Abstract
Age-related macular degeneration (AMD) is a multi-factorial disease that is the leading cause of irreversible and severe vision loss in the developed countries. It has been suggested that the pathogenesis of dry AMD involves impaired protein degradation in retinal pigment epithelial cells (RPE). RPE cells are constantly exposed to oxidative stress that may lead to the accumulation of damaged cellular proteins, DNA and lipids and evoke tissue deterioration during the aging process. The ubiquitin-proteasome pathway and the lysosomal/autophagosomal pathway are the two major proteolytic systems in eukaryotic cells. NRF-2 (nuclear factor-erythroid 2-related factor-2) and PGC-1α (peroxisome proliferator-activated receptor gamma coactivator-1 alpha) are master transcription factors in the regulation of cellular detoxification. We investigated the role of NRF-2 and PGC-1α in the regulation of RPE cell structure and function by using global double knockout (dKO) mice. The NRF-2/PGC-1α dKO mice exhibited significant age-dependent RPE degeneration, accumulation of the oxidative stress marker, 4-HNE (4-hydroxynonenal), the endoplasmic reticulum stress markers GRP78 (glucose-regulated protein 78) and ATF4 (activating transcription factor 4), and damaged mitochondria. Moreover, levels of protein ubiquitination and autophagy markers p62/SQSTM1 (sequestosome 1), Beclin-1 and LC3B (microtubule associated protein 1 light chain 3 beta) were significantly increased together with the Iba-1 (ionized calcium binding adaptor molecule 1) mononuclear phagocyte marker and an enlargement of RPE size. These histopathological changes of RPE were accompanied by photoreceptor dysmorphology and vision loss as revealed by electroretinography. Consequently, these novel findings suggest that the NRF-2/PGC-1α dKO mouse is a valuable model for investigating the role of proteasomal and autophagy clearance in the RPE and in the development of dry AMD.
Abbreviations: ER, endoplasmic reticulum; AMD, age-related macular degeneration; RPE, retinal pigment epithelium; PUFAs, polyunsaturated fatty acids; ROS, reactive oxygen species; NRF-2, nuclear factor-erythroid 2-related factor-2; ARE, antioxidant response element; Keap1, kelch-like ECH-associated protein 1; PGC-1, peroxisome proliferator-activated receptor gamma coactivator-1; PRC, PGC-1-related coactivator; SOD2, superoxide dismutase 2; TRX1, thioredoxin; AMPK, AMP-activated protein kinase; SIRT1, sirtuin 1; UPS, ubiquitin-proteasome system; p62/SQSTM1, sequestosome 1; LC3, microtubule-associated protein 1A/1B light chain 3; dKO, double knockout; KO, knockout; WT, wild type; 4-HNE, 4-hydroxynonenal); ROI, region of interest; TEM, transmission electron microscopy; ATF4, activating transcription factor 4; GRP78, glucose-regulated protein 78; Iba-1, ionized calcium binding adaptor molecule 1; ERG, electroretinography; Rd8, retinal degeneration 8; MRI, magnetic resonance imaging; mtDNA, mitochondrial DNA
Keywords: Aging, Autophagy, Degeneration, Oxidative stress, Protein aggregation, Proteasome
Highlights
-
•
NRF-2/PGC-1α dKO mouse model shows a dry AMD-like phenotype.
-
•
Loss of NRF-2/PGC-1α genes increased oxidative and ER stress in RPE cells.
-
•
High oxidative stress was associated with impaired autophagy and proteasomal clearance.
-
•
The pathology becomes manifest as an age-related loss of photoreceptor function.
1. Introduction
Chronic oxidative and endoplasmic reticulum (ER) stress, impaired autophagy, mitochondrial dysfunction and inflammation are strongly linked to age-related macular degeneration (AMD) [1], [2], [3], [4], [5], [6]. One clinical hallmark of AMD is the degeneration of retinal pigment epithelial (RPE) cells, a process that associates with the accumulation of oxidative stress–derived lysosomal lipofuscin, impairing lysosomal degradation, and the presence of extracellular protein/lipid deposits (drusen) between the basal lamina of the RPE and the inner collagenous layer of the Bruch's membrane [7], [8], [9], [10], [11]. AMD can be subdivided into dry and wet forms with 80% and 20% prevalences, respectively. There are no effective treatments for dry AMD.
The quiescent RPE cells in the macula are constantly subjected to high oxidative stress due to the diurnal digestion of high concentrations of polyunsaturated fatty acids (PUFAs) originating from the phagocytosis of the retinal outer segments, exposure to the oxygen-rich choriocapillary circulation, and the light-induced production of reactive oxygen species (ROS) by cellular pigment granules [7]. The mitochondria also produce ROS as a by-product of respiration. During the aging of RPE cells, the cellular capacity to neutralize ROS diminishes due to a decreased production of ROS-scavengers and other antioxidants [12], [13].
In RPE cells, the NRF-2/ARE (nuclear factor-erythroid 2-related factor-2/antioxidant response element) pathway plays the major role in the oxidative stress regulation [14], [15]. In response to oxidative stress, the NRF-2 is released from NRF-2-Keap1 (kelch-like ECH-associated protein 1) complex allowing the translocation of NRF-2 from cytosol into the nucleus and the expression of several antioxidant defense system and autophagy -related genes and proteasomal subunits [16], [17], [18], [19], [20]. Mice models with NRF-2 deficiency have been shown to be more susceptible to oxidative damage and protein aggregation than WT (wild type) mice and to negatively regulate autophagy-related genes [21], [22].
