Significance
Bacterial pathogens must sense cues in the host environment and adapt with appropriate transcriptional responses to cause infections. Although identifying global transcriptional changes in a pathogen during infections would reveal these mechanisms of success, such work in human infections is difficult and rare. Here we use samples from a controlled human model of enterotoxigenic Escherichia coli (ETEC) infection to identify ETEC’s global transcriptional response to the human host. We found ETEC senses environmental oxygen to coordinate virulence gene expression during human infections via the transcriptional regulator FNR, and that FNR regulates biofilm formation in ETEC. This work identified a global regulator of virulence genes in ETEC where toxin and adhesin expression is coordinated with pathogen proximity to the epithelium.
Keywords: ETEC, enterotoxigenic E. coli, FNR, toxin, cfa
Abstract
Enterotoxigenic Escherichia coli (ETEC) is a global diarrheal pathogen that utilizes adhesins and secreted enterotoxins to cause disease in mammalian hosts. Decades of research on virulence factor regulation in ETEC has revealed a variety of environmental factors that influence gene expression, including bile, pH, bicarbonate, osmolarity, and glucose. However, other hallmarks of the intestinal tract, such as low oxygen availability, have not been examined. Further, determining how ETEC integrates these signals in the complex host environment is challenging. To address this, we characterized ETEC’s response to the human host using samples from a controlled human infection model. We found ETEC senses environmental oxygen to globally influence virulence factor expression via the oxygen-sensitive transcriptional regulator fumarate and nitrate reduction (FNR) regulator. In vitro anaerobic growth replicates the in vivo virulence factor expression profile, and deletion of fnr in ETEC strain H10407 results in a significant increase in expression of all classical virulence factors, including the colonization factor antigen I (CFA/I) adhesin operon and both heat-stable and heat-labile enterotoxins. These data depict a model of ETEC infection where FNR activity can globally influence virulence gene expression, and therefore proximity to the oxygenated zone bordering intestinal epithelial cells likely influences ETEC virulence gene expression in vivo. Outside of the host, ETEC biofilms are associated with seasonal ETEC epidemics, and we find FNR is a regulator of biofilm production. Together these data suggest FNR-dependent oxygen sensing in ETEC has implications for human infection inside and outside of the host.
Enterotoxigenic Escherichia coli (ETEC) is a global Gram-negative diarrheal pathogen of the small intestine that infects up to 200 million people worldwide each year (1). ETEC infections result in ∼100,000 deaths annually and rely on a trio of classical ETEC virulence factors to cause disease (2–4). The prototypical ETEC strain H10407 harbors heat-labile (LT) and heat-stable (ST) enterotoxins along with the colonization factor antigen I (CFA/I) adhesin (5). The production of CFA/I is controlled by the transcriptional regulator CfaD, which activates the expression of the cfaABCE operon on virulence plasmid p948 (6). The outer membrane usher protein CfaC and periplasmic chaperon CfaA assemble the extracellular CfaB pillus with the CfaE adhesin tip (7, 8). Robust CFA/I adhesin production by strain H10407 occurs in CFA media, a namesake standard ETEC complex growth media.
Alongside the CFA/I adhesin, H10407 also harbors the LT enterotoxin and two homologs of the ST enterotoxin denoted as Sta1 and Sta2 (3, 9, 10). The enzymatic activity of these enterotoxins results in water efflux from host intestinal cells which produces profuse diarrhea (11, 12). sta1 and sta2 are found on plasmids p666 and p948, respectively. Despite their conserved functional role, sta1 and sta2 have different promoters and their expression can be inversely regulated (13). Recent work has shown the cAMP receptor protein (CRP), in response to changes in extracellular glucose concentrations, can oppositely regulate sta1 and sta2 promoter::lacZ fusion reporter constructs in E. coli K-12 (13). The significance of differential regulation between two ST toxin promoters remains unclear, although it demonstrates how a single environmental factor can have a complex effect on virulence factor expression in ETEC. LT toxin is a heterohexameric, A-B subunit enterotoxin encoded by the eltAB operon on virulence plasmid p666 (14). It is functionally and structurally similar to the diarrhea-inducing cholera toxin produced by Vibrio cholerae (14, 15).
Although ETEC H10407 contains all of these classical virulence factors, only one adhesin and one enterotoxin is required for symptomatic disease (16). ETEC can also contain a large group of nonclassical (17) virulence factors that have been shown to contribute either directly or indirectly to virulence-related phenotypes in E. coli. Notably, genes etpABC (18) and tibAB (19) were originally described in ETEC, and have been shown to enable host cell adhesion in vivo and host cell adhesion/invasion in vitro, respectively.
Decades of research on classical virulence factor gene regulation in ETEC has demonstrated how over 20 environmental conditions influence expression of individual virulence genes, including pH, bile, bicarbonate, osmolarity, and glucose (6, 13, 20). Understanding how ETEC responds to these signals in the mammalian intestinal tract for effective colonization and transmission is challenging, particularly considering just one of these factors can alter expression of virulence factors in opposing ways. For example, not only does glucose oppositely regulate ST homologs, but it can also oppositely regulate ST and LT toxins using promoter::lacZ fusion assays (13). To our knowledge, there are no reports of a common regulation for all classical virulence factors simultaneously.
Successful intestinal colonization requires appropriate bacterial transcriptional adaptation to the host environment. We therefore examined the ETEC transcriptome in human infection samples to understand how ETEC responds to the host in vivo. The host-specific response has important implications for therapy design, as it can reveal that high-priority drug and vaccine targets may have unexpected expression patterns in vivo (21). Here we used samples from a controlled human infection model to examine the global transcriptome of ETEC H10407 directly in human infection samples (ClinicalTrials.gov Identifier NCT01922856).
We performed RNA sequencing directly on the stool samples of five infected volunteers in the absence of therapeutic treatment. Many studies have investigated the expression of individual ETEC genes in infected stool samples; however, variation between wild-type isolates can make interpretation of these data challenging. In this study, each volunteer was infected simultaneously from the same H10407 inoculum. This allowed us to study biological replicates of human infection without confounding differences between strains or inoculums, and further, we performed a global analysis of ETEC gene expression directly in the infected samples using next-generation high-throughput RNA sequencing.
We were surprised to find the LT toxin genes and CFA/I adhesin gene expression were down-regulated in infected stool samples compared with laboratory growth in CFA media. After considering the stool samples represented the anaerobic large intestinal environment, and that little work has been done to characterize ETEC in low-oxygen conditions, we examined the ETEC transcriptome during growth in microaerobic and anaerobic atmospheres. Besides the anaerobic large intestinal environment, anaerobic conditions in the proximal small intestine are fortified by bacterial overgrowth (22) and the distal small intestinal lumen naturally has near-zero levels of oxygen (23). Remarkably, we discovered anaerobic in vitro growth fully replicated the virulence gene expression pattern seen in the stool samples, indicating environmental oxygen played an important role in understanding virulence factor expression in vivo. Anaerobic growth in vitro results in a significant decrease in expression of cfaD, the cfa operon, and the eltAB operon, and an increase in sta2 expression compared with aerobic growth. We went on to determine that the effect of oxygen on virulence gene expression is dependent on the oxygen-sensitive transcriptional regulator fumarate nitrate reduction (FNR) regulator (24, 25). Deletion of fnr in H10407 resulted in a significant increase in expression of all classical virulence factor genes, including both sta1 and sta2, indicating FNR activity results in a global repression of virulence gene expression in ETEC.
