T1320 and PKCε is not immediately clear, as NEO1 phosphorylation is reduced by the HFD but restored by PKCε knockdown (category VII).
Fig. 2.
siRNA screen uncovers previously unknown activators and inhibitors of insulin signaling. A total of 125 phosphoproteins was targeted with siRNA and stimulated with 1 or 10 nM insulin. Cells were fixed and stained with a phosphospecific antibody against AKT S473 phosphorylation; staining was quantified by single-cell image analysis as described in SI Appendix, Fig. S2. (A) Heatmap of siRNA screen with 1 nM insulin stimulation (three replicates). Heatmap intensity presented as the percentage of AKT S473 phosphorylation signal relative to the RISC-free siRNA control set at 100%. (B) Heatmap of siRNA screen with 10 nM insulin stimulation (three replicates). Heatmap intensities reflect the relative percent effect in AKT S473 phosphorylation of each individual siRNA, compared with the RISC-free control siRNA set as 0% effect and the INSR control siRNA set as 100% effect. (C) Representative images from the 1 nM insulin stimulation (green, FITC-AKT pS473; blue, DAPI). RISC-free and INSR control siRNAs are shown with Grb2 and Neo1 siRNAs, showing increase in AKT pS473 staining with gene knockdown. (D) Representative images from the 10 nM insulin stimulation siRNA screen (green, FITC-AKT pS473; blue, DAPI). (E) Model of the modulation of canonical insulin signaling by the HFD and PKCε. PKCε may influence insulin signaling through phosphoproteins modulated by the PKCε ASO alone (categories II and VIII) or by the PKCε ASO and HFD (categories III and VII). Insulin signaling may also be influenced by proteins whose phosphorylation is up- or down-regulated by the HFD alone (categories IV and VI); however, these proteins are unlikely to play a role in the rescue of insulin sensitivity by PKCε knockdown. CHD7 and DGKD are represented twice [and designated by an asterisk (*)] as we observed multiple phosphorylation events on these proteins that showed different fold-change values upon different treatments. Phosphoproteins that were shown by siRNA screen to activate or inhibit AKT S473 phosphorylation are shown in blue and yellow, respectively.
In the screen for activators, we observed several proteins known to promote insulin signaling, including IRS1 and IRS2. We also observed that RPS6 knockdown had an effect similar to knockdown of INSR (108% effect for RPS6, with 100% effect set by the INSR control siRNA) and greater than the IRS1/2 knockdowns (82% and 93% effect for IRS1 and IRS2 siRNAs, respectively; P < 0.05, one-way ANOVA) (Fig. 2D and SI Appendix, Table S1). This effect was not quite as strong, but still apparent, when validated by Western blot (SI Appendix, Fig. S4). This result suggests that the RPS6 protein is a strong activator of insulin signaling; given that RPS6 phosphorylation correlates with insulin resistance in the basal state in vivo and is rescued by PKCε knockdown, aberrant phosphorylation of RPS6 by DAG-activated PKCε may be an additional driver of insulin resistance perhaps by lowering the fold change upon insulin stimulation.
In addition to RPS6 as an activator of insulin signaling, we observed several other proteins that have not previously been implicated as activators of insulin signaling (Fig. 2 B, D, and E and SI Appendix, Table S1) with consistent results by imaging and Western blot. ATP7B, BDH1, LAMTOR1, NRIP1, PRKCB, RIPK3, and SEC16B were identified as activators of insulin signaling that may contribute to the rescue of insulin sensitivity by PKCε knockdown, as phosphorylation of these phosphoproteins was altered by PKCε knockdown but not the HFD (category II/VIII, Fig. 2E). ATN1, CHD7, DCAF5, EIF3A, KIF20A, and TOM1 were identified as activators of insulin signaling whose phosphorylation is modulated by the HFD independent of PKCε knockdown (category IV/VI, Fig. 2E) and are thus unlikely to be involved in HFD-induced insulin resistance through PKCε, although they may contribute to the insulin-resistant phenotype through other mechanisms (Fig. 2 B, D, and E) (31, 33–35).
The activator diacylglycerol kinase delta (DGKD) is interesting in that we observed multiple phosphorylation events on this protein; however, these events appeared in different categories, suggesting differential regulation by the HFD (DGKD S690, category VI) and by PKCε (DGKD S665, category II). It is possible that the PKCε knockdown-driven reduction in phosphorylation on DGKD S665 may compensate for or override the HFD-driven changes in promoting insulin sensitivity, or that the changes are mediated solely through DGKD S690, which was just barely below the twofold cutoff for category III in comparison 2. DGKD has been shown to phosphorylate DAGs and convert them into phosphatidic acid, regulating the balance of these two important signaling lipids (36); it has also been shown that DGKD is regulated by conventional PKCs (cPKCs) (PKCα, β, γ) in Cos-7 cells (fibroblasts) (37) and that DGKD deficiency can lead to insulin resistance in skeletal muscle (38). We observed that knocking down DGKD decreased the AKT pS473 signal upon insulin stimulation; we hypothesize that this manipulation increased levels of DAGs and activated PKCε, which resulted in the decreased AKT pS473 signal (observed in both the 10 and 1 nM insulin screens), although we cannot rule out other direct or indirect effects through protein–protein interactions or the potential reduction of phosphatidic acid. It is also possible that, in liver and liver cell lines, DGKD itself is regulated by PKCε rather than cPKCs, creating a feedback loop. These hypotheses will be tested in future work.