The peroxisome proliferator-activated receptor gamma coactivator-1 (PGC-1) family, consisting of PGC-1α, PGC-1β and PRC (PGC-1-related coactivator), regulates a network governing the transcriptional control of mitochondrial biogenesis and respiratory function as well as targeting mitochondrial antioxidant defense system [23], [24], [25]. The role of PGC-1α is to serve as a switch between mitochondrial biogenesis and oxidative damage by controlling the mitochondrial levels of ROS. Loss-of-function studies of PGC-1α have shown burst of ROS and an increase in mitochondrial damage and degradation whereas gain-of-function enhanced mitochondrial biogenesis and the expression of mitochondrial antioxidant defense system-related genes, such as SOD2 (superoxide dismutase 2) and TRX1 (thioredoxin) [23], [25]. However, both pathways, mitochondrial biogenesis and ROS control, strive for the preservation of mitochondrial homeostasis. Post-translational modification of the energy sensors, AMPK (AMP-activated protein kinase) and SIRT1 (sirtuin 1) that induce autophagy, are known to regulate PGC-1α activities [26], [27], [28]. In RPE cells, PGC-1α has been shown to drive mitochondrial biogenesis as well as activating the antioxidant defense system [29], [30].
A high ROS production combined with impaired antioxidant systems results in detrimental protein aggregation [8], [31]. The ubiquitin-proteasome system (UPS) and the lysosomal/autophagosomal degradation system share the major responsibility of maintaining cellular proteostasis [8], [32]. Both systems recognize and actively select the material destined for degradation and recycling in cells. The p62/SQSTM1 (sequestosome 1) has a multifactorial role since it acts as a bridge shuttling proteins between UPS and autophagy clearance as well as regulating the antioxidant response in RPE cells during oxidative stress [9], [33], [34]. Proteins assigned to the degradation are often ubiquitinated and directed to the proteasomal degradation. However, the rate of protein degradation by proteasomes is limited, and when UPS is overwhelmed, for example under heavy oxidative stress, proteins form aggregates which cannot be removed by proteasomes. Autophagy is specialized for removing protein aggregates and large cell organelles, such as mitochondria. p62/SQSTM1 recognizes ubiquitinated perinuclear protein aggregates and the p62/SQSTM1 tagged material is then isolated from the cytosol in p62/SQSTM1-LC3 (microtubule-associated protein 1A/1B light chain 3) interaction-guided autophagosome formation [35]. In the final step of the autophagy process, a lysosome is fused to the autophagosome resulting in the formation of the autolysosome followed by the degradation of its contents including ubiquitin, p62/SQSTM1 and LC3. Thereby, these proteins can be exploited as biomarkers of protein aggregation and autophagy activity [35], [36]. Moreover, p62/SQSTM1 interacts with the NRF-2/ARE pathway by disrupting the NRF-2-Keap1 complex leading to nuclear localization of NRF-2 [37].
The large and increasing number of individuals affected by AMD means that this represents an urgent global health emergency requiring effective primary prevention strategies as well as improvements in the current treatments for this disease. Therefore, new studies contributing to a better understanding of the mechanisms regulating antioxidant defense system, proteostasis, and mitochondrial function may lead to breakthroughs in AMD research. Since the NRF-2 and PGC-1α transcription factors have a key role in cellular detoxification and mitochondrial function in response to oxidative stress, we generated and characterized the NRF-2/PGC-1α double knockout (dKO) mouse model to study retinal changes that occur in degradative pathways and document features previously associated with AMD.
2. Material & methods
2.1. Animals
All animal protocols were approved by the Animal Experiment Board of Finland and conducted in compliance with the European Community Council Directives 2010/63/EU and ARVO statement for the Use of Animals in Ophthalmic and Vision Research. Mice were group-housed in the Laboratory Animal Centre of University of Eastern Finland, Kuopio, Finland on a 12-h light/dark cycle and given food and water ad libitum.
The mice strains deficient in NRF-2 or PGC-1α (C57BL/6J background) used in this study have been described earlier [38], [39]. To obtain mice lacking both NRF-2 and PGC-1α, homozygous-null NRF-2 and PGC-1α strains were crossed first, and the resultant double heterozygotes were crossed together. Female and male KO (knockout) and dKO mice at six weeks, three months and one year of age were used for this study, together with age-matched WT (wild type) controls.
2.2. The genotyping of NRF-2 and PGC-1α knockout mice
The genomic mouse DNA was extracted from small ear samples by heating the tissues for 1 h at 95 °C in 100 μl of 25 mM NaOH + 0.2 mM EDTA. The solutions were neutralized by adding 100 μl of 40 mM Tris-HCl pH 7.5, and centrifuged for 10 min at 2000 rpm. The upper halves of the supernatants (100 μl) were collected to be used directly as templates in the PCR reactions.
The genotypes of PGC-1α were detected with a 4-primer PCR. The primer sequences were: WTA, 5′-CCA GTT TCT TCA TTG GTG TG; WTB, 5′-ACC TGT CTT TGC CTA TGA TTC; KOA, 5′-TCC AGT AGG CAG AGA TTT ATG AC; KOB, 5′-CCA ACT GTC TAT AAT TCC AGT TC. The NRF-2 genotypes were studied with the following three primers: LacZ, 5′-GCG GAT TGA CCG TAA TGG CAT AGG; Nrf2–5′, 5′-TGG ACG GGA CTA TTG AAG GCT G; Nrf2–3’, GCC GCC TTT TCA GTA GAT GGA CG.
