Abstract
Therapies against hepatitis B virus (HBV) have improved in recent decades; however, the development of individualized treatments has been limited by the lack of individualized infection models. In this study, we used human induced pluripotent stem cell (hiPSC) to generate a functional liver organoid (LO) that inherited the genetic background of the donor, and evaluated its application in modeling HBV infection and exploring virus–host interactions. To establish a functional hiPSC-LO, we cultured hiPSC-derived endodermal, mesenchymal, and endothelial cells with a chemically defined medium in a three-dimensional microwell culture system. Based on cell-cell interactions, these cells could organize themselves and gradually differentiate into a functional organoid, which exhibited stronger hepatic functions than hiPSC derived hepatic like cell (HLC). Moreover, the functional LO demonstrated more susceptibility to HBV infection than hiPSC-HLC, and could maintain HBV propagation and produce infectious virus for a prolonged duration. Furthermore, we found that virus infection could cause hepatic dysfunction of hiPSC-LOs, with down-regulation of hepatic gene expression, induced release of early acute liver failure markers, and altered hepatic ultrastructure. Therefore, our study demonstrated that HBV infection in hiPSC-LOs could recapitulate virus life cycle and virus induced hepatic dysfunction, suggesting that hiPSC-LOs may provide a promising individualized infection model for the development of individualized treatment for hepatitis.
Keywords: Liver organoid, hiPSC, Hepatitis B virus, Virus-host interactions
1. Introduction
Although vaccines and therapies against hepatitis B virus (HBV) have improved in recent decades, an estimated 257 million people are still living with hepatitis B virus infection with markedly heterogeneous outcomes [1, 2]. Some individuals have self-limiting, symptom-free infection, whereas others develop liver cirrhosis and/or hepatocellular carcinoma [2]. This drastic heterogeneity in outcome cannot be justified completely by the contributing factors, such as viral genotype diversity, environmental and demographic variables, and differences in age of patient with infection [3]. Evidence has indicated that the genetic background of the host might play an important role in virus-induced outcomes [4, 5]. Although various models have been developed for HBV infection, these models do not represent individualized genetic backgrounds, which limits their application in understanding the potential impact of the host's genetic makeup during virus infection. Since long, primary human hepatocytes (PHH) have been a valuable model for HBV infection studies [6]; however, donor shortage and variable quality limit their applications. More recently, the Na+-taurocholate cotransporting polypeptide (NTCP) was identified as a HBV entry receptor, and the exogenous expression of NTCP could successfully induce susceptibility to HBV infection in non-susceptible hepatocarcinoma cells [7]. However, the genetic background of these hepatocarcinoma cells could not broadly represent the human population. Hepatocytes from chimeric mice also face similar problems [8]. Therefore, the generation of new hepatocytes, with patient-specific genetic background and susceptibility to HBV infection, is paramount for individualized hepatitis studies.
In the recent decade, the ability of human induced pluripotent stem cell (hiPSC) to differentiate into various terminal cell types, inheriting the donor's genetic background, has been successfully tested and proved [9]. This has created a unique opportunity to accurately model diseases and develop new treatments by using hiPSCs. Studies have reported that hiPSC-derived hepatic-like cells (hiPSC-HLC) could exhibit and inherit hepatic characteristics similar to those of the donor [10, 11]. HBV infection have also been reported in hiPSC-HLC [[12], [13], [14]]. However, these HLCs displayed low hepatic function that was easily lost, greatly limiting its application in the investigation of virus-host interactions [[12], [13], [14], [15]]. The liver is known to have a complex and highly organized architecture consisting of numerous cell types, with hepatic differentiation being accurately controlled by its interactions with non-parenchymal cells during liver organogenesis [16]. Therefore, reconstruction of these interactions might be a plausible approach to promote hepatic differentiation, long-term maintenance of hepatic functions, and generation of an effective infection model to understand virus-host interactions.
The liver develops from specific hepatic endoderm in a microenvironment surrounded by mesenchymal and endothelial progenitor cells located in the septum transversum. The mesenchymal and endothelial cell microenvironment has been considered a key element to initiate hepatic differentiation and promote liver development [17, 18]. By recapitulating this microenvironment, we have previously generated a self-organized hiPSC liver organoid (LO) that could grow into a vascularized and functional tissue post-transplantation [19, 20]. However, the self-organized organoid had a few drawbacks such as limited hepatic function before transplantation, and a huge size (approximate diameter of 3000–5000 μm) that inhibited its ability to differentiate well in vitro [19, 20]. In the current study, we aimed to generate a functional hiPSC liver organoid that could act as a reliable and feasible ex vivo infection model for hepatitis studies.
2. Materials & methods
2.1. Cell culture
The TkDA3 human iPSC clone used in this study was kindly provided by K. Eto and H. Nakauchi. Undifferentiated iPSCs were maintained on a growth factor-reduced Matrigel (BD Biosciences, San Diego, CA)-coated dish with mTeSR1 medium (Stem Cell Technologies, Vancouver, BC, Canada). HUVECs and human bone marrow (BM)-MSCs were maintained in endothelial cell growth medium (Lonza, Walkersville, MD) and mesenchymal cell growth medium (Lonza), respectively.
Cryopreserved PHHs (lot number: AKB (PHH-1) and TLY (PHH-2)) were puchased from Bioreclamation IVT (Baltimore, MD, USA) and thawed according to the manufacturer's instruction. The PHHs were cultured in Williams E medium supplemented with 5% FBS, 1 μM Dexamethasone, 100 IU/mL Penicillin, 100 μg/mL Streptomycin, 4 μg/mL Human Recombinant Insulin, 2 mM GlutaMAXTM, and 15 mM HEPES pH 7.4. 24 h later, PHHs were used for HBV infection, Q-PCR anaysis and ELISA analysis.
Phoenix human hepatocytes (Phoenix-HHs) were isolated from humanized mice (PhoenixBio Co., Ltd., Higashihiroshima, Japan), without cryopreservation. Phoenix-HHs were cultured with hepatic growth medium (PhoenixBio). After 24 h of culture, Phoenix-HHs were used for HBV infection.
The HepG2-TET-NTCP cells were generated by Kei Miyakawa and Akihide Ryo as previous report [21]. HepG2.2.15.7 cells were obtained from Takaji Wakita [22], which are a HepG2.2.15 clone producing a higher level of HBV. The HepG2-TET-NTCP cells and HepG2.2.15.7 cells were maintained on collagen-I coated dishes with DMEM/F-12 (Life Technologies, Gaithersburg, MD), 2 mM GlutaMAX (Life Technologies), 10% fetal bovine serum (Life Technologies), 10 mM HEPES (Life Technologies) and 5 μg/mL insulin (Sigma–Aldrich, St. Louis, MO). All cells were maintained at 37 °C in a humidified incubator with 5% CO2.
2.2. Cell differentiation and organoid generation
To differentiate HLCs and LOs from hiPSC, we first differentiated endoderm from hiPSC according to a previously reported protocol [23]. Then hiPSC-endoderm was then cultured and differentiated into HLC as described previously [23].
To generate hiPSC-LOs, 2.5 × 105 hiPSC endoderm cells, 1.75 × 105 HUVECs, and 2.5 × 104 human BM-MSCs were co-cultured in a 3D microwell plate (Elplasia, Kuraray, Tokyo, Japan) with a serum-free differentiation (SFD) medium [24]. All cells were maintained at 37 °C in a humidified incubator with 5% CO2. After 15 days of differentiation, hiPSC-LOs were used for HBV infection experiments as well as other analysis.
To generate HepG2-TET-NTCP organoids, 2.5 × 105 HepG2-TET-NTCP cells, 1.75 × 105 HUVECs, and 2.5 × 104 human BM-MSCs were co-cultured in DMEM/12: EGM = 1:1 with 1 mM GlutaMAX, 5% fetal bovine serum, 5 mM HEPES and 2.5 μg/mL insulin in a 3D microwell plate. After 24 h culture with or without DOX, HepG2-TET-NTCP organoids were used for HBV infection experiments as well as other analysis.
2.3. HBV preparation, infection and inhibition assays
HBV stocks were derived from supernatants of HepG2.2.15.7 cells, which were stably transfected with a complete HBV genome (genotype D) as described previously [21]. HiPSCs-LO, hiPSCs-HLC, HepG2-tet-NTCP organoids, and PHHs were infected with HBV [500 genome equivalents (GEq)/cell or 5000 GEq/cell] in the presence of 4% polyethylene glycol 8000 in 24-well plates. After 10 days post infection or 20 days post infection, cultured cells were then harvested. pg RNA was quantified by SYBR Green (Takara Bio, Otsu, Japan) with primers listed in Table S1. The expression of pg RNA was normalized against expression of β-ACTB (Thermo Fisher Scientific, Waltham, MA).
In inhibition assay, hiPSC-LOs were infected with HBV at 5000 GEq/cell, and 100 nM Myrcludex was added into the medium 2 h before infection; 1.8 mM Entecavir was added to the medium during infection; 1000 IU/mL IFN-α (Sigma–Aldrich) and 1000 IU/mL IFN-γ (Sigma–Aldrich) were added to the medium during infection.
2.4. Intracellular vDNA and cccDNA isolation and quantification
Infected cells were collected after infection. Total DNA in the cells were purifed using DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany). The concentration of were determined by naondrop spectrophotometer (Thermo Fischer Scientific, Waltham, MA), and the DNA concentration were adjust to 20 ng/μl for further experiment. 2 μl adjusted DNA sample were used to quatify vDNA by SYBR Green with a standard curve done by using plasmid pUC-HBV over a range of 107–101 copies. To quanty cccDNA copies, adjusted DNA sample 25 μl of 20 ng/μl of HBV DNA samples were treated with plasmid-safe DNase at 37 °C for 1 h according to the manuscript. After digestion, the plasmid safe DNase were inactivated at 70 °C for 30 min. 2 μl sample were used to quantify cccDNA copies by SYBR Green with a standard curve done by using plasmid pUC-HBV over a range of 107–101 copies. The primer used for HBV DNA and cccDNA quantification were list in Table S1.
2.5. Supernatant vDNA isolation and quantification
Supernatant from infected cells were collected. 200 μl supernatant were mix with 200 μl AL buffere and 20 μl proteinase K. Total DNA in the supernatant were then purifed using DNeasy Blood & Tissue Kit (QIAGEN) and DNA was eluted with 30 μL of elusion buffer. 2 μl DNA sample were used to quatify supernatant DNA by SYBR Green with a standard curve done by using plasmid pUC-HBV over a range of 107–101 copies. The primer used for HBV DNA quantification were list in Table S1.
2.6. RNA isolation and quantitative real-time polymerase chain reaction
Total RNA was isolated using a PureLink viral RNA/DNA mini kit (Thermo Fisher Scientific, Carlsbad, CA). RNA (1 μg) was used as a template for single-strand cDNA synthesis with a high-capacity cDNA reverse transcription kit (Thermo Fisher Scientific) according to the manufacturer's instructions. Q-PCR was performed with cDNA using specific primers and Universal Probe Library (UPL) probes. The primers and UPL probes used in this study are listed in Table S1. All data were calculated using the ΔΔCT method with β-ACTB (Thermo Fisher Scientific) as a normalization control.
2.7. ALB assay, urea production, CYP3A, LDH release, and ALT assay
Human ALB was measured using a human ALB enzyme-linked immunosorbent assay kit (Bethyl Laboratories, Montgomery, TX). Urea production was assayed using a QuantiChrom urea assay kit (BioAssay System, Hayward, CA), CYP3A activity was detected using a P450-Glo CYP3A4 assay kit (Promega, Madison, WI, USA), LDH release was detected using the LDH Cytotoxicity Detection Kit (Roche, Roche, Boehringer Mannheim, Germany), and ALT was detected using FUJI DRI-CHEM SLIDE GPT/ALT-PIII (Tokyo, Japan) according to the manufacturer's instructions. The number of hiPSC-HLCs, and PHHs cells were analyzed by Incell analyzer 2000 (GE Healthcare, Cardiff, United Kingdom) with Hoechst 33342 staining. To calculate the cell number in LOs, total DNA of the cell number counted hiPSC-HLCs and hiPSC-LOs were extracted using DNeasy Blood & Tissue Kit, and DNA were eluted with 50 μL of elusion buffer. Then the concentration of those DNA were determined by naondrop spectrophotometer. The total cell number in hiPSC-LOs were calculated by following formula:
2.8. Indocyanine green uptake and release
ICG dry− powder (Akorn, Buffalo Grove, IL) (10 mg) was dissolved in 10 mL of hepatocyte culture medium (HCM; Lonza) to obtain a 1 mg/mL stock. hiPSC-LOs were incubated with ICG in suspension or plated for 4 h at 37 °C in a humidified incubator with 5% CO2. Then, hiPSC-LOs were washed three times with phosphate-buffered saline (PBS) and incubated in fresh HCM for another 5 h to determine the ICG release.
2.9. Tissue processing and immunofluorescence
hiPSC-LOs were embedded in optimal cutting temperature compound (Sakura Finetek Japan., Co., Ltd., Tokyo, Japan), and 7-μm sections were prepared and mounted on MAS-GP type A-coated slides (Matsunami, Osaka, Japan). Sections were fixed in a 4% paraformaldehyde solution in PBS for 10 min, washed three times with PBS, and blocked for 60 min with 10% ECL prime blocking agent in PBS containing 0.3% Triton X-100, followed by three washes with PBS. Then, the sections were incubated with HBc (Dako, Japan), HBs (Bio-rad, CA, USA), ALB (Bethyl Laboratories) and NTCP antibodies in the blocking buffer at 4 °C overnight. The sections were washed three times with PBS and incubated with a fluorescence-conjugated secondary antibody for another 60 min at room temperature. Finally, the sections were washed three times in PBS and covered with a mounting solution containing 4′,6-diamidino-2-phenylindole. Fluorescence was detected with a Zeiss Axio Imager M1 microscope. The NTCP antibody was produced by Kei Miyakawa and Akihide Ryo and tested using HepG2-TET-NTCP cells and organoids with or without doxycycline treatment (Fig. S2A and S2B). The HBc antibody were tested in infected PHHs (Fig. S2D). To quantification of HBc positive cells, we first count the ALB positive cells according to ALB and nuclear staining, then count the HBc positive cells among this ALB positive cells. The percentage of ALB+HBc+ in infected hiPSC-LOs were calculated by following formula:
2.10. Transmission electron microscopy
Samples in culture medium were fixed with an equal amount of 4% paraformaldehyde and 4% glutaraldehyde in 0.1 M phosphate buffer at 4 °C for 1 h, followed by incubation with 2% glutaraldehyde in 0.1 M phosphate buffer at 4 °C overnight. The fixed samples were postfixed with 2% osmium tetroxide, dehydrated through a graded series of ethyl alcohol, and embedded in a fresh 100% resin. Ultrathin sections (70 nm) were cut with an ultramicrotome (Ultracut UCT, Leica, Vienna, Austria) and stained with 2% uranyl acetate. Then, the sections were washed with distilled water and stained with a lead stain solution. Grids were observed under a transmission electron microscope (JEM-1400Plus, JEOL, Ltd., Tokyo, Japan) at an acceleration voltage of 80 kV. Digital images were taken with a CCD camera (VELETA, Olympus Soft Imaging Solutions GmbH, Munster, Germany).
2.11. Statistics
Values are expressed as the mean ± standard error of the mean (SEM). The statistical significance of differences was evaluated by the Mann-Whitney U test when two groups were compared or by one-way ANOVA and Bonferroni's multiple comparison tests when multiple groups were compared. A p < .05 was considered statistically significant, Statistical analysis was performed using GraphPad Prism. Additional experimental procedures are listed in Supplemental information Materials & Methods.
3. Results
3.1. In vitro generation of functional liver organoids from hiPSC
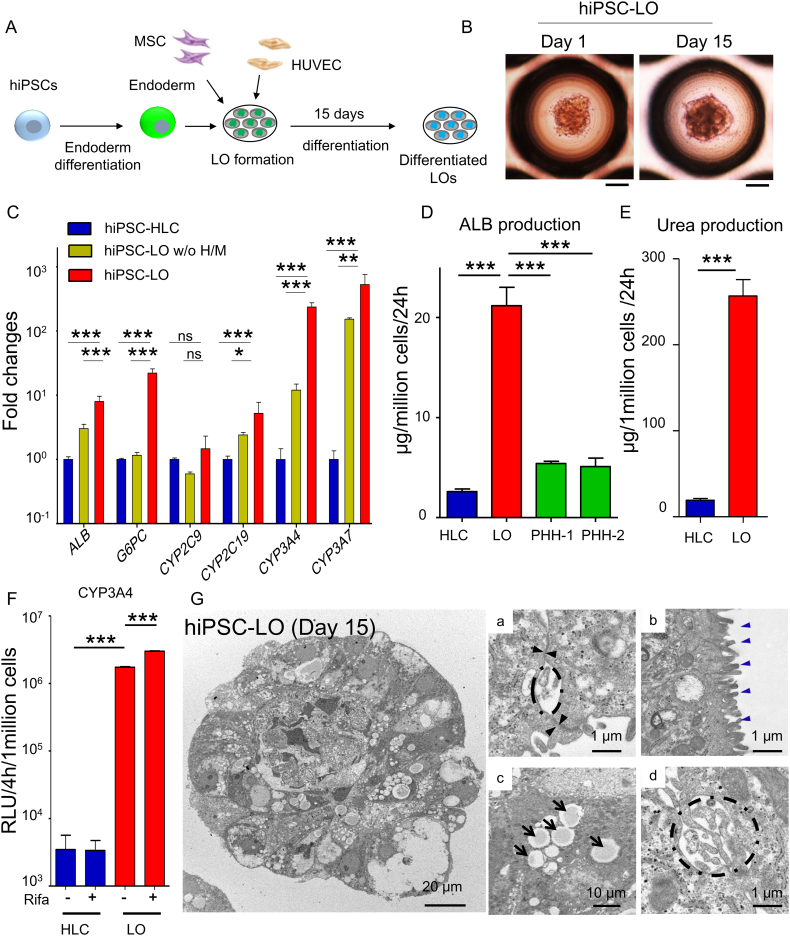
To generate functional liver organoids, we first differentiated hiPSCs into endoderm that expressed lineage specific hepatic makers (Fig. S1A and S1B), and then co-cultured the hiPSC-endoderm with human umbilical vein endothelial cells (HUVECs) and mesenchymal stem cells (MSCs) in a 3D microwell culture system (Fig. 1A). After 24 h, all three co-cultured cells were found to self-organize into organoids having a uniform size (205.1 ± 37.5 μm, n = 532) (Fig. S1C). Quantitative polymerase chain reaction (Q-PCR) analysis was performed on hiPSC-LO after 15 days of differentiation (Fig. 1B). The results revealed that hiPSC-LO had differentiated into hepatic lineages with expression of specific hepatic functional genes (Fig. 1C and Fig. S1D). We also found that the expression of these genes in differentiated LOs was significantly higher than that in hiPSC-HLCs (Fig. 1C and Fig. S1D). In addition, the omission of HUVECs and MSCs in hiPSC-LOs (hiPSC-LO w/o H/M) could delay hepatic lineage differentiation with lower expression of hepatic genes (Fig. 1C), indicating that the endothelial/mesenchymal environment was important to promote hepatic differentiation in LOs.
Fig. 1.
Generation of functional liver organoids from hiPSCs.
(A) Schematic representation of generation and differentiation of liver organoids (LOs) from hiPSCs. After 15 days of differentiation, differentiated LOs were collected and analyzed. (B) Morphology of hiPSC-LOs at day 1 and day 15. Scale bar, 100 μm. (C) Q-PCR analysis showing expression of hepatic genes: ALB, G6PC, CYP2C9, CYP2C19, CYP3A4, and CYP3A7 in hiPSC-HLCs (n = 4), differentiated hiPSC-LOs w/o H/M (deletion of HUVEC and MSC at LOs generation) (n = 4), and differentiated hiPSC-LOs (n = 4). (D) ELISA for ALB secretion of hiPSC-HLCs (n = 9), differentiated hiPSC-LOs (n = 11), PHH-1(24 h cultured, n = 3), and PHH-2 (24 h cultured, n = 3). (E) Quantification of urea production in hiPSC-HLCs (n = 8) and differentiated hiPSC-LOs (Day 15, n = 8). (F) Quantification of CYP3A activity in differentiated hiPSC-HLCs (n = 4) and hiPSC-LOs (n = 4), with or without 25 μM rifampicin treatment. (G) Transmission electron microscopy analysis showing ultrastructure of differentiated hiPSC-LO. (a) Tight junction (black arrowhead), (b) Microvilli (blue arrowhead), (c) Lipid droplets (black arrow), and (d) Bile canaliculi (dotted line). *p < .05, **p < .01, ***p < .001; ns, not significant.
Hepatic function analysis indicated that differentiated hiPSC-LOs show about 4–8-fold higher ALB (albumin) secretion (Fig. 1D and Fig. S1E), and 12-fold higher urea production than hiPSC-HLCs (Fig. 1E). Moreover, hiPSC-LOs displayed approximately 500-fold higher CYP (cytochrome P450) 3A4 activity than hiPSC-HLCs, and the CYP3A4 activity of hiPSC-LOs could be further induced by rifampicin (Fig. 1F). We further explored the ultrastructure of the differentiated hiPSC-LO with transmission electron microscopy (TEM). TEM data revealed that hepatic lineages in organoids had typical hepatic features, such as tight junctions, microvilli on the cell membrane, lipid droplets in the cytoplasm, and bile capillaries between hepatic cells (Fig. 1G). These differentiated organoids also had the ability to uptake and release indocyanine green (ICG) (Fig. S1F), a general characteristic of the liver. Collectively, these results demonstrate the successful generation of a well-differentiated hiPSC-LO with enhanced hepatic function and liver-specific features by recapitulating multiple cellular interactions in an ex vivo culture system.
3.2. Susceptibility to HBV infection in hiPSC liver organoids
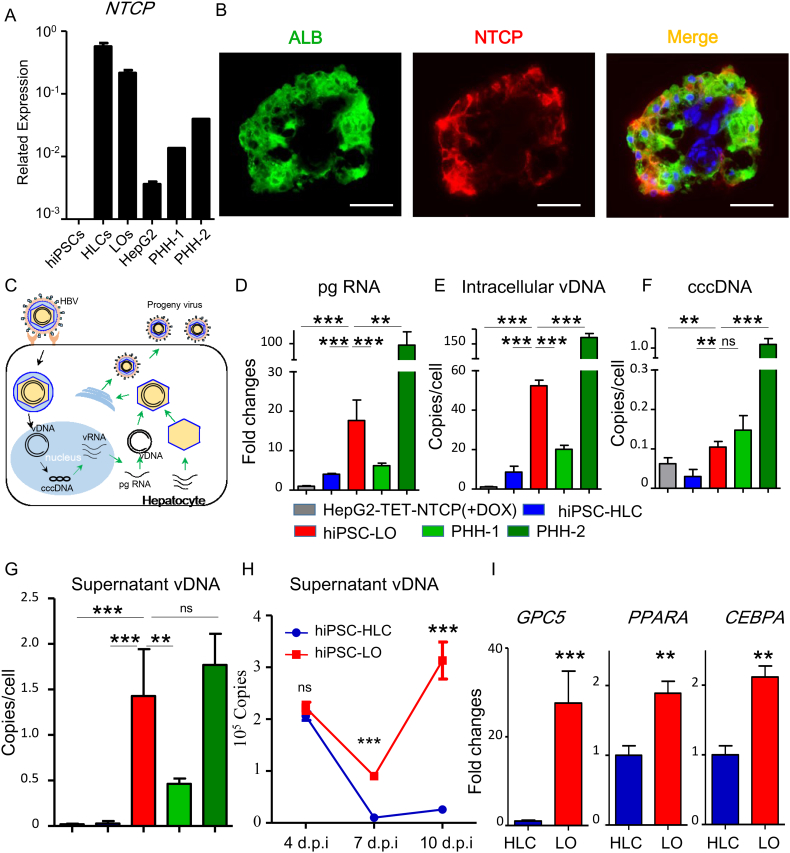
To evaluate whether the differentiated hiPSC-LO could be used as a potential hepatic source for HBV infection, we detected the expression of NTCP in hiPSC-LO using the q-PCR assay. Analysis showed that the expression of NTCP could be detected and was comparatively higher in hiPSC-LOs, than in the PHHs (Fig. 2A). Immunofluorescence analysis also confirmed NTCP positive hepatic lineages in the hiPSC-LO (Fig. 2B). We then tested the infection susceptibility of hiPSC-LO with HBV produced from HepG2.2.15.7 cells, and compared its susceptibility with hiPSC-HLCs and PHHs through the expression of pregenomic (pg) RNA, intercellular viral DNA (vDNA), covalently closed circular DNA (cccDNA), and supernatant vDNA at 10 days post infection (dpi) (Fig. 2C). In agreement with previous reports [[12], [13], [14]], hiPSC-HLCs could be infected by HBV with the detection of viral RNA and DNA (Fig. 2D-G). Further, infected hiPSC-LOs showed much higher expression of pg RNA, and increased copy numbers of intercellular vDNA, cccDNA, and supernatant vDNA than infected hiPSC-HLCs (Fig. 2D-G and Fig. S3A). Moreover, the expression levels of pg RNA and the copy numbers of intercellular vDNA, cccDNA, and supernatant vDNA of infected hiPSC-LOs were intermediate between that of two different infected PHHs (Fig. 2D-G), indicating that the virus susceptibility of hiPSC-LO was comparable to PHHs. We monitored the dynamic virus release in infected hiPSC-LOs during the infection. From 7 dpi to 10 dpi, a significant release of viruses from the infected hiPSC-LOs was observed, but not from infected hiPSC-HLCs (Fig. 2H). Additionally, the expression of known infection promoting factors, GPC5 (glypican 5), PPARA (peroxisome proliferator-activated receptor alpha), and CEBPA (CCAAT/enhancer-binding protein alpha) [[25], [26], [27]], was found to be higher in hiPSC-LO than in hiPSC-HLC (Fig. 2I). These results indicated that differentiated hiPSC-LO could be a robust infection model.
Fig. 2.
Susceptibility of hiPSC-derived liver organoids to HBV infection.
(A) Q-PCR analysis of NTCP expression in hiPSCs, hiPSC-HLCs (HLCs) (n = 4), differentiated hiPSC-LOs (LOs) (n = 4), HepG2 cells (n = 4), PHH-1, and PHH-2. (B) Immunofluorescence analysis of ALB and NTCP in differentiated hiPSC-LOs. Green, ALB; Red, NTCP. Scale bars, 50 μm. (C) Schematic representation of HBV life cycle in infected hepatocyte. (D-G) Q-PCR quantification of HBV pregenomic RNA (pgRNA) (D), intracellular virus DNA (vDNA) (E), covalently closed circular DNA (cccDNA) (F), and supernatant vDNA (G) in HepG2-TET-NTCP (+DOX) (n = 4), hiPSC-HLCs (n = 8), hiPSC-LOs (n = 8), PHH-1(n = 4), and PHH-2 (n = 4) at 10 days post infection (dpi) with 500 genome equivalent (GEq)/cell infection, pgRNA expression in infected HepG2-TET-NTCP (+DOX) as a control. (H) Q-PCR quantification of supernatant vDNA in hiPSC-LOs and hiPSC-HLCs at 4, 7 and 10 dpi (n = 4). (F) The expression of GPC5, PPARA, and CEBPA in hiPSC-HLCs (n = 4) and hiPSC-LOs (n = 4) was detected by Q-PCR, hiPSC-HLCs were used as a control. *p < .05, **p < .01, ***p < .001; ns, not significant.
3.3. Long term maintenance of hepatic function and virus propagation in hiPSC liver organoids
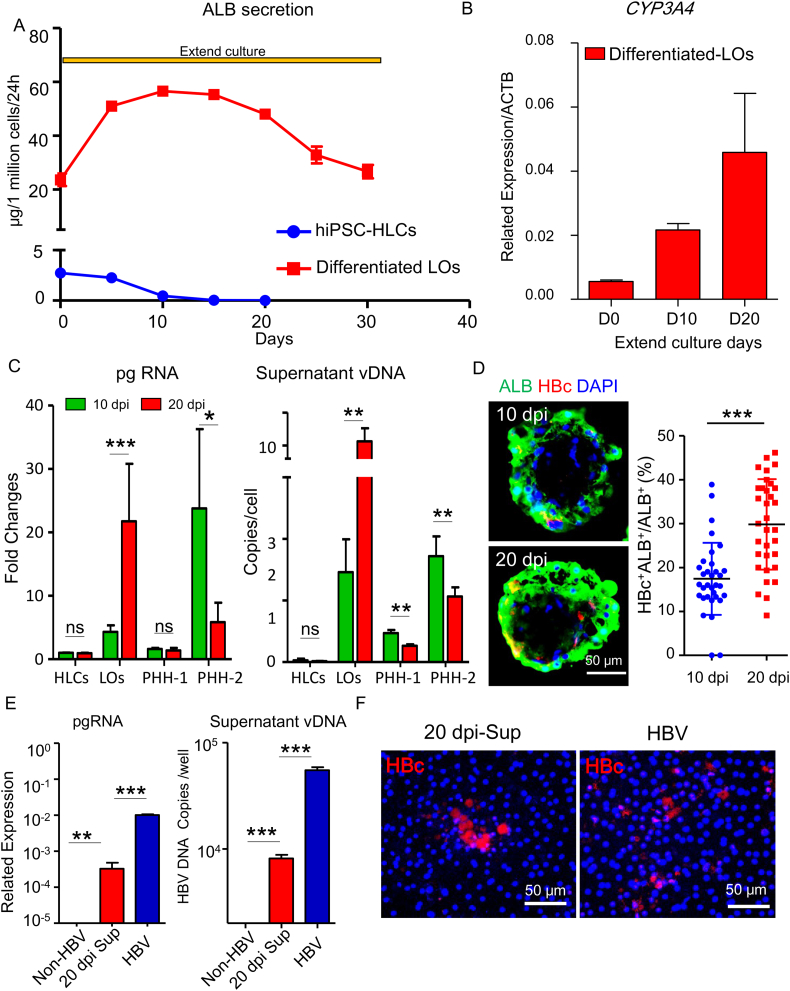
Next, we evaluated the ability of differentiated hiPSC-LO to maintain its hepatic function and its potential to be a long-term infection model. Dynamic monitoring showed that ALB secretion steadily increased in differentiated hiPSC-LO, and could be sustained for 20 days (Fig. 3A). In contrast, a sharp decline of ALB secretion was observed in hiPSC-HLCs after only 5 days of extended culture (Fig. 3A). The expression of CYP3A4 in hiPSC-LO also constantly increased during this period (Fig. 3B). These results demonstrate that the hiPSC-LOs displayed prolonged maintenance of hepatic functions, and have the potential to be a long-term infection model.
Fig. 3.
Long-term maintenance of hepatic function and HBV propagation in hiPSC liver organoids.
(A) ELISA for ALB secretion of hiPSC-HLCs and differentiated hiPSC-LOs in extended culture (n = 5, each). (B) Q-PCR analysis of CYP3A4 expression of hiPSC-LOs during extended culture, n = 3. (C) Quantification of HBV pgRNA (left) and supernatant vDNA (right) in hiPSC-HLCs (n = 4), hiPSC-LOs (n = 8), PHH-1 (n = 4) and PHH-2 (n = 4) at 10 and 20 dpi by Q-PCR. 10 dpi of hiPSC-HLCs was used as a control. (D) Immunofluorescence analysis of ALB and HBc in infected hiPSC-LOs at 10 and 20 dpi. Infection rates were calculated based on the number of HBc and ALB double positive cells (ALB+HBc+) relative to that of total ALB-positive cells (ALB+), n = 24 liver organoids. (E) Quantification of HBV pgRNA and supernatant vDNA in PXB-HH infected with supernatant of infected hiPSC-LOs (20 dpi) and HBV derived from HepG2 2.15.7 (n = 6 each) at 10 dpi, non-infected PXB-HH was used as a negative control (F) Immunofluorescence analysis of HBc in PXB-HH infected with supernatant of infected hiPSC-LOs (20 dpi) and HBV derived from HepG2 2.15.7 at 10 dpi. *p < .05, **p < .01, ***p < .001, ns, not significant.
We further investigated viral replication and propagation in hiPSC-LOs by increasing the infection time from 10 days to 20 days. Compared with 10 dpi infected LOs, pgRNA expression and supernatant vDNA copy numbers markedly increased in 20 dpi infected LOs (Fig. 3C). Such an increase was not observed in infected hiPSC-HLCs and PHHs (Fig. 3C). Immunofluorescence analysis also showed that the proportion of HBV core antigen (HBc) positive hepatic lineages (HBc+ALB+) increased in hiPSC-LOs prolonging the infection time (Fig. 3D). To further investigate whether infectious progeny viruses could be produced from infected hiPSC-LOs, we collected the supernatant from infected hiPSC-LOs at 20 dpi (20 dpi-Sup, from day 17 to day 20) and examined its infectivity in PXB-HHs. Notably, the progeny virus displayed infectivity, confirmed by pgRNA quantification in PXB-HHs (Fig. 3E). Furthermore, the progeny virus could complete their life cycle based on detectable vDNA in the supernatant (Fig. 3E) and intracellular HBc staining (Fig. 3F). These results indicate that this organoid system could serve as a long-term ex vivo infection model.
3.4. Induction of hepatic dysfunction in hiPSC liver organoids with HBV infection
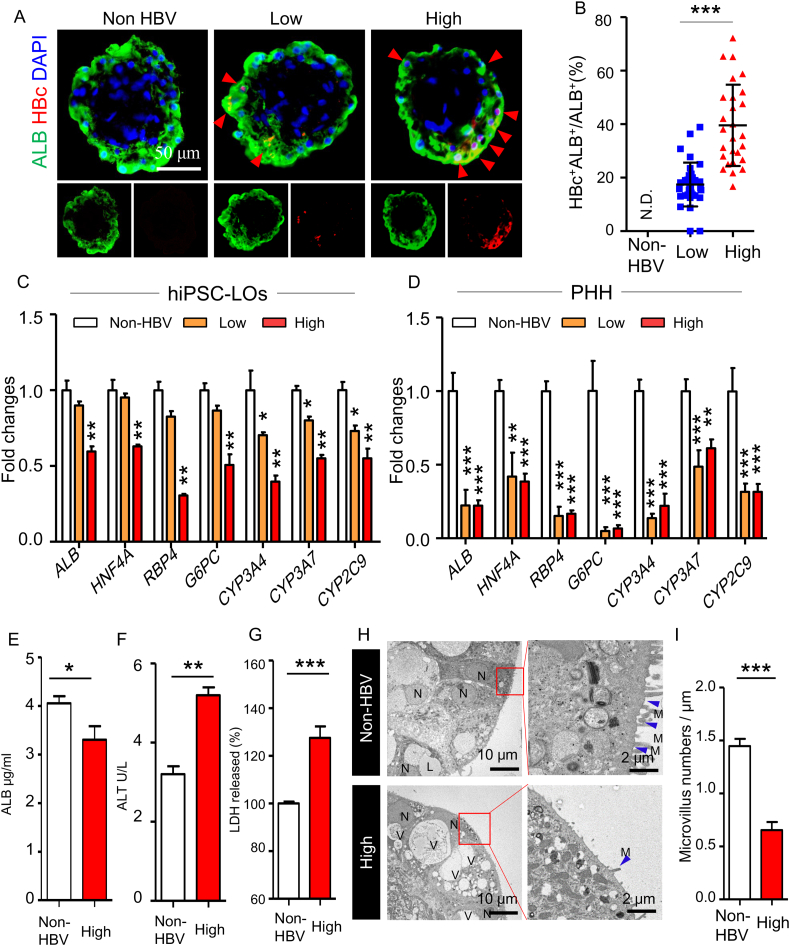
The prolonged maintenance of hepatic function in differentiated hiPSC-LOs could be helpful in understanding the host consequences caused by virus infection without any concerns about inherent reduction in hepatic function. To explore the host consequences of viral infection, we infected the differentiated hiPSC-LOs with different doses of the virus: 0 GEq (genome equivalent)/cell (non-HBV), 500 GEq/cell (low dose), and 5000 GEq/cell (high dose) [28, 29], and analyzed the changes in infected hiPSC-LOs. The increased amount of virus could markedly promote the infection in hiPSC-LOs (Fig. 4A, B and S4A). Further, expression analysis established that virus infection could dose-dependently impair the expression of CYP3A4, CYP3A7, and CYP2C9 (Fig. 4C). Additionally, a high-dose infection could down-regulate the expression of ALB, G6PC (glucose-6-phosphatase catalytic-subunit), HNF4A (hepatocyte nuclear factor 4 alpha), and RBP4 (retinol binding protein 4) (Fig. 4C). To confirm that the impaired expression of hepatic genes was a result of virus infection, we used myrcludex (a HBV entry inhibitor) and entecavir (an anti-HBV nucleos(t)ide) to inhibit virus infection. Upon treatment with these drugs, the expression of pg RNA, and copy number of intracellular vDNA and supernatant vDNA were significantly reduced (Fig. S4B), and the expression of hepatic functional genes was enhanced in infected LOs (Fig. S4C). The infection impaired hepatic gene expression was also detected in infected PHHs (Fig. 4D), but not in infected HepG2-TET-NTCP organoids (Fig. S4E). Moreover, a decreased ALB secretion was observed in high-dose infected LOs (Fig. 4E). To further confirm that viruses caused hepatic dysfunction, we measured the level of aminotransferase (ALT) and lactate dehydrogenase (LDH) that act as markers for early acute liver failure [30], in the supernatant of infected LOs, and found an increased level of ALT and LDH (Fig. 4F and G). TEM analysis indicated that, compared with non-infected LOs, the infected LOs had increased number of vacuoles in the hepatocyte cytoplasm (Fig. 4H), which occupied a significant proportion of the cytoplasm and pushed the nucleus into the cell periphery (Fig. 4H). Additionally, infected LOs showed reduced membrane microvilli (Fig. 4H and I), a characteristic of fibrosing liver disease [31]. HBV infection has been considered as a driving force for epithelial-mesenchymal transition (EMT) in liver cancer development [32]. In infected LOs, we found the expression of EMT markers SNAI2 (Snail Family Transcriptional Repressor 2) and TWIST1 (Twist Family BHLH Transcription Factor 1) was significantly up-regulated (Fig. S4F). These findings indicate that this organoid infection system could recapitulate virus induced hepatic dysfunction.
Fig. 4.
Induction of hepatic dysfunction in hiPSC liver organoids with HBV infection.
(A) Immunofluorescence analysis of HBc and ALB in hiPSC-LOs at doses of 0 (Non-HBV), 500 (low dose), and 5000 (high dose) GEq/cell infection at 10 dpi. (B) The percentage of ALB+HBc+ cells relative to that of total ALB+ cells, n = 22 liver organoids. (C) Q-PCR quantification of hepatic genes ALB, HNF4A, G6PC, RBP4, CYP3A4, CYP3A7, and CYP2C9 in non-, low-dose and high-dose HBV-infected hiPSC-LOs at 10 dpi, n = 4. (D) Q-PCR quantification of hepatic genes ALB, HNF4A, G6PC, RBP4, CYP3A4, CYP3A7, and CYP2C9 in non-, low-dose and high-dose HBV-infected PHH-2 at 10 dpi, n = 3. (E) ELISA analysis of ALB secretion in non- and high-dose HBV-infected hiPSC-LOs, n = 5. (F) Quantification of ALT in the supernatant of non- and high-dose HBV-infected hiPSC-LOs at 10 dpi, n = 5. (G) Detection of LDH in the supernatant of non- and high-dose HBV-infected hiPSC-LOs at 10 dpi, n = 5, non-HBV group used as a control. (H) Transmission electron microscopy analysis of the ultrastructure of hiPSC-LOs with non- and high-dose HBV infection at 10 dpi, N: nuclear; V: vacuole; L: lipid drop; blue arrow: microvilli. (I) Numbers of microvilli in non- and high-dose HBV infected hiPSC-LOs, n = 5.
During virus infection, the host innate immune cells recruited into the infection site produce interferons (IFNs) that help inhibit and eliminate the virus [33]. To mimic this immune defense, we treated infected hiPSC-LOs with IFNα, and found that IFNα could induce transcription of antiviral genes, including VIPERIN (virus inhibitory protein, endoplasmic reticulum-associated, interferon-inducible), ISG15 (interferon stimulated gene 15), ISG20 (interferon stimulated gene 20), and MX2 (Myxovirus Resistance Protein 2) (Fig. S5A). Upon IFNα and IFNγ treatment, virus replication was significantly suppressed with down-regulated expression of pgRNA (Fig. S5B), and a marked inhibition in expression of hepatic genes in the infected hiPSC-LOs (Fig. S5E). These observations suggest that the innate immune activation could effectively inhibit virus replication but would induce additional hepatic injury in hiPSC-LOs.
4. Discussion
In the current study, we developed a novel method to generate functional liver organoids with a 3D microwell system (Table S2). In this new culture system, liver organoids with small diameters (approximately 200 μm) were generated, with improved nutrient permeation and absorption, resulting in effective hepatic differentiation and susceptibility to HBV infection. The susceptibility of hiPSC-LOs to infection was comparable to cryopreserved PHHs. Moreover, this organoid system can mimic the virus induced hepatic dysfunction, indicating that infection in hiPSC-LOs might be able to recapitulate in vivo virus - host interactions.
With exogenous expression of human NTCP, different donor derived human hepatocarcinoma cells could gain susceptibility to HBV infection, but their susceptibility varied among cells [34]. Meanwhile, mouse hepatocytes overexpressing human NTCP were not susceptible to HBV infection [35]. These studies have confirmed that NTCP is a necessary factor but not sufficient for HBV infection; other human liver specific factors would also be necessary in the regulation of HBV life cycle, and these factors may be related to the host's genetic background. To test this idea, we compared the HBV infection in LOs generated from different genetic background and observed different levels of susceptibility among them (Fig. S3B). Furthermore, we found that hiPSC-LOs and hiPSC-HLCs generated from same genetic background had comparable expression of NTCP, but had significant difference in virus susceptibility, suggesting that hepatic lineages in distinct differentiation stages might express different levels of liver-specific factors important for virus infection. Thus, further detailed analysis of the differences between single donor-derived LO and HLC would help in the identification of potential risk and resistance factors for infection. Our novel functional hiPSC-LO provides a new avenue to investigate the role of the host genetic background in HBV infection and prognosis of individual infection and has the potential to be used in personalized hepatitis treatment.
HBV was not considered as a cytopathic virus because it was believed that the virus itself might not cause the hepatocellular damage in acute and chronic HBV infection [36]; some studies have also stated that HBV infection could not induce hepatic apoptosis [37, 38]. However, these models likely display cancer cell characteristics and unstable hepatic function, due to which they probably could not reproduce normal viral cytopathic effects. Besides, subsequent studies have observed that HBV infection could induce hepatic damage in patients and mouse models with severe immunosuppression [39, 40], suggesting that HBV may indeed cause cytopathic effects on hepatocytes in vivo, which might be taken care of by rapid host immune responses. In this study, we accidentally found that HBV could induce hepatic dysfunction of hiPSC-LOs with significantly reduced hepatic function, induced release of hepatic injury markers, and altered hepatic ultrastructure. Functional LOs generated without immune cells, when infected, displayed an impaired hepatic function, further favoring the idea that HBV might be a cytopathic virus. Although the mechanisms underlying the cytopathic effects of the virus have not yet been defined, the accumulation of HBV proteins has been reported to result in the appearance of cellular vacuolization in hepatocytes [39], and to induce stress and increase reactive oxygen species (ROS) in the endoplasmic reticulum [41]. The increased ROS might further induce autophagy and disrupt membrane lipids to alter hepatic ultrastructure [42, 43]. In this study, we found that HBV infection did not induce cell apoptosis or lysis of hiPSC-LOs (Fig. S4D), supporting the view that HBV might be a cytopathic virus, but not a cytolytic one.
The highly efficient and fast hepatic innate immune response to pathogen is initiated through IFNs [44]. Inhibition of IFN activation could induce robust replication of HBV in PHHs [13], but this HBV-activated IFN response in hepatocytes was not detected in hiPSC-LOs (Fig. S5F) or in other infected hepatic cells [45]. This loss of self-defense might advance the stealthy replication of HBV in hiPSC-LOs. Although studies have suggested that IFNs inhibit HBV replication in a non-cytolytic manner [46, 47], our results suggest that IFNs could efficiently inhibit viral replication, but could also inhibit hepatic function in the infected hiPSCs-LOs. This aggravated hepatic injury might be a side effect of IFNs. This IFN-mediated liver injury has also been observed in mice models and clinical trials [48, 49]. We were unable to observe IFN induced cccDNA degradation in infected LOs, possibly because of the low APOBEC3A expression in hiPSC-LOs (Fig. S5C and S5D) [47]. These observations suggest that the organoids may still have some characteristics different from adult hepatocytes. Thus, further efforts are required to generate adult liver-like organoids from hiPSCs.
In summary, we have generated functional hiPSC-LOs that can efficiently recapitulate host-virus interactions by mimicking the virus life cycle and display HBV-induced hepatic dysfunction, indicating that this LO may be a reliable and feasible personalized infection model for individualized hepatitis study and treatments.
The following are the supplementary data related to this article.
List of quantitative RT-PCR primers and probes for human genes.
The difference of Liver organoids generation protocols in this study and in previous reports.
Supplementary material
Funding sources
This work was supported by the grants from the JST Research Center Network for Realization of Regenerative Medicine: 17bm0304002h0105, the Japan Agency for Medical Research and Development: 17fk0310103j0001, and the Japan Society for the Promotion of Scienc KAKENHI (Grant-in-Aid for JSPS Research Fellow):18F18101. Y.-Z. N was supported by JSPS International Fellowships for Research in Japan (P18101).
Conflicts of interest
The authors declare no competing financial interests.
Author contributions
Y.-Z.N., Y.-W.Z., and H.T. designed the study, analyzed the data, and prepared the manuscript; Y.-Z.N. and K.M. performed the experiments; S.M., R.-R.Z., Y.U., K.S., T.T., and A.R. were involved in study design and supervised experiments; all authors discussed the results and commented on the manuscript.
Research in context
Evidence suggests that the genetic background of patients with Hepatitis B influences the markedly heterogeneous outcomes seen across these patients. However, very few infection models exist that can represent a patient's genetic background. Our new technique makes this possible by generation of functional liver organoids using human induced pluripotent stem cells (hiPSCs). The hiPSC-derived functional liver organoids can be a robust and long-term HBV infection model, which recapitulates viral lifecycle and virus-induced hepatic dysfunction. It provides a promising approach to understand the precise roles of genetic background in virus-induced host outcomes and develop personalized medicine for hepatitis B patients.
Acknowledgments
We thank Mayu Miyamoto for technical support and assistance with HBV preparation.
Contributor Information
Yun-Wen Zheng, Email: ywzheng@md.tsukuba.ac.jp.
Hideki Taniguchi, Email: rtanigu@med.yokohama-cu.ac.jp.
References
- 1.Revill P., Testoni B., Locarnini S., Zoulim F. Global strategies are required to cure and eliminate HBV infection. Nature Reviews. Gastroenterology & Hepatology. 2016;13:239–248. doi: 10.1038/nrgastro.2016.7. [DOI] [PubMed] [Google Scholar]
- 2.Zeng Z., Guan L., An P., Sun S., O'brien S.J., Winkler C.A. A population-based study to investigate host genetic factors associated with hepatitis B infection and pathogenesis in the Chinese population. BMC Infectious Diseases. 2008;8:1. doi: 10.1186/1471-2334-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thursz M., Yee L., Khakoo S. Understanding the host genetics of chronic hepatitis B and C. Seminars in Liver Disease. 2011;31:115–127. doi: 10.1055/s-0031-1276642. [DOI] [PubMed] [Google Scholar]
- 4.Burgner D., Jamieson S.E., Blackwell J.M. Genetic susceptibility to infectious diseases: big is beautiful, but will bigger be even better? The Lancet Infectious Diseases. 2006;6:653–663. doi: 10.1016/S1473-3099(06)70601-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin T.M., Chen C.J., Wu M.M., Yang C.S., Chen J.S., Lin C.C. Hepatitis B virus markers in Chinese twins. Anticancer Research. 1989;9:737–741. [PubMed] [Google Scholar]
- 6.Galle P.R., Hagelstein J., Kommerell B., Volkmann M., Schranz P., Zentgraf H. In vitro experimental infection of primary human hepatocytes with hepatitis B virus. Gastroenterology. 1994;106:664–673. doi: 10.1016/0016-5085(94)90700-5. [DOI] [PubMed] [Google Scholar]
- 7.Yan H., Zhong G., Xu G., He W., Jing Z., Gao Z., Huang Y., Qi Y., Peng B., Wang H., Fu L., Song M., Chen P., Gao W., Ren B., Sun Y., Cai T., Feng X., Sui J., Li W. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife. 2012;1 doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishida Y., Yamasaki C., Yanagi A., Yoshizane Y., Fujikawa K., Watashi K. Novel robust in vitro hepatitis B virus infection model using fresh human hepatocytes isolated from humanized mice. The American Journal of Pathology. 2015;185:1275–1285. doi: 10.1016/j.ajpath.2015.01.028. [DOI] [PubMed] [Google Scholar]
- 9.Stadtfeld M., Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes & Development. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rashid S.T., Corbineau S., Hannan N., Marciniak S.J., Miranda E., Alexander G. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. The Journal of Clinical Investigation. 2010;120:3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takayama K., Morisaki Y., Kuno S., Nagamoto Y., Harada K., Furukawa N. Prediction of interindividual differences in hepatic functions and drug sensitivity by using human iPS-derived hepatocytes. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:16772–16777. doi: 10.1073/pnas.1413481111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaneko S., Kakinuma S., Asahina Y., Kamiya A., Miyoshi M., Tsunoda T. Human induced pluripotent stem cell-derived hepatic cell lines as a new model for host interaction with hepatitis B virus. Scientific Reports. 2016;6:29358. doi: 10.1038/srep29358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shlomai A., Schwartz R.E., Ramanan V., Bhatta A., De Jong Y.P., Bhatia S.N. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:12193–12198. doi: 10.1073/pnas.1412631111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia Y., Carpentier A., Cheng X., Block P.D., Zhao Y., Zhang Z. Human stem cell-derived hepatocytes as a model for hepatitis B virus infection, spreading and virus-host interactions. Journal of Hepatology. 2017;66:494–503. doi: 10.1016/j.jhep.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu Y., Liu H., Ikeda Y., Amiot B.P., Rinaldo P., Duncan S.A. Hepatocyte-like cells differentiated from human induced pluripotent stem cells: relevance to cellular therapies. Stem Cell Research. 2012;9:196–207. doi: 10.1016/j.scr.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Si-Tayeb K., Lemaigre F.P., Duncan S.A. Organogenesis and development of the liver. Developmental Cell. 2010;18:175–189. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 17.Watt A.J., Zhao R., Li J., Duncan S.A. Development of the mammalian liver and ventral pancreas is dependent on GATA4. BMC Developmental Biology. 2007;7:37. doi: 10.1186/1471-213X-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaret K.S. Regulatory phases of early liver development: paradigms of organogenesis. Nature Reviews. Genetics. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 19.Takebe T., Sekine K., Enomura M., Koike H., Kimura M., Ogaeri T. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 20.Takebe T., Zhang R.R., Koike H., Kimura M., Yoshizawa E., Enomura M. Generation of a vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature Protocols. 2014;9:396–409. doi: 10.1038/nprot.2014.020. [DOI] [PubMed] [Google Scholar]
- 21.Miyakawa K., Matsunaga S., Watashi K., Sugiyama M., Kimura H., Yamamoto N. Molecular dissection of HBV evasion from restriction factor tetherin: a new perspective for antiviral cell therapy. Oncotarget. 2015;6:21840–21852. doi: 10.18632/oncotarget.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogura N., Watashi K., Noguchi T., Wakita T. Formation of covalently closed circular DNA in Hep38.7-Tet cells, a tetracycline inducible hepatitis B virus expression cell line. Biochemical and Biophysical Research Communications. 2014;452:315–321. doi: 10.1016/j.bbrc.2014.08.029. [DOI] [PubMed] [Google Scholar]
- 23.Kajiwara M., Aoi T., Okita K., Takahashi R., Inoue H., Takayama N. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12538–12543. doi: 10.1073/pnas.1209979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nie Y.Z., Zheng Y.W., Ogawa M., Miyagi E., Taniguchi H. Human liver organoids generated with single donor-derived multiple cells rescue mice from acute liver failure. Stem Cell Research & Therapy. 2018;9:5. doi: 10.1186/s13287-017-0749-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi B.H., Park G.T., Rho H.M. Interaction of hepatitis B viral X protein and CCAAT/ enhancer-binding protein alpha synergistically activates the hepatitis B viral enhancer II/pregenomic promoter. The Journal of Biological Chemistry. 1999;274:2858–2865. doi: 10.1074/jbc.274.5.2858. [DOI] [PubMed] [Google Scholar]
- 26.Verrier E.R., Colpitts C.C., Bach C., Heydmann L., Weiss A., Renaud M. A targeted functional RNA interference screen uncovers glypican 5 as an entry factor for hepatitis B and D viruses. Hepatology. 2016;63:35–48. doi: 10.1002/hep.28013. [DOI] [PubMed] [Google Scholar]
- 27.Yu X., Mertz J.E. Critical roles of nuclear receptor response elements in replication of hepatitis B virus. Journal of Virology. 2001;75:11354–11364. doi: 10.1128/JVI.75.23.11354-11364.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortega-Prieto A.M., Skelton J.K., Wai S.N., Large E., Lussignol M., Vizcay-Barrena G. 3D microfluidic liver cultures as a physiological preclinical tool for hepatitis B virus infection. Nature Communications. 2018;9:682. doi: 10.1038/s41467-018-02969-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winer B.Y., Huang T.S., Pludwinski E., Heller B., Wojcik F., Lipkowitz G.E. Long-term hepatitis B infection in a scalable hepatic co-culture system. Nature Communications. 2017;8:125. doi: 10.1038/s41467-017-00200-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kotoh K., Enjoji M., Kato M., Kohjima M., Nakamuta M., Takayanagi R. A new parameter using serum lactate dehydrogenase and alanine aminotransferase level is useful for predicting the prognosis of patients at an early stage of acute liver injury: a retrospective study. Comparative Hepatology. 2008;7:6. doi: 10.1186/1476-5926-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedman S.L. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. The Journal of Biological Chemistry. 2000;275:2247–2250. doi: 10.1074/jbc.275.4.2247. [DOI] [PubMed] [Google Scholar]
- 32.Lau C.C., Sun T., Ching A.K., He M., Li J.W., Wong A.M. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell. 2014;25:335–349. doi: 10.1016/j.ccr.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 33.Shin E.C., Sung P.S., Park S.H. Immune responses and immunopathology in acute and chronic viral hepatitis. Nature Reviews. Immunology. 2016;16:509–523. doi: 10.1038/nri.2016.69. [DOI] [PubMed] [Google Scholar]
- 34.Ni Y., Lempp F.A., Mehrle S., Nkongolo S., Kaufman C., Falth M. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology. 2014;146:1070–1083. doi: 10.1053/j.gastro.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 35.Li H., Zhuang Q., Wang Y., Zhang T., Zhao J., Zhang Y. HBV life cycle is restricted in mouse hepatocytes expressing human NTCP. Cellular & Molecular Immunology. 2014;11:175–183. doi: 10.1038/cmi.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chisari F.V., Ferrari C. Hepatitis B virus immunopathogenesis. Annual Review of Immunology. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 37.Schulze-Bergkamen H., Untergasser A., Dax A., Vogel H., Buchler P., Klar E. Primary human hepatocytes–a valuable tool for investigation of apoptosis and hepatitis B virus infection. Journal of Hepatology. 2003;38:736–744. doi: 10.1016/s0168-8278(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 38.Sells M.A., Chen M.L., Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foo N.C., Ahn B.Y., Ma X., Hyun W., Yen T.S. Cellular vacuolization and apoptosis induced by hepatitis B virus large surface protein. Hepatology. 2002;36:1400–1407. doi: 10.1053/jhep.2002.36819. [DOI] [PubMed] [Google Scholar]
- 40.Meuleman P., Libbrecht L., Wieland S., De Vos R., Habib N., Kramvis A., Roskams T., Leroux-Roels G. Immune suppression uncovers endogenous cytopathic effects of the hepatitis B virus. Journal of virology. 2006;80:2797–2807. doi: 10.1128/JVI.80.6.2797-2807.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsieh Y.H., Su I.J., Wang H.C., Chang W.W., Lei H.Y., Lai M.D. Pre-S mutant surface antigens in chronic hepatitis B virus infection induce oxidative stress and DNA damage. Carcinogenesis. 2004;25:2023–2032. doi: 10.1093/carcin/bgh207. [DOI] [PubMed] [Google Scholar]
- 42.Birben E., Sahiner U.M., Sackesen C., Erzurum S., Kalayci O. Oxidative stress and antioxidant defense. World Allergy Organization Journal. 2012;5:9–19. doi: 10.1097/WOX.0b013e3182439613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang S.W., Ducroux A., Jeang K.T., Neuveut C. Impact of cellular autophagy on viruses: insights from hepatitis B virus and human retroviruses. Journal of Biomedical Science. 2012;19:92. doi: 10.1186/1423-0127-19-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crispe I.N. Hepatocytes as immunological agents. Journal of Immunology. 2016;196:17–21. doi: 10.4049/jimmunol.1501668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng X., Xia Y., Serti E., Block P.D., Chung M., Chayama K. Hepatitis B virus evades innate immunity of hepatocytes but activates cytokine production by macrophages. Hepatology. 2017;66:1779–1793. doi: 10.1002/hep.29348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guidotti L.G., Rochford R., Chung J., Shapiro M., Purcell R., Chisari F.V. Viral clearance without destruction of infected cells during acute HBV infection. Science. 1999;284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- 47.Lucifora J., Xia Y., Reisinger F., Zhang K., Stadler D., Cheng X. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221–1228. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kakimi K., Lane T.E., Wieland S., Asensio V.C., Campbell I.L., Chisari F.V. Blocking chemokine responsive to gamma-2/interferon (IFN)-gamma inducible protein and monokine induced by IFN-gamma activity in vivo reduces the pathogenetic but not the antiviral potential of hepatitis B virus-specific cytotoxic T lymphocytes. The Journal of Experimental Medicine. 2001;194:1755–1766. doi: 10.1084/jem.194.12.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vento S., DI Perri G., Garofano T., Cosco L., Concia E., Ferraro T. Hazards of interferon therapy for HBV-seronegative chronic hepatitis. Lancet. 1989;2:926. doi: 10.1016/s0140-6736(89)91595-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of quantitative RT-PCR primers and probes for human genes.
The difference of Liver organoids generation protocols in this study and in previous reports.
Supplementary material