Surprisingly, in addition to the previously unknown regulators of insulin signaling we have described, we identified PKCε itself as an activator of insulin signaling. Rat hepatoma cells transfected with PKCε siRNA showed an effect of 77% of that of the INSR control siRNA, suggesting that knockdown of PKCε in this model impairs insulin signaling. This result suggests that, in the normal state, PKCε may be required for proper insulin signaling; however, in the lipid-laden liver, DAG-activated PKCε becomes inhibitory and knocking down the kinase becomes protective.
Large-Scale Validation of PKCε Substrates.
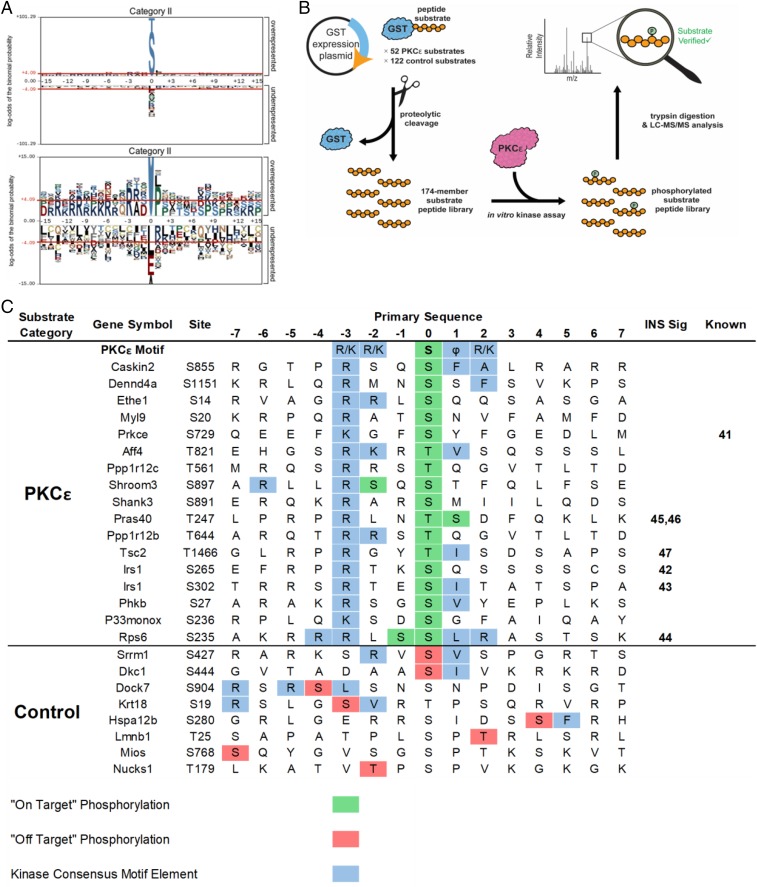
To identify the kinases driving the phosphosite changes observed in each category, we performed motif analysis using pLogo (39), which aligns the central phosphorylated residue with 15 flanking amino acids from the native protein context on either side and identifies enriched residues at each position. As predicted, we observed a PKC-like motif (RxxS/T) (Fig. 3A and SI Appendix, Fig. S5A) in category II, with a strong −3R motif and a hydrophobic residue at +1 (L, M, V) corresponding to a PKCε preference (40). We also observed an RxxS/T motif in category VIII, but without the +1 hydrophobic preference, suggesting compensation for PKCε knockdown by other PKCs (SI Appendix, Fig. S5B). Conversely, the changes in the phosphoproteome caused by the HFD showed a strong cyclin-dependent kinase motif (SPxKKK) and are the focus of a separate study (SI Appendix, Fig. S5B). No significant motif enrichments were observed in categories I, III, IX, and VII due to their relatively low number of phosphosites.
Fig. 3.
Motif analysis and mass spectrometry-based kinase assays identify previously unknown PKCε substrates. (A) Motif analysis of phosphopeptides from category II revealed a PKC-like motif (RxxS/T). Residues shaded in gray are fixed, while the size of the residue correlates with the enrichment of that residue at a position. Residues above the red lines are statistically significant (P < 0.05). (B) Workflow for kinase–substrate relationship determination using a substrate peptide display library. Substrates were selected based on phosphoproteomic data and PKC motif analysis, expressed as a C-terminal fusion to GST in E. coli, purified, and cleaved from the GST. The peptide substrate display library was incubated with PKCε and digested with trypsin; peptide substrates that were phosphorylated were identified by LC-MS/MS followed by database searching. (C) PKCε substrates identified by substrate peptide display library. The amino acid sequence of each substrate peptide is shown with the site of in vivo phosphorylation at position 0 with 7 aa flanking. “On-target” phosphorylation (in green) indicates the site of in vivo and in vitro phosphorylation matched. “Off-target” phosphorylation (in red) indicates phosphorylation by PKCε at unintended sites. PKCε motif elements are shown in blue. References are included for known members of the insulin signaling pathway (INS Sig) and for known kinase–phosphosite relationships (Known).