Both reactions included 4 μl of DNA extracted from mouse ears, reaction buffer, 100 μM of each dNTPs, 1.5 mM of MgCl2, 1 μM of each primers, and 1.2 U of DreamTaq DNA polymerase (Thermo Fisher Scientific Waltham, MA USA), in a volume of 30 μl. For the PGC-1α, samples were denatured at 95 °C for 5 min, followed by 39 cycles at 95 °C for 30 s, 58 °C for 30 s, 72 °C for 30 s, and a final extension at 72 °C for 7 min. For the NRF-2 genotyping, the reaction conditions were: denaturation at 95 °C for 5 min, followed by 35 cycles at 95 °C for 30 s, at 59 °C for 30 s, and at 72 °C for 45 s, and a final extension at 72 °C for 7 min. All PCR products were separated using 1% agarose (Seakem LE, Cambrex) gel electrophoresis in standard Tris-acetate-EDTA buffer, visualized in UV light with ethidium bromide, and photographed. The amplicon sizes the for wild type allele of PGC-1α are 600 bp and for the KO allele 400 bp. Their sizes for the NRF-2 alleles are 700 and 400 bp, respectively (Supplementary Fig. 1).
2.3. Immunomapping for detection of protein aggregation, autophagy and oxidative stress markers: Ubiquitin, p62/SQSTM1, Beclin-1, LC3 and 4-HNE
The mice were sacrificed with cervical dislocation and the eyes were immediately carefully enucleated and placed in PBS (pH 7.4). Before dehydrating in graded series of ethanol and embedding in paraffin, the eyes were fixed in 4% paraformaldehyde in 0.1 M phosphate buffer for 24–48 h. Five μm thick parasagittal serial sections were cut from embedded blocks with a microtome (SM2000 R, Leica, Heidelberg, Germany). After a random selection of slides from each of the individual specimens, the sections were dewaxed and rehydrated and the labeling of the retina was performed according to previously published methods, with minor modifications [36]. Briefly, the sections were treated with blocking solution (#IHC-101B Bethyl laboratories, Montgomery, TX, USA) for 30 min at room temperature to prevent the possibility of non-specific binding of the primary antibodies; the samples were then immunostained with different primary antibodies as follows: anti-ubiquitin (1/200; Dako, Glostrup, Denmark), anti-SQSTM1/p62 (1/100; Abcam, Cambridge, UK), anti-Beclin 1 (1/160; Novus Biologicals, CO, USA), anti 4-HNE (4-hydroxynonenal) (1/200; LifeSpan BioSciences, Inc, Seattle, WA, USA), anti LC3B (1/200; Cell Signaling Technology, Danvers, MA, USA) overnight, at 4 °C. After rinsing the primary antibodies in TBS, the samples were then incubated with a secondary antibody anti mouse Alexa Fluor 594 (1/500; Thermo Fisher Scientific Waltham, MA USA) for 3 h at room temperature and rinsed with TBS. A fluorescent nuclear marker DAPI (4′,6-diamidino-2-phenylindole dihydrochloride, #D9542, Sigma, St. Louis, MO, USA) diluted 1:10 000 in TBS was added and incubated for 30 min at room temperature. The sections were rinsed with TBS and covered with Mowiol mounting media. Immunofluorescence was evaluated independently of randomly selected sections by three researchers and no signal was recorded from technical negative controls (TBS instead of primary antibody). After the staining, the samples were analyzed as described above, photomicrographs were taken in a Zeiss AX10 Imager A2 (Zeiss, Göttingen, Germany) microscope. Images were captured sequentially from the green, red, and far-red channels on slices using a 63× oil immersion objective (NA:1.42, Plan Apochromat). The microscope settings were identical for all scans and kept constant during imaging. Representative scans were taken with a Jenoptik ProgRes C5 (Zeiss, Göttingen, Germany) digital camera mounted onto the microscope. In all imaging procedures, gamma adjustment was made on the whole image in order to maintain appropriate contrast and images were processed using Adobe Photoshop for documentation. The control sections originated from the same eyes and the validity of the sequential staining was then verified with the same protocol as described above except that one of the primary antibodies was omitted. The negative control samples displayed only a sub-minimal autofluorescence signal.
2.4. Spectral imaging analysis
The immunohistochemical results were examined by three independent observers searching for immunoreactive RPE cells. The different markers distribution pattern were semi-quantitatively analyzed by computer imaging as follows. High power view RGB 8-bit images of single RPE cells were generated with a Jenoptik ProgRes C5 as it described above in fluorescent mode, using a 63× objective lens. Blue pseudocolor of the nuclei was assigned by the investigators to help segmentation processes of RPE cell nuclei, respectively. For quantitation of the different protein aggregation and autophagy related (Ubiquitin, p62/SQSTM1, Beclin-1, LC3B) marker expression and oxidative stress marker (4-HNE), 10 RPE cells per animal of interest (n = 3 WT and n = 3 NRF-2/PGC-1α dKO) were manually designated as regions of interest (ROI). Special care was taken to select two representative areas per section for collecting RPEs from each individual samples close to the vicinity of optic nerve. Sampling for RPE image analysis was performed without external knowledge on samples genotype to be able to compare without influences from testers' preferences or expectations. A spectral imaging containing the characteristic wavelength emission of Alexa Fluor 594 was created by sampling pure RED spectra from RGB images; these were converted to black and white images, i.e. only the positive signal of interest was analyzed further. Via this grayscale conversion, the original, true RED color spectral information was automatically converted into one grayscale image. This process is based on the calculation of the average intensity per channel, therefore one can keep the original staining intensity information. Furthermore, this conversion makes it possible to focus only on the RED channel of interest. Using the open source Image J software (http//:imagej.nih.gov; NIH, USA), we quantified the immunostaining in the ROI, the software measured the amount of target signal (intensity of B&W pseudo-color) within each ROI, by measuring the density per pixel and returning a nominal value between 0 (the limit of noise level i.e. data point from a blank sample /=negative immunohistochemical reaction/ was determined below 20) and 255 (white maximal staining) for that pixel. The integrated average gray level used in our further analysis was the sum of all the separate pixels in the examined area. Final data were displayed as average gray value/cell. With this setup described briefly above, we believe that the risk of interpretation bias is avoided. However, obtaining accurate results with this semi-quantitative-immunohistochemical analysis depends upon the specificity of the antibodies used.
2.5. TEM analysis
Prior to transmission electron microscopy (TEM), the mice eyes were pre-fixed with 3% paraformaldehyde in 0.075 M cacodylate buffer (pH 7.2) with 1% glutaraldehyde for 5–7 days at + 4 °C. After 2 × 10 min washing in 0.075 M cacodylate buffer, the samples were post-fixed in 1% osmium tetraoxide and 1.5% potassium-ferrocyanide in 0.1 M cacodylate buffer (pH 7.2) for 2 h, and again washed with cacodylate buffer for 2 × 10 min and 5 min in distilled water prior to standard ethanol dehydration. Subsequently, the samples were infiltrated and embedded in LX-112 resin (Ladd Research, Williston, VT, USA). Polymerization was carried out at 60 °C for 48–72 h. One-micron semi-thin sections were cut with a Reichert Ultracut E microtome (Leica Microsystems Inc, IL, USA), stained with 1% toluidine blue, and examined with a light microscope to find the localization of interest for further TEM sectioning. Next, the 70 nm thick sections (cut with Leica EM UC7, Wetzlar, Germany) were stained with 1% uranyl acetate for 30 min and with lead citrate for 2 min. The sections were examined with a JEM-2100F TEM (Jeol, Tokyo, Japan) at 200 kV. Lipofuscin-like aggregates and autophagic compartments were manually counted by three different individuals. Similarly as with the histological samples, representative areas per sample were selected for collecting RPEs from each individual sample close to the vicinity of the optic nerve. Six cells were randomly selected from each group (WT and dKO mice aged 12 months, n = 3) for counting.
2.6. Vacuole area fraction analysis
The analysis technique used in this study objectively measured the morphometric parameter of the vacuoles at the basal cytoplasmic site of RPE from TEM images, to characterize area fractions of vacuoles in the WT and dKO RPE at 1 year of age. Using the open source Image J software (http//:imagej.nih.gov; NIH, USA), the software measured the size of vacuoles and data were expressed according to equal size ROI (200 µm2).
2.7. Detection of lipofuscin-like granules in the RPE cells
During the regular dewaxing and rehydration, the processed WT and dKO samples of interest were washed with 0.3% w/v Sudan Black B (SBB, cat. no. 3545-12, EMD Chemicals, Gibbstown, NJ, USA) in 70% ethanol at 37 °C for 20 min, rinsed with PBS 3 times 10 min and then the sections were mounted with Mowiol mounting Media (Sigma, St. Louis, MO, USA) [40]. The samples were examined in a laser scanning confocal microscope (Zeiss AX10 Imager A2, Zeiss, Göttingen, Germany) with sequential scanning and detection of dot-like lipofuscin granules, respectively, followed by merging and saving of the images.
2.8. Apoptosis assay
TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) assay was performed using the ApopTag® Peroxidase in situ Apoptosis Detection Kit (Millipore S.A.S., Molsheim, France) according to the manufacturer's instructions in one-year-old samples. TUNEL stained sections were washed in PBS and counterstained with DAPI. The sections were dehydrated and mounted. Positive and negative controls were used to control staining quality.
2.9. Immunohistochemistry of ER stress markers
One-year-old WT and dKO retinas were stained in triplicate with rabbit anti-ATF4 (activating transcription factor 4, Abcam Cambridge, MA) and rabbit anti-GRP78 (glucose-regulated protein 78, Abcam Cambridge, MA) antibodies, separately. Immunohistochemistry was performed on paraffin embedded sections after antigen retrieval using an Invitrogen AEC red (Carlsbad, CA) substrate kit. For accurate comparisons, all sections were stained simultaneously and slides were developed under the same conditions. Imaging was performed with the Leica-Aperio CS digital ScanScope (Leica Microsystems, Buffalo, IL) at 40× magnification. When measuring positive pixels, images were digitally scribed using Aperio ScanScope software to create a ROI that encircled the entire cross-sectional profiles of the retina on each slide. The positive pixel count algorithm Aperio v.9.1 was used to determine stained vs. unstained pixel counts for areas within each ROI.
2.10. Flat mount and RPE size analysis
Mice eyes were collected and fixed in 2% PFA for 2 h. The samples were then processed for the RPE/choroid flatmount preparation using a protocol previously described [34], [35]. Briefly, the anterior segment of the eye, lens and the neuronal retina were removed under a dissecting microscope. Four to five vertical cuts were made from the edge of the RPE/choroid/sclera cup to the equator. The samples were then washed and treated with 1% triton X-100 for 2 h, followed by incubation with rabbit anti-mouse Iba-1 (ionized calcium binding adaptor molecule 1, Abcam, Cambridge, MA) 1:100, at 4 °C for 16 h, followed by FITC-conjugated (fluorescein isothiocyanate) goat anti-rabbit IgG (1:200, Abcam) and Alexa Fluor 568 Phalloidin (1:100, Life Technologies) at room temperature for 2 h. The samples were washed and flatmounted on a glass slide for confocal microscopy investigation (Eclipse TE200-U; Nikon UK Ltd.). Z-stack confocal images of RPE flatmounts were reconstructed using the NIS Element (Nikon) software. The border of each RPE cell was outlined based on the Phalloidin (F-actin) staining, and the size of each RPE cell was measured manually using the same software.
2.11. ERG recordings
We tested the retinal function in three-month-old (referred to as “young”; WT, n = 6; dKO, n = 6) and one-year-old mice (referred to as “old”; WT, n = 6; dKO, n = 7) by ERG (electroretinography, Thorlabs PM100D, Thorlabs, NJ, USA). The ERG method has been described in detail previously [41]. The animals were dark-adapted overnight for at least 12 h. They were anesthetized with isoflurane (induction: concentration 4–4.5%; maintenance: concentration ~ 1.2%) and placed into a customized stereotaxic frame and kept warm with a physiological heating pad at 38 °C. The corneal ERG electrode was a silver wire constructed as a semi-circular loop. One stainless steel subcutaneous needle was attached into the cheek (reference) and another into the lower back (common ground). All animal manipulations were performed under a dim red light and animals were further dark-adapted in a completely dark room for 5 min. The dark-adapted light stimulation was performed in an ascending series with respect to the stimulus intensity (3.60, −2.90, −2.10, −1.65, −0.45, 0.50, 1.00, 1.60 and 2.00 log cd·s/m2), and the inter-stimulus interval (ISI) was increased accordingly (2 s, 4 s, 4 s, 10 s, 10 s, 10 s, 20 s, 50 s and 50 s, respectively). Finally, a paired-flash paradigm was used to isolate the cone component from the mixed rod-cone waveform [42]. A strong flash (2.00 log cd·s/m2) was delivered to evoke transient rod saturation and 400 ms later a second probe flash (2.00 log cd·s/m2) was delivered to generate a cone-specific response. Rod-saturation at 400 ms after flash was assumed based on a previous ex vivo ERG saturation test [41], where the strongest flash was estimated to yield ~ 14,000 photoisomerizations (*R) per mouse rod. The light intensity was quantified with an energy meter (Thorlabs PM100D, Thorlabs, NJ, USA). In the waveform analysis, the baseline for ERG response was taken as an average amplitude between −100 and 0 ms before the stimulus onset. The a-wave amplitude was calculated from baseline to the trough of the first negative deflection after the stimulus onset. The b-wave was calculated from the a-wave trough to the peak of the first major positive wave. The ´cone´ response amplitude was calculated between the largest negativity and the major positive wave peaking around 40 ms after the probe flash.
2.12. Statistical analysis
All statistical analysis was performed using SPSS statistics software (SPSS Inc., Chicago, Ill, USA). p-values < 0.05 were considered significant. Spectral imaging analysis: The statistical analysis of the data was conducted with a one-way analysis of variance (ANOVA), followed by Games-Howell post hoc test (n = 30). TEM analysis: The significance of differences between WT and dKO groups were analyzed with Student's t-test (n = 18). Vacuole area fraction analysis: The statistical analysis of the data was conducted with a Student's t-test (n = 25). Immunohistochemistry of ER stress markers: The average cell numbers from 5 different regions were calculated. Student's t-test was applied to assess statistical differences between WT and dKO groups (n = 4–7). Flat mount and RPE size analysis: Statistical significance was analyzed with Student's t-test (n = 4). ERG recordings: a- and b-wave amplitude and b-a ratio statistical analyses were performed with a two-way repeated measured ANOVA using the genotype as the between-subject factor and stimulus intensity as the within-subjects factor, followed by Bonferroni post hoc test. The cone-flash amplitude was analyzed with a regular two-way ANOVA.
3. Results
We investigated the effects of NRF-2/PGC-1α dKO on macroscopic anatomy and age-related RPE degeneration using light, confocal, and electron microscopy, as well as performing a functional electroretinography analysis (Supplementary Figs. 2 and 3). To ensure that there were no complications due to the spontaneous Rd8 (retinal degeneration 8) frameshift mutation c.3481delC in Crb1 (crumbs homolog 1) resulting retinal degeneration [43], [44], screening for this mutation was performed for mouse lines used in this study: WT, NRF-2 KO, PGC-1α KO and NRF-2/PGC-1α dKO. Our analysis revealed that these mice strains were negative for the Rd8 mutant allele (Supplementary Fig. 4).
3.1. NRF-2/PGC-1α gene inactivation evokes severe axial skeleton and eye morphology alterations
The radiographs reveal the abnormally small axial skeleton already present in six-week-old dKO mice (Supplementary Fig. 3). In comparison with WT controls, the dKO mice had lighter body weights and expressed prominent dysmorphic features, such as a shorter axial skeleton, smaller limbs and head parameters at all ages studied (Supplementary Fig. 3). The fur of the dKO mice was also more greyish than in the corresponding single KOs or WT mice. The macrophotography and micro-MRI (magnetic resonance imaging) revealed a smaller eyeball size in the dKOs.
3.2. NRF-2/PGC-1α dKO mice display increased protein aggregation and oxidative stress markers in RPE cells
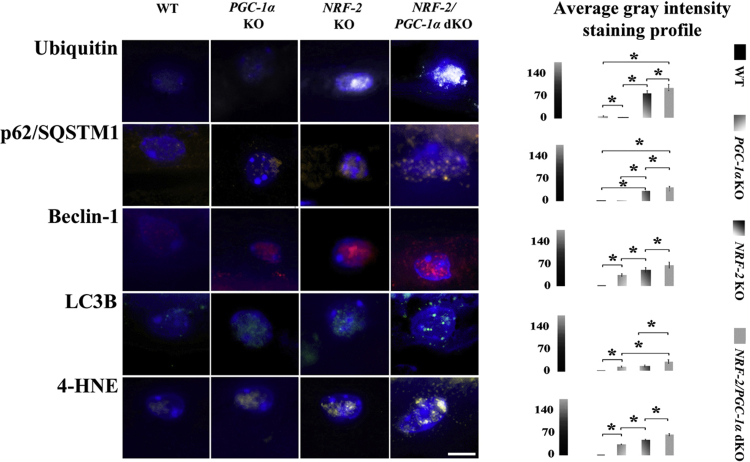
Chronic oxidative stress and impaired autophagy are strongly linked to RPE degeneration and the protein aggregation observed in AMD [1], [3]. The content and localization of protein aggregates-conjugated marker ubiquitin, autophagy regulator Beclin-1, autophagy markers p62/SQSTM1 and LC3B, and oxidative stress marker 4-HNE (product of lipid peroxidation) were evaluated in RPE cells by immunostainings and confocal microscopy in one-year-old WT, PGC-1α KO, NRF-2 KO and NRF-2/PGC-1α dKO mice (Fig. 1).
Fig. 1.
NRF-2/PGC-1αdKO mice display increased protein aggregation, autophagy and oxidative stress markers in RPE cells. Representative images and comparative computer-aided densitometric assay focusing on ubiquitin (white), p62/SQSTM1 (brown), Beclin-1 (red), LC3B (green) and 4-HNE (yellow) immunofluorescences in RPE cells (DAPI-stained nuclei/blue) of one-year-old wild type (WT), PGC-1α KO, NRF-2 KO and NRF-2/PGC-1α dKO mice. The scale bar indicates 5 µm. The gray level intensities of WT, PGC-1α KO, NRF-2 KO and NRF-2/PGC-1α dKO mice represent average intensities. Combined results from three different animals per genotype and n = 30. *p < 0.001 one-way ANOVA followed by Games-Howell post hoc test (n = 30). Results are expressed as means ± SD.
Removal of PGC-1α gene resulted in higher levels of Beclin-1 (p < 0.001), LC3B (p < 0.001) and 4-HNE (p < 0.001) compared to the WT mice. However, the marker of protein aggregation, ubiquitin, decreased significantly and the p62/SQSTM1 levels remained stable indicating an increase in oxidative stress but, as expected, functional UPS/autophagy appears capable to maintain mitochondrial/protein homeostasis. NRF-2 KO resulted in higher expression levels of all markers studied compared to the WT (p < 0.001). The marker of oxidative stress (4-HNE) as well as autophagy markers Beclin-1 and p62/SQSTM1 and protein aggregation marker ubiquitin were significantly elevated in NRF-2 KO compared to PGC-1α KO mice suggesting stronger oxidative stress, insufficient proteasome function and autophagic clearance. NRF-2/PGC-1α dKO mice had the highest accumulation of all markers studied suggesting the highest degree oxidative stress and more severe defect in the UPS/autophagy clearance.
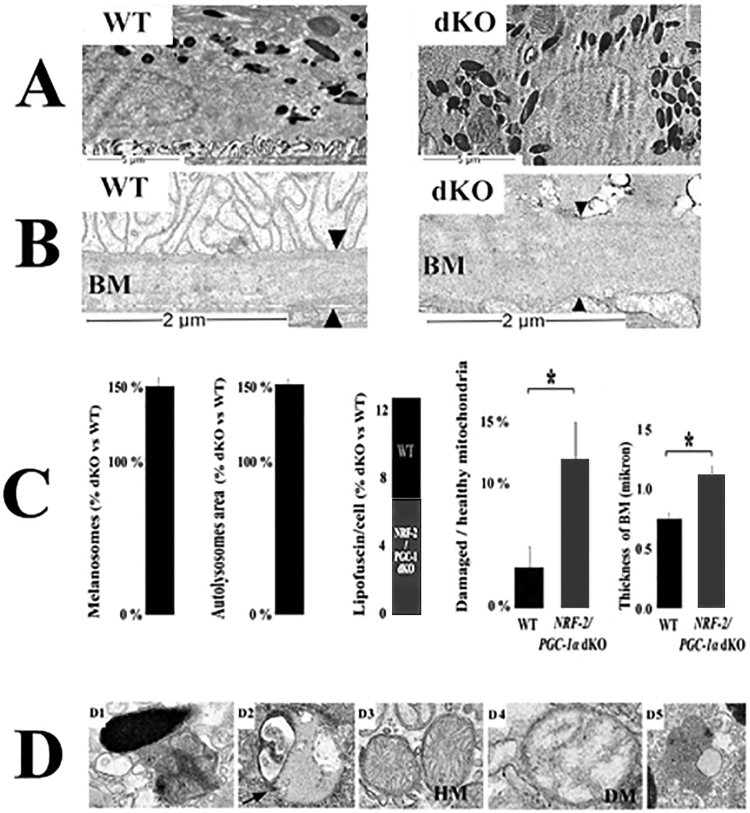
3.3. Ultrastructural analysis indicates an increase in autolysosomes, damaged mitochondria and melanosomes in NRF-2/PGC-1α dKO mice
Since insufficient autophagic clearance and high oxidative stress were detected in dKO mice, we assessed ultrastructural changes of WT and dKO RPE cells from the TEM images (Fig. 2). Compared to intact normal cell structures in WT RPE cells, the examination of dKO revealed the presence of larger autolysomes, higher ratio of damaged mitochondria (p < 0.05), thicker Bruch's membrane (p < 0.05), the loss of basal infoldings and moderate increase in lipofuscin particles. Interestingly, the number of melanosomes was higher in dKO mice. Moreover, Bruch's membrane elastic and collagenous layers were not often observed, and increased thickness of Bruch's membrane was detected along with an accumulation of electron dense amorphous material in dKO animals.
Fig. 2.
Transmission electron microscopy (TEM) analysis revealed dry AMD-like pathology in one-year-oldNRF-2/PGC-1αdKO mice. Representative TEM image of WT RPE indicates normal cellular organelles, while in one year old dKO RPE loss of basal infoldings and thickened Bruchs's membrane (arrowheads) were detected (A and B). (C) Increased amount of (D1) melanosomes, (D2) autolysosomes, (D3, D4) damaged mitochondria and (D5) lipofuscin were detected in dKO RPE. The scale bars indicate 2 µm. WT = wild type; dKO = double knockout; BM = Bruch's membrane; HM = healthy mitochondria, DM = damaged mitochondria Data are quantified as % ratio of dKO versus WT ± SD. Thickness of Bruch's membrane (BM) microns ± SD. *p < 0.05, Student's t-test (n = 18).
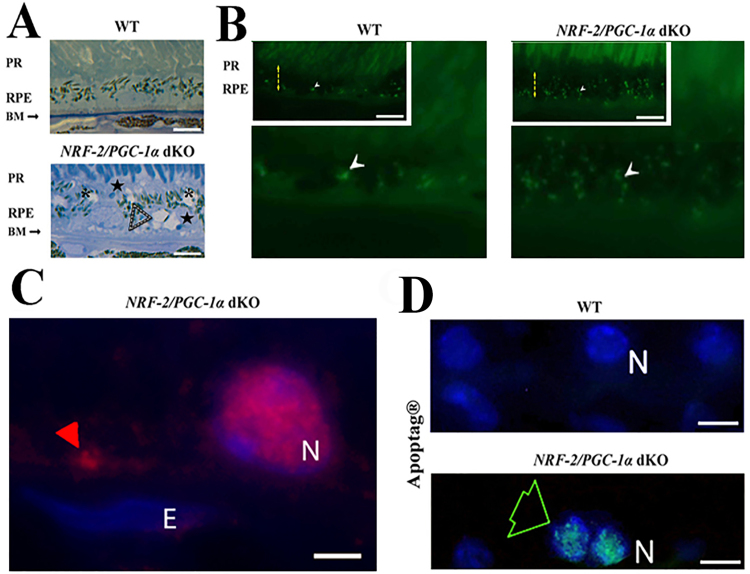
3.4. Micro-structural changes in the RPE and Bruch's membrane of NRF-2/PGC-1α dKO mice
Our aim was then to evaluate whether the NRF-2/PGC-1α dKO tissue samples showed AMD-like pathological features at the histological level. Analysis of the epoxy semi-thin and wax sections revealed Bruch's membrane dysmorphology, increased accumulation of lipofuscin-like particles and cytoplasmic alteration in the dKO RPE (Fig. 3A). Drusen-like deposits between RPE and Bruch's membrane were positive for ubiquitin staining (Fig. 3B and C). The intracellular cystic degeneration of RPE, nuclear debris and apoptosis (Fig. 3D) were observed in dKO RPE cells.
Fig. 3.
The pathological changes of RPE in NRF-2/PGC-1α double knockout (dKO) mice. (A) The light microscopic images of WT and dKO samples of the toluidine blue-stained epoxy section from one-year-old mouse. The white dashed arrowheads indicate the dome-shaped extracellular deposits between the RPE and Bruch's membrane (upper panels). The cystic cytoplasmic alterations were observed in dKO samples (asterisks). (B) Excessive accumulation of lipofuscin-like material (white arrowhead) in the dKO RPE. In the inserts of Fig. B On the insert of figure B, the yellow dashed arrows indicate the RPE layer. The scale bar indicates 10 µm. (C) The red arrowhead indicates the ubiquitin positivity of drusen-like deposits in the close vicinity of Bruch's membrane. (D) Restricted apoptosis was detected within some cells in the RPE layer (green arrow) of the dKO retina with Apoptag® kit. PR: photoreceptor layer; RPE: retinal pigment epithelium; BM: Bruch's membrane. The scale bar indicates 5 µm. E (endothelial cell nucleus); N (RPE nucleus).
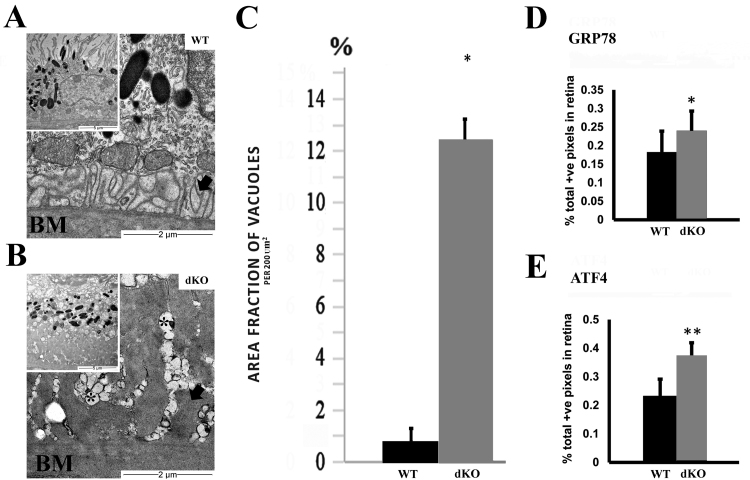
3.5. NRF-2/PGC-1α dKO mice RPE cells display increased ER stress markers and loss of basal infoldings
The involvement of ER stress was assessed by ultrastructural analysis and staining with anti-GRP78 and anti-ATF4. Ultrastructural analysis revealed statistically significant ER vacuolization as a marker of ER damage in dKO RPE cells compared to corresponding WT RPE cells (Fig. 4A-C). The retinas from one-year-old dKO exhibited more intense staining for GRP78 (p < 0.05) and ATF4 (p < 0.01) in the RPE layer when compared to the corresponding tissues in WT controls (Fig. 4D-E).
Fig. 4.
NRF-2/PGC-1α dKO mice display increased endoplasmic reticulum (ER) stress in RPE cells. (A) Representative transmission electron microscopy (TEM) image of WT RPE indicates normal cellular structures and melanosomes. (B) Increased number of melanosomes, the basal laminar deposits (arrow), the loss of basal infoldings and ER damage (asterisks) were detected in dKO samples. (C) Increase of the ER vacuolization (% per 200 µm2) was recorded in dKO RPE compared to aged matched WT samples. Increased immunohistochemical staining intensity for (D) GRP78 and (E) ATF4 was detected in RPE layer of dKO mice compared to WT. The scale bar indicates 10 µm (A, B) and 2 µm (C, D). *p < 0.05, **p < 0.01, Students t-test. Data are mean ± SD (A-D n = 4–7; E n = 25).
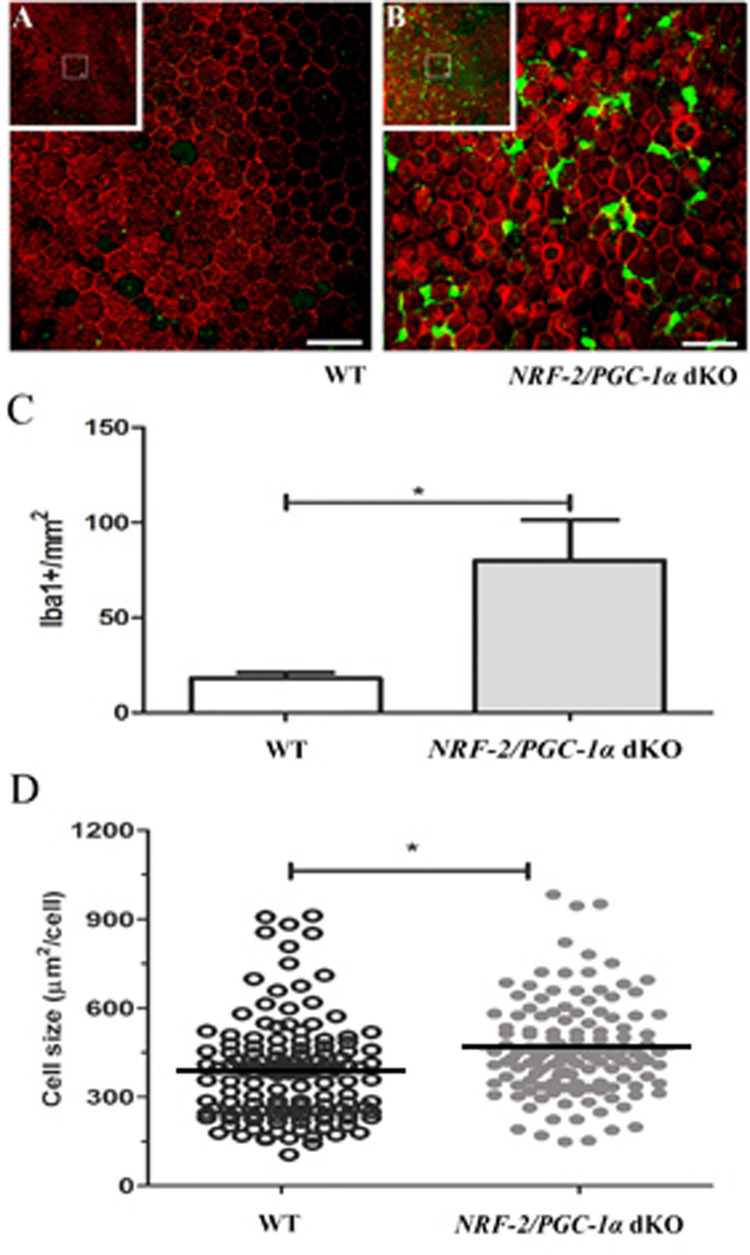
3.6. Iba-1 is upregulated and the size of RPE cell is increased in NRF-2/PGC-1α dKO mice
The innate immune system is activated in a number of degenerative and inflammatory retinal disorders, including AMD. Retinal mononuclear phagocytes are also present in most mouse models of retinal disease [45]. To evaluate whether Iