Abstract
Background
People with HIV (PWH) demonstrate increased cardiovascular disease (CVD), due in part to increased immune activation, inflammation, and endothelial dysfunction.
Methods
In a randomized trial (INTREPID), 252 HIV-infected participants with dyslipidemia and no history of coronary artery disease were randomized (1:1) to pitavastatin 4 mg vs. pravastatin 40 mg for 52 weeks. Using a proteomic discovery approach, 92 proteins biomarkers were assessed using Proximity Extension Assay technology to determine the effects of statins on key atherosclerosis and CVD pathways among PWH. 225 participants had specimens available for biomarker analysis pre- and post-baseline.
Findings
The mean age was 49.5 ± 8.0 (mean ± SD), LDL-C 155 ± 25 mg/dl and CD4 count 620 ± 243 cell/mm3. Among all participants, three proteins significantly decreased: tissue factor pathway inhibitor [TFPI; t-statistic = −6.38, FDR p-value<0.0001], paraoxonase 3 [PON3; t-statistic = −4.64, FDR p-value = 0.0003], and LDL-receptor [LDLR; t-statistic = −4.45, FDR p-value = 0.0004]; and two proteins significantly increased galectin-4 [Gal-4; t-statistic = 3.50, FDR p-value = 0.01] and insulin-like growth factor binding protein 2 [IGFBP-2; t-statistic = 3.21, FDR p-value = 0.03]. The change in TFPI was significantly different between the pitavastatin and pravastatin groups. Among all participants, change in TFPI related to the change in LDL-C (r = 0.43, P < 0.0001) and change in Lp-PLA2 (r = 0.29, P < 0.0001).
Interpretation
Using a proteomics approach, we demonstrated that statins led to a significant reduction in the levels of TFPI, PON3, and LDLR and an increase in Gal-4 and IGFBP-2, key proteins involved in coagulation, redox signaling, oxidative stress, and glucose metabolism. Pitavastatin led to a greater reduction in TFPI than pravastatin. These data highlight potential novel mechanisms of statin effects among PWH.
Fund
This work was supported by an investigator-initiated grant to S.K.G. from KOWA Pharmaceuticals America, Inc. and the National Institutes of Health [P30 DK040561; Nutrition Obesity Research Center at Harvard]. M.T. was support by National Institutes of Health [5KL2TR001100-05; Harvard Catalyst KL2 grant].
Keywords: HIV, Statin, Atherosclerosis, Proteomics, Cardiovascular disease
Graphical abstract
Highlights
-
•
Among PWH, statins significantly decreased three proteins [tissue factor pathway inhibitor (TFPI), paraoxonase 3 (PON3), and LDL-receptor (LDLR)].
-
•
Among PWH, statins significantly increased galectin-4 (Gal-4) and insulin-like growth factor binding protein 2 (IGFBP-2).
-
•
The proteins significantly affected by statin therapy are involved in important pathways in atherosclerosis and cardiovascular disease.
-
•
The change in TFPI was directly related to the change in LDL-C and a systemic marker of arterial inflammation (Lp-PLA2).
Research in context.
Evidence before this study
Prior epidemiological studies have demonstrated that people living with HIV (PWH) have an increased risk of atherosclerotic cardiovascular disease (CVD). Persistent immune activation, endothelial dysfunction, and increased arterial inflammation have been implicated in the pathophysiology of atherosclerotic CVD in HIV. Randomized controlled trials evaluating the lipid lowering effect of statins among people living with HIV have examined the effect of statins on several protein biomarkers using single-assay radioimnunoassays. These prior studies, however, have been limited in their ability to evaluate potential pleotropic statin effects among individuals with HIV, which is of clinical significance given the multiple contributors to increased atherosclerotic CVD in this population. Prior proteomics studies of statin effects have been performed but are very small and none have been conducted in HIV, a population in whom statins hold a potentially critical role to reduce inflammation.
Added value of this study
We performed the largest study to date evaluating the change in proteins after statin therapy among individuals with HIV using a novel proteomic approach — Proximity Extension Assay (PEA) technology. This proteomic approach permitted simultaneous assessment of the effects of statins on over 90 proteins in our study population. The statins investigated – pitavastatin and pravastatin – have been shown to have less drug-drug interactions with anti-retroviral therapy compared to other statins and therefore, are clinically relevant and important among PWH. Through this evaluation, we identified 5 proteins which significantly changed (false discovery rate p-value <0.05) with statin therapy – tissue factor pathway inhibitor, paraoxonase 3, and LDL-receptor, galectin-4 and insulin-like growth factor binding protein 2. These proteins notably play important roles in various pathways central to atherogenesis and atherosclerotic CVD, including tissue-factor mediate coagulation, oxidative stress, redox signal pathways and glucose metabolism.
Implications of all the available evidence
Our findings identified several pathways through which statins may affect CVD risk among PWH. Given that PWH have increased cardiovascular disease risk even after controlling for traditional risk factors, understanding the effect of statins on different cardiovascular pathways is critical to understanding the potential role of statins in preventing and treating atherosclerotic CVD among PWH.
Alt-text: Unlabelled Box
1. Introduction
People living with HIV(PWH) demonstrate increased cardiovascular disease (CVD) rates which remain increased controlling for traditional risk factors. [1], [2] Persistent immune activation, arterial inflammation, endothelial dysfunction and increased coagulation among PWH on anti-retroviral therapy (ART) are thought to contribute to increased CVD among PWH [3]. Strategies to reduce and/or prevent atherosclerosis are critical to decreasing CVD mortality in this population.
Statins may be a useful strategy to reduce and/or prevent atherosclerotic CVD among PWH [4]. [5] Atorvastatin, for example, has been shown to reduce atherosclerotic plaque among HIV-infected participants [5], whereas rosuvastatin has been shown to reduce the progression of carotid intima-media thickness (cIMT) [4]. We previously described the effects of two statins known to have few drug-drug interactions with anti-retroviral therapy (ART), pitavastatin and pravastatin, on LDL and effects on a limited number of inflammatory markers among dyslipidemic HIV-infected participants in INTREPID (HIV-infected patieNts and TREatment with PItavastatin vs. pravastatin for Dyslipidemia) [6,7], a large double-blind, parallel-group study. Larger reductions in the pitavastatin group were shown for lipoprotein associated phospholipase A2 (Lp-PLA2), oxidized LDL (oxLDL), and soluble CD14 (sCD14) [7], a biomarker that has been associated with the progression of atherosclerosis [8] and mortality [9] in HIV.
This prior work assessed only specific biomarkers, using traditional single-assay radioimmunoassay (RIA) and other immunoassay techniques. To advance discovery of important pathways affected by statin therapy, we first used a Proximity Extension Assay in a small study of atorvastatin in HIV and identified multiple pathways of interest [10]. We now extend this approach to the much larger INTREPID trial, to compare the effects of pitavastatin and pravastatin on over 90 proteins, simultaneously. To our knowledge, this is the largest proteomics discovery analysis to date, investigating statin effects on key atherosclerosis and CVD pathways in HIV. The proteomic technique utilized in this current analysis offers a high-throughput, sensitive and specific method to evaluate proteins that may be associated with atherosclerotic disease in HIV [10,11]. We hypothesized that statins would have novel effects on heretofore unknown, but relevant, atherosclerosis pathways in HIV. Moreover, given our prior data [7], we hypothesized that pitavastatin would result in significantly greater changes in relevant cardiovascular pathways among PWH.
2. Methods
2.1. Study design
INTREPID was a randomized, double-blind, double-dummy, active-controlled, parallel-group, superiority trial performed from 23 February 2011 to 29 March 2013 [6]. The study was conducted at 45 sites in the United States and Puerto Rico, and the primary aim was to compare the effect of pitavastatin 4 mg daily vs. pravastatin 40 mg daily on LDL-C reduction in individuals with HIV and dyslipidemia for 12 weeks, followed by a 40-week safety extension period. Primary results for this study at Week 12 and Week 52, including data on safety, have been reported [6]. Written informed consent was obtained from all participants prior to enrollment and approval was obtained by the institutional review board at each participating study site. The trial is registered on ClinicalTrials.gov (NCT01301066).
Male or female participants, with documented HIV infection, between the ages of 18 and 70, with a CD4-cell count >200 cells/ml for >3 months, and HIV-1 RNA <200 copies/ml, prescribed ART for >6 months, and without change in ART within 3 months prior to randomization, were eligible. Participants on darunavir were excluded due to a significant drug-drug interaction between darunavir and pravastatin. Participants with conditions causing secondary dyslipidemia, history of coronary heart disease or a coronary heart disease equivalent, prior or current muscular or neuromuscular disease, or active systemic infections were excluded. Participants receiving statins were eligible after a 4-week washout period. After a wash-out period of at least 4 weeks and a 4-week diet stabilization period (previously described) [6], lipid eligibility was determined based on a LDL-C value of at least 130 mg/dl but <220 mg/dl and triglyceride value <400 mg/dl.
Participants were randomized 1:1 to active pitavastatin 4 mg once daily and a matching pravastatin placebo vs. pravastatin 40 mg once daily and a matching pitavastatin placebo. Randomization was performed using a central interactive voice response system as previously described [6], and study investigators, site staff, and participants were masked to treatment codes.
2.2. Procedures
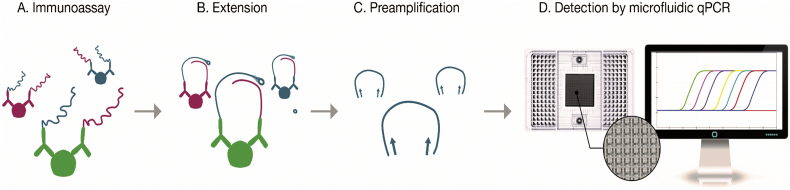
Study visits occurred every 4 weeks for the first 12 weeks with subsequent visits occurring quarterly through Week 52. Measurements of the creatinine, lipid panel, and CD4 cell count, were performed using standard techniques. HIV-RNA was assessed using Cobas AmpliPrep and COBAS TaqMan HIV-1 Test, version 2.0 (Roche Molecular Diagnostics, Pleasanton, CA, USA; lower limit of detection of 20 copies per ml). Plasma sCD163, hs-IL6, sCD14, MCP-1, Lp-PLA2, and oxLDL were quantified as previously detailed elsewhere [7]. EDTA plasma samples collected at baseline and at the end of study visit were sent for analysis of 92 proteins. Analysis of 92 proteins comprising the Olink Cardiovascular III panel (http://www.olink.com/products/cvd-iii-panel/) was performed using PEA (Proximity Extension Assay) technology. The PEA technique allows simultaneous assessment of proteins using oligonucleotide-labeled antibody probe pairs that bind to each protein within the sample (Fig. 1). The PEA technique also permits accurate assessment of protein levels with repeated measurements by requiring both dual recognition of correctly matched antibody pairs and DNA-barcoding from sequence-specific oligonucleotides to generate a signal [12]. The assay measures fold change in log 2 units. For this analysis, the raw data are converted into a t-statistic which can be compared across assays. The coefficient of variation of Olink's Cardiovascular III panel proteins is demonstrated in Supplemental Table 1. Detailed information on the PEA technique and the Cardiovascular III Panel are found on http://www.olink.com/products/document-download-center/. In terms of validation to immunoassays, we previously assessed a few protein biomarkers using standard immunoassays among participants within the INTREPID trial [7]; and among the proteins previously assessed, two were also evaluated using PEA technology as part of this current study. Significant correlations were shown for soluble scavenger receptor cysteine-rich type 1 protein M130 (sCD163) and monocyte chemotactic protein 1 (MCP-1) comparing PEA vs. the standardized assay for these proteins (both P < .0001).
Fig. 1.
Overview of the Proximity Extension Assay demonstrating the (a) immunoassay, (b) extension, (c) preamplification, (d) and detection steps used for proteomic analyses of protein biomarkers in this study. [12] Image courtesy of Olink Proteomics AB. (a) For this immunoassay, pairs of specific antibodies labeled with DNA oligonucleotides bind to their target-specific antigens (protein biomarkers) in a solution. Antigen binding, in turn, brings the PEA probes in close proximity. (b) Pair-wise binding of matching PEA probes occurs through hybridization and upon addition of a DNA polymerase results in extension of the two oligonucleotides to form DNA barcodes that will now serve as a reporter of the protein biomarkers. (c) The addition of primers results in preamplication of the new DNA sequences. (d) Microfluidic qPCR is used for amplification and quantification of these new DNA barcodes.
2.3. Statistical analysis
The initial objective of INTREPID was to compare the between-group difference in the percent change in fasting serum LDL-C between two treatment groups — pitavastatin and pravastatin [6]. In this post-hoc analysis, changes in the level of 92 proteins before and after statin therapy were assessed among all participants first pooling data from both groups and then among each treatment group individually. Secondary objectives included the assessment of between-group difference in the change of the protein biomarkers assessed before and after statin therapy. All available data were included in an intent to treat analysis. A total of 35 participants did not complete the study to Week 52 but had interim data available for this analysis [median follow up duration: 12.1 (4.4, 12.4) weeks (Median, IQR)] and therefore were included in this intent to treat analysis. Baseline demographic data, HIV-specific parameters, and markers of immune activation and arterial inflammation are presented as mean ± standard deviation and the means of each statin group were compared using a student's t-test for continuous variables and χ2 test for categorical variables. Among all study participants and for each study group, a t-statistic for the treatment effect of the statin(s) and the corresponding p-value was calculated. This analysis was repeated for the pitavastatin group and pravastatin group, respectively. Between group differences in mean change of these proteins in the pitavastatin vs pravastatin groups were determined using the independent samples t-test for each protein. Due to the large number of proteins assessed, false discovery rate p-values (FDR p-values) were determined [13]. Statistically significant change in the level of a protein was defined by a false discovery rate p-value <0.05. Tests for interaction were performed to determine if use of specific ART classes, e.g. protease-inhibitor use, non-nucleoside reverse transcriptase inhibitor use (NNRTI), and integrase-inhibitor use, modified the effect of statins on proteins assessed. Bivariate analyses between two variables were performed using a Pearson correlation coefficient. With 225 evaluable participants, the current proteomics discovery analysis had 90% power to detect a 0.3 SD change, based on estimated SD of 1.0, with a false discovery rate of 0.05 using a two-sided one-sample t-test. Statistical analyses were performed using SAS software (version 9.4; SAS Institute) and SAS JMP software (version 11.0; SAS Institute).
3. Results
3.1. Study participants
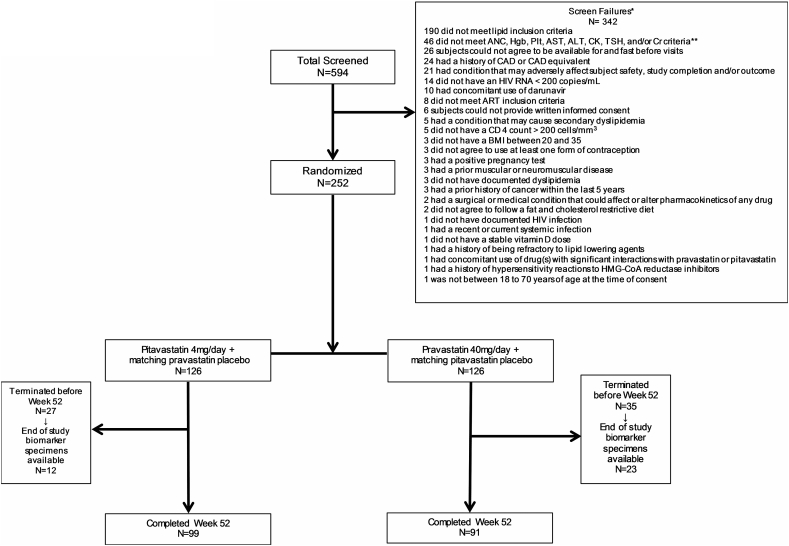
A total of 252 participants were deemed eligible and randomly assigned to receive pitavastatin (126 participants) or pravastatin (126 participants). Paired samples were available for proteomic analyses from 190 participants completing to Week 52 and from 35 additional participants with specimens available from an interim visit (n = 225; Fig. 2). Of the 225 participants with samples before and after treatment for this analysis, 114 were randomized to pravastatin and 111 to pitavastatin.
Fig. 2.
Consort Diagram.
Out of 594 participants screened, 342 participants were screen failures. Two hundred and fifty-two participants were enrolled and randomized.
*Twenty-seven participants who were screen failures had at least two or more criteria for inclusion that were not met and/or exclusion criteria that were met.
**Other Inclusion criteria: absolute neutrophil count >750 cells/ml, hemoglobin at least 9.0 g/dl for female participants and at least 10.0 g/dl for male participants, platelets at least 100,000/ml, ALT and AST 2.5 times the upper limit of normal (ULN) or less (note participants coinfected with hepatitis B or C were required to have ALT and AST 1.5 times the ULN or less), fasting glucose 125 mg/dl or less, CK 3 times the ULN or less (if a transient increase in CK level was suspected due to exercise or trauma, CK may have been repeated at screening after an ‘exercise washout’ at the discretion of the Investigator), serum creatinine 1.3 times the ULN or less and estimated glomerular filtration rate (eGFR) at least 60 ml/min per 1.73 m2 based on the modification of diet in renal disease equation at http://www.nephron.com/MDRD_GFR.cgi (if a creatinine level was suspected to be temporarily increased due to factors such as dehydration, creatinine testing may have been repeated and the eGFR may have been recalculated at screening at the discretion of the Investigator), and TSH <1.5 times the ULN.
Abbreviations: ANC, absolute neutrophil count; hgb, hemoglobin; plt, platelet; AST, aspartate aminotransferase; ALT, alanine transferase; CK, creatinine kinase; TSH, thyroid-stimulating hormone; Cr, creatinine; CAD, coronary heart disease; HIV RNA, Human Immunodeficiency Virus ribonucleic acid; ART, antiretroviral therapy; BMI, body mass index; HMG-CoA, 3-hydroxy-3-methylglutaryl-coenzyme A
3.2. Baseline demographics
The mean age for study participants was 49.5 ± 8.0 [mean ± standard deviation (SD)]. Eighty-eight percent were male and 26% were Hispanic or Latino. The mean Framingham Risk Score was 6.5 ± 4.9% and mean LDL-C level was 155 ± 25 mg/dl. With regards to HIV-specific parameters, the mean CD4 lymphocyte count was 620 ± 243 cells/mm3 and log HIV viral load was 1.1 ± 0.2 copies. Baseline demographics did not significantly differ between groups other than for a small difference in CD4 lymphocyte count (Table 1). Eighty-four out of the 225 participants (37%) were on a PI-based therapy, 120 out of the 225 participants (53%) were on an NNRTI-based therapy, two out of the 225 participants were on both a protease inhibitor and NNRTI, and the remainder of participants were either on an integrase-inhibitor-based therapy or NRTIs only. Baseline demographics did not differ among the 225 participants included in this study and the 27 participants who were not included because they did not have available blood samples for proteomic analysis (Supplemental Table 2). Prior analysis of INTREPID study participants demonstrated a mean duration of HIV infection of 12.6 ± 7.5 years [6].
Table 1.
Baseline demographics and immune markers.
| Pitavastatin N = 111 | Pravastatin N = 114 | |
|---|---|---|
| Age | 50.0 ± 7.6 | 49.1 ± 8.4 |
| Sex, % female | 14 (16/111) | 11 (12/114) |
| Race | ||
| White % | 85 (94/111) | 76 (87/114) |
| Black % | 12 (14/111) | 19 (21/114) |
| Asian % | 1 (1/111) | 1 (1/114) |
| Other % | 2 (2/111) | 4 (4/114) |
| Hispanic % | 24 (27/111) | 28 (32/114) |
| Hepatitis B/C | 8 (9/111) | 11 (12/114) |
| eGFR (ml/min/1.73m2) | 80 ± 16 | 81 ± 16 |
| Total Cholesterol (mg/dl) | 239 ± 32 | 238 ± 31 |
| HDL-C (mg/dl) | 49 ± 15 | 49 ± 12 |
| LDL-C (mg/dl) | 156 ± 27 | 154 ± 24 |
| Triglycerides (mg/dl) | 177 ± 95 | 169 ± 70 |
| BMI (kg/m2) | 27.4 ± 4.6 | 28.3 ± 5.0 |
| Framingham Risk Score % | 6.7 ± 5.1 | 6.3 ± 4.7 |
| Log HIV-1 Viral Load (copies/ml) | 1.1 ± 0.3 | 1.1 ± 0.2 |
| CD4 count (cells/mm3)⁎ | 663 ± 264 | 576 ± 212 |
| sCD163 (ng/ml) | 1114 ± 567 | 1006 ± 393 |
| hsIL-6 (pg/ml) | 1.7 ± 2.8 | 1.7 ± 2.3 |
| MCP-1 (pg/ml) | 158.7 ± 67.0 | 151.8 ± 67.8 |
| sCD14 (ng/ml) | 2085 ± 1285 | 1893 ± 1031 |
| oxLDL (U/l) | 78.3 ± 21.9 | 77.8 ± 18.9 |
| Lp-PLA-2 (ng/ml) | 193.8 ± 61.6 | 186.9 ± 71.7 |
| hsCRP (mg/dl) | 4.3 ± 8.7 | 5.5 ± 14.8 |
Normally distributed data are reported as mean ± standard deviation.
No significant differences between baseline values unless indicated.
Abbreviations: eGFR, estimated glomerular filtration rate; HDL-C, high density lipoprotein cholesterol; LDL-C, low density lipoprotein cholesterol; BMI, body mass index; HIV-1, Human Immunodeficiency Virus-1; sCD163, soluble CD163; hsIL-6, high sensitivity interleukin-6, MCP-1, monocyte chemoattractant protein-1; sCD14, soluble CD14; oxLDL, oxidized low density lipoprotein; Lp-PLA2, lipoprotein-associated phospholipase A2; hs-CRP, high sensitivity c-reactive protein.
P-Value = 0.008
3.3. Change in protein biomarkers from baseline
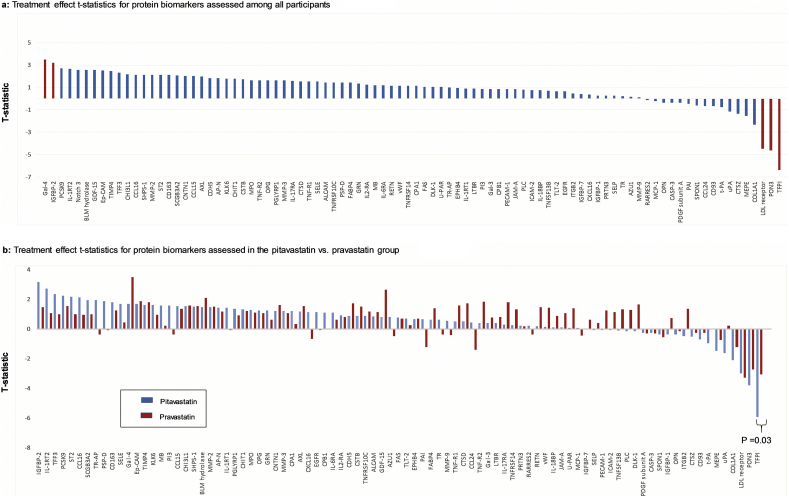
The overall pattern of effects within each treatment group was generally concordant in that proteins most often changed in the same direction in response to each statin. A small minority of discordant responses were seen, but changes between statin arms for these proteins did not achieve statistical significance (Supplemental Table 3). Therefore, the primary analysis pooled the data from each group to obtain an overall effect. In this intent to treat analysis, three proteins, tissue factor pathway inhibitor (TFPI), paraoxonase 3 (PON3), and LDL-receptor (LDLR) decreased, (Fig. 3a; Table 2a) and two proteins, galectin-4 (Gal-4) and insulin-like growth factor binding protein 2 (IGFBP-2), increased with statin therapy using a FDR p-value <0.05. Additional proteins, including proprotein convertase subtilisin/kexin type 9 (PCSK9), met nominal P-value thresholds, but only trended toward significance with FDR p-value ≤0.1 (Table 2a). The t-statistics for the remainder proteins which were nonsignificant (absolute t-statistic <2) are demonstrated in Supplemental Table 4. Among all participants, no significant modification of statin effects on protein levels was seen with use of protease-inhibitors, NNRTI's, and integrase-inhibitors for those proteins meeting FDR P-values <0.05 (Supplemental Tables 6a, 6b, and 6c).
Fig. 3.
Waterplot diagrams demonstrating t-statistic on y-axis and protein biomarkers on x-axis among (a) all participants and (b) among the pitavastatin group and pravastatin group.
(a) Among all participants, 2 proteins significantly increased (defined as a false discovery rate p-value <0.05): Gal-4 and IGFBP-2 and 3 proteins significantly decreased: TFPI, PON3, and LDLR. Protein biomarkers that significantly changed with statin therapy are shaded in red.
Abbreviations: Gal-4, galectin-4; IGFBP-2, insulin-like growth factor-binding protein 2; PCSK-9, proprotein convertase subtilisin/kexin type 9; IL-1RT2, interleukin-1 receptor type 2; Notch 3, neurogenic locus notch homolog protein 3; BLM hydrolase, bleomycin hydrolase; GDF-15, growth/differentiation factor 15; Ep-CAM, epithelial cell adhesion molecule; TIMP4, metalloproteinase inhibitor 4; TFF3, trefoil factor 3; CHI3L1, chitinase-3-like protein 1; CCL16, c-c motif chemokine 16; SHPS-1, tyrosine-protein phosphatase non-receptor type substrate 1; CD163, scavenger receptor cysteine-rich type 1 protein M130; SCGB3A2, secretoglobin family 3A member 2; CNTN1, contactin-1; CCL15, c-c motif chemokine 15; AXL, tyrosine-protein kinase receptor UFO; CDH-5, cadherin-5; AP-N, aminopeptidase N; KLK6, kallikrein-6; CHIT1, chitotriosidase-1; CSTB, cystatin-B; MPO, myeloperoxidase; TNF-R2; tumor necrosis factor receptor 2; OPG, osteoprotegerin; PGLYRP1, peptidoglycan recognition protein 1; MMP-3, matrix metalloproteinase-3; IL-17RA, interleukin-17 receptor a; CTSD, cathepsin D; TNF-R1, tumor necrosis factor receptor 1; SELE, E-selectin; ALCAM, CD166 antigen; TNFRSF10C, tumor necrosis factor receptor superfamily member 10C; PSP-D, pulmonary surfactant-associated protein D; FABP4, fatty acid-binding protein, adipocyte; GRN, granulins; IL2-RA, interleukin-2 receptor subunit alpha; MB, myoglobin; IL-6RA, interleukin-6 receptor subunit alpha; RETN, resistin; vWF, von Willebrand factor; TNFRSF14, tumor necrosis factor receptor superfamily member 14; CPA1, carboxypeptidase A1; FAS, tumor necrosis factor receptor superfamily member 6; DLK-1, protein delta homolog 1; U-PAR, urokinase plasminogen activator surface receptor; TR-AP, tartrate-resistant acid phosphatase type 5; EPHB4, ephrin type-B receptor 4; IL-17RA, interleukin-17 receptor a; LTBR, lymphotoxin-beta receptor; PI3, elafin; Gal-3, galectin-3; CPB1, carboxypeptidase B; PECAM-1, platelet endothelial cell adhesion molecule; JAM-a, junctional adhesion molecule a; PLC, perlecan; ICAM-2, intercellular adhesion molecule 2; IL-18BP, interleukin-18-binding protein; TNFSF13B, tumor necrosis factor ligand superfamily member 13B; TLT-2, trem-like transcript 2 protein; EGFR, epidermal growth factor receptor; ITGB2, integrin beta-2; IGFBP-7, insulin-like growth factor-binding protein 7; CXCL16, c-x-c motif chemokine 16; IGFBP-1, insulin-like growth factor-binding protein 1; PRTN3, myeloblastin; SELP, P-selectin; TR, transferrin receptor protein 1; AZU1, azurocidin; MMP-9, matrix metalloproteinase-9; RARRES2; retinoic acid receptor responder protein 2; MCP-1, monocyte chemotactic protein 1; OPN, osteopontin; CASP-3, caspase-3; PDGF subunit a, platelet-derived growth factor subunit a; PAI, plasminogen activator inhibitor 1; SPON1, spondin-1; CCL24, c-c motif chemokine 24; CD93, complement component C1q receptor; t-PA, tissue-type plasminogen activator; uPA; urokinase-type plasminogen activator; CTSZ, cathepsin Z; MEPE, matrix extracellular phosphoglycoprotein; TFPI, tissue factor pathway inhibitor
(b) In comparing the treatment effects for protein biomarkers between the pitavastatin group and the pravastatin group, TFPI was the only protein biomarker which demonstrated a significant difference in treatment effects between these two groups (pitavastatin vs. pravastatin: −5.92 vs.-3.06, P = 0.03). The pitavastatin t-statistic treatment effect on each protein biomarker are shaded in blue whereas the pravastatin t-statistic treatment effect on each protein biomarker are shaded in red. Proteins are ordered based upon their pitavastatin t-statistic treatment effect.
Abbreviations: IGFBP-2, insulin-like growth factor-binding protein 2; IL-1RT2, interleukin-1 receptor type 2; TFF3, trefoil factor 3; PCSK-9, proprotein convertase subtilisin/kexin type 9; ST2, ST2 protein; CCL16, c-c motif chemokine 16; SCGB3A2, secretoglobin family 3A member 2; TR-AP, tartrate-resistant acid phosphatase type 5; PSP-D, pulmonary surfactant-associated protein D; Notch 3, neurogenic locus notch homolog protein 3; CD163, scavenger receptor cysteine-rich type 1 protein M130; SELE, E-selectin; Gal-4, galectin-4; Ep-CAM, epithelial cell adhesion molecule; TIMP4, metalloproteinase inhibitor 4; KLK6, kallikrein-6; MB, myoglobin; PI3, elafin; CCL15, c-c motif chemokine 15; CHI3L1, chitinase-3-like protein 1; SHPS-1, tyrosine-protein phosphatase non-receptor type substrate 1; BLM hydrolase, bleomycin hydrolase; MMP-2, matrix metalloproteinase-2; AP-N, aminopeptidase N; IL-1RT1, interleukin-1 receptor type 1; PGLYRP1, peptidoglycan recognition protein 1; CHIT1, chitotriosidase-1; MPO, myeloperoxidase; OPG, osteoprotegerin; GRN, granulins; CNTN1, contactin-1; MMP-3, matrix metalloproteinase-3; CPA1; carboxypeptidase A1; AXL, tyrosine-protein kinase receptor UFO; CXCL16, c-x-c motif chemokine 16; EGFR, epidermal growth factor receptor; CPB1, carboxypeptidase B; IL-6RA, interleukin-6 receptor subunit alpha; IL2-RA, interleukin-2 receptor subunit alpha; CDH-5, cadherin-5; CSTB, cystatin-B; TNFRSF10C, tumor necrosis factor receptor superfamily member 10C; ALCAM, CD166 antigen; GDF-15, growth/differentiation factor 15; AZU1, azurocidin; FAS, Tumor necrosis factor receptor superfamily member 6;; TLT-2, trem-like transcript 2 protein; EPHB4, ephrin type-B receptor 4; PAI, plasminogen activator inhibitor 1; FABP4, fatty acid-binding protein, adipocyte; TR, transferrin receptor protein 1; MMP-9, matrix metalloproteinase-9; TNF-R1, tumor necrosis factor receptor 1; CTSD, cathepsin D; CCL24, c-c motif chemokine 24; TNF-R2; tumor necrosis factor receptor 2; Gal-3, galectin-3; LTBR, lymphotoxin-beta receptor; IL-17RA, interleukin-17 receptor a; TNFRSF14, tumor necrosis factor receptor superfamily member 14; PRTN3, myeloblastin; RARRES2; retinoic acid receptor responder protein 2; RETN, resistin; vWF, von Willebrand factor; IL-18BP, interleukin-18-binding protein; JAM-a, junctional adhesion molecule a; U-PAR, urokinase plasminogen activator surface receptor; MCP-1, monocyte chemotactic protein 1; IGFBP-7, insulin-like growth factor-binding protein 7; SELP, P-selectin; PECAM-1, platelet endothelial cell adhesion molecule; ICAM-2, intercellular adhesion molecule 2; TNFSF13B, tumor necrosis factor ligand superfamily member 13B; PLC, perlecan; DLK-1, protein delta homolog 1; PDGF subunit a, platelet-derived growth factor subunit a; CASP-3, caspase-3; SPON1, spondin-1; IGFBP-1, insulin-like growth factor-binding protein 1; OPN, osteopontin; ITGB2, integrin beta-2; CTSZ, cathepsin Z; CD93, complement component C1q receptor; t-PA, tissue-type plasminogen activator; MEPE, matrix extracellular phosphoglycoprotein; uPA; urokinase-type plasminogen activator; COL1A1, collagen alpha-1(I) chain; LDL receptor; low-density lipoprotein receptor; PON3, paraoxonase 3; TFPI, tissue factor pathway inhibitor
Table 2.
Difference in protein biomarker levels before and after statin therapy.
| 2a. All participants | |||
|---|---|---|---|
| Protein | T-statistic | P-Value | False discovery rate p-Value |
| TFPI | −6.38 | <0.0001 | <0.0001 |
| PON3 | −4.64 | <0.0001 | 0.0003 |
| LDLR | −4.45 | <0.0001 | 0.0004 |
| Gal-4 | 3.50 | 0.0006 | 0.01 |
| IGFBP-2 | 3.21 | 0.002 | 0.03 |
| PCSK9 | 2.72 | 0.007 | 0.10 |
| IL-1RT2 | 2.66 | 0.008 | 0.10 |
| Notch 3 | 2.57 | 0.01 | 0.10 |
| GDF-15 | 2.56 | 0.01 | 0.10 |
| BLM hydrolase | 2.56 | 0.01 | 0.10 |
| Ep-CAM | 2.52 | 0.01 | 0.10 |
| TIMP4 | 2.45 | 0.01 | 0.11 |
| COL1A1 | −2.32 | 0.02 | 0.14 |
| TFF3 | 2.31 | 0.02 | 0.14 |
| CHI3L1 | 2.19 | 0.03 | 0.16 |
| SHPS-1 | 2.13 | 0.03 | 0.16 |
| CCL16 | 2.13 | 0.03 | 0.16 |
| ST2 | 2.12 | 0.03 | 0.16 |
| CD163 | 2.12 | 0.03 | 0.16 |
| MMP-2 | 2.12 | 0.04 | 0.16 |
| SCGB3A2 | 2.09 | 0.04 | 0.16 |
| CNTN1 | 2.02 | 0.04 | 0.18 |
| CCL15 | 2.02 | 0.04 | 0.18 |
|
2b. Pitavastatin group | |||
| TFPI | −5.92 | <0.0001 | <0.0001 |
| PON3 | −3.79 | 0.0002 | 0.01 |
| IGFBP-2 | 3.15 | 0.002 | 0.06 |
| LDLR | −2.99 | 0.003 | 0.08 |
| IL-1RT2 | 2.73 | 0.007 | 0.13 |
| TFF3 | 2.36 | 0.02 | 0.30 |
| PCSK9 | 2.26 | 0.03 | 0.33 |
| ST2 | 2.16 | 0.03 | 0.35 |
| CCL16 | 2.14 | 0.03 | 0.35 |
| COL1A1 | −2.01 | 0.05 | 0.39 |
|
2c. Pravastatin group | |||
| Gal-4 | 3.50 | 0.0007 | 0.06 |
| LDLR | −3.29 | 0.001 | 0.06 |
| TFPI | −3.06 | 0.003 | 0.08 |
| PON3 | −2.74 | 0.007 | 0.16 |
| GDF-15 | 2.66 | 0.009 | 0.16 |
| BLM hydrolase | 2.10 | 0.04 | 0.53 |
Among all participants, protein biomarkers with a p-value of <0.05 and an absolute t-statistic >2 are shown. Of these protein biomarkers, 5 protein biomarkers had a false discovery rate p-value that was <0.05 (TFPI, PON3, LDLR, Gal-4, and IGFBP-2) and are shaded. Another 6 proteins trended toward a significant change with a false discovery rate p-value ≤0.10.
Abbreviations: TFPI, tissue factor pathway inhibitor; PON 3, paraoxonases 3; LDLR, low density lipoprotein receptor; Gal-4, galectin-4; IGFBP-2, insulin-like growth factor binding protein 2; PCSK9 proprotein convertase subtilisin/kexin type 9; IL-1RT2, interleukin-1 receptor, type II; Notch 3, neurogenic locus notch homolog protein 3; GDF-15, growth/differentiation factor 15; BLM hydrolase; bleomycin hydrolase; Ep-CAM, epithelial cell adhesion molecule; TIMP4; metalloproteinase inhibitor 4; COLIA1, collagen, type 1, alpha 1; TFF3, trefoil factor 3; CHI3LI, chitinase-3 like protein 1; SHPS-1, Src homology 2 (SH2) domain-containing protein tyrosine phosphatase substrate 1; CCL16, Chemokine (C-C motif) ligand 16; ST, ST2 protein; CD163, cluster of differentiation 163; MMP-2, matrix metalloproteinase-2; SCGB3A2, secretoglobulin family 3A member 2; CNTN1, contactin 1; CCL15, chemokine (C-C motif) ligand 15.
Among the pitavastatin group, protein biomarkers with a p-value of <0.05 and an absolute t-statistic >2 are shown. Of these protein biomarkers, 2 protein biomarkers had a false discovery rate p-value that was <0.05 (TFPI and PON3) and are shaded, whereas 2 other protein biomarkers trended toward a significant change with a false discovery rate p-value <0.10.
Abbreviations: TFPI, tissue factor pathway inhibitor; PON 3, paraoxonases 3; IGFBP-2, insulin-like growth factor binding protein 2; LDLR, low density lipoprotein receptor; IL-1RT2, interleukin-1 receptor, type II; TFF3, trefoil factor 3; PCSK9 proprotein convertase subtilisin/kexin type 9; ST2, ST2 protein; CCL16, Chemokine (C-C motif) ligand 16; COLIA1, collagen, type 1, alpha 1.
In the pravastatin group, protein biomarkers with a p-value of <0.05 and an absolute t-statistic >2 are shown. None of the protein biomarkers had a false discovery rate p-value of <0.05. There were 3 protein biomarkers which trended toward a significant change with a false discovery rate p-value <0.10.
Abbreviations: Gal-4, galectin-4; LDLR, low density lipoprotein receptor; TFPI, tissue factor pathway inhibitor; PON 3, paraoxonases 3; GDF-15, growth/differentiation factor 15; BLM hydrolase; bleomycin hydrolase.
3.4. Change in protein biomarkers from baseline in the pitavastatin group compared to the pravastatin group
Although the overall pattern of effects was similar for both statins studied, there were some specific differences with respect to relevant proteins. Within the pitavastatin group, TFPI and PON3 were significantly reduced using a FDR p-value <0.05(Table 2b), whereas no proteins met this threshold within the pravastatin group (Table 2c). The change (i.e., t-statistic for the treatment effect) in TFPI was significantly greater among the pitavastatin vs. pravastatin group (Fig. 3b; pitavastatin vs. pravastatin: −5.92 vs.-3.06, P = 0.03).
3.5. Predictive markers of change in proteins after statin therapy
In evaluating the relationship between baseline parameters and the change in the proteins among all participants, only baseline Framingham Risk Score was predictive of change in PON3 (r = 0.17, P = 0.01; Supplemental Table 5a). Within the pitavastatin group, the change in TFPI was related to baseline log HIV-1 viral load (r = 0.20, P = 0.03; Supplemental Table 5b) and inversely related to baseline Lp-PLA2 levels (r = −0.21, P = 0.032). None of the selected baseline parameters predicted change in these proteins within the pravastatin group (Supplemental Table 5c).
We also evaluated the change in proteins with change in LDL-C, and change in other relevant biomarker data available from prior analyses using traditional assays in the study participants [7]. Among all participants, the change in TFPI was directly related to the change in LDL-C (r = 0.43, P < 0.0001; Table 3a) and the change in Lp-PLA2 (r = 0.29, P < 0.0001). The change in LDL-C was also directly related to the change in PON3 (r = 0.20, P = 0.004) and change in LDLR (r = 0.27, P < 0.0001; Table 3a). Within the pitavastatin group, the change in LDL-C was directly related to the change in TFPI (r = 0.54, P < 0.0001; Table 3b), PON3 (r = 0.34, P = 0.0008), and LDLR (r = 0.42, P < 0.0001). Within the pravastatin group, the change in LDL-C was also directly related to the change in TFP1 (r = 0.26, P = 0.008; Table 3c).
Table 3.
Correlation Analysis between Changes in LDL-C and Immune Markers and Changes in Protein Biomarkers.
| 3a. All participants | |||||
|---|---|---|---|---|---|
| delta TFPI | delta PON3 | delta LDLR | delta Gal-4 | delta IGFBP-2 | |
| delta LDL-C | r = 0.43 | r = 0.20 | r = 0.27 | r = −0.05 | r = −0.26 |
| P < 0.0001 | P = 0.004 | P < 0.0001 | P = 0.50 | P = 0.0002 | |
| delta sCD163 | r = 0.01 | r = −0.11 | r = 0.06 | r = 0.04 | r = −0.12 |
| P = 0.83 | P = 0.12 | P = 0.39 | P = 0.60 | P = 0.08 | |
| delta hsIL-6 | r = 0.06 | r = 0.007 | r = 0.01 | r = 0.04 | r = 0.10 |
| P = 0.40 | P = 0.92 | P = 0.87 | P = 0.57 | P = 0.15 | |
| delta MCP -1 | r = −0.14 | r = −0.11 | r = −0.006 | r = −0.07 | r = 0.07 |
| P = 0.05 | P = 0.11 | P = 0.93 | P = 0.31 | P = 0.32 | |
| delta sCD14 | r = 0.06 | r = −0.11 | r = −0.0009 | r = 0.05 | r = −0.02 |
| P = 0.39 | P = 0.11 | P = 0.99 | P = 0.49 | P = 0.78 | |
| delta oxLDL | r = 0.04 | r = −0.13 | r = 0.10 | r = −0.02 | r = −0.21 |
| P = 0.52 | P = 0.06 | P = 0.13 | P = 0.82 | P = 0.002 | |
| delta Lp-PLA2 | r = 0.29 | r = 0.02 | r = 0.25 | r = 0.03 | r = −0.19 |
| P < 0.0001 | P = 0.75 | P = 0.0003 | P = 0.66 | P = 0.006 | |
| delta hsCRP | r = 0.03 | r = 0.006 | r = −0.10 | r = 0.07 | r = −0.001 |
| P = 0.72 | P = 0.94 | P = 0.16 | P = 0.32 | P = 0.99 | |
| 3b. Pitavastatin group | |||||
|---|---|---|---|---|---|
| delta TFPI | delta PON3 | delta LDLR | delta Gal-4 | delta IGFBP-2 | |
| delta LDL-C | r = 0.54 | r = 0.34 | r = 0.42 | r = −0.06 | r = −0.14 |
| P < 0.0001 | P = 0.0008 | P < 0.0001 | P = 0.57 | P = 0.17 | |
| delta sCD163 | r = 0.03 | r = −0.08 | r = 0.02 | r = 0.03 | r = −0.08 |
| P = 0.73 | P = 0.40 | P = 0.82 | P = 0.76 | P = 0.45 | |
| delta hsIL-6 | r = 0.004 | r = −0.07 | r = −0.04 | r = 0.07 | r = 0.04 |
| P = 0.97 | P = 0.46 | P = 0.65 | P = 0.49 | P = 0.65 | |
| delta MCP -1 | r = −0.13 | r = −0.15 | r = 0.15 | r = −0.17 | r = −0.04 |
| P = 0.19 | P = 0.14 | P = 0.14 | P = 0.09 | P = 0.68 | |
| delta sCD14 | r = 0.008 | r = −0.11 | r = −0.04 | r = 0.003 | r = 0.005 |
| P = 0.94 | P = 0.29 | P = 0.71 | P = 0.97 | P = 0.96 | |
| delta oxLDL | r = 0.06 | r = −0.11 | r = 0.16 | r = −0.002 | r = −0.21 |
| P = 0.52 | P = 0.26 | P = 0.12 | P = 0.99 | P = 0.03 | |
| delta Lp-PLA2 | r = 0.43 | r = 0.10 | r = 0.43 | r = 0.07 | r = −0.09 |
| P < 0.0001 | P = 0.33 | P < 0.0001 | P = 0.46 | P = 0.39 | |
| delta hsCRP | r = 0.01 | r = −0.18 | r = −0.12 | r = 0.02 | r = −0.008 |
| P = 0.90 | P = 0.09 | P = 0.23 | P = 0.85 | P = 0.94 | |
| 3c. Pravastatin group | |||||
|---|---|---|---|---|---|
| delta TFPI | delta PON3 | delta LDLR | delta Gal-4 | delta IGFBP-2 | |
| delta LDL-C | r = 0.26 | r = 0.05 | r = 0.16 | r = −0.10 | r = −0.37 |
| P = 0.008 | P = 0.61 | P = 0.11 | P = 0.31 | P = 0.0001 | |
| delta sCD163 | r = −0.05 | r = −0.16 | r = 0.12 | r = 0.03 | r = −0.17 |
| P = 0.64 | P = 0.11 | P = 0.23 | P = 0.76 | P = 0.08 | |
| delta hsIL-6 | r = 0.08 | r = 0.05 | r = 0.05 | r = 0.01 | r = 0.14 |
| P = 0.44 | P = 0.63 | P = 0.62 | P = 0.89 | P = 0.16 | |
| delta MCP-1 | r = −0.14 | r = −0.07 | r = −0.16 | r = 0.05 | r = 0.16 |
| P = 0.16 | P = 0.45 | P = 0.11 | P = 0.64 | P = 0.09 | |
| delta sCD14 | r = 0.07 | r = −0.13 | r = 0.04 | r = 0.08 | r = −0.03 |
| P = 0.50 | P = 0.17 | P = 0.70 | P = 0.42 | P = 0.77 | |
| delta oxLDL | r = −0.008 | r = −0.20 | r = 0.02 | r = −0.06 | r = −0.25 |
| P = 0.94 | P = 0.04 | P = 0.86 | P = 0.52 | P = 0.01 | |
| delta Lp-PLA2 | r = 0.10 | r = −0.07 | r = 0.08 | r = −0.05 | r = −0.27 |
| P = 0.30 | P = 0.48 | P = 0.43 | P = 0.62 | P = 0.005 | |
| delta hsCRP | r = 0.05 | r = 0.06 | r = −0.12 | r = 0.12 | r = −0.001 |
| P = 0.65 | P = 0.55 | P = 0.26 | P = 0.26 | P = 0.99 | |
Correlation Analysis between Changes in LDL-C and Immune Markers and Changes in Protein Biomarkers among (a) All Participants, (b) Pitavastatin group, and (c) Pravastatin group.
Correlation between the protein biomarkers which significantly changed with statin therapy and LDL-C and systemic markers of immune activation are shown. Significant correlations (p < 0.05).
Abbreviations: TFPI, tissue factor pathway inhibitor; PON 3, paraoxonases 3; LDLR, low density lipoprotein receptor; Gal-4, galectin-4; IGFBP-2, insulin-like growth factor binding protein 2; LDL-C, low density lipoprotein cholesterol; sCD163, soluble CD163; hsIL-6, high sensitivity interleukin-6, MCP-1, monocyte chemoattractant protein-1; sCD14, soluble CD14; oxLDL, oxidized low density lipoprotein; Lp-PLA2, lipoprotein-associated phospholipase A2; hs-CRP, high sensitivity c-reactive protein.
4. Discussion
Very limited studies have begun to assess the pleiotropic effects of statins in human disease models using a proteomic approach [[14], [15], [16], [17]]. HIV is a useful model to assess the effects of statins on key pathways of immune activation, endothelial dysfunction, and inflammation. This study provides new biological insights into the effects of statins leveraging a large randomized trial broadly investigating the effects of statins on cardiovascular and atherogenesis protein biomarkers in HIV. To accomplish this, we used a high throughput proteomics method and PEA technology, to simultaneously assess the levels of over 90 protein biomarkers and identified significant changes in several notable protein biomarkers with statin therapy. Of note, the statins utilized in this study – pitavastatin and pravastatin – have fewer drug-drug interactions with ART compared to other statins such as simvastatin and atorvastatin, which are metabolized predominately by the CYP450 3A4 isoenzyme [18]. As such, these statins - pitavastatin and pravastatin - are ideally suited for prevention and/or treatment of CVD and dyslipidemia among PWH on ART, lending clinical relevance to our findings. Moreover, these particular statins may aggravate glucose parameters less than other statins, suggesting potential utility in non-HIV populations. [[19], [20], [21], [22]]
In our study, TFPI levels were significantly decreased with statin therapy; and the reduction in TFPI was significantly greater in the pitavastatin group compared to the pravastatin group. In a prior study among participants living with HIV by deFilippi, et al., the effects of atorvastatin versus placebo on protein biomarkers were investigated using PEA technology. In that study, TFPI was also significantly reduced with atorvastatin versus placebo [10]. Moreover, data from the current study are consistent with a very small prior study demonstrating lovastatin reduced TFPI in non-HIV-infected subjects [23]. TFPI, of note, is a natural inhibitor of tissue factor-mediated coagulation which is synthesized predominately by the vascular endothelium [24,25]. Tissue factor-mediated coagulation plays a central role in acute thrombosis associated with the disruption of atherosclerotic plaque [26]. Moreover, animal studies also suggest that TFPI plays a role in atherogenesis in addition to thrombosis [24]. While most of the TFPI protein can be found bound to the endothelium, the remainder 15–20% can be found in the plasma, either attached to lipoproteins or unbound [27]. It is the unbound TFPI which has physiologically active, anti-coagulant properties [28]. The importance of TFPI is highlighted by the fact that complete deficiency in this protein has never been described in humans and complete deficiency of TFPI in animal models results in embryonic lethality [24]. As such, given the physiologic importance of TFPI, particularly in atherosclerosis, significant changes in the level of this protein biomarker may have clinically relevant implications in the treatment and/or prevention of CVD among PWH.
Increased TFPI levels have been described in various conditions including smoking, diabetes, male gender, increasing age, atherosclerosis, and HIV [29]. And notably, activated monocytes expressing tissue factor have been implicated in the pathogenies of plaque rupture in HIV [30,31]. Increased TFPI levels have been described in individuals with coronary heart disease, with highest levels in those with higher CAD severity. As such, it could be that increased TFPI levels in the peripheral circulation among individuals with atherosclerosis are compensatory or conversely a marker of endothelial dysfunction.
Since plasma TFPI is primarily bound to apolipoprotein B (apo B), reduction via statins might be expected to lower lipoprotein-bound TFPI [23,32]. Indeed, in the current study, the degree of TFPI reduction was highly related to the decrease in LDL-C. By reducing LDL-C, statins could alter the balance between free and bound TFPI, and increase free TFPI, due to a reduction in the major carrier apo B. Future studies assessing statin effects on TFPI activity at the endothelial surface will provide additional insights to this important question.
We previously have described increased arterial inflammation in PWH and how this could also contribute to the increased risk of CVD among PWH [[33], [34], [35]]. Within the INTREPID trial, in turn, we previously demonstrated significant reduction with statin therapy of a systemic marker of arterial inflammation — Lp-PLA2 [7] — which has been associated with increased CVD in the general population [36] and among PWH [37,38]. Interestingly, in this current study, there was a significant relationship between the reduction of TFPI and the reduction of Lp-PLA2 with statin therapy. This relationship highlights the potential interplay between coagulation and inflammation that has been described in various inflammatory states and now requires further investigation among PWH [39].
In addition to the effects of statins on coagulation pathways, we also demonstrated a significant change among PWH with statin therapy in protein biomarkers involved in oxidative stress and redox signaling pathways. PON3, for example, was significantly reduced by statin therapy and this reduction remained significant with analysis of participants in the pitavastatin arm only. PON3 notably can be found on the surface of HDL particles where it is involved in the oxidative modification of LDL-C in addition to its role in monocyte activation [40]. In a cross-sectional study evaluating PWH as well as individuals without HIV, systemic levels of PON3 were notably three times higher in the HIV group [41]. Given the role of PON3 in oxidation, increased levels among PWH may be a marker of oxidative stress. Numerous studies have demonstrated that HIV infection induces oxidative stress through increases in reactive oxygen species triggered directly by several HIV-1 proteins [42]; and oxidative stress through various mechanisms, in turn, plays an important role in atherogenesis [43]. Alternatively, reduction of PON3 may lead to increases in oxidized LDL, though our data did not show a significant pattern in this regard.
Statin therapy in our study also led to a significant change in levels of another protein biomarker, Gal-4, which may also affect atherogenesis. Gal-4, of note, plays an important role in stabilizing lipid rafts [44], which in turn has downstream effects in redox signaling pathways [45]. No prior studies, however, have examined levels of Gal-4 among PWH and/or among individuals with atherosclerosis. Taken together, the significant change in systemic levels of protein markers such as PON3 and Gal-4 with statin therapy among PWH seen in this study highlight potential effects of statins on oxidative stress and redox signaling pathways which are known to play a significant role in atherogenesis [43].
This study demonstrated a decrease in LDLR in response to statin therapy. While statins are known to upregulate LDLR on the hepatic cell surface due to cholesterol depletion, they may simultaneously increase surface recycling of LDLR as well as degradation, due to an increase in PCSK9. Indeed, trends toward increased PCSK9 with statin therapy were shown in the current study. Although the net effect on circulating LDLR resulting from these effects of statin therapy is not known, animal studies have suggested that statins may reduce the half-life of the LDLR [46]. Further research on this, particularly regarding statin effects on PCSK9 in HIV, is needed.
Statin therapy, and most notably pitavastatin, increased insulin like growth factor binding protein-2 (IGFBP-2) in this study. IGFBP-2, through induction of GLUT-4 translocation, contributes to improved insulin sensitivity [47]. This effect on IGFBP-2 may help to explain the neutral effects of pitavastatin on glucose parameters as shown in INTREPID [6] and is consistent with effects of pitavastatin shown in preclinical studies on GLUT-4 translocation.
INTREPID included dyslipidemic individuals with HIV. This study is therefore highly relevant for those HIV patients most in need of statin therapy, but different results might be seen among HIV-infected individuals with lower LDL-C levels. Moreover, we studied two specific statins of importance to the HIV field, given their general lack of significant interactions with ART. Our data may not apply to other statins, but we do note similarities to prior studies that reinforce our results, for example effects on TFPI noted with atorvastatin in a much smaller study of HIV patients with normal LDL-C. Changes in systemic levels of proteins may not reflect the activity of each protein at the tissue level or the biologically active form of the protein. Despite these limitations, the study has many strengths. The study was large enough to allow us to contrast effects across key statins clinically relevant to the management of CVD in HIV. Moreover, we used a highly stringent false discovery rate p-values to determine treatment effects and a high-throughput assessment of protein biomarker levels with demonstrated sensitivity and specificity.
In conclusion, this study is the largest study evaluating the change in protein biomarkers after statin therapy among individuals with HIV. Endothelial dysfunction [3,48] as well as increased systemic immune activation and arterial inflammation [33] are all felt to be significant contributors to increased CVD among PWH. Through simultaneous assessment of systemic protein levels in these pathways, using PEA technology, we identified key protein biomarkers which significantly changed after statin therapy. Additional studies are now needed to explore the potential mechanisms of statin effects on these pathways among PWH, both in terms of pathophysiology and potential optimization of treatment effects. Moreover, the clinical significance of changes in these pathways identified in this novel discovery approach need to be determined. For example, key pathways identified in this study will also inform analyses in the large REPRIEVE trial (NCT02344290), and embedded mechanistic sub-study investigating statin effects to prevent primary CVD events in HIV. Finally, similar proteomic approaches should now be performed in large, randomized studies among non-HIV populations to further extend these findings.
The following are the supplementary data related to this article.
Cardiovascular Disease Panel III Coefficient of Variation for Proximity Extension Assay
Baseline Demographics for Participants Included and Not Included in Study Analysis
Mean Change and T-statistic for Treatment Effect in the Pitavastatin Group and the Pravastatin Group and in the Pitavastatin Group vs. the Pravastatin Group
Difference in Protein Biomarker Levels Before and After Statin Therapy (All Participants)
Correlation Analysis between Changes Protein Biomarkers and Baseline Demographics. 5a. All Participants. 5b. Pitavastatin Group. 5c. Pitavastatin Group.
Modification of Statin Effects on Protein Levels by Protease Inhibitor Use
Modification of Statin Effects on Protein Levels by NNRTI Use
Modification of Statin Effects on Protein Levels by Integrase-inhibitor Use
Author contributions
MT was involved in data collection, data analysis, data interpretation, figure and table preparation, reference search, writing, and editing the manuscript; KVF was involved in data collection and editing the manuscript; LS was involved in data collection, data analysis, figure and table preparation, and editing the manuscript; MVZ, JL, and CD were involved in editing the manuscript. CAS was involved in study design, data collection, data interpretation, writing, and editing the manuscript; HL was involved in data analysis, data interpretation, writing, and editing the manuscript; IG was involved in data collection, data analysis, data interpretation, and editing the manuscript. MAT and JAA were involved in parent study design, served as investigators, and were involved in data interpretation, writing, and editing the manuscript; SKG was involved in study design, data collection, data analysis, data interpretation, reference search, figure and table preparation, writing, and editing the manuscript.
Declaration of interests
MVZ has participated in a scientific advisory board meeting for Roche Diagnostics and has received investigator-initiated research funding to her institution from Gilead, unrelated to the present manuscript. JL has served as served as a consultant for Gilead Sciences and Viiv Healthcare, both unrelated to the manuscript. CD unrelated to this manuscript receives research funding from Roche Diagnostics, has consulted for Abbott Diagnostics, FujiRebio, Metanomics, Ortho Diagnostics and Siemens Healthcare, received royalties from UpToDate. CAS is an employee of KOWA Pharmaceuticals America, Inc. IG is an employee of Olink Proteomics. MAT has received research support to her institution from KOWA Pharmaceuticals to conduct the parent study. She has received unrelated research support to her institution from Bristol Myers Squibb, CytoDyn Inc., Gilead Sciences, GlaxoSmithKline, Merck Sharp Dohme, Roche Laboratories, Taimed Inc., and ViiV Healthcare. JAA received grants from KOWA Pharmaceuticals to conduct the parent study; she also received grants from Gilead Sciences and Glaxo-Smith-Kline/ViiV Healthcare, and received scientific advisory board personal fees from Gilead, Janssen, Merck, and Glaxo-Smith-Kline/ViiV Healthcare, all unrelated to the present study. SKG has received research support to his institution from KOWA Pharmaceuticals. Unrelated to this manuscript he has consulted for Theratechnologies, Gilead and Navidea and received research support from Gilead and Theratechnologies. MT, KVF, LS, and HL have no disclosures to report.
Funding
This work was supported by an investigator-initiated grant to S.K.G. from KOWA Pharmaceuticals America, Inc. and the National Institutes of Health [P30 DK040561; Nutrition Obesity Research Center at Harvard]. M.T. was support by National Institutes of Health [5KL2TR001100-05; Harvard Catalyst KL2 grant].
Role of sponsor
The Sponsor funded the study but had no role in the analysis of the data nor in the decision to publish the data.
References
- 1.Triant V.A., Lee H., Hadigan C., Grinspoon S.K. Increased acute myocardial infarction rates and cardiovascular risk factors among patients with human immunodeficiency virus disease. J. Clin. Endocrinol. Metab. 2007 Jul.;92(7):2506–2512. doi: 10.1210/jc.2006-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freiberg M.S., Chang C.C., Kuller L.H., Skanderson M., Lowy E., Kraemer K.L. HIV infection and the risk of acute myocardial infarction. JAMA Intern. Med. 2013 Apr. 22;173(8):614–622. doi: 10.1001/jamainternmed.2013.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beltran L.M., Rubio-Navarro A., Amaro-Villalobos J.M., Egido J., Garcia-Puig J., Moreno J.A. Influence of immune activation and inflammatory response on cardiovascular risk associated with the human immunodeficiency virus. Vasc. Health Risk Manag. 2015;11:35–48. doi: 10.2147/VHRM.S65885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Longenecker C.T., Sattar A., Gilkeson R., McComsey G.A. Rosuvastatin slows progression of subclinical atherosclerosis in patients with treated HIV infection. AIDS. 2016 Sep 10;30(14):2195–2203. doi: 10.1097/QAD.0000000000001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lo J., Lu M.T., Ihenachor E.J., Wei J., Looby S.E., Fitch K.V. Effects of statin therapy on coronary artery plaque volume and high-risk plaque morphology in HIV-infected patients with subclinical atherosclerosis: A randomised, double-blind, placebo-controlled trial. Lancet HIV. 2015 Feb;2(2):e52–e63. doi: 10.1016/S2352-3018(14)00032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aberg J.A., Sponseller C.A., Ward D.J., Kryzhanovski V.A., Campbell S.E., Thompson M.A. Pitavastatin versus pravastatin in adults with HIV-1 infection and dyslipidaemia (INTREPID): 12 week and 52 week results of a phase 4, multicentre, randomised, double-blind, superiority trial. Lancet HIV. 2017 Jul.;4(7) doi: 10.1016/S2352-3018(17)30075-9. (e284-e94) [DOI] [PubMed] [Google Scholar]
- 7.Toribio M., Fitch K.V., Sanchez L., Burdo T.H., Williams K.C., Sponseller C.A. Effects of pitavastatin and pravastatin on markers of immune activation and arterial inflammation in HIV. AIDS. 2017 Mar 27;31(6):797–806. doi: 10.1097/QAD.0000000000001427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Longenecker C.T., Jiang Y., Orringer C.E., Gilkeson R.C., Debanne S., Funderburg N.T. Soluble CD14 is independently associated with coronary calcification and extent of subclinical vascular disease in treated HIV infection. AIDS. 2014 Apr 24;28(7):969–977. doi: 10.1097/QAD.0000000000000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandler N.G., Wand H., Roque A., Law M., Nason M.C., Nixon D.E. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J. Infect. Dis. 2011 Mar 15;203(6):780–790. doi: 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Defilippi C., Lo J., Christenson R., Grundberg I., Stone L., Zanni M.V. Novel mediators of statin effects on plaque in HIV: A proteomics approach. AIDS. 2018 Apr 24;32(7):867–876. doi: 10.1097/QAD.0000000000001762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lind L., Arnlov J., Lindahl B., Siegbahn A., Sundstrom J., Ingelsson E. Use of a proximity extension assay proteomics chip to discover new biomarkers for human atherosclerosis. Atherosclerosis. 2015 Sep;242(1):205–210. doi: 10.1016/j.atherosclerosis.2015.07.023. [DOI] [PubMed] [Google Scholar]
- 12.Assarsson E., Lundberg M., Holmquist G., Bjorkesten J., Thorsen S.B., Ekman D. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE. 2014;9(4) doi: 10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Royal Stat. Society 1995. 1995;57(1):289–300. [Google Scholar]
- 14.Bhandari S., Gupta P., Quinn P., Sandhu J., Hakimi A., Jones D. Pleiotropic effects of statins in hypercholesterolaemia: a prospective observational study using a lipoproteomic based approach. Lancet. 2015 Feb 26;385(Suppl. 1):S21. doi: 10.1016/S0140-6736(15)60336-1. [DOI] [PubMed] [Google Scholar]
- 15.Thongtang N., Diffenderfer M.R., Ooi E.M.M., Barrett P.H.R., Turner S.M., Le N.A. Metabolism and proteomics of large and small dense LDL in combined hyperlipidemia: Effects of rosuvastatin. J. Lipid Res. 2017 Jul;58(7):1315–1324. doi: 10.1194/jlr.M073882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barderas M.G., Tunon J., Darde V.M., De la Cuesta F., Jimenez-Nacher J.J., Tarin N. Atorvastatin modifies the protein profile of circulating human monocytes after an acute coronary syndrome. Proteomics. 2009 Apr;9(7):1982–1993. doi: 10.1002/pmic.200700583. [DOI] [PubMed] [Google Scholar]
- 17.Gordon S.M., McKenzie B., Kemeh G., Sampson M., Perl S., Young N.S. Rosuvastatin alters the proteome of high density lipoproteins: Generation of alpha-1-antitrypsin enriched particles with anti-inflammatory properties. Mol. Cell. Proteomics. 2015 Dec;14(12):3247–3257. doi: 10.1074/mcp.M115.054031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chauvin B., Drouot S., Barrail-Tran A., Taburet A.M. Drug-drug interactions between HMG-CoA reductase inhibitors (statins) and antiviral protease inhibitors. Clin. Pharmacokinet. 2013 Oct;52(10):815–831. doi: 10.1007/s40262-013-0075-4. [DOI] [PubMed] [Google Scholar]
- 19.Sugiyama S., Fukushima H., Kugiyama K., Maruyoshi H., Kojima S., Funahashi T. Pravastatin improved glucose metabolism associated with increasing plasma adiponectin in patients with impaired glucose tolerance and coronary artery disease. Atherosclerosis. 2007 Oct;194(2):e43–e51. doi: 10.1016/j.atherosclerosis.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 20.Mita T., Watada H., Nakayama S., Abe M., Ogihara T., Shimizu T. Preferable effect of pravastatin compared to atorvastatin on beta cell function in Japanese early-state type 2 diabetes with hypercholesterolemia. Endocr. J. 2007 Jun;54(3):441–447. doi: 10.1507/endocrj.k06-198. [DOI] [PubMed] [Google Scholar]
- 21.Mita T., Nakayama S., Abe H., Gosho M., Iida H., Hirose T. Comparison of effects of pitavastatin and atorvastatin on glucose metabolism in type 2 diabetic patients with hypercholesterolemia. J. Diabet. Investig. 2013 May 6;4(3):297–303. doi: 10.1111/jdi.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park J.B., Jung J.H., Yoon Y.E., Kim H.L., Lee S.P., Kim H.K. Long-term Effects of high-doSe pitavaStatin on diabetogenicity in comparison with atorvastatin in patients with metabolic syndrome (LESS-DM): study protocol for a randomized controlled trial. Trials. 2017 Oct 27;18(1):501. doi: 10.1186/s13063-017-2229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen J.B., Huseby N.E., Sandset P.M., Svensson B., Lyngmo V., Nordoy A. Tissue-factor pathway inhibitor and lipoproteins. Evidence for association with and regulation by LDL in human plasma. Arterioscler. Thromb. 1994 Feb;14(2):223–229. doi: 10.1161/01.atv.14.2.223. [DOI] [PubMed] [Google Scholar]
- 24.Winckers K., ten Cate H., Hackeng T.M. The role of tissue factor pathway inhibitor in atherosclerosis and arterial thrombosis. Blood Rev. 2013 May;27(3):119–132. doi: 10.1016/j.blre.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Lupu C., Lupu F., Dennehy U., Kakkar V.V., Scully M.F. Thrombin induces the redistribution and acute release of tissue factor pathway inhibitor from specific granules within human endothelial cells in culture. Arterioscler. Thromb. Vasc. Biol. 1995 Nov;15(11):2055–2062. doi: 10.1161/01.atv.15.11.2055. [DOI] [PubMed] [Google Scholar]
- 26.Ardissino D., Merlini P.A., Ariens R., Coppola R., Bramucci E., Mannucci P.M. Tissue-factor antigen and activity in human coronary atherosclerotic plaques. Lancet. 1997 Mar 15;349(9054):769–771. doi: 10.1016/S0140-6736(96)11189-2. [DOI] [PubMed] [Google Scholar]
- 27.Werling R.W., Zacharski L.R., Kisiel W., Bajaj S.P., Memoli V.A., Rousseau S.M. Distribution of tissue factor pathway inhibitor in normal and malignant human tissues. Thromb. Haemost. 1993 Apr 1;69(4):366–369. [PubMed] [Google Scholar]
- 28.Bajaj M.S., Birktoft J.J., Steer S.A., Bajaj S.P. Structure and biology of tissue factor pathway inhibitor. Thromb. Haemost. 2001 Oct;86(4):959–972. [PubMed] [Google Scholar]
- 29.Mitchell C.T., Kamineni A., Palmas W., Cushman M. Tissue factor pathway inhibitor, vascular risk factors and subclinical atherosclerosis: the multi-ethnic study of atherosclerosis. Atherosclerosis. 2009 Nov.;207(1):277–283. doi: 10.1016/j.atherosclerosis.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Funderburg N.T., Mayne E., Sieg S.F., Asaad R., Jiang W., Kalinowska M. Increased tissue factor expression on circulating monocytes in chronic HIV infection: relationship to in vivo coagulation and immune activation. Blood. 2010 Jan 14;115(2):161–167. doi: 10.1182/blood-2009-03-210179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Funderburg N.T., Zidar D.A., Shive C., Lioi A., Mudd J., Musselwhite L.W. Shared monocyte subset phenotypes in HIV-1 infection and in uninfected subjects with acute coronary syndrome. Blood. 2012 Nov 29;120(23):4599–4608. doi: 10.1182/blood-2012-05-433946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen J.B., Huseby K.R., Huseby N.E., Ezban M., Nordoy A. Tissue factor pathway inhibitor in complex with low density lipoprotein isolated from human plasma does not possess anticoagulant function in tissue factor-induced coagulation in vitro. Thromb. Res. 1997 Mar 1;85(5):413–425. doi: 10.1016/s0049-3848(97)00029-7. [DOI] [PubMed] [Google Scholar]
- 33.Subramanian S., Tawakol A., Burdo T.H., Abbara S., Wei J., Vijayakumar J. Arterial inflammation in patients with HIV. JAMA. 2012 Jul 25;308(4):379–386. doi: 10.1001/jama.2012.6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zanni M.V., Toribio M., Robbins G.K., Burdo T.H., Lu M.T., Ishai A.E. Effects of antiretroviral therapy on immune function and arterial inflammation in treatment-naive patients with human immunodeficiency virus infection. JAMA Cardiol. 2016 Jul 1;1(4):474–480. doi: 10.1001/jamacardio.2016.0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zanni M.V., Toribio M., Wilks M.Q., Lu M.T., Burdo T.H., Walker J. Application of a novel CD206+ macrophage-specific arterial imaging strategy in HIV. J. Infect. Dis. 2017 Apr 15;215(8):1264–1269. doi: 10.1093/infdis/jix095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson A., Gao P., Orfei L., Watson S., Di Angelantonio E., Kaptoge S. Lipoprotein-associated phospholipase a(2) and risk of coronary disease, stroke, and mortality: collaborative analysis of 32 prospective studies. Lancet. 2010 May 1;375(9725):1536–1544. doi: 10.1016/S0140-6736(10)60319-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross Eckard A., Longenecker C., Jiang Y., Debanne S., Labbato D., Storer N. Lipoprotein-associated phospholipase a and cardiovascular disease risk in HIV infection. HIV Med. 2014 Oct;15(9):537–546. doi: 10.1111/hiv.12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mangili A., Ahmad R., Wolfert R.L., Kuvin J., Polak J.F., Karas R.H. Lipoprotein-associated phospholipase A2, a novel cardiovascular inflammatory marker, in HIV-infected patients. Clin. Infect. Dis. 2014 Mar;58(6):893–900. doi: 10.1093/cid/cit815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levi M., van der Poll T., Buller H.R. Bidirectional relation between inflammation and coagulation. Circulation. 2004 Jun 8;109(22):2698–2704. doi: 10.1161/01.CIR.0000131660.51520.9A. [DOI] [PubMed] [Google Scholar]
- 40.Kowalska K., Socha E., Milnerowicz H. Review: the role of paraoxonase in cardiovascular diseases. Ann. Clin. Lab. Sci. 2015 Spring;45(2):226–233. [PubMed] [Google Scholar]
- 41.Aragones G., Garcia-Heredia A., Guardiola M., Rull A., Beltran-Debon R., Marsillach J. Serum paraoxonase-3 concentration in HIV-infected patients. Evidence for a protective role against oxidation. J. Lipid Res. 2012 Jan.;53(1):168–174. doi: 10.1194/jlr.P018457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivanov A.V., Valuev-Elliston V.T., Ivanova O.N., Kochetkov S.N., Starodubova E.S., Bartosch B. Oxidative stress during HIV infection: Mechanisms and consequences. Oxidative Med. Cell. Longev. 2016;2016 doi: 10.1155/2016/8910396. (8910396) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Catalgol B., Kartal Ozer N. Lipid rafts and redox regulation of cellular signaling in cholesterol induced atherosclerosis. Curr. Cardiol. Rev. 2010 Nov;6(4):309–324. doi: 10.2174/157340310793566181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lemaire-Ewing S., Lagrost L., Neel D. Lipid rafts: a signalling platform linking lipoprotein metabolism to atherogenesis. Atherosclerosis. 2012 Apr;221(2):303–310. doi: 10.1016/j.atherosclerosis.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 45.Zhang A.Y., Yi F., Zhang G., Gulbins E., Li P.L. Lipid raft clustering and redox signaling platform formation in coronary arterial endothelial cells. Hypertension. 2006 Jan;47(1):74–80. doi: 10.1161/10.1161/01.HYP.0000196727.53300.62. [DOI] [PubMed] [Google Scholar]
- 46.Ness G.C., Zhao Z., Lopez D. Inhibitors of cholesterol biosynthesis increase hepatic low-density lipoprotein receptor protein degradation. Arch. Biochem. Biophys. 1996 Jan 15;325(2):242–248. doi: 10.1006/abbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 47.Assefa B., Mahmoud A.M., Pfeiffer A.F.H., Birkenfeld A.L., Spranger J., Arafat A.M. Insulin-like growth factor (IGF) binding protein-2, independently of IGF-1, induces GLUT-4 translocation and glucose uptake in 3T3-L1 adipocytes. Oxidative Med. Cell. Longev. 2017;2017 doi: 10.1155/2017/3035184. (3035184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duprez D.A., Neuhaus J., Kuller L.H., Tracy R., Belloso W., De Wit S. Inflammation, coagulation and cardiovascular disease in HIV-infected individuals. PLoS ONE. 2012;7(9) doi: 10.1371/journal.pone.0044454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cardiovascular Disease Panel III Coefficient of Variation for Proximity Extension Assay
Baseline Demographics for Participants Included and Not Included in Study Analysis
Mean Change and T-statistic for Treatment Effect in the Pitavastatin Group and the Pravastatin Group and in the Pitavastatin Group vs. the Pravastatin Group
Difference in Protein Biomarker Levels Before and After Statin Therapy (All Participants)
Correlation Analysis between Changes Protein Biomarkers and Baseline Demographics. 5a. All Participants. 5b. Pitavastatin Group. 5c. Pitavastatin Group.
Modification of Statin Effects on Protein Levels by Protease Inhibitor Use
Modification of Statin Effects on Protein Levels by NNRTI Use
Modification of Statin Effects on Protein Levels by Integrase-inhibitor Use