Our phosphoproteomics approach and motif analysis identified a list of potential PKCε substrates that fit with a canonical motif and responded to direct PKCε knockdown in vivo. To provide more direct evidence of a PKCε kinase/substrate relationship in a high-throughput and site-specific manner, we deployed a recently developed peptide substrate expression platform for in vitro LC-MS/MS coupled kinase assays (Fig. 3B). Fifty-two potential PKCε substrates were selected from categories II and III (peptides showing a greater than twofold change in PKCε knockdown regardless of diet), which fit a generic PKC motif (RxxS/T). In addition to these target substrate peptides, we included two groups of control phosphopeptides; one group of 72 peptides from category VI that conforms to a different kinase consensus motif (+1P) and one group of 50 peptides from category V that showed no fold change and did not contain a discernable kinase motif. This method is similar to the recently published SERIOHL-KILR method (41), except (i) we used rat instead of human and (ii) we made a targeted, rather than unbiased, library of substrate peptides.
PKCε was able to phosphorylate 17 of the 52 potential substrate library peptides in at least three of four replicates (Fig. 3C and SI Appendix, Table S2). Substrates were considered “on-target” when phosphorylation occurred at the same site as in vivo studies and matched the PKCε motif. Only 9 of 132 control peptides were phosphorylated (1 outside the −7 to +7 window in Fig. 3C) and the majority of these phosphorylated control peptides contained elements of a PKC-consensus motif (−3R/−2R, +1 hydrophobic) but most were “off-target” since they did not match in vivo sites. Bona fide on-target substrates included a known PKCε autophosphorylation site S729 (42); however, the remainder of the substrates that we observed were either previously unknown, or known substrates at different positions than previously known. Significantly, these other sites included the p70S6K sites S265 and S302 on IRS1 (43, 44) (homologous to human S270 and S307) and S235/S236 on RPS6 (45), as well as the AKT sites T247 on PRAS40 (46, 47) (T246 in human) and T1466 on TSC2 (T1462 in human) (48) (Fig. 3C). PKCε, p70S6K, and AKT are all members of the AGC family of kinases and share an RxxS/T motif preference. Although p70S6K and AKT additionally prefer −5R while PKCε prefers +1 hydrophobic and +2R/K (39), these results suggest functional signaling crosstalk between PKCε and p70S6K and/or AKT.
PKCε Can Cross Talk with p70S6K Substrates.
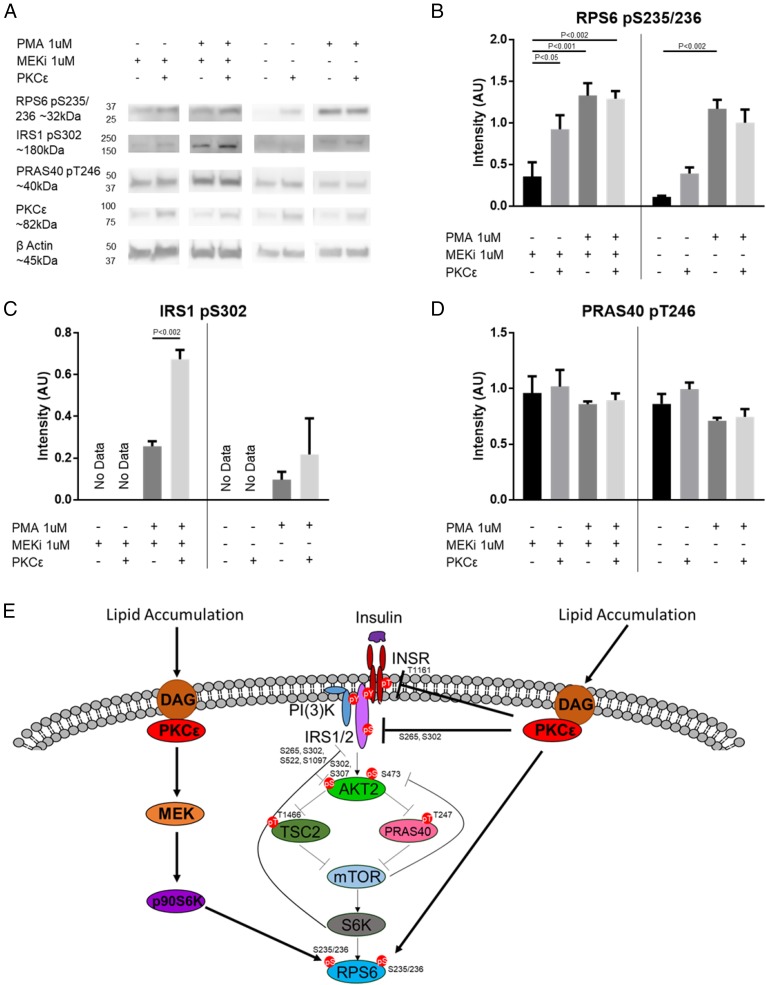
p70S6K and AKT are key kinases in the insulin signaling cascade and are known to regulate a variety of processes including apoptosis, proliferation, cytoskeletal organization, protein synthesis, mRNA splicing, transcription, metabolism, and negative feedback of insulin signaling (49, 50). Our in vivo phosphoproteomics data suggested cross talk between these kinases and PKCε, and our in vitro data provided additional support to this hypothesis. To further examine this cross talk in cells, we used p70SK1/p70S6K2 double-knockout (DKO) mouse embryonic fibroblasts (MEFs) where PKCε activity toward p70S6K substrates could be tested in the absence of p70S6K activity. Surprisingly, DKO cells have persistent phosphorylation at RPS6 S235/236 despite lacking p70S6K (51). This phosphorylation has been explained by a MAPK–p90S6K pathway downstream of PKC activation (45, 51) and is consistent with the role of PKCs in MAPK pathway activation (52). To account for this additional S6K pathway, we used the potent and specific MEK inhibitor trametinib (MEKi) to block the MAPK–p90S6K pathway in the p70SK1/p70S6K2 DKO MEFs to examine PKCε cross talk in a S6K-null background.
In the S6K-null background (DKO MEKi+), we saw that PKCε transfection significantly increased RPS6 S235/236 phosphorylation (Fig. 4 A and B), supporting direct phosphorylation of RPS6 by PKCε. Low levels of RPS6 S235/236 phosphorylation were observed in our null background, and we assume that this is due to native PKC isoforms. Western blots confirmed low levels of native PKCε expression, and phorbol 12-myristate 13-acetate (PMA) stimulation increased RPS6 phosphorylation in nontransfected (NT) cells. While overexpression of PKCε alone increases RPS6 S235/236 phosphorylation, we did not see further PMA stimulation in PKCε-transfected cells. Baseline IRS1 S302 phosphorylation was below the limit of quantitation in DKO MEKi+ cells with or without PKCε overexpression. However, we observed a significant increase in IRS1 S302 phosphorylation with PMA treatment and PKCε overexpression (Fig. 4C). PMA activation of native PKCs alone in untransfected cells, with or without MEKi, could not reproduce this effect. These data support direct phosphorylation of IRS1 S302 by PKCε; however, unlike RPS6 phosphorylation, PMA stimulation is required. Even though this result does not rule out the possibility that PKCε transfection itself primes another kinase or PKC isoform to respond to PMA and phosphorylate IRS1 S302, we believe that direct phosphorylation by PKCε is the simplest interpretation of this result. In contrast to RPS6 S235/236 and IRS1 S302 phosphorylation, phosphorylation on PRAS40 was not altered by PKCε transfection, stimulation with PMA, or MEKi treatment (Fig. 4D and SI Appendix, Fig. S6A), suggesting that these sites are not regulated by PKCε in S6K-null cells. Measuring the phosphorylation at TSC2 T1466 was inconclusive because of a poorly performing phosphoantibody.
Fig. 4.
PKCε can cross talk with p70S6K substrates. (A) Western blot of p70S6K1/p70S6K2 DKO cells with or without WT PKCε transfection, treated with or without 1 μM MEKi, followed by treatment with or without 1 μM PMA. Densitometry for RPS6 S235/236 phosphorylation (different exposures for −MEKi and +MEKi) (B), IRS1 S302 phosphorylation (C), or PRAS40 T246 phosphorylation (D). n = 3. Bands are normalized to β-actin. Vertical bars separate +MEKi and −MEKi. P values for two-way ANOVA with Tukey’s multiple-comparisons test are as indicated. Full Western blots with selected lanes indicated in SI Appendix, Fig. S6B. (E) Model of PKCε modulation of canonical insulin signaling. In addition to modulating the INSR itself and other known members of the insulin signaling pathway, PKCε may influence insulin signaling through phosphoproteins modulated by the PKCε ASO alone (categories II and VIII) or by the PKCε ASO and HFD (categories III and VII) that were shown by siRNA screen to activate (blue) or inhibit (yellow) AKT S473 phosphorylation; direct targets of PKCε are shown by the solid lines.
PKCε phosphorylation of canonical p70S6K substrates in vitro and in S6K-null cells supports the in vivo phosphoproteomics data, suggesting that PKCε can cross talk with p70S6K signaling pathways (Fig. 4E). Although PKCε appeared to cross talk with canonical AKT sites in vivo and in vitro, we could not find further support for this in cell lines; thus, these and other sites not confirmed as direct PKCε substrates required further investigation.
Discussion
While INSR T1160 phosphorylation by PKCε is an important mechanistic link between increased DAGs and hepatic insulin resistance (8), this single phosphorylation event is unlikely to encapsulate the full effect of PKCε on the signaling and physiology of HFD-induced hepatic insulin resistance. We therefore adopted a systems approach to understand how PKCε drives HFD-induced hepatic insulin resistance; by comparing phosphoproteomes among normal, insulin-resistant, and PKCε-deficient livers, we gain additional insight into the many signaling pathways that may be driving these states.
We also used an siRNA-based functional screen to determine which differentially regulated phosphoproteins could directly influence canonical insulin signaling. This screen identified many phosphoproteins spread across phosphorylation categories that, when knocked down in a rat hepatoma cell line, affect insulin signaling as measured by AKT pS473 staining. These proteins include RPS6, NEO1, and DGKD whose phosphorylation was regulated by both the HFD and PKCε (although in the case of RPS6 and NEO1 the same site was observed, while in DGKD two different sites were regulated); changes in the phosphorylation of these proteins may contribute to insulin resistance. The activators of insulin signaling ATP7B, BDH1, LAMTOR1, NRIP1, PRKCB, RIPK3, and SEC16B may also be involved in the restoration of insulin sensitivity by PKCε knockdown but are unlikely to drive insulin resistance as their phosphorylation was only altered by the PKCε knockdown but not the HFD. Together, these expand the network by which PKCε may promote insulin resistance or sensitivity beyond just phosphorylation of the insulin receptor.
In addition, we developed a method to screen potential kinase–substrate relationships in higher throughput than previously possible, leading to the identification of PKCε substrates in the setting of HFD-induced insulin resistance. Given the ability of LC-MS/MS to localize phosphorylated residues, and that our substrate library peptides map back to peptides observed in vivo, site-specific kinase–substrate relationships validated in this manner are extremely accurate at the peptide level and provide a platform to screen many potential substrates in high-throughput. Additional validation with full-length substrate protein and/or coimmunoprecipitation with the narrowed list of potential substrates could further strengthen these results.
Curiously, we did observe several previously described PKCε phosphosites in our study, such as ALDOA S39, ALDOA S46, ETFDH S550, and SDHA S22, that did not have significant fold change with the HFD or PKCε knockdown and were therefore not included in the substrate peptide display library. However, the majority of known PKCε substrates from previous studies (17, 53) were not observed in our study; this result suggests that differences in cell type/tissue and PKCε activation do affect the relevant PKCε network. In addition, it is possible that limiting our library to peptides containing a RxxS/T motif excluded legitimate PKCε substrates from our analysis. For example, we observed PRKD3 S735 in category II; this site has been described as a PKCε/PKCδ substrate but was not included in our potential substrate library because it lacks a RxxS/T motif. The sites that were observed in DGKD (S665 and S690) were also not included in the PKCε substrate library because they lacked the RxxS/T motif as well. Expanding our approach to all peptides in a given category would overcome this limitation and will be addressed in future studies.
We have also shown in this study that PKCε can phosphorylate IRS1 and RPS6 in cross talk with p70S6K, but that PKCε does not appear to cross talk with AKT in cells. Although several studies have implicated the canonical p70S6K substrate IRS1 S302 as a mediator of insulin resistance [reviewed by Copps and White (54)] and have suggested that reduced phosphorylation at this site is responsible for protection against lipid- and age-induced insulin resistance in p70S6K-null mice (55), an IRS1 S302A knockin mouse model demonstrated that this mutation by itself was not sufficient to alter normal insulin signaling or feedback regulation by p70S6K (56). However, it is possible that this site works in concert with other phosphosites targeted by p70S6K and PKCε to drive negative feedback and insulin resistance. These sites include IRS1 S265, which was also phosphorylated by PKCε in vivo and in vitro but not validated by Western blot due to lack of a commercially available phosphospecific antibody, and IRS1 S1100 (human S1101), which has been shown to impair insulin signaling (57, 58), but was just slightly below the strict twofold change cutoff in our study (1.84-fold change) in the PKCε knockdown condition and thus excluded from our initial analysis. Investigating the role of these sites individually and in combination could determine how or if they contribute to insulin resistance.
PKCε may also drive insulin resistance through RPS6 phosphorylation. RPS6 phosphorylation has been implicated in the regulation of glucose homeostasis and cell size in addition to its canonical role in regulating protein translation (59); mice with whole-body mutation of S6K target serine residues on RPS6 to alanine show improved peripheral insulin sensitivity through an unknown mechanism despite hyperglycemia (which is mainly driven by a pancreatic beta-cell defect) (59). Even though the physiologic outcome of baseline PKCε (or PKCε–MAPK–p90S6K) phosphorylation of RPS6 in DKO cells is unknown, our data show that phosphorylation on RPS6 in vivo correlates with PKCε activation by DAG and is rescued by PKCε knockdown. Given that we, through a siRNA screen, and others, through S-to-A knockin models (59), have shown that RPS6 can regulate insulin signaling and sensitivity, it is probable that PKCε phosphorylation of RPS6 also contributes to insulin resistance. Further study is required to determine the mechanism by which RPS6 influences insulin signaling and sensitivity, and how phosphorylation in normal and disease states regulates those processes. Our data suggest a model whereby PKCε cross talk with p70S6K results in the inappropriate phosphorylation of p70S6K substrates, leading to the activation of negative-feedback mechanisms that reduce insulin signaling and drive insulin resistance (Fig. 4E).
Overall, these results highlight the need to investigate entire signaling networks in disease states using systems biology and omics techniques to understand how the entire system responds to perturbation and how these responses result in the complex changes in physiology that are observed. Using these approaches in this study, we have broadened our understanding of the proteins involved in mediating insulin signaling and insulin resistance, uncovered future lines of investigation into how insulin signaling and resistance are regulated, and provided potential protein targets for the treatment of T2D.
Materials and Methods
Animals.
All experimental protocols involving animals were reviewed and approved by the Institutional Animal Care and Use Committee of Yale University School of Medicine before study initiation. Rats were treated with ASO targeting a scramble sequence or PKCε for 4 wk by i.p. injection; before experiments, rats were either maintained on regular chow or switched to HFD with sucrose water for 3 d. HFD feeding, ASO treatment, and phenotyping have been described previously (7, 60, 61). Detailed methods are available in SI Appendix, Supplemental Materials and Methods.
Phosphoproteomics.
Phosphoproteomic analysis of protein extracts was performed as reported earlier (62) with the following changes. Dimethyl labeling of peptides was performed before phosphopeptide enrichment with TiO2. Furthermore, we introduced an additional ERLIC fractionation step to further separate TiO2-enriched phosphopeptides. In our hands, this combination of phosphopeptide enrichment by TiO2 and fractionation by ERLIC provides superior performance compared with ERLIC fractionation alone (63). Detailed protein extraction, digestion, and analysis methods are available in SI Appendix, Supplemental Materials and Methods; full results are available in Dataset S2.
siRNA Screen.
All screening was performed at and in collaboration with the Yale Center for Molecular Discovery. Dharmacon siGENOME rat siRNAs were obtained from GE Healthcare as SMART pools of four siRNAs per gene. Individual siRNA sequences are available in Dataset S4. Rat hepatoma cells were treated with 20 nM individual SMART pools for 48 h, serum starved for 12 h, and then treated with 1 or 10 nM insulin. Following treatment, cells were washed, fixed, permeabilized, and stained with an antibody against AKT pS473 (Cell Signaling Technologies), followed by Alexa Fluor 488 (Molecular Probes). Cells were imaged using the InCell 2200 Imaging System (GE Corporation) and analyzed using InCell Analyzer software. Detailed transfection, imaging, and image analysis methods are available in SI Appendix, Supplemental Materials and Methods.
Kinase Substrate Validation.
Of the 203 phosphosites observed in categories II and III, we selected the 52 peptide sequences that had −3R for this library. We also selected the 72 peptides out of the 269 observed in category VI that had +1P as a control group. In category V, we selected 50 peptides that had 0-fold change in both comparisons, and that had no elements of either the PKC motif (−3R) or the +1P motif surrounding the central phosphorylated residue to serve as additional controls. Predicted kinase substrate phosphosites were encoded as peptides at least 21 aa long. The predicted phosphorylatable Ser or Thr residue was positioned in the middle of the peptide (position 0) flanked on the N and C termini by the 10 aa occurring in the native sequence of the corresponding rat protein (or more out to the next native tryptic site). Peptides were expressed as fusions to GST in C321.ΔA.ΔserB Escherichia coli, after which they were purified from the bacteria, cleaved from GST by PreScission protease treatment, and separated from the cleaved GST using molecular-weight cutoff spin columns. Peptides were incubated in vitro with PKCε, followed by tryptic digestion and LC-MS/MS analysis. Detailed peptide construction, cloning, expression, purification, in vitro kinase reaction, and proteomic workup methods are available in SI Appendix, Supplemental Materials and Methods; library members are available in Dataset S5.
Additional Reagents and Methods.
Additional reagents, cell lines, and methods are available in SI Appendix, Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We acknowledge Ionis Pharmaceuticals for providing the PKCε ASO, Drs. Carol Mercer and Sara Kozma for the p70S6K DKO cells, and Dr. Terence Wu in the West Campus Analytical Core for help in maintaining instruments. J.R. is supported by National Institutes of Health Grants R01GM117230, P01DK017433, and P30DK45735. G.I.S. is supported by National Institutes of Health Grants R01DK40936, R01DK113984, and P30DK45735. B.M.G. and K.W.B. are supported by National Science Foundation Graduate Research Fellowship Grant DGE1122492. M.C.P. is supported by National Institutes of Health Medical Scientist Training Program Grants T32GM007205 and F30DK104596.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.F.W. is a guest editor invited by the Editorial Board.
Data deposition: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository, https://www.ebi.ac.uk/pride/archive/, (dataset indentifier PXD010209).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804379115/-/DCSupplemental.
References
- 1.Perry RJ, Samuel VT, Petersen KF, Shulman GI. The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature. 2014;510:84–91. doi: 10.1038/nature13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petersen MC, Vatner DF, Shulman GI. Regulation of hepatic glucose metabolism in health and disease. Nat Rev Endocrinol. 2017;13:572–587. doi: 10.1038/nrendo.2017.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumashiro N, et al. Cellular mechanism of insulin resistance in nonalcoholic fatty liver disease. Proc Natl Acad Sci USA. 2011;108:16381–16385. doi: 10.1073/pnas.1113359108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magkos F, et al. Intrahepatic diacylglycerol content is associated with hepatic insulin resistance in obese subjects. Gastroenterology. 2012;142:1444–1446.e2. doi: 10.1053/j.gastro.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ter Horst KW, et al. Hepatic diacylglycerol-associated protein kinase Cε translocation links hepatic steatosis to hepatic insulin resistance in humans. Cell Rep. 2017;19:1997–2004. doi: 10.1016/j.celrep.2017.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samuel VT, et al. Mechanism of hepatic insulin resistance in non-alcoholic fatty liver disease. J Biol Chem. 2004;279:32345–32353. doi: 10.1074/jbc.M313478200. [DOI] [PubMed] [Google Scholar]
- 7.Samuel VT, et al. Inhibition of protein kinase Cepsilon prevents hepatic insulin resistance in nonalcoholic fatty liver disease. J Clin Invest. 2007;117:739–745. doi: 10.1172/JCI30400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen MC, et al. Insulin receptor Thr1160 phosphorylation mediates lipid-induced hepatic insulin resistance. J Clin Invest. 2016;126:4361–4371. doi: 10.1172/JCI86013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kikkawa U, Takai Y, Tanaka Y, Miyake R, Nishizuka Y. Protein kinase C as a possible receptor protein of tumor-promoting phorbol esters. J Biol Chem. 1983;258:11442–11445. [PubMed] [Google Scholar]
- 10.Aziz MH, et al. Protein kinase Cvarepsilon mediates Stat3Ser727 phosphorylation, Stat3-regulated gene expression, and cell invasion in various human cancer cell lines through integration with MAPK cascade (RAF-1, MEK1/2, and ERK1/2) Oncogene. 2010;29:3100–3109. doi: 10.1038/onc.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durgan J, Parker PJ. Regulation of the tumour suppressor Fbw7α by PKC-dependent phosphorylation and cancer-associated mutations. Biochem J. 2010;432:77–87. doi: 10.1042/BJ20100799. [DOI] [PubMed] [Google Scholar]
- 12.Meshki J, Caino MC, von Burstin VA, Griner E, Kazanietz MG. Regulation of prostate cancer cell survival by protein kinase Cepsilon involves bad phosphorylation and modulation of the TNFalpha/JNK pathway. J Biol Chem. 2010;285:26033–26040. doi: 10.1074/jbc.M110.128371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niture SK, Gnatt A, Jaiswal AK. Oncogene PKCε controls INrf2-Nrf2 interaction in normal and cancer cells through phosphorylation of INrf2. J Cell Sci. 2013;126:5657–5669. doi: 10.1242/jcs.133819. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Liu GS, Cohen MV, Mochly-Rosen D, Downey JM. Protein kinase C-epsilon is responsible for the protection of preconditioning in rabbit cardiomyocytes. J Mol Cell Cardiol. 1999;31:1937–1948. doi: 10.1006/jmcc.1999.1026. [DOI] [PubMed] [Google Scholar]
- 15.Ping P, Zhang J, Pierce WM, Jr, Bolli R. Functional proteomic analysis of protein kinase C epsilon signaling complexes in the normal heart and during cardioprotection. Circ Res. 2001;88:59–62. doi: 10.1161/01.res.88.1.59. [DOI] [PubMed] [Google Scholar]
- 16.Mayr M, et al. Proteomic and metabolomic analysis of cardioprotection: Interplay between protein kinase C epsilon and delta in regulating glucose metabolism of murine hearts. J Mol Cell Cardiol. 2009;46:268–277. doi: 10.1016/j.yjmcc.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Budas G, et al. Identification of EPKC targets during cardiac ischemic injury. Circ J. 2012;76:1476–1485. doi: 10.1253/circj.cj-11-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards TS, et al. Protein kinase C spatially and temporally regulates gap junctional communication during human wound repair via phosphorylation of connexin43 on serine368. J Cell Biol. 2004;167:555–562. doi: 10.1083/jcb.200404142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stawowy P, et al. Protein kinase C epsilon mediates angiotensin II-induced activation of beta1-integrins in cardiac fibroblasts. Cardiovasc Res. 2005;67:50–59. doi: 10.1016/j.cardiores.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Alvarez-Arias DA, Campbell KS. Protein kinase C regulates expression and function of inhibitory killer cell Ig-like receptors in NK cells. J Immunol. 2007;179:5281–5290. doi: 10.4049/jimmunol.179.8.5281. [DOI] [PubMed] [Google Scholar]
- 21.Ong ST, et al. Phosphorylation of Rab5a protein by protein kinase CE is crucial for T-cell migration. J Biol Chem. 2014;289:19420–19434. doi: 10.1074/jbc.M113.545863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajagopal S, et al. Site-specific regulation of CAV2.2 channels by protein kinase C isozymes betaII and epsilon. Neuroscience. 2009;159:618–628. doi: 10.1016/j.neuroscience.2008.12.047. [DOI] [PubMed] [Google Scholar]
- 23.Yang L, et al. Protein kinase C isoforms differentially phosphorylate Cav1.2 α1c. Biochemistry. 2009;48:6674–6683. doi: 10.1021/bi900322a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao BM, et al. Proteomic analysis of livers from fat-fed mice deficient in either PKCδ or PKCε identifies Htatip2 as a regulator of lipid metabolism. Proteomics. 2014;14:2578–2587. doi: 10.1002/pmic.201400202. [DOI] [PubMed] [Google Scholar]
- 25.Schäfer A, et al. The epoxyeicosatrienoic acid pathway enhances hepatic insulin signaling and is repressed in insulin-resistant mouse liver. Mol Cell Proteomics. 2015;14:2764–2774. doi: 10.1074/mcp.M115.049064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sah SP, Singh B, Choudhary S, Kumar A. Animal models of insulin resistance: A review. Pharmacol Rep. 2016;68:1165–1177. doi: 10.1016/j.pharep.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 27.Turner N, et al. Distinct patterns of tissue-specific lipid accumulation during the induction of insulin resistance in mice by high-fat feeding. Diabetologia. 2013;56:1638–1648. doi: 10.1007/s00125-013-2913-1. [DOI] [PubMed] [Google Scholar]
- 28.Aebersold R, Goodlett DR. Mass spectrometry in proteomics. Chem Rev. 2001;101:269–295. doi: 10.1021/cr990076h. [DOI] [PubMed] [Google Scholar]
- 29.Al-Hasani H, et al. Identification of Ser-1275 and Ser-1309 as autophosphorylation sites of the insulin receptor. FEBS Lett. 1997;400:65–70. doi: 10.1016/s0014-5793(96)01342-7. [DOI] [PubMed] [Google Scholar]
- 30.Tennagels N, Telting D, Parvaresch S, Maassen JA, Klein HW. Identification of Ser1275 and Ser1309 as autophosphorylation sites of the human insulin receptor in intact cells. Biochem Biophys Res Commun. 2001;282:387–393. doi: 10.1006/bbrc.2001.4589. [DOI] [PubMed] [Google Scholar]
- 31.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 32.The UniProt Consortium UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Francis GA, Fayard E, Picard F, Auwerx J. Nuclear receptors and the control of metabolism. Annu Rev Physiol. 2003;65:261–311. doi: 10.1146/annurev.physiol.65.092101.142528. [DOI] [PubMed] [Google Scholar]
- 34.Seth A, et al. The transcriptional corepressor RIP140 regulates oxidative metabolism in skeletal muscle. Cell Metab. 2007;6:236–245. doi: 10.1016/j.cmet.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fritah A, et al. Absence of RIP140 reveals a pathway regulating glut4-dependent glucose uptake in oxidative skeletal muscle through UCP1-mediated activation of AMPK. PLoS One. 2012;7:e32520. doi: 10.1371/journal.pone.0032520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imai S, Sakane F, Kanoh H. Phorbol ester-regulated oligomerization of diacylglycerol kinase delta linked to its phosphorylation and translocation. J Biol Chem. 2002;277:35323–35332. doi: 10.1074/jbc.M202035200. [DOI] [PubMed] [Google Scholar]
- 37.Imai S, Kai M, Yamada K, Kanoh H, Sakane F. The plasma membrane translocation of diacylglycerol kinase delta1 is negatively regulated by conventional protein kinase C-dependent phosphorylation at Ser-22 and Ser-26 within the pleckstrin homology domain. Biochem J. 2004;382:957–966. doi: 10.1042/BJ20040681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chibalin AV, et al. Downregulation of diacylglycerol kinase delta contributes to hyperglycemia-induced insulin resistance. Cell. 2008;132:375–386. doi: 10.1016/j.cell.2007.12.035. [DOI] [PubMed] [Google Scholar]
- 39.O’Shea JP, et al. pLogo: A probabilistic approach to visualizing sequence motifs. Nat Methods. 2013;10:1211–1212. doi: 10.1038/nmeth.2646. [DOI] [PubMed] [Google Scholar]
- 40.Nishikawa K, Toker A, Johannes FJ, Songyang Z, Cantley LC. Determination of the specific substrate sequence motifs of protein kinase C isozymes. J Biol Chem. 1997;272:952–960. doi: 10.1074/jbc.272.2.952. [DOI] [PubMed] [Google Scholar]
- 41.Barber KW, et al. Kinase substrate profiling using a proteome-wide serine-oriented human peptide library. Biochemistry. 2018;57:4717–4725. doi: 10.1021/acs.biochem.8b00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cenni V, et al. Regulation of novel protein kinase C epsilon by phosphorylation. Biochem J. 2002;363:537–545. doi: 10.1042/0264-6021:3630537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, Gao Z, Yin J, Quon MJ, Ye J. S6K directly phosphorylates IRS-1 on Ser-270 to promote insulin resistance in response to TNF-α signaling through IKK2. J Biol Chem. 2008;283:35375–35382. doi: 10.1074/jbc.M806480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah OJ, Hunter T. Turnover of the active fraction of IRS1 involves raptor-mTOR- and S6K1-dependent serine phosphorylation in cell culture models of tuberous sclerosis. Mol Cell Biol. 2006;26:6425–6434. doi: 10.1128/MCB.01254-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roux PP, et al. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem. 2007;282:14056–14064. doi: 10.1074/jbc.M700906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 47.Sancak Y, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 48.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 49.Tavares MR, et al. The S6K protein family in health and disease. Life Sci. 2015;131:1–10. doi: 10.1016/j.lfs.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 50.Hers I, Vincent EE, Tavaré JM. Akt signalling in health and disease. Cell Signal. 2011;23:1515–1527. doi: 10.1016/j.cellsig.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 51.Pende M, et al. S6K1−/−/S6K2−/− mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ueda Y, et al. Protein kinase C activates the MEK-ERK pathway in a manner independent of Ras and dependent on Raf. J Biol Chem. 1996;271:23512–23519. doi: 10.1074/jbc.271.38.23512. [DOI] [PubMed] [Google Scholar]
- 53.Hornbeck PV, et al. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Copps KD, White MF. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia. 2012;55:2565–2582. doi: 10.1007/s00125-012-2644-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Um SH, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 56.Copps KD, Hancer NJ, Qiu W, White MF. Serine 302 phosphorylation of mouse insulin receptor substrate 1 (Irs1) is dispensable for normal insulin signaling and feedback regulation by hepatic S6 kinase. J Biol Chem. 2016;291:8602–8617. doi: 10.1074/jbc.M116.714915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tremblay F, et al. Identification of IRS-1 Ser-1101 as a target of S6K1 in nutrient- and obesity-induced insulin resistance. Proc Natl Acad Sci USA. 2007;104:14056–14061. doi: 10.1073/pnas.0706517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, et al. Protein kinase C theta inhibits insulin signaling by phosphorylating IRS1 at Ser(1101) J Biol Chem. 2004;279:45304–45307. doi: 10.1074/jbc.C400186200. [DOI] [PubMed] [Google Scholar]
- 59.Ruvinsky I, et al. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19:2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cantley JL, et al. CGI-58 knockdown sequesters diacylglycerols in lipid droplets/ER-preventing diacylglycerol-mediated hepatic insulin resistance. Proc Natl Acad Sci USA. 2013;110:1869–1874. doi: 10.1073/pnas.1219456110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 62.Kettenbach AN, Sano H, Keller SR, Lienhard GE, Gerber SA. SPECHT—single-stage phosphopeptide enrichment and stable-isotope chemical tagging: Quantitative phosphoproteomics of insulin action in muscle. J Proteomics. 2015;114:48–60. doi: 10.1016/j.jprot.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alpert AJ. Electrostatic repulsion hydrophilic interaction chromatography for isocratic separation of charged solutes and selective isolation of phosphopeptides. Anal Chem. 2008;80:62–76. doi: 10.1021/ac070997p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.