We also found that H10407 produces significantly more biofilm when grown anaerobically compared with aerobic growth, and that a Δfnr H10407 strain produces significantly more biofilms than wild type under anaerobic conditions. In afflicted areas, ETEC biofilm formation on abiotic surfaces is correlated with seasonal epidemics (26), which suggests FNR-dependent oxygen sensing may contribute to human ETEC infection both inside and outside of the host.
Overall, by examining the transcriptome of ETEC in infection samples from a controlled human infection model, this study identified an environmental factor and corresponding transcriptional regulator that influence global virulence and biofilm regulation. This work suggests the low-oxygen environment of the intestinal lumen likely represses ETEC virulence gene expression in vivo—particularly when bacterial overgrowth fortifies anaerobic conditions in the proximal small intestine (22, 27), in the distal small intestine where luminal oxygen levels are near zero (23), and in the anaerobic large intestine during transmission (23). However, oxygen seepage from intestinal epithelial cells is sensed by E. coli via FNR in vivo, and the highest oxygen concentrations exist nearest the epithelium (28, 29). Therefore, ETEC virulence gene expression is likely repressed in the low-oxygen lumen and is relieved when the oxygen-intolerant FNR is inactivated near the epithelium.
Controlled Human Infection Samples and RNA Sequencing
Transcriptional adaptation to the host environment is essential for intestinal pathogens to colonize and cause disease. To understand how ETEC adapts to the human host, we characterized the transcriptome of ETEC H10407 directly in infected diarrhea samples taken from volunteers in a controlled human infection model (ClinicalTrials.gov Identifier NCT01922856). The same inoculum was used to infect volunteers at 1.2 × 107 colony forming units (cfu) each. The infected samples used in this study came from volunteers who were included as naïve, unvaccinated infection controls in the infection model. These volunteers were not prescribed a fixed diet during their infection.
RNA extracted from stool mostly represents RNA derived from host cells, the native microbiome, and food waste. To specifically identify ETEC transcripts in the dataset, reads were trimmed to 21 bases long and mapped with 100% exact homology to the H10407 reference genome as previously described for similarly complex samples (21). To be sure commensal E. coli were not influencing our RNA-sequencing data, we measured the ratio of mapped ETEC chromosomal transcripts to transcripts that mapped to virulence plasmids. If a volunteer had a prominent commensal E. coli colonization, which would likely produce transcripts that are highly homologous to ETEC chromosomal transcripts, we expected ETEC chromosomal transcripts would be overrepresented compared with ETEC-specific virulence plasmid transcripts. We did not find this to be the case for any of our sequenced samples.
Environmental Oxygen Explains in Vivo Virulence Gene Expression
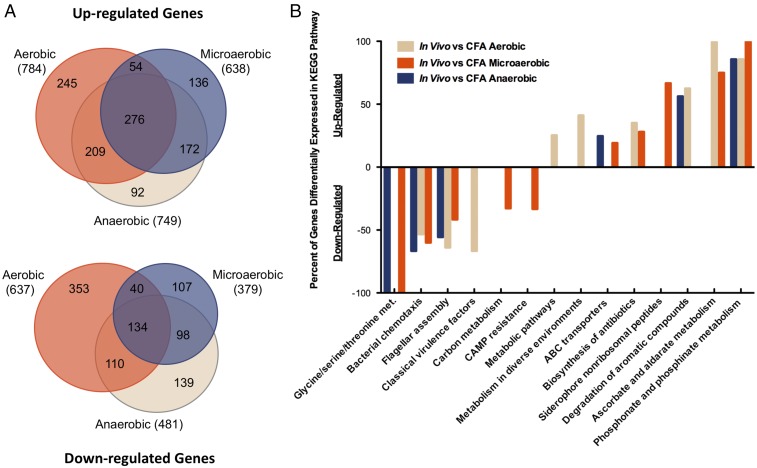
We determined the ETEC transcriptome directly in five infected diarrhea samples, each from different volunteers, which together as a sample group represent our in vivo transcriptome. A total of 784 genes were up-regulated and 637 genes were down-regulated in vivo compared with aerobic midlog phase growth in CFA broth (Fig. 1A and Datasets S1 and S2). Given the close association with the host, we hypothesized classical virulence factor genes would be up-regulated in the stool vs. in vitro growth. Surprisingly, classical virulence factors were largely down-regulated in stool compared with standard laboratory growth, including cfaD, the cfa operon, and eltAB (Figs. 1B and 2). Stool is frequently used as a proxy for understanding the state of the intestinal environment, and a hallmark of the intestinal tract is low availability of oxygen (29). The difference between environmental atmospheres was a major discrepancy between our aerobically grown laboratory comparison and ETEC in infected stool. Despite the in vivo relevance of growth in limited oxygen atmospheres, we found little published research describing ETEC in such conditions. To address this, we grew biological triplicates of ETEC H10407 in microaerobic (85% N, 10% CO2, 5% O2) and anaerobic (95% N, 5% H, 0% O2) atmospheres to midlog phase with shaking (SI Appendix, Fig. S1) and compared these low-oxygen transcriptomes to the in vivo stool transcriptomes (Figs. 1 and 2 and Dataset S1).
Fig. 1.
ETEC gene regulation in vivo. (A) The number of up-regulated or down-regulated genes in infected stool compared with midlog phase in vitro CFA broth growth in the indicated atmospheres. Microaerobic atmosphere was 85% nitrogen, 10% CO2, 5% O2. Anaerobic atmosphere was 95% nitrogen, 5% hydrogen, 0% oxygen. (B) KEGG orthology pathways statistically significantly (P < 0.05) represented in genes differentially regulated in vivo (fold change > |5|, FDR P < 0.05) using a hypergeometric test. Classical virulence factors were manually curated into our KEGG pathway list to be included in the analysis. In vivo samples represent five infected diarrhea samples from different human volunteers and in vitro conditions each represent three independent biological replicates.
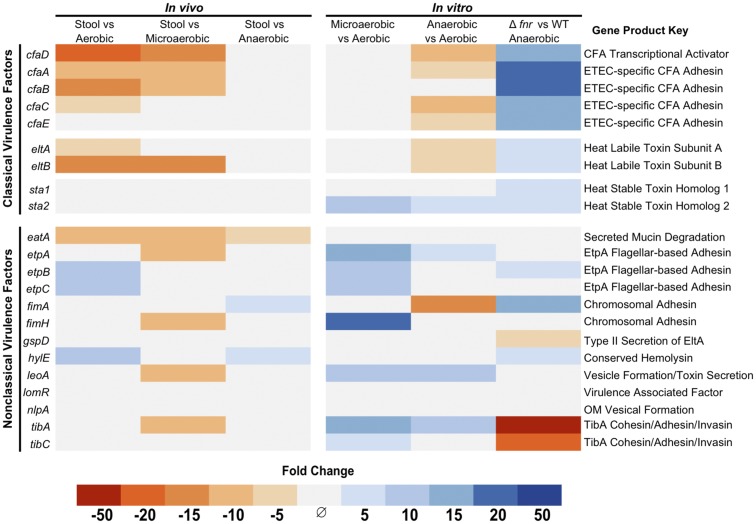
Fig. 2.
