Abstract
CMTM6, a previously uncharacterized protein, was identified as a critical regulator of PD-L1, which is reported as an immune checkpoint inhibitor, to modulate the T cell activities both in vitro and in vivo of other tumors. However, the role of CMTM6 has so far remained unclear in glioma. To investigate the role of CMTM6 in gliomas, we analyzed the transcriptome level, genomic profiles and its relationship with clinical practice. 1862 glioma samples with transcriptome data were enrolled in this study, including CGGA RNA-seq, TCGA RNA-seq, CGGA-microarray, GSE16011 and IVY GBM databases. Clinical information and genomic profiles containing somatic mutations and DNA copy numbers were also obtained. We found that CMTM6 expression was highly correlated with major clinical and molecular characteristics. Cases with high CMTM6 expression were more likely to be predicted as malignant entities and frequent with genomic aberrations of driver oncogenes. Moreover, gene ontology analysis based on significantly correlated genes of CMTM6 expression exhibited that CMTM6 was associated with immune responses and inflammatory activities. CMTM6 was synthetic with other immune checkpoint inhibitors. Additionally, CMTM6 was involved in immune functions via modulating T-lymphocyte-mediated anti-tumor immunity. Finally, high CMTM6 expression was associated with reduced survival time and may serve as a strong indicator of poor prognosis in gliomas. In brief, High level of CMTM6 expression is closely related to high malignant gliomas. Meanwhile, CMTM6 plays an important role in regulating T cell activation and antitumor responses. Therefore, CMTM6 is a promising target for developing immunotherapy of gliomas.
Keywords: Glioma, CMTM6, Inflammatory activity, Immune response, Prognosis, Epigenetics
Highlights
-
•
High malignancy of gliomas, such as high grade, IDH wildtype, or Mesenchymal subtype gliomas, exhibit high CMTM6 expression.
-
•
Somatic mutations and copy number alterations of the tumor-associated genes are prevalent in gliomas with high CMTM6 expression.
-
•
CMTM6 is involved in the immune response and inflammatory activities in gliomas.
-
•
CMTM6 is a potential predictor of poor prognosis in glioma patients.
Research in context.
CMTM6, a type-3 transmembrane protein of previously unknown functions, was recently identified as a critical regulator of PD-L1 in the tumor cells. However, the role of CMTM6 has so far remained unclear in human cancer tissue. To investigate the relationship between CMTM6 expression and clinical practice, we analyzed the molecular and clinical characteristics of CMTM6 expression in large sample-sized glioma. High level of CMTM6 expression was associated with malignant entities. Different CMTM6 expression also indicated diverse genomic aberrations. Moreover, gene ontology analysis revealed that CMTM6 played an important role in the regulation of immune responses and inflammatory activations, especially in modulating suppression of T cell immunity to antitumor. Furthermore, patients with high CMTM6 expression had shorter median survival. These findings suggest that CMTM6 can be served as a novel potential therapeutic target and may facilitate the improvement of the current therapies.
Alt-text: Unlabelled Box
1. Introduction
Gliomas are the most common primary malignant tumors of the central nervous system (CNS) in adults [1]. Despite advances in comprehensive therapy, such as neurosurgical resection, adjuvant radiotherapy, and alkylating agent temozolomide chemotherapy, patients who suffer from gliomas still have a short median survival time, due to the aggressiveness of tumors, resistance to treatments, and recurrence over time [2]. In the past decade, studies on the anticancer immune responses for other tumors have promoted clinical advances in the limited success of conventional therapies [3]. Meanwhile, the discovery of CNS lymphatic system has provided a new theoretical basis and opportunity for brain tumor immunotherapy [4].
Patients have been observed to benefit upon blockage of the interaction between programmed death-1 (PD-1) and ligand-1 (PD-L1) to inhibit the suppression of T cell immune responses [5]. Meanwhile, clinical trials of PD-1/PD-L1 targeting in glioma have been initiated [6]. However, the rate of objective response has been <13% - 56% in the therapies of other tumors with PD-1/PD-L1 inhibitors, while the rate of complete response has been 1% - 16% [[7], [8], [9], [10], [11], [12], [13]]. Hence, the acknowledgment of the molecular regulation of PD-L1 expression is limited.
Recently, two studies reported that CKLF-like MARVEL transmembrane domain containing 6 (CMTM6), a previously unknown protein at the plasma membrane, was identified as a critical regulator of the PD-L1 protein [14, 15]. CMTM6 promotes PD-L1 expression in tumor cells against T cells [15]. On the contrary, the depletion of CMTM6 relieves T cell immunosuppression [14]. CMTM6, as a new immune checkpoint in tumor-induced immunosuppression, exhibits new avenues to overcome tumor immune escape.
Previous studies published that CMTM6 suppresses T cell activation and the anti-tumor responses in vitro and in mouse melanoma models [14, 15]. However, they did not show the capacitation of CMTM6 expression in neither the whole WHO Grade gliomas nor even the brain tumor. To clarify the CMTM6 status in all gliomas, we gathered RNA-seq data of 325 glioma samples from Chinese Glioma Genome Atlas (CGGA) dataset. Moreover, our findings were verified in RNA-seq data from TCGA network, CGGA microarray, and GSE16011 microarray. In addition, genomic profiles containing somatic mutations and DNA copy numbers were also obtained. This study is the first integrative study characterizing CMTM6 expression in whole grade glioma both molecularly and clinically.
2. Materials and methods
2.1. Data collection
This study was approved by Capital Medical University Institutional Review Board (IRB). In CGGA dataset, we collected transcriptome sequencing data of 325 samples generated by Illumina HiSeq platform. Clinical and molecular information was obtained from the CGGA database. The methodology for detecting IDH mutation status was described in the previous study [16]. The Cancer Genome Atlas (TCGA) mRNAseq data of 698 gliomas, ranging from WHO grade II to grade IV, and 5 normal brain tissues were downloaded from public databases (https://portal.gdc.cancer.gov/). 301 glioma cases were obtained from CGGA microarray dataset, and 276 cases were from GSE16011 cohort. RNA-seq data for specific tumor anatomic structure in GBM, identified by H&E staining, was from Ivy Glioblastoma Atlas Project (ttp://glioblastoma.alleninstitute.org/).
2.2. Bioinformatic analysis
648 cases with somatic mutations and 690 cases with somatic copy number alternations (CNAs), which corresponded to the cases with RNA-seq data, were downloaded from TCGA database. Copy number alternations associated with CMTM6 expression were analyzed by GISTIC 2.0 [17]. The thresholded copy number at alteration peaks were from GISTIC analysis (all_lesions.cof_99.txt file) [18]. The biological functions of the related genes were explored by GO analysis in DAVID Bioinformatics Resources 6.8 [19]. Gene set variation analysis (GSVA) analysis was described in the previous study [16]. After Spearman correlation analysis, gene ontology (GO) analysis of the most correlated genes was constructed by Heatmap. The GO geneset was downloaded from AmiGO 2 Web portal (http://amigo.geneontology.org/amigo/landing). Inflammatory related metagenes were described as before [20, 21]. Metagene expression values were performed by calculating the mean of the normalized expression values of all genes in the respective cluster, as described in a previous study [21].
2.3. Statistical analysis
Correlations between continuous variables were evaluated by Spearman correlation analysis. The Student t-test, one-way ANOVA, or Pearson's Chi-squared test was used to assess differences in variables between groups. The survival distributions were described by the Kaplan – Meier survival curve and the log-rank test was used to test the statistical significance. The prognostic value of CMTM6 was estimated by Univariate and multivariate Cox proportional hazard model analysis. Patients with missing information were excluded from the corresponding analysis. All statistical analyses were performed using IBM SPSS Statistical software (version 24.0, Armonk, NY: IBM Corp) and R project (version 3.4.1, https://www.r-project.org/). A p-value <.05 is considered as significant. All statistical tests were two-sided.
3. Results
3.1. Associations of CMTM6 expression with clinical and molecular characteristics in gliomas
CMTM6 expression in gliomas was dramatically higher than in normal brain tissues (Supplementary Fig. S1, Supplementary Tables S1 and S2). Gliomas from the RNA-seq and microarray were arranged in order of increasing CMTM6 expression (Fig. 1A). We found a positive correlation between CMTM6 expression and age at diagnosis in CGGA dataset, TCGA dataset, and CGGA microarray, while a negative association was shown between CMTM6 expression and KPS score in TCGA dataset and CGGA microarray (Fig. 1A). Although no significant difference was detected between primary gliomas and recurrent gliomas, even in glioblastoma mutilforme (GBM), CMTM6 expression was higher in primary GBM than in secondary GBM in CGGA-microarray cohort (Supplementary Fig. S1B and S1C). Due to the histopathological heterogeneity of glioma, the RNA-sequencing data of glioma were analyzed according to WHO grade system, histology, and IDH mutation status. The expression of CMTM6 was the highest in the glioblastoma (WHO grade IV) compared with WHO grade II and grade III glioma (Fig. 1B). Besides, CMTM6 expression increased in higher histopathologic malignancies (Fig. 1C). Meanwhile, according to 2016 WHO classification of tumors in the central nervous system, we divided gliomas into five group including lower-grade glioma (LGG) oligodendroglioma (LGG-Oligo, IDH-mutant LGG with TERT promoter mutation or 1p/19q codeletion), LGG astrocytoma (LGG-Astro, IDH-mutant LGG without TERT promoter mutation or 1p/19q codeletion and with ATRX mutation), LGG with wild-type IDH status (LGG IDH-wildtype), GBM with mutant IDH status (GBM IDH-mutant), and GBM with wild-type IDH status (GBM IDH-wildtype) [1, 22]. CMTM6 expression was highest in GBM IDH-mutant but lowest in LGG-Oligo (Supplementary Fig. S1D). Additionally, the higher expression level of CMTM6 was detected in the gliomas with a wild-type IDH gene (Fig. 1D; Supplementary Fig. S1E), which plays an essential role in the progression of glioma. Furthermore, we found that the IDH wild-type glioma showed a distinct pattern expression level of CMTM6 from IDH mutation glioma in the different grade (Supplementary Figs. S1F). These findings indicated that the high expression of CMTM6 predicted a high malignant glioma. Then, we also quantified CMTM6 expression in RNA-seq data for specific tumor anatomic structure, identified by H&E staining, from Ivy Glioblastoma Atlas Project (http://glioblastoma.alleninstitute.org/). High levels of CMTM6 expression in GBM was enriched in the hyperplastic blood vessels and microvascular proliferation (Fig. 1E), which are important in the progression of tumors.
Fig. 1.
The landscape of clinical and molecular features in associations with CMTM6 expression. A. Datasets were arranged in order of increasing CMTM6 expression. The relationships between CMTM6 expression and clinical characteristics were evaluated. ns, *, and *** indicate no significant difference, p < .05, and p < .001, respectively. B. The expression levels of CMTM6 increased by the WHO grade. ***, p < .001. C. The expression levels of CMTM6 increased by the histopathologic classification. ***, p < .001. D. CMTM6 expression was upregulated in the IDH wild-type gliomas compared with the IDH mutant gliomas. ***, p < .001. E. CMTM6 expression was detected in the different location of GBM in IVY GBM database. ***, p < .001. F. The CMTM6 expression pattern in the TCGA molecular subtype. ROC curves predicted CMTM6 as a biomarker of mesenchymal subtype glioma. ***, p < .001.
Molecular classification provides a fresh perspective to define different patient outcomes based on transcriptomic and genomic dimensions [23]. As a result, we investigated the distribution of CMTM6 expression in different molecular subclasses of glioma according to the definition of TCGA network. As shown in Figs. 1F, CMTM6 was dramatically upregulated in mesenchymal subtype in CGGA dataset as well as in TCGA dataset, compared with other subtypes. Furthermore, ROC curve was used to evaluate the discrimination ability of CMTM6 expression for Mesenchymal subtype in all grade glioma. Surprisingly, the area under the curve of CMTM6 expression were 88.7% and 89.4% in CGGA cohort and TCGA cohort, respectively (Figs. 1F). These results suggested that CMTM6 may play an important role in the progression of glioma. Meanwhile, CMTM6 may serve as a biomarker for the Mesenchymal subtype.
3.2. CMTM6 expression associates with distinct patterns of genomic alterations
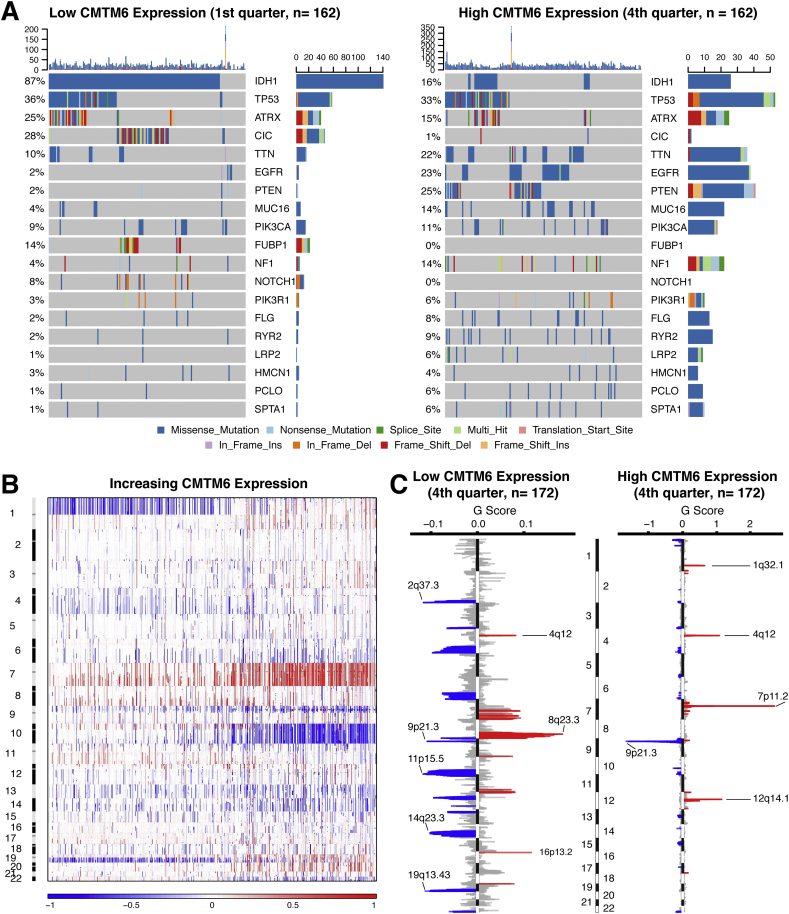
To explore the molecular mechanisms in gliomas, somatic mutations and copy number alterations from TCGA database were analyzed. A positive correlation was detected between CMTM6 expression somatic mutations (R = 0.2449, P < .001, Supplementary Fig. S2A). Based on the order of increasing CMTM6 expression, cases were classified into two, three, or four groups. Parallel analyses were performed in each kind of grouping to enhance the credibility of our findings. After comparing the frequency of mutations between cases with low and high CMTM6 expression, more somatic mutations were revealed in cases with high CMTM6 expression (1st vs. 2nd half, 8660 vs. 12,154 mutations; 1st vs 3rd tertile, 5504 vs. 8703 mutations; 1st vs. 4th, 4179 vs. 6847 mutations). Mutations in IDH1, CIC, FUBP1, and NOTCH1 were significantly enriched in the cases with low CMTM6 expression (Fig. 2A; Supplementary Fig. S3; Supplementary Table S3). Meanwhile, high frequency of mutations in EGFR, PTEN, and NF1 was observed in gliomas with high CMTM6 expression (Fig. 2A; Supplementary Fig. S3; Supplementary Table S3). A significant difference of mutations in TTN, MUC16, FLG, RYR2, LRP2, and SPTA1 was also detected in various conditions of CMTM6 expression.
Fig. 2.
Distinct genomic profiles associated with CMTM6 expression. A. Differential somatic mutations were detected in gliomas with low and high CMTM6 expression. B. The overall CNAs profile in order of increasing CMTM6 expression. C. GISTIC 2.0 amplifications and deletions in gliomas with low and high CMTM6 expression. Chromosomal locations of peaks of significantly recurring focal amplification (red) and deletions (blue) were presented.
Then, somatic copy number alternations were investigated between cases with low CMTM6 expression and cases with high CMTM6 expression. A positive correlation was also observed between CMTM6 expression and CNAs (R = 0.2127, P < .001, Supplementary Fig. S2B). Besides, more segment count of CNAs was found in cases with high CMTM6 expression (1st vs. 2nd half, 40,215 vs. 57,547 CNAs; 1st vs 3rd tertile, 25,644 vs. 38,003 CNAs; 1st vs. 4th, 18,672 vs. 28,064 CNAs). As shown in Fig. 2B, an amplification of Chr 7 accompanied with a deletion of Chr 10 was enriched in gliomas with high CMTM6 expression. Whereas, the incidence of the 1p/19q codeletion, which is a genomic hallmark in oligodendroglioma [1], reduced along with increasing of CMTM6 expression in gliomas (Fig. 2B). GISTIC 2.0 analysis of all gliomas identified 29 significantly reoccurring focal amplifications and 38 deletion events (Supplementary Dataset S1). In the cases with high CMTM6 expression, focal amplification peaks, containing well-characterized driver oncogenes such as PIK3C2B (1q32.1), PDGFRA (4q12), EGFR (7p11.2), and CDK4 (12q14.1), were observed accompanied by a 9p21.3 (CDKN2A and CDKN2B) focal deletion peak (Fig. 2C; Supplementary Fig. S3). Although a 4q12 peak was also detected in the cases with low CMTM6 expression, the G score was obviously lower than the gliomas with high CMTM6 expression. Meanwhile, significant amplifications showed peaks in 8q23.3 and 16q13.2, while the frequently deleted genomic regions were 2q37.3, 9p21.3, 11p15.5, and 19q13.43 (Fig. 2C, Supplementary Fig. S3).
3.3. CMTM6 related biological processes
According to the aforementioned results, CMTM6 may play an essential role in the biological functions of glioma. To clarify the biological roles of CMTM6 expression in glioma, we selected the 1020 genes and 1690 genes, which were strongly correlated with CMTM6 by Pearson correlation analysis (Pearson |R| > 0.5), in CGGA dataset and TCGA dataset, respectively, to perform a subsequent analysis. Then, the biological functions of the related genes were explored by GO analysis in DAVID Bioinformatics Resources 6.8 [19]. We found that the genes, which were positively correlated with CMTM6 expression, were mostly involved in inflammatory response and immune response, when the gene functions were sorted by p-value in increasing order in CGGA database (Fig. 3A). Meanwhile, these results were also verified in the TCGA database (Fig. 3B). In both CGGA cohort and TCGA cohort, the negatively related genes were involved in normal biological processes such as neurotransmitter secretion, memory, and others (Supplement Figs. S4A and B). In addition, the significantly related genes shared by two datasets were chosen to draw the heat maps (Supplement Figs. S4C and D). As mentioned earlier, the expression of CMTM6 was high in GBM and mesenchymal subtype glioma, and we used the GBM cases to make further investigation. As shown in Supplement Fig. S5, related genes were still involved in inflammatory response and immune response.
Fig. 3.
Gene ontology analysis for CMTM6 in glioma. A, B. Gene ontology analysis of positively related biological process showed that CMTM6 was mostly in inflammatory response and immune response in CGGA and TCGA datasets. C, D. Heatmaps showed that most immune-related genes were positively correlated with CMTM6 expression in CGGA and TCGA datasets, while a small number of genes were negatively associated.
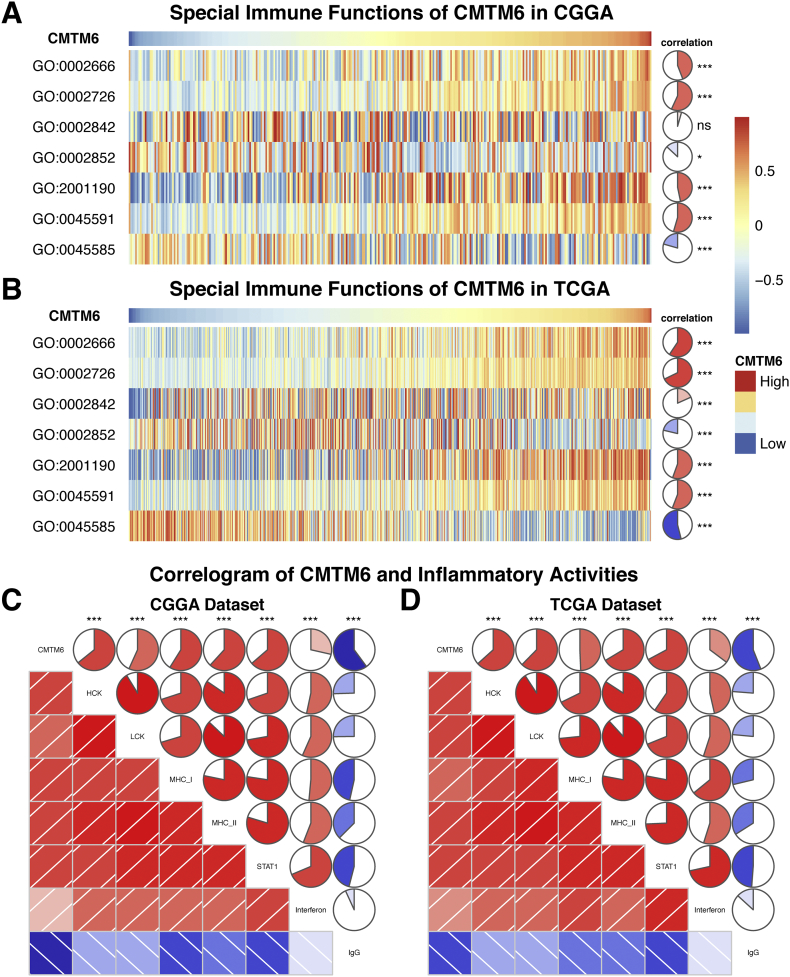
3.4. CMTM6 is related to immune response
To clarify the role of CMTM6 in the immune response in glioma, we found the gene sets related to immune responses from the AmiGo 2 Web portal as described previously [16]. Then, we chose 314 genes which were most relevant to CMTM6 (Pearson |R| > 0.4) to draw the heat maps. Among these genes, 291 genes were positively correlated with CMTM6 expression, while the number of negatively related genes was 23 (Figs. 3C and D). Thus, in glioma CMTM6 is positively correlated with most relevant immune responses while negatively correlated with few relevant immune responses.
3.5. CMTM6 is synergistic with other checkpoint members in tumor-induced immune response
In the recent study, CMTM6 was identified as a critical regulator of PD-L1 [14, 15], which is involved in tumor immunity escapes. Here, PD-L1 is one of the ligands of PD-1, while PD-L2 is the other ligand. CD80, expressed on the surface of activated CD8+ T cells, interacts with PD-L1 to limit antitumor CD8+ T cell response. Therefore, to test this relationship, we performed Pearson correlation analysis with CMTM6, PD-L1, PD-L2, PD-1, and CD80 expression in both CGGA dataset and TCGA dataset. In whole grade gliomas of CGGA cohort, CMTM6 showed high correlation with PD-L2 and CD80, followed by PD-1 and PD-L1 (Supplementary Fig. S6A). We also detected the correlation in low-grade glioma and glioblastoma of CGGA dataset (Supplementary Fig. 6B and C). Furthermore, we observed similar results with whole grade gliomas, which were also verified in TCGA database (Supplementary Fig. S6D, E, and F). This data suggested that CMTM6 was involved in the regulation of PD-1/PD-L1 pathway.
Current studies show that combination therapy with inhibition of immune checkpoints increases the clinical benefit, and pharmacological treatments that target the immune checkpoints molecules are being extensively evaluated in preclinical or clinical studies [24, 25]. To explore the relationship between CMTM6 and immune checkpoints, we also enrolled the additional genes of immune checkpoints, such as CTLA-4, TIM-3, LAG-3, and others, into the analysis. As shown in Fig. 4, CMTM6 demonstrates a high correlation with TIM-3 and H7-H3 in both whole grade gliomas and low-grade gliomas, and CMTM6 expression is tightly associated with TIM-3 in glioblastomas.
Fig. 4.
Correlation of immune checkpoint members in whole gliomas (A, D), low-grade gliomas (B, E), and GBM (C, F).
3.6. CMTM6 is associated with T cell immunity in glioma
CMTM6 is involved in the progression of melanoma, lung cancer, and thyroid cancer cells in the co-culture experiments by inhibiting T cells [14, 15], but it has remained unclear whether CMTM6 plays a same role in the gliomas. To evaluate the relationship between CMTM6 and T cell immune response in gliomas, we performed GSVA analysis (Fig. 5A and B). We found that CMTM6 was positively correlated with positive regulation of T cell tolerance induction, positive regulation of T cell cytokine production, positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell, and positive regulation of regulatory T cell differentiation. Meanwhile, CMTM6 was negatively associated with regulation of T cell mediated cytotoxicity directed against tumor cell target and positive regulation of cytotoxic T cell differentiation. These results were verified mutually in CGGA and TCGA cohorts. Since CMTM6 is a positive regulator of PD-L1 [15], it is possible that CMTM6 may play an important role in inhibiting T cell anti-tumor immunity in gliomas through its positive regulation of PD-L1.
Fig. 5.
CMTM6 related T cell immunity and inflammatory activities in gliomas. A, B. The relationship between CMTM6 and T cell immunity in CGGA and TCGA dataset. ns, *, and *** indicate no significant difference, p < .05, and p < .001, respectively. C, D. The relationship between CMTM6 and inflammatory activities in CGGA and TCGA dataset. ***, p < .001. GO:0002666, positive regulation of T cell tolerance induction. GO:0002726, positive regulation of T cell cytokine production. GO:0002842, positive regulation of T cell mediated immune response to tumor cell. GO:0002852, regulation of T cell mediated cytotoxicity directed against tumor cell target. GO:2001190, positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell. GO:0045591, positive regulation of regulatory T cell differentiation. GO:0002842, positive regulation of cytotoxic T cell differentiation.
3.7. CMTM6 is relevant to inflammatory activities in gliomas
Because CMTM6 was also involved in the inflammatory response in gliomas, we analyzed seven metagenes by the method as previously described to clarify the role of CMTM6 in the inflammatory response [14, 21]. As shown in Fig. 5C and D, CMTM6 expression was positively correlated with HCK, LCK, MHC-I, MHC-II, STAT1, and interferon, while negatively associated with IgG in both CGGA and TCGA databases. These findings indicated that CMTM6 was upregulated in the activating of macrophages, signaling transduction of T cells, and antigen-presenting cells, but negatively associated with B lymphocytes related metagenes. Furthermore, we also confirmed that CMTM6 played important immune and inflammatory functions in gliomas.
3.8. CMTM6 predicts worse survival in gliomas
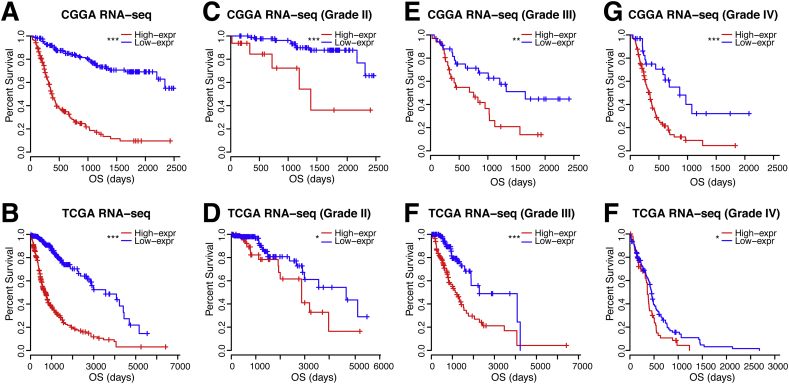
Since CMTM6 was negatively associated with T cell immune response, we further investigated the prognostic value of CMTM6 by Kaplan – Meier analysis and Cox proportional hazard model analysis. As shown in Fig. 6, patients with high CMTM6 expression tumors experienced significantly shorter survival in whole gliomas. Moreover, similar results were also detected in glioma patients of WHO Grade II, WHO Grade III, and WHO Grade IV in both CGGA and TCGA cohorts (Figs. 6). Survival curves were also depicted in different histopathologic classification (Supplementary Fig. S7), IDH mutant status (Supplementary Fig. S8), and 2016 WHO molecular classification (Supplementary Fig. S9). Patients with gliomas in a high level of CMTM6 expression show inferior outcome, although some subgroups failed to acquire the statistical significance. Furthermore, to explore the clinical prognostic significance of CMTM6 in gliomas, Cox regression analysis was performed. In the Univariate analysis, it showed that CMTM6, age at diagnosis, high WHO Grade, malignant histopathology, and IDH mutations were significantly associated with overall survival in both CGGA and TCGA databases (Table 1). Importantly, CMTM6 was still a significant predictor in both cohorts, after adjusting the age and gender in the multivariate analysis. These results revealed that CMTM6 may facilitate to serve as an indicator for the poor prognosis in glioma due to suppression of T cell immune response to antitumor.
Fig. 6.
Clinical outcome in patients with gliomas in low and high CMTM6 expression. Kaplan – Meier survival analysis was performed. ns, *, and *** indicate no significant difference, p < .05, and p < .001, respectively.
Table 1.
Univariate and multivariate cox analyses in gliomas.
| Factor | CGGA RNA-seq set |
TCGA RNA-seq set |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Univariate |
Multivariate |
Univariate |
Multivariate |
|||||||
| sample | P value | HR | P value | HR | sample | P value | HR | P value | HR | |
| Age | ||||||||||
| Increasing years | 310 | <0.001 | 1.038 | 0.961 | 1.000 | 678 | <0.001 | 1.066 | <0.001 | 1.036 |
| Gender | ||||||||||
| Male vs. Female | 310 | 0.345 | 0.847 | 678 | 0.061 | 1.262 | ||||
| WHO Grade | ||||||||||
| Grade II | 105 | Ref. | Ref. | Ref. | Ref. | 222 | Ref. | Ref. | Ref. | Ref. |
| Grade III | 67 | <0.001 | 5.865 | <0.001 | 4.150 | 242 | <0.001 | 2.986 | 0.003 | 1.961 |
| Grade IV | 138 | <0.001 | 14.710 | <0.001 | 49.383 | 167 | <0.001 | 18.593 | <0.001 | 4.988 |
| Histopathology | a | a | ||||||||
| Oligodendroglioma | 39 | Ref. | Ref | Ref. | Ref. | 180 | Ref. | Ref. | Ref. | Ref. |
| Oligoastrocytoma | 67 | 0.007 | 7.294 | 0.012 | 6.591 | 116 | 0.262 | 1.322 | 0.043 | 1.670 |
| Astrocytoma | 66 | <0.001 | 13.909 | 0.002 | 10.598 | 168 | 0.008 | 1.774 | 0.102 | 1.444 |
| Glioblastoma | 138 | <0.001 | 44.882 | b | 167 | <0.001 | 12.726 | b | ||
| TCGA molecular classification | ||||||||||
| Neural | 76 | ref | Ref | Ref. | Ref. | 134 | Ref. | Ref. | Ref. | Ref. |
| Proneural | 99 | 0.009 | 2.182 | 0.273 | 1.477 | 345 | 0.424 | 0.843 | 0.450 | 0.837 |
| Classical | 70 | <0.001 | 6.194 | 0.466 | 1.282 | 98 | <0.001 | 5.326 | 0.258 | 0.733 |
| Mesenchymal | 65 | <0.001 | 10.793 | 0.169 | 1.733 | 114 | <0.001 | 6.023 | 0.699 | 0.895 |
| Radiation | ||||||||||
| Yes vs. No | 287 | <0.001 | 0.429 | <0.001 | 0.454 | |||||
| Chemotherapy | ||||||||||
| Yes vs. No | 279 | 0.079 | 0.852 | |||||||
| IDH status | ||||||||||
| Mutation vs. wild-type | 310 | <0.001 | 0.244 | 0.240 | 0.702 | 668 | <0.001 | 0.117 | <0.001 | 0.375 |
| CMTM6 | ||||||||||
| Increasing expression | 310 | <0.001 | 1.105 | 0.013 | 1.047 | 678 | <0.001 | 1.088 | 0.030 | 1.029 |
Degree of freedom reduced because of constant or linearly dependent covariates.
Constant or Linearly Dependent Covariates Glioblastoma = Grade IV.
4. Discussion
Glioma is the most prevalent and lethal primary brain tumors in adults. As a result of the limited improvements of outcome in conventional treatment, including surgical resection, radiotherapy, and chemotherapy, new therapeutic methods are urgently needed. As a novel therapeutic approach in the last decade, immunotherapy, especially the targeting PD-1/PD-L1, has achieved marked success in preclinical or clinical trials of many malignant tumors, such as those of lung cancer [9], breast cancer [26], renal cancer [13], melanoma [7], and head and neck cancer [10]. Additionally, blocking of PD-1/PD-L1 in glioma has been initiated in clinical studies [6]. However, the response rate of treatment via blocking PD-1/PD-L1 has low in the other tumors [[7], [8], [9], [10], [11], [12], [13]], due to less understanding of the molecular mechanism of regulation of PD-1/PD-L1. Recently, two papers have indicated that CMTM6 is a critical regulator of PD-L1 [14, 15]. However, the status of CMTM6 in clinical samples of gliomas and the relationship between CMTM6 and immune responses have been unclear. Identification of the molecular and clinical correlation between CMTM6 expression and immune activities in gliomas will facilitate the establishment of a new therapeutic target and may improve the current therapies.
Here, we analyzed the CMTM6 expression in glioma. The high levels of CMTM6 showed an association with malignant entities, such as high WHO Grade, the IDH wildtype status in gliomas. Besides, CMTM6 was highly enriched in mesenchymal subtype gliomas. Mesenchymal subtype is characterized by mesenchymal differentiation with NF1 mutations, presenting immunosuppression and aggression [23, 27]. Meanwhile, Immunosuppressive cytokines and immune checkpoint inhibitor are enriched in mesenchymal subtype [28]. Several studies reported that high PD-L1 expression was observed in the mesenchymal subtype [20, 29, 30]. Therefore, in mesenchymal subtype glioma, CMTM6 may be activated and be involved in the immunosuppressive microenvironment via its positive regulation of PD-L1 [14]. This is the first study to present the expression pattern of CMTM6 in gliomas according to WHO Grade system, TCGA subtypes, or 2016 WHO molecular classifications. Furthermore, unique genomic landscapes have been performed to evaluate the tumor progression as an early event initiating pathogenesis [22, 31]. In this study, we investigated the distinct genomic alternations in order of increasing CMTM6 expression. We observed the events of somatic mutations and CNAs were positively associated with CMTM6 expression. The oncogenic drivers, such as EGFR, PDGFRA, PIK3C2B, and CDK4 were detected high amplification peaks in the cases with high CMTM6 expression [32, 33]. Meanwhile, a deletion peak of CDKN2A and CDKN2B, tumor suppressor genes, was also observed in the gliomas with high CMTM6 expression [34]. In addition, genomic alternations and heterogeneity may contribute to transforming the tumor microenvironment, promoting the progression of tumor and the resistance to therapies [33]. These findings suggested that CMTM6 expression was associated with the malignant biological process. Exploring and revealing the mechanism of CMTM6 in glioma may develop the therapeutic approaches to overcome this disease.
In the gene ontology analysis of the biological functions of CMTM6, we found that CMTM6 plays an important role in inflammatory activities and immune responses in whole gliomas, especially in glioblastoma. The correlation between CMTM6 and other immune checkpoints also suggested that CMTM6 may play a suppressive role in the anti-tumor immune responses. Moreover, CMTM6 was expressed synergistically with PD-1, PD-L1, PD-L2, and CD80, which is involved in the regulation of PD-1/PD-L1 pathway. From previous works, CMTM6 has been shown to enhance the PD-L1 protein stability and protect PD-L1 from being targeted for lysosomal degradation [14, 15]. PD-1, as a major negative immune regulator, is involved in controlling T cell activation, T cell exhaustion, T cell tolerance, and resolution of inflammation [35]. Meanwhile, PD-L1 expression is significantly upregulated in high-grade gliomas [20]. High levels of PD-L1 expression on tumors limits the host immune responses via ligation of PD-1 on cytotoxic T cells [36]. However, depletion of CMTM6, via the reduction of PD-L1, significantly enhanced T cell activation ex vivo and in vivo [14]. In our depth analysis of special immune functions of CMTM6 in gliomas from clinical databases, we found that CMTM6 was positively associated with regulation of T cell tolerance induction, T cell cytokine production, and regulatory T cell differentiation, while negatively correlated with regulation of cytotoxic T cell differentiation. Collectively, the special effect of CMTM6 demonstrate that CMTM6 functions as an important immune checkpoint inhibitor by modulating T-lymphocyte-mediated anti-tumor immunity. In addition, CMTM6 was also involved in inflammatory reaction to promote the progression in gliomas, via activation of tumor-associated macrophage and dysfunctional T cells [16, 21, 37]. Thus, CMTM6 was suggested to be a potential target of the immunotherapy for gliomas.
Critically, a high level of CMTM6 was associated with a poor prognosis. High CMTM6 expression predicted significantly worse survival in each database. Importantly, this prognostic value was also observed in GBM patients, which were referred to poor outcome. Meanwhile, we found that the CMTM6 expression was shown positively related to T cell tolerance induction and regulatory T cell differentiation. Besides, T cell exhaustion and an excessive number of regulatory T cells contribute to the immunosuppression of GBM patients and lead to a poor prognosis and shorter survival [37, 38]. As a result, CMTM6 may serve as a potential prognostic predictor for glioma patients.
Cancer immunotherapy has shown the potential benefits of cancer therapy, such as advanced metastatic cancer, specifically melanoma and renal cell carcinoma [39]. After immune checkpoint inhibitors achieve excellent results initial preclinical proof-of-principle studies, the strategy moved to the clinical evaluation [39, 40]. Besides, the antibody, targeting cytotoxic T lymphocyte antigen 4 (CTLA-4), has been approved for clinical treatment of cancer [41]. In addition, the clinical trials of immune checkpoint blockades for gliomas, such as anti-PD-1 (NCT02359565), anti-PD-L1 (NCT02794883), anti-CTLA-4 (NCT03233152), and anti-lymphocyte activation gene 3 protein (LAG3, NCT02658981), are ongoing. Moreover, the combination of immune checkpoint inhibitors has shown higher response rates and longer survival in patients with advanced melanoma, even brain metastases [7, 42]. As a key regulator of PD-1/PD-L1 pathways, our findings indicated that CMTM6 showed a desirable correlation with other immune checkpoints. Thus, it is suggested that combined anti-CMTM6 with other immune checkpoint blockades may be a novel approach for glioma treatments. However, we recognize that the current evidence is correlative. Future studies are necessary to demonstrate that inhibiting CMTM6 can reduce the tumor growth and extend the survival of gliomas.
Our findings indicated that CMTM6 may serve as an important biomarker in gliomas including the tumor mutational profile, genomic, histologic and clinical features. The biological significances of CMTM6 were investigated by the use of large sample-sized glioma cohorts. As a key regulator of T lymphocyte-mediated anti-tumor immunity, CMTM6 expression in cellular components within the tumor microenvironment and the immunoregulatory role of CMTM6 need to be addressed in further studies. Among limitations, the elevated expression induction of CMTM6 can help to explain why high-grade glioma are refractory to immunotherapy, but the mechanism by which CMTM6 is upregulated in malignant glioma remains to be understood. CMTM6 is a new gene, little is known about its transcriptional or non-transcriptional regulation.
Taken together, through transcriptomic and genomic profiling data, we found that CMTM6 was upregulated in high malignant glioma, and was synergistic with other immune checkpoint inhibitors. Moreover, CMTM6 was involved in T cell activation and antitumor immunity. Finally, CMTM6 was revealed to be correlated with the clinical outcome of glioma patients. These findings establish CMTM6 as a novel potential target to enhance the anticancer therapies.
Funding sources
This work was supported by the Capital Health Research and Development of Special (2014-2-1072) and Beijing Municipal Natural Science Foundation (7142054).
Declaration of interest
No potential conflicts of interest were disclosed.
Author contributions
Conception and design: XG, CZ, JZ, WJ. Development of methodology: XG, CZ, QS. Acquisition of data: XG, CZ, JZ. Analysis and interpretation of data: XG, JZ, QS. Writing, review and/or revision of the manuscript: XG, CZ, GS, WJ. Administrative, technical, or material support: XG, CZ, QS. Study supervision: WJ. All authors read and approved the final manuscript.
Acknowledgments
We gratefully acknowledge contributions from Chinese Glioma Genome Atlas network and TCGA Research Network. The first author is particularly indebted to Profs. Wang Jia and Dandan Sun for help during the study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.08.012.
Appendix A. Supplementary data
Supplementary figures
Supplementary Fig. S1 Clinical and molecular features in associations with CMTM6 expression. A. CMTM6 expression in normal brain tissues and gliomas. ***, p < .001. B. MTM6 expression in normal brain tissues and gliomas. C. CMTM6 expression in primary GBM, recurrent GBM, and secondary GBM. ***, p < .001. D. CMTM6 expression in 2016 WHO molecular classification. *, and *** indicate p < .05, and p < .001, respectively. E. ROC curve analysis showed that CMTM6 had 78.2% and 86.7% sensitivity and specificity to predict IDH wild-type state glioma in CGGA and TCGA database respectively. F. CMTM6 expression across IDH state in different WHO grades. *, and *** indicate p < .05, and p < .001, respectively.
Supplementary Fig S2 Correlation between CMTM6 expression and somatic mutations and CNAs. High CMTM5 expression predicted more somatic mutations (A) and CNAs (B).
Supplementary Fig S3 Distinct somatic mutation and CNAs profiles associated with CMTM6 expression.
Supplementary Fig S4 Gene ontology analysis for CMTM6 in glioma. A, B. Gene ontology analysis of CMTM6 negatively related biological process in CGGA and TCGA datasets. C, D. Heatmaps of gene ontology analysis for CMTM6 in gliomas in CGGA and TCGA datasets.
Supplementary Fig S5 Gene ontology analysis for CMTM6 in GBM. A, B. Gene ontology analysis showed that CMTM6 was mostly in inflammatory response and immune response in GBM.
Supplementary Fig S6 Correlation of PD-L1 homologs and family members in whole gliomas (A, D), low-grade gliomas (B, E), and GBM (C, F).
Supplementary Fig S7 Clinical outcome in different histopathologic classification. ns, *, and *** indicate no significant, p < .05, and p < .005, respectively.
Supplementary Fig S8 Clinical outcome in different IDH mutant status. *, and *** indicate p < .05, and p < .005, respectively.
Supplementary Fig S9 Clinical outcome in 2016 WHO molecular classification. ns and *** indicate no significant and p < .005, respectively.
Supplementary tables
Table S1 Clinical and molecular characteristics of patients included in this study.
Table S2 The information of the location in IVY GBM dataset.
Table S3 Genomic aberrations with different frequency between cases with lower and higher CMTM6 expression
Supplementary dataset
The alteration of peaks of CNAs in gliomas with different CMTM6 expression.
References
- 1.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Jiang T., Mao Y., Ma W., Mao Q., You Y., Yang X. CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2016;375(2):263–273. doi: 10.1016/j.canlet.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 3.Preusser M., Lim M., Hafler D.A., Reardon D.A., Sampson J.H. Prospects of immune checkpoint modulators in the treatment of glioblastoma. Nat Rev Neurol. 2015;11(9):504–514. doi: 10.1038/nrneurol.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Louveau A., Smirnov I., Keyes T.J., Eccles J.D., Rouhani S.J., Peske J.D. Structural and functional features of central nervous system lymphatic vessels. Nature. 2015;523(7560):337–341. doi: 10.1038/nature14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou W., Wolchok J.D., Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016;8(328):328rv4. doi: 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue S., Hu M., Iyer V., Yu J. Blocking the PD-1/PD-L1 pathway in glioma: a potential new treatment strategy. J Hematol Oncol. 2017;10(1):81. doi: 10.1186/s13045-017-0455-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolchok J.D., Chiarion-Sileni V., Gonzalez R., Rutkowski P., Grob J.J., Cowey C.L. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2017;377(14):1345–1356. doi: 10.1056/NEJMoa1709684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellmunt J., de Wit R., Vaughn D.J., Fradet Y., Lee J.L., Fong L. Pembrolizumab as second-line therapy for advanced urothelial carcinoma. N Engl J Med. 2017;376(11):1015–1026. doi: 10.1056/NEJMoa1613683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reck M., Rodriguez-Abreu D., Robinson A.G., Hui R., Csoszi T., Fulop A. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375(19):1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 10.Ferris R.L., Blumenschein G., Jr., Fayette J., Guigay J., Colevas A.D., Licitra L. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. 2016;375(19):1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nghiem P.T., Bhatia S., Lipson E.J., Kudchadkar R.R., Miller N.J., Annamalai L. PD-1 blockade with pembrolizumab in advanced Merkel-cell carcinoma. N Engl J Med. 2016;374(26):2542–2552. doi: 10.1056/NEJMoa1603702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brahmer J., Reckamp K.L., Baas P., Crino L., Eberhardt W.E., Poddubskaya E. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373(2):123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Motzer R.J., Escudier B., McDermott D.F., George S., Hammers H.J., Srinivas S. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373(19):1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burr M.L., Sparbier C.E., Chan Y.C., Williamson J.C., Woods K., Beavis P.A. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549(7670):101–105. doi: 10.1038/nature23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mezzadra R., Sun C., Jae L.T., Gomez-Eerland R., de Vries E., Wu W. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549(7670):106–110. doi: 10.1038/nature23669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li G., Wang Z., Zhang C., Liu X., Cai J., Wang Z. Molecular and clinical characterization of TIM-3 in glioma through 1,024 samples. Oncoimmunology. 2017;6(8) doi: 10.1080/2162402X.2017.1328339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Research Network, Electronic address wbe, Cancer Genome Atlas Research N Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169(7):1327–1341. doi: 10.1016/j.cell.2017.05.046. [e23] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z., Zhang C., Liu X., Wang Z., Sun L., Li G. Molecular and clinical characterization of PD-L1 expression at transcriptional level via 976 samples of brain glioma. Oncoimmunology. 2016;5(11) doi: 10.1080/2162402X.2016.1196310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rody A., Holtrich U., Pusztai L., Liedtke C., Gaetje R., Ruckhaeberle E. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res. 2009;11(2):R15. doi: 10.1186/bcr2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang C., Cheng W., Ren X., Wang Z., Liu X., Li G. Tumor purity as an underlying key factor in glioma. Clin Cancer Res. 2017;23(20):6279–6291. doi: 10.1158/1078-0432.CCR-16-2598. [DOI] [PubMed] [Google Scholar]
- 23.Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao X., Subramanian S. Oncogenic pathways that affect antitumor immune response and immune checkpoint blockade therapy. Pharmacol Ther. 2018;181:76–84. doi: 10.1016/j.pharmthera.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Sharma P., Allison J.P. The future of immune checkpoint therapy. Science (New York, NY) 2015;348(6230):56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 26.Brahmer J.R., Tykodi S.S., Chow L.Q., Hwu W.J., Topalian S.L., Hwu P. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phillips H.S., Kharbanda S., Chen R., Forrest W.F., Soriano R.H., Wu T.D. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 28.Doucette T., Rao G., Rao A., Shen L., Aldape K., Wei J. Immune heterogeneity of glioblastoma subtypes: extrapolation from the cancer genome atlas. Cancer Immunol Res. 2013;1(2):112–122. doi: 10.1158/2326-6066.CIR-13-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berghoff A.S., Kiesel B., Widhalm G., Rajky O., Ricken G., Wohrer A. Programmed death ligand 1 expression and tumor-infiltrating lymphocytes in glioblastoma. Neuro Oncol. 2015;17(8):1064–1075. doi: 10.1093/neuonc/nou307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heiland D.H., Haaker G., Delev D., Mercas B., Masalha W., Heynckes S. Comprehensive analysis of PD-L1 expression in glioblastoma multiforme. Oncotarget. 2017;8(26):42214–42225. doi: 10.18632/oncotarget.15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki H., Aoki K., Chiba K., Sato Y., Shiozawa Y., Shiraishi Y. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47(5):458–468. doi: 10.1038/ng.3273. [DOI] [PubMed] [Google Scholar]
- 32.Nobusawa S., Lachuer J., Wierinckx A., Kim Y.H., Huang J., Legras C. Intratumoral patterns of genomic imbalance in glioblastomas. Brain Pathol (Zurich, Switzerland) 2010;20(5):936–944. doi: 10.1111/j.1750-3639.2010.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson D.R., Wu Y.M., Lonigro R.J., Vats P., Cobain E., Everett J. Integrative clinical genomics of metastatic cancer. Nature. 2017;548(7667):297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krimpenfort P., Ijpenberg A., Song J.Y., van der Valk M., Nawijn M., Zevenhoven J. p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature. 2007;448(7156):943–946. doi: 10.1038/nature06084. [DOI] [PubMed] [Google Scholar]
- 35.Baumeister S.H., Freeman G.J., Dranoff G., Sharpe A.H. Coinhibitory pathways in immunotherapy for cancer. Annu Rev Immunol. 2016;34:539–573. doi: 10.1146/annurev-immunol-032414-112049. [DOI] [PubMed] [Google Scholar]
- 36.Smyth M.J., Ngiow S.F., Ribas A., Teng M.W. Combination cancer immunotherapies tailored to the tumour microenvironment. Nat Rev Clin Oncol. 2016;13(3):143–158. doi: 10.1038/nrclinonc.2015.209. [DOI] [PubMed] [Google Scholar]
- 37.Weller M., Roth P., Preusser M., Wick W., Reardon D.A., Platten M. Vaccine-based immunotherapeutic approaches to gliomas and beyond. Nat Rev Neurol. 2017;13(6):363–374. doi: 10.1038/nrneurol.2017.64. [DOI] [PubMed] [Google Scholar]
- 38.Mirzaei R., Sarkar S., Yong V.W. T cell exhaustion in glioblastoma: intricacies of immune checkpoints. Trends Immunol. 2017;38(2):104–115. doi: 10.1016/j.it.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 39.Ribas A., Wolchok J.D. Cancer immunotherapy using checkpoint blockade. Science (New York, NY) 2018;359(6382):1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sahin U., Tureci O. Personalized vaccines for cancer immunotherapy. Science (New York, NY) 2018;359(6382):1355–1360. doi: 10.1126/science.aar7112. [DOI] [PubMed] [Google Scholar]
- 41.Patel S.A., Minn A.J. Combination Cancer therapy with immune checkpoint blockade: mechanisms and strategies. Immunity. 2018;48(3):417–433. doi: 10.1016/j.immuni.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Long G.V., Atkinson V., Lo S., Sandhu S., Guminski A.D., Brown M.P. Combination nivolumab and ipilimumab or nivolumab alone in melanoma brain metastases: a multicentre randomised phase 2 study. Lancet Oncol. 2018;19(5):672–681. doi: 10.1016/S1470-2045(18)30139-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Supplementary Fig. S1 Clinical and molecular features in associations with CMTM6 expression. A. CMTM6 expression in normal brain tissues and gliomas. ***, p < .001. B. MTM6 expression in normal brain tissues and gliomas. C. CMTM6 expression in primary GBM, recurrent GBM, and secondary GBM. ***, p < .001. D. CMTM6 expression in 2016 WHO molecular classification. *, and *** indicate p < .05, and p < .001, respectively. E. ROC curve analysis showed that CMTM6 had 78.2% and 86.7% sensitivity and specificity to predict IDH wild-type state glioma in CGGA and TCGA database respectively. F. CMTM6 expression across IDH state in different WHO grades. *, and *** indicate p < .05, and p < .001, respectively.
Supplementary Fig S2 Correlation between CMTM6 expression and somatic mutations and CNAs. High CMTM5 expression predicted more somatic mutations (A) and CNAs (B).
Supplementary Fig S3 Distinct somatic mutation and CNAs profiles associated with CMTM6 expression.
Supplementary Fig S4 Gene ontology analysis for CMTM6 in glioma. A, B. Gene ontology analysis of CMTM6 negatively related biological process in CGGA and TCGA datasets. C, D. Heatmaps of gene ontology analysis for CMTM6 in gliomas in CGGA and TCGA datasets.
Supplementary Fig S5 Gene ontology analysis for CMTM6 in GBM. A, B. Gene ontology analysis showed that CMTM6 was mostly in inflammatory response and immune response in GBM.
Supplementary Fig S6 Correlation of PD-L1 homologs and family members in whole gliomas (A, D), low-grade gliomas (B, E), and GBM (C, F).
Supplementary Fig S7 Clinical outcome in different histopathologic classification. ns, *, and *** indicate no significant, p < .05, and p < .005, respectively.
Supplementary Fig S8 Clinical outcome in different IDH mutant status. *, and *** indicate p < .05, and p < .005, respectively.
Supplementary Fig S9 Clinical outcome in 2016 WHO molecular classification. ns and *** indicate no significant and p < .005, respectively.
Supplementary tables
Table S1 Clinical and molecular characteristics of patients included in this study.
Table S2 The information of the location in IVY GBM dataset.
Table S3 Genomic aberrations with different frequency between cases with lower and higher CMTM6 expression
Supplementary dataset
The alteration of peaks of CNAs in gliomas with different CMTM6 expression.