Oxygen and FNR-dependent virulence gene expression in ETEC. Differentially regulated (fold change > |3|, FDR-corrected P < 0.05) ETEC virulence genes in vivo compared with in vitro growth in different atmospheric oxygen concentrations. In vitro anaerobic growth replicated the in vivo expression pattern of classical ETEC virulence factors. In vivo samples represent five infected diarrhea samples from different human volunteers and in vitro conditions each represent a group of three independent biological replicates.
Remarkably, growth under in vitro anaerobic conditions led to similar virulence factor expression profiles as seen in stool samples, with no statistically significant difference in expression of any classical virulence factor between the two conditions (Fig. 2 and Dataset S1). This suggests understanding ETEC’s response to environmental oxygen is important for understanding ETEC virulence gene regulation in vivo. Notably, the expression of both ST toxin genes did not differ in vivo compared with any environmental oxygen concentration. This lack of statistical significance likely reflects variation in the expression of sta genes between the complex patient samples (SI Appendix, Fig. S2). However, in controlled in vitro growths, sta2 gene expression is influenced by environmental oxygen while sta1 is not (Fig. 2). Although highly homologous, the differences in nucleotide sequence between sta1 and sta2 ensured their expression could be independently determined when using our mapping parameters, and contrasting regulation between these two genes has been previously established in promoter::lacZ fusion assays (13). Overall, we found that simply altering environmental oxygen alone can greatly influence virulence factor expression in ETEC, with CFA, ST, and LT toxin genes all showing altered expression in response to changes in environmental oxygen (Fig. 2).
FNR Transcriptional Regulator Globally Influences Virulence Factor Expression
To better understand how ETEC sensing of environmental oxygen is linked with virulence gene expression, we considered the oxygen-sensitive transcription factor FNR. FNR is a well-characterized dimeric transcription factor which can be directly inactivated by oxygen (24, 25). In oxygenated conditions, an exposed Fe-S cluster in FNR is oxidized which eliminates the ability of FNR monomers to dimerize. The activity of antioxidant enzymes such as IscS can reverse this process, restoring FNR protein activity (30). In this way, FNR activity is directly sensitive to environmental oxygen in a reversible manner.
We therefore hypothesized the anaerobic virulence factor repression was dependent on FNR activity. To test this, we used recombineering to produce a Δfnr H10407 strain and compared the transcriptomes of three biological replicates of wild type and Δfnr H10407 grown anaerobically to midlog phase in CFA broth. We found a global increase of ETEC virulence factor expression in the Δfnr background (Fig. 2 and Dataset S1). Interestingly, both sta genes were up-regulated in this background, indicating that FNR can influence their expression despite limited changes between aerobic and anaerobic growth in wild-type bacteria.
FNR Binds to Classical Virulence Factor Promoter Regions
To examine whether FNR was directly influencing virulence factor gene expression, we used the online database Prodoric (31) to determine whether there were FNR binding sites in the upstream regions of virulence genes. We found multiple predicted FNR binding sites in the promoter regions of all classical virulence genes (SI Appendix, Figs. S3–S6). Using electrophoretic mobility shift assays, we found FNR was capable of binding to the promoter regions of cfaD, eltA, sta1, and sta2 in vitro (SI Appendix, Figs. S3–S6). This is consistent with the Δfnr RNA-sequencing data and direct FNR repression of classical virulence genes and operons.
However, FNR has a well-defined role in regulating low-oxygen metabolism genes and therefore has a broad influence on cell physiology (SI Appendix, Fig. S7) (24). Consequently, it is possible that FNR is additionally altering virulence factor expression by influencing other known or unknown regulatory factors, although our data suggest this is unlikely. Sensing of glucose concentrations via the CRP transcription factor was recently shown to influence transcription of promoter::lacZ fusions, and in enterohemorrhagic E. coli (EHEC), CRP expression increases when grown anaerobically (32). However, we did not find differential expression of CRP (ETEC_3608) when wild-type ETEC or Δfnr was grown anaerobically (Dataset S1).
Osmolarity was also shown to regulate expression of virulence promoter::lacZ fusions via the genome-binding protein HNS (13), and HNS was significantly down-regulated threefold during anaerobic growth compared with aerobic growth (Dataset S1). HNS has been shown to influence FNR and CRP binding to promoter regions (24) and directly represses eltAB, sta1, and sta2 promoter::lacZ reporter fusion expression in an E. coli K-12 background (13). Therefore, decreased expression of HNS during anaerobic growth could relieve this direct repression and result in increased expression of toxin genes. However, only sta2 showed increased expression during anaerobic growth, and HNS expression was not altered in the Δfnr background compared with wild type. Interestingly, previous work with promoter::lacZ fusions showed sta2 was also the only toxin gene to be repressed by both direct HNS-dependent repression and HNS-dependent inhibition of promoter activation. This stronger reliance on HNS regulation may explain why sta2 was the only virulence factor to have increased expression while HNS expression is decreased. Previous work also showed HNS repression of an eltAB operon promoter::lacZ fusion in a K-12 background. However, despite a threefold decrease in HNS expression during anaerobic growth, we did not see relief of this repression in ETEC, and our data show the down-regulation of eltAB is FNR dependent.
Beyond sensing oxygen, glucose, and osmolarity, ETEC is likely integrating other in vivo signals to regulate virulence factor expression, including previously described bile and bicarbonate, and interaction with epithelial cells in vitro (33). Our data show that without additional manipulations, simply changing environmental oxygen influences adhesin, LT, and ST toxin gene expression, and that deletion of the oxygen-sensitive FNR transcription factor results in a global increase in expression of all classical virulence factor genes.
FNR Is a Regulator of ETEC Biofilm Formation
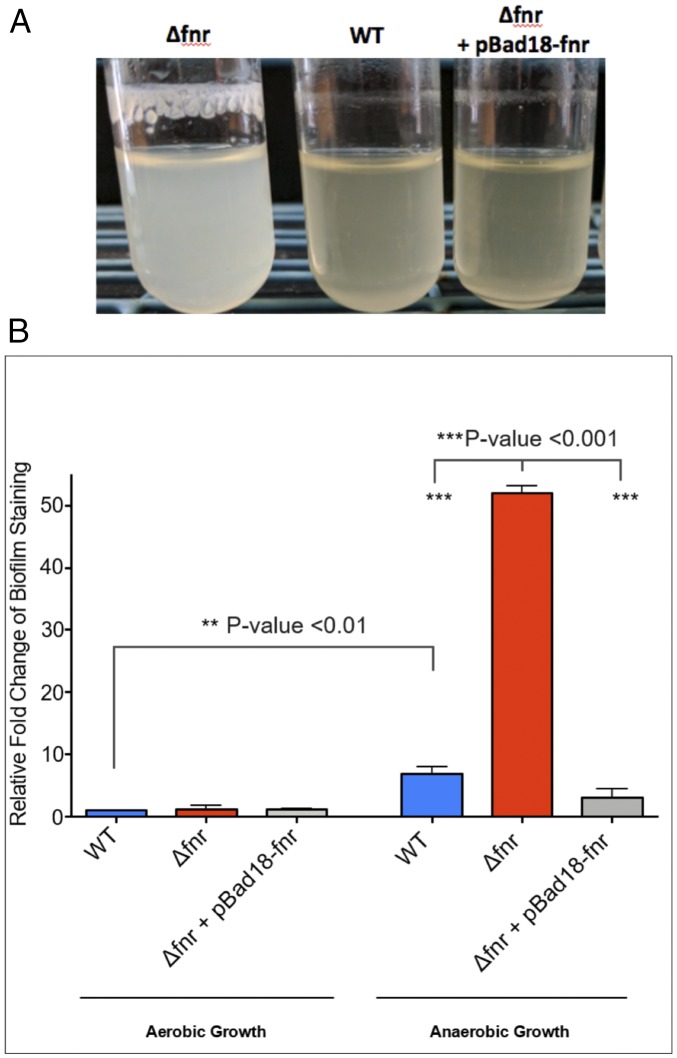
Biofilm production provides bacteria protection from environmental stressors inside and outside of the host and has been well described in many pathogens (34). In ETEC, biofilm production on abiotic surfaces within the homes of endemic areas is correlated with seasonal ETEC epidemics (26). While characterizing ETEC in low-oxygen conditions, we unexpectedly observed Δfnr H10407 spontaneously produced prominent biofilms during anaerobic overnight growth in CFA broth at 37 °C, despite vigorous shaking (Fig. 3). Quantitation of biofilm production via crystal violet staining revealed biofilm production increased 10-fold compared to the wild type, and this phenotype was complemented when FNR expression was restored via expression from an inducible plasmid. More broadly, we found wild-type ETEC H10407 biofilm production increased 5-fold when grown anaerobically compared with aerobic growth.
Fig. 3.
FNR-regulated biofilm formation in ETEC. (A) A picture of spontaneously occurring biofilm production when H10407 is grown anaerobically at 37 °C with shaking. Image is representative of more than 10 independent replicates. An fnr mutant produces more biofilm than wild type and this phenotype can be complemented by expressing the fnr gene from the pBad18 plasmid in the fnr:kan mutant background. All depicted growth included 0.2% arabinose for consistency with the induced complemented strain. (B) Quantification of ETEC biofilm formation exemplified in Fig. 5A using optical density readings of crystal violet-stained biofilms as described in Materials and Methods. All growth represents three independent biological replicates grown with 0.2% arabinose for consistency with the induced complementation strain. The relative amounts of biofilm produced by various ETEC H10407 strains in the indicated conditions are shown compared with wild-type ETEC H10407 biofilm production, which is set as 1 as a reference. Statistical significance was determined by one-way ANOVA with a Tukey multiple comparison test (P values <0.05 were considered significant) and error bars represent the SEM.
As expected, the FNR mutant produces wild-type levels of biofilm when grown aerobically. However, anaerobic biofilm production in the fnr mutant was increased 50-fold compared with the wild-type strain grown aerobically. Together, these data depict an interesting dynamic where ETEC can readily grow as a biofilm in an anaerobic environment, yet activation of FNR in this condition limits the extent of biofilm production.
Bacteria rely on a variety of mechanisms to form biofilms in response to changes in their environment (35). To identify FNR-influenced genes that contribute to anaerobic biofilm formation in ETEC, we sought to identify up-regulated genes in Δfnr H10407 that have been previously associated with biofilm formation. The ETEC TibAB adhesin has been shown to contribute to cohesion and biofilm formation (19), but unexpectedly, these genes are resoundingly down-regulated in the Δfnr strain (Fig. 2), suggesting they are unlikely to be responsible for increased biofilm formation. Alternatively, the main structural subunit of the Fim adhesin, fimA, was up-regulated >10-fold. Fim-based adhesion in ETEC occurs in vivo and in vitro via a specific binding of the adhesive tip protein FimH to d-mannose (36, 37). Therefore, exogenous d-mannose quenches Fim adhesive ability and disrupts biofilm formation, and sensitivity to d-mannose is frequently used as the standard to examine Fim-dependent biofilm formation or cellular agglutination (37–39). To see whether d-mannose-dependent adhesion was contributing to biofilm formation in Δfnr H10407, we measured biofilm production in the presence of increasing concentrations of d-mannose. A concentration-dependent significant decrease in biofilm production was seen in the presence of d-mannose, suggesting the Fim adhesin contributes to biofilm formation in ETEC (SI Appendix, Fig. S8).
ETEC Transcriptome in Human Infection Samples
Beyond virulence factors, we defined ETEC genes and pathways that were differentially regulated in vivo based on comparisons to different environmental oxygen conditions. Over 1,000 genes are differentially regulated in vivo compared with the atmospheric conditions we tested (Fig. 1A and Datasets S1 and S2). In vitro microaerobic growth most closely matched the infected stool transcriptome, while aerobic growth was the most different (Fig. 1A and Datasets S1 and S2). A statistical analysis of pathways that are differentially regulated in vivo, per environmental oxygen comparison, show virulence factor expression differences were highly dependent on atmospheric oxygen growth while chemotaxis and motility pathways were not (Fig. 1B).
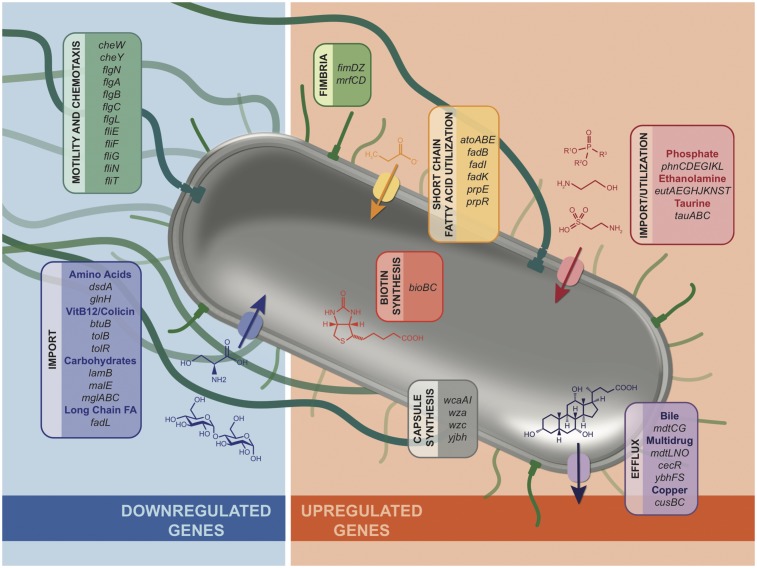
Despite the differences between atmospheres, 361 genes were differentially regulated in vivo independently of environmental oxygen changes (Fig. 1A and Dataset S2). Shared functional roles clearly emerged while examining these data by hand (Fig. 4). We found that many nutrient acquisition pathways that have been important for intestinal colonization in small animal models are significantly differentially regulated in human infection samples. For example, short chain fatty acid utilization has been shown to be crucial for colonization and/or survival of some intestinal microbes (40), and we see short chain fatty acid utilization genes in ETEC are strongly up-regulated in human infection. Congruently, the long chain fatty acid import protein fadL is down-regulated in vivo.
Fig. 4.
ETEC genes that showed differential gene expression in vivo despite the in vitro atmosphere comparisons (fold change > |3|, FDR-corrected P < 0.05), manually curated to highlight genes with clear functional roles. All genes listed showed significant gene expression changes in vivo compared with each of three in vitro atmospheric types of growth (aerobic, microaerobic, and anaerobic). Therefore, other in vivo signals likely influence the differential expression of these genes. See Dataset S1 for all changes in gene expression.
Similarly, striking regulation is seen in ethanolamine utilization, with up-regulation of nine genes involved in uptake and utilization of ethanolamine during human infection. Host intestinal epithelial cell turnover releases ethanolamine into the intestinal lumen, and in EHEC and Salmonella, ethanolamine is used as a nitrogen source to create a competitive advantage over the microbiota to establish an infection (41, 42). These data jointly suggest ETEC may be utilizing ethanolamine during human infection in a similar manner. Biotin synthesis was also up-regulated in vivo, and in Salmonella, biotin synthesis is up-regulated upon epithelial cell infection, and deletion of the biotin synthesis gene bioB resulted in attenuated mouse colonization (43).
Beyond nutrient acquisition genes, many genes that play a role in resistance to antibiotics or host factors are differentially regulated in vivo (Fig. 4). These include the mucin degradation gene eatA; fimbrial genes fimD, fimZ, mrfC, and mrfD; and invasion/secretion-associated genes invH, mrfC and mrfD, and cexE. Genes involved in multidrug efflux and resistance were also up-regulated, including bile acid and novobiocin/fosfomycin resistance genes mdtC/mdtG, sulfa drug resistance genes mdtN and mdtO, and cefoperazone and chloramphenicol resistance genes ybhF and ybhS.
Common Gene Regulation Between Uropathogenic E. coli and ETEC During Human Infection
Few transcriptomes of bacterial pathogens in human infection samples exist. An exemplary dataset is that of uropathogenic E. coli (UPEC) (44), where an infected urine sample typically represents a monoculture of urinary tract-infecting bacteria. This minimally complex sample enables efficient sequencing and analysis of in vivo pathogen transcriptomes. Interestingly, many of the up-regulated ETEC genes in infected stool were also up-regulated in infected urine samples collected from human UPEC infections (44). Notably, ethanolamine utilization, phosphonate utilization, taurine import, colanic acid synthesis, and copper efflux were all major up-regulated pathways despite the differences between these pathogens and site of infection. As most E. coli urinary tract infections are established by E. coli found in fecal flora (45, 46), these data suggest a common transcriptional response to the host across E. coli strains and infection sites in humans.
Discussion
We examined ETEC H10407 directly in human infection samples from a controlled human infection model and found atmospheric oxygen influences virulence gene expression via the FNR transcription factor. Previously, ETEC has not been well characterized in low-oxygen conditions and the role of FNR in ETEC virulence and physiology was unknown.
We found CFA/I adhesin genes, including the master regulator cfaD, and the LT toxin gene eltAB, are down-regulated when ETEC is grown anaerobically. Consistent with FNR-dependent repression, these genes are up-regulated in a Δfnr strain, including both ST genes in H10407, sta1 and sta2. This strikingly shows that a single regulator in ETEC can globally influence virulence factor expression, and in vitro, FNR can bind to the promoter regions of each of these genes. Similarly, anaerobic growth in vitro, which represents the near-zero oxygen availability status of the distal small intestine and anaerobic large intestine, as well as bacterial overgrowth in the proximal small intestine, mimicked the virulence gene expression profile seen in stool samples.
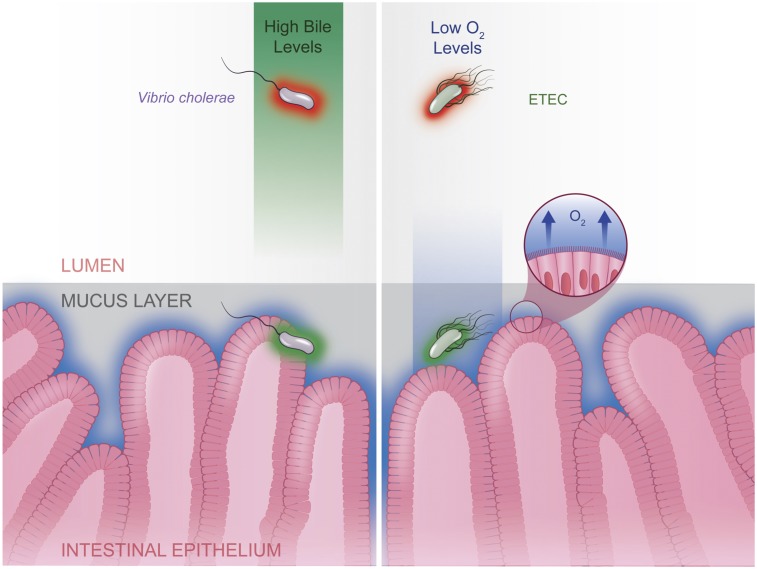
Low-oxygen availability is a hallmark of the intestinal environment, and in vivo, E. coli can respond to their proximity to the epithelium by sensing oxygen seepage from host cells (28). Together these data suggest ETEC virulence factor expression may be repressed by FNR in the low-oxygen lumen, and that repression is relieved when ETEC nears the intestinal epithelium and oxygen inactivates FNR (Fig. 5). Convergent evolution across intestinal pathogens appears to have favored similar methods of host-cell proximity sensing. For example, the diarrheal pathogen V. cholerae, also senses a host cue (bile) to repress cholera toxin expression in the lumen (47), which is then relieved when in close proximity to host cells (Fig. 5).
Fig. 5.
A model of oxygen-dependent ETEC H10407 virulence gene regulation. The luminal environment inhibits virulence factor expression (red) which is relieved (green) within close proximity to epithelial cells. Our study suggests ETEC directly senses environmental oxygen via the oxygen-sensitive transcription factor FNR, which results in repressed virulence gene expression in low-oxygen environments like the intestinal lumen. When ETEC nears the intestinal epithelium, where oxygen seepage from host cells creates a zone of increased oxygen concentrations, FNR is inactivated, which relieves virulence gene repression nearest to the host epithelium. Other intestinal pathogens, such as V. cholerae, use a similar proximity-sensitive approach to regulating virulence genes within the host. In V. cholerae, bile has been shown to repress cholera toxin expression, and similar to oxygen, bile exists as a gradient within the intestinal environment. In both cases, concentrations of host-derived factors result in repression of virulence gene expression in the lumen and relief of the repression when near the target epithelium.
FNR and the ability to switch between aerobic and anaerobic metabolism is known to be essential for E. coli colonization (48), and other intestinal pathogens also utilize FNR to regulate select colonization factors (28). The FNR regulon has been well defined in laboratory E. coli K-12 and interplays with other regulator proteins such as HNS and the related CRP transcriptional regulator which responds to cAMP in low-glucose conditions (24). Recent work has demonstrated the activity of both HNS and CRP contribute to virulence gene expression in aerobic conditions using promoter::lacZ fusion assays in K-12 background (13), and certainly, all may play a role in the low-glucose, low-oxygen environment of the intestinal tract.
Beyond classical virulence factor expression changes, we found many cellular processes were differentially regulated in vivo. These include ethanolamine utilization, short chain fatty acid utilization, phosphonate utilization, and copper efflux. Notably, previous work demonstrated these same processes were also up-regulated in human UPEC infections. Further, all of these have been shown to contribute to fitness in small animal models of infection, and here we found they respond transcriptionally to the human host during ETEC infection.
Besides offering insight into ETEC gene regulation in vivo, these data have implications for studies that examine ETEC virulence factor expression in collected infected samples (49, 50). Our data suggest stool samples should be maintained or preserved with consideration for atmospheric oxygen to be sure differences in virulence factor expression between samples or strains cannot be attributed to differences in oxygen exposure after collection.
During our characterization of ETEC in low-oxygen conditions we noticed a striking oxygen-sensitive biofilm production phenotype. Only a few studies have investigated ETEC biofilm formation, although ETEC biofilms in home water reservoirs in endemic areas are correlated with seasonal epidemics (26). Here we show robust ETEC biofilms can be produced overnight when Δfnr H10407 is grown anaerobically in CFA media. Together, this demonstrates an interesting FNR-dependent link between ETEC’s lifestyle in the environment and in the host. This study represents the joint efforts between clinicians and researchers to define an in vivo perspective of a worldwide human pathogen.
Materials and Methods
ETEC Human Infection Model.
Volunteers for enterotoxigenic E. coli H10407 infection (ClinicalTrials.gov Identifier NCT01922856, Navy Medical Research Center IRB NMRC.2013.0011) were healthy adults from the mid-Atlantic region of the United States. Volunteers signed informed consent forms before participating and allowed for future use of samples produced during the study. Volunteers were either male or female between 18 and 50 y of age, were in general good health, without significant medical history or previous exposure to ETEC or V. cholerae, did not have an abnormal stool patterns, and did not regularly use laxatives or antacids. On the day of the challenge, volunteers drank 120 mL of bicarbonate buffer, followed by 30 mL of bicarbonate buffer containing 1.2 × 107 cfu of virulent ETEC H10407. Inoculum bacteria were initially grown overnight at 37 °C on 3 CFA agar plates supplemented with bile salt, and then suspended in PBS. PBS resuspension was kept on ice until being incorporated into the final bicarbonate buffer inoculum given to volunteers. Inoculum concentration was initially estimated via optical density 600nm readings on the day of the challenge and then confirmed using colony forming unit enumeration on LB agar plates after overnight incubation at 37 °C.
Infected Sample Preparation for RNA Sequencing.
ETEC H10407-infected diarrhea was weighed and added 1:1 in RNAlater reagent as quickly as possible after production and frozen at −80 °C. We worked with only infected diarrhea samples from volunteers who were not vaccinated against ETEC, were not receiving antibiotic treatment at the time of production, did not have a restricted diet, and were not receiving prescribed rehydration therapy. Preserved infected samples were thawed in a secondary container on ice and total RNA was extracted via TRIzol-chloroform phase separation. DNA was removed (Turbo DNA-Free DNase kit, Ambion) and rRNA was depleted (Ribo-Zero human and bacterial rRNA removal kits, Illumina). Libraries were built for Illumina next-generation sequencing (Ultra Directional RNA Library Prep Kit for Illumina, New England Biolabs).
In Vitro ETEC Sample Preparation for RNA Sequencing.
Enterotoxigenic E. coli strain H10407 was grown at 37 °C in either standard aerobic atmospheric conditions, or in an atmosphere-controlled chamber for low-oxygen growth (Coy Laboratory). The microaerobic atmosphere used was 85% nitrogen, 10% carbon dioxide, and 5% oxygen, while the anaerobic atmosphere was 85% nitrogen, 10% carbon dioxide, and 5% hydrogen. For bacterial growth, CFA agar plates were streaked from a frozen stock of ETEC H10407 and grown overnight at 37 °C in the indicated atmosphere. Single colonies were used to inoculate 5-mL aliquots of CFA broth in triplicate and grown with shaking in the indicated atmosphere. For low-oxygen atmosphere growth, CFA broth aliquots were equilibrated with the atmosphere overnight with shaking at 37 °C before inoculation. For RNA sequencing, all cultures were grown in triplicate to midlog phase in the respective atmosphere, harvested via centrifugation within the respective atmosphere, and resuspended in TRIzol reagent within the respective atmosphere for immediate RNA extraction. DNA was then removed (Turbo DNA-Free DNase Kit, Ambion), rRNA was removed (Ribo-Zero Bacterial rRNA Removal Kit, Illumina), and the samples were prepared for Illumina sequencing using the TruSeq Stranded mRNA Library Kit (Illumina).
Illumina RNA Sequencing and Analysis.
Infected samples are complex and ETEC RNA represents a small portion of total sample RNA. To identify ETEC-specific transcripts, we trimmed sample reads to 21 base pairs in length and then mapped them with 100% exact sequence homology to the ETEC H10407 reference genome using CLC Genomics WorkBench software, as has been previously described with similarly complex samples (21). At least 3 million reads were mapped to the H10407 reference genome per sample. Five infected diarrhea samples each from different volunteers were sequenced and analyzed individually, and then together as a group represented our in vivo condition for analysis. For in vitro samples, reads were trimmed and mapped in an identical manner, with at least 15 million reads mapping to the reference genome, and all in vitro conditions represent a group of three biological replicates.
CLC Genomics WorkBench (Qiagen) was used to determine differential gene expression fold changes between conditions and a false discovery rate (FDR)-corrected P value was used to determine statistical significance. We considered a significant change in expression between groups to be a weighted proportions fold change > |3| with a FDR P value of <0.05.
To determine whether any native commensal E. coli were influencing the mapped reads from in vivo samples, we compared the ratio of chromosomally mapped reads vs. reads mapped to ETEC-specific virulence plasmids. We assumed commensal E. coli genomes would share extensive homology to the ETEC chromosome, and therefore if commensal E. coli were influential, chromosomal reads would be overrepresented compared with reads mapped to ETEC virulence plasmids. We did not find chromosomal reads to be overrepresented, and therefore we did not see a large representation of non-ETEC reads mapping to the ETEC reference genome.
ETEC Recombineering and Complementation.
To generate a knockout of fnr in ETEC H10407, Lambda Red-mediated gene replacement was used as previously described in E. coli (51). Briefly, the kanamycin cassette from the pkD4 plasmid was PCR amplified with 50 base pair flanking regions, which were homologous to the immediate up- and downstream regions of the chromosomally encoded fnr gene. This linear PCR product was cleaned using the QIAquick PCR Purification Kit and electroporated into H10407 containing the pkD46 plasmid, which produces the Lambda Red recombinase and mediates the homologous recombination of the fnr locus with linear PCR product. Recombinant bacteria were selected for on kanamycin LB agar, and fnr:kan mutants were verified by PCR, Sanger sequencing, and by observing an expected growth defect in low-oxygen conditions compared with wild type (SI Appendix, Fig. S1). fnr:kan was maintained on agar plates and in liquid medium in the presence of kanamycin.
The fnr complementation plasmid (pBad18-fnr) is an arabinose-inducible pBad18 plasmid carrying a chloramphenicol resistance cassette and was built using Gibson assembly. pBad18-fnr was electroporated into the fnr:kan H10407 genetic background. Complementation was determined by PCR verification of the plasmid, alongside rescue of the fnr:kan anaerobic growth defect (SI Appendix, Fig. S1) as well as rescue of the increased biofilm formation phenotype (Fig. 3). fnr:kan pBad18-fnr was maintained in the presence of kanamycin and chloramphenicol and with 0.2% arabinose when indicated.
Electrophoretic Mobility Shift Assays.
FNR activity is sensitive to oxygen, making most standard in vitro assays, including purification, difficult. To address this, we used a well-characterized FNR variant, (FNRD154A)2, which is physically linked as an active dimer and is therefore active in aerobic environments. This variant has been validated for use in electrophoretic mobility shift assays (EMSAs) and purified (FNRD154A)2-His6 was generously provided to us by Lisa Kay Nolan’s laboratory, University of Georgia, Athens, GA and prepared as previously described (39). Purified protein was stored at −80 °C in binding buffer (20 mM Tris HCL, 50 mM NaCl, 40 mM EDTA, 4 mM DTT, 10% glycerol, pH 6.8).
All assays were carried out as previously described (39) by mixing increasing amounts of (FNRD154A)2-His6 with the PCR-amplified promoter regions of virulence factor genes as the DNA probe. PCR-amplified promoter regions were size selected using a Zymoclean DNA Gel Recovery Kit (Zymogen) and then cleaned using the DNA Clean and Concentrator Kit (Zymogen). Binding reactions of (FNRD154A)2-His6 to DNA probes were performed in binding buffer mentioned above with 0.5 mg/mL BSA, in 25 μL total volume, at 37 °C for 30 min. Concentrations of protein and probe used are detailed per assay in SI Appendix, Figs. S3–S6 for clarity. Reaction mixtures were run in a 6% polyacrylamide DNA retardation gel (Invitrogen) in 0.5 TBE buffer (Invitrogen) at 200 V for 45 min, and gels were stained with ethidium bromide in 0.5 TBE buffer for 10 min at room temperature before being exposed to UV light for imaging.
Biofilm Production and Measurement.
We found ETEC H10407 spontaneously grew biofilms on glass test tubes when grown anaerobically at 37 °C in 5 mL of CFA broth with vigorous shaking. To quantitate biofilm production, growth media were removed from test tubes using a serological pipette, and then 0.1% filter sterilized crystal violet stain was added to the tube to completely submerge the biofilm, and the biofilm was incubated for 20 min at room temperature. The stain was removed via serological pipette, and then stained biofilms were carefully washed at least three times with PBS to be sure there was no residual stain remaining in the tube. Stained and washed biofilms were air dried at room temperature for 30 min and then solubilized in 95% ethanol via pipetting. Dilutions of the solubilized stain were pipetted into a 96-well clear bottom plate, and then the optical density at 595 nm was determined for each sample using 95% ethanol as a blank. All conditions were tested in biological triplicate. Statistical significance was determined across samples using a one-way ANOVA followed by Tukey’s multiple comparison test, and error bars represent the SEM.
Supplementary Material
Acknowledgments
We thank our friend, Dr. Patricia Guerry of the Naval Medical Research Center, for helping to establish collaboration and insightful conversations and Lisa Kay Nolan and Nicolle Barbieri for their generous gift of (FNRD154A)2-His6 and their EMSA insight. This work was supported by the National Institutes of Health (NIH) Grants AI064184 and AI076322 (to M.S.T.). The views expressed herein are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, the Department of Defense, or the US Government. F.M.P., C.K.P, M.S.R, R.L.G., M.G.P., and S.J.S. are employees of the US Government, and this work was performed as part of their official duties. Title 17 U.S.C. §105 provides that “Copyright protection under this title is not available for any work of the United States Government.” Title 17 U.S.C. §101 defines a US Government work as a work prepared by a military service member or employee of the US Government as part of that person’s official duties.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequencing data are publicly available for download at the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) at SRA accession SRP151249.
See Commentary on page 9342.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808982115/-/DCSupplemental.
References
- 1.Gupta SK, et al. Part III. Analysis of data gaps pertaining to enterotoxigenic Escherichia coli infections in low and medium human development index countries, 1984–2005. Epidemiol Infect. 2008;136:721–738. doi: 10.1017/S095026880700934X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hosangadi D, Smith PG, Giersing BK. Considerations for using ETEC and Shigella disease burden estimates to guide vaccine development strategy. Vaccine. 2017 doi: 10.1016/j.vaccine.2017.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harro C, et al. Refinement of a human challenge model for evaluation of enterotoxigenic Escherichia coli vaccines. Clin Vaccine Immunol. 2011;18:1719–1727. doi: 10.1128/CVI.05194-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DuPont HL, et al. Pathogenesis of Escherichia coli diarrhea. N Engl J Med. 1971;285:1–9. doi: 10.1056/NEJM197107012850101. [DOI] [PubMed] [Google Scholar]
- 5.Evans DG, Silver RP, Evans DJ, Jr, Chase DG, Gorbach SL. Plasmid-controlled colonization factor associated with virulence in Esherichia coli enterotoxigenic for humans. Infect Immun. 1975;12:656–667. doi: 10.1128/iai.12.3.656-667.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hodson C, et al. Control of virulence gene expression by the master regulator, CfaD, in the prototypical enterotoxigenic Escherichia coli strain, H10407. Front Microbiol. 2017;8:1525. doi: 10.3389/fmicb.2017.01525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y-F, et al. Structure of CFA/I fimbriae from enterotoxigenic Escherichia coli. Proc Natl Acad Sci USA. 2009;106:10793–10798. doi: 10.1073/pnas.0812843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao R, Liu Y, Savarino SJ, Xia D. Off-pathway assembly of fimbria subunits is prevented by chaperone CfaA of CFA/I fimbriae from enterotoxigenic E. coli. Mol Microbiol. 2016;102:975–991. doi: 10.1111/mmi.13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozaki H, et al. Molecular structure of the toxin domain of heat-stable enterotoxin produced by a pathogenic strain of Escherichia coli. A putative binding site for a binding protein on rat intestinal epithelial cell membranes. J Biol Chem. 1991;266:5934–5941. [PubMed] [Google Scholar]
- 10.Yamamoto T, Tamura T, Yokota T. Primary structure of heat-labile enterotoxin produced by Escherichia coli pathogenic for humans. J Biol Chem. 1984;259:5037–5044. [PubMed] [Google Scholar]
- 11.Sack RB, Froehlich JL. Antigenic similarity of heat-labile enterotoxins from diverse strains of Escherichia coli. J Clin Microbiol. 1977;5:570–572. doi: 10.1128/jcm.5.6.570-572.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sack DA, Merson MH, Wells JG, Sack RB, Morris GK. Diarrhoea associated with heat-stable enterotoxin-producing strains of Escherichia coli. Lancet. 1975;2:239–241. doi: 10.1016/s0140-6736(75)90958-7. [DOI] [PubMed] [Google Scholar]
- 13.Haycocks JRJ, Sharma P, Stringer AM, Wade JT, Grainger DC. The molecular basis for control of ETEC enterotoxin expression in response to environment and host. PLoS Pathog. 2015;11:e1004605. doi: 10.1371/journal.ppat.1004605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sixma TK, et al. Refined structure of Escherichia coli heat-labile enterotoxin, a close relative of cholera toxin. J Mol Biol. 1993;230:890–918. doi: 10.1006/jmbi.1993.1209. [DOI] [PubMed] [Google Scholar]
- 15.De Haan L, Hirst TR. Cholera toxin: A paradigm for multi-functional engagement of cellular mechanisms (review) Mol Membr Biol. 2004;21:77–92. doi: 10.1080/09687680410001663267. [DOI] [PubMed] [Google Scholar]
- 16.Levine MM, et al. Diarrhea caused by Escherichia coli that produce only heat-stable enterotoxin. Infect Immun. 1977;17:78–82. doi: 10.1128/iai.17.1.78-82.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Del Canto F, et al. Distribution of classical and nonclassical virulence genes in enterotoxigenic Escherichia coli isolates from Chilean children and tRNA gene screening for putative insertion sites for genomic islands. J Clin Microbiol. 2011;49:3198–3203. doi: 10.1128/JCM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy K, et al. Enterotoxigenic Escherichia coli EtpA mediates adhesion between flagella and host cells. Nature. 2009;457:594–598. doi: 10.1038/nature07568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherlock O, Vejborg RM, Klemm P. The TibA adhesin/invasin from enterotoxigenic Escherichia coli is self recognizing and induces bacterial aggregation and biofilm formation. Infect Immun. 2005;73:1954–1963. doi: 10.1128/IAI.73.4.1954-1963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicklasson M, Sjöling Å, von Mentzer A, Qadri F, Svennerholm A-M. Expression of colonization factor CS5 of enterotoxigenic Escherichia coli (ETEC) is enhanced in vivo and by the bile component Na glycocholate hydrate. PLoS One. 2012;7:e35827. doi: 10.1371/journal.pone.0035827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crofts AA, et al. Campylobacter jejuni transcriptional and genetic adaptation during human infection. Nat Microbiol. 2018;3:494–502. doi: 10.1038/s41564-018-0133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorbach SL, Tabaqchali S. Bacteria, bile, and the small bowel. Gut. 1969;10:963–972. doi: 10.1136/gut.10.12.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman ES, et al. Microbes vs. chemistry in the origin of the anaerobic gut lumen. Proc Natl Acad Sci USA. 2018;115:4170–4175. doi: 10.1073/pnas.1718635115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myers KS, et al. Genome-scale analysis of escherichia coli FNR reveals complex features of transcription factor binding. PLoS Genet. 2013;9:e1003565. doi: 10.1371/journal.pgen.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volbeda A, Darnault C, Renoux O, Nicolet Y, Fontecilla-Camps JC. The crystal structure of the global anaerobic transcriptional regulator FNR explains its extremely fine-tuned monomer-dimer equilibrium. Sci Adv. 2015;1:e1501086. doi: 10.1126/sciadv.1501086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmed D, et al. Presence of enterotoxigenic Escherichia coli in biofilms formed in water containers in poor households coincides with epidemic seasons in Dhaka. J Appl Microbiol. 2013;114:1223–1229. doi: 10.1111/jam.12109. [DOI] [PubMed] [Google Scholar]
- 27.El Aidy S, van den Bogert B, Kleerebezem M. The small intestine microbiota, nutritional modulation and relevance for health. Curr Opin Biotechnol. 2015;32:14–20. doi: 10.1016/j.copbio.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Marteyn B, et al. Modulation of Shigella virulence in response to available oxygen in vivo. Nature. 2010;465:355–358. doi: 10.1038/nature08970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng L, Kelly CJ, Colgan SP. Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A review in the theme: Cellular responses to hypoxia. Am J Physiol Cell Physiol. 2015;309:C350–C360. doi: 10.1152/ajpcell.00191.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwartz CJ, Djaman O, Imlay JA, Kiley PJ. The cysteine desulfurase, IscS, has a major role in in vivo Fe-S cluster formation in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:9009–9014. doi: 10.1073/pnas.160261497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eckweiler D, Dudek C-A, Hartlich J, Brötje D, Jahn D. PRODORIC2: The bacterial gene regulation database in 2018. Nucleic Acids Res. 2018;46:D320–D326. doi: 10.1093/nar/gkx1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurabayashi K, Tanimoto K, Tomita H, Hirakawa H. Cooperative actions of CRP-cAMP and FNR increase the fosfomycin susceptibility of enterohaemorrhagic Escherichia coli (EHEC) by elevating the expression of glpT and uhpT under anaerobic conditions. Front Microbiol. 2017;8:426. doi: 10.3389/fmicb.2017.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kansal R, et al. Transcriptional modulation of enterotoxigenic Escherichia coli virulence genes in response to epithelial cell interactions. Infect Immun. 2013;81:259–270. doi: 10.1128/IAI.00919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar A, Alam A, Rani M, Ehtesham NZ, Hasnain SE. Biofilms: Survival and defense strategy for pathogens. Int J Med Microbiol. 2017;307:481–489. doi: 10.1016/j.ijmm.2017.09.016. [DOI] [PubMed] [Google Scholar]
- 35.Flemming H-C, et al. Biofilms: An emergent form of bacterial life. Nat Rev Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 36.Puorger C, Vetsch M, Wider G, Glockshuber R. Structure, folding and stability of FimA, the main structural subunit of type 1 pili from uropathogenic Escherichia coli strains. J Mol Biol. 2011;412:520–535. doi: 10.1016/j.jmb.2011.07.044. [DOI] [PubMed] [Google Scholar]
- 37.Sheikh A, et al. Highly conserved type 1 pili promote enterotoxigenic E. coli pathogen-host interactions. PLoS Negl Trop Dis. 2017;11:e0005586. doi: 10.1371/journal.pntd.0005586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schembri MA, Klemm P. Biofilm formation in a hydrodynamic environment by novel fimh variants and ramifications for virulence. Infect Immun. 2001;69:1322–1328. doi: 10.1128/IAI.69.3.1322-1328.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbieri NL, et al. FNR regulates the expression of important virulence factors contributing to the pathogenicity of avian pathogenic Escherichia coli. Front Cell Infect Microbiol. 2017;7:265. doi: 10.3389/fcimb.2017.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luethy PM, et al. Microbiota-derived short-chain fatty acids modulate expression of Campylobacter jejuni determinants required for commensalism and virulence. MBio. 2017;8:e00407-17. doi: 10.1128/mBio.00407-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson CJ, Clark DE, Adli M, Kendall MM. Ethanolamine signaling promotes Salmonella niche recognition and adaptation during infection. PLoS Pathog. 2015;11:e1005278. doi: 10.1371/journal.ppat.1005278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bertin Y, et al. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ Microbiol. 2011;13:365–377. doi: 10.1111/j.1462-2920.2010.02334.x. [DOI] [PubMed] [Google Scholar]
- 43.Denkel LA, Rhen M, Bange F-C. Biotin sulfoxide reductase contributes to oxidative stress tolerance and virulence in Salmonella enterica serovar Typhimurium. Microbiology. 2013;159:1447–1458. doi: 10.1099/mic.0.067256-0. [DOI] [PubMed] [Google Scholar]
- 44.Subashchandrabose S, et al. Host-specific induction of Escherichia coli fitness genes during human urinary tract infection. Proc Natl Acad Sci USA. 2014;111:18327–18332. doi: 10.1073/pnas.1415959112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreno E, et al. Relationship between Escherichia coli strains causing acute cystitis in women and the fecal E. coli population of the host. J Clin Microbiol. 2008;46:2529–2534. doi: 10.1128/JCM.00813-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamamoto S, et al. Genetic evidence supporting the fecal-perineal-urethral hypothesis in cystitis caused by Escherichia coli. J Urol. 1997;157:1127–1129. [PubMed] [Google Scholar]
- 47.Gupta S, Chowdhury R. Bile affects production of virulence factors and motility of Vibrio cholerae. Infect Immun. 1997;65:1131–1134. doi: 10.1128/iai.65.3.1131-1134.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones SA, et al. Respiration of Escherichia coli in the mouse intestine. Infect Immun. 2007;75:4891–4899. doi: 10.1128/IAI.00484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Begum YA, et al. In situ analyses directly in diarrheal stool reveal large variations in bacterial load and active toxin expression of enterotoxigenic Escherichia coli and Vibrio cholerae. mSphere. 2018;3:e00517-17. doi: 10.1128/mSphere.00517-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sjöling A, et al. In vivo expression of the heat stable (estA) and heat labile (eltB) toxin genes of enterotoxigenic Escherichia coli (ETEC) Microbes Infect. 2006;8:2797–2802. doi: 10.1016/j.micinf.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 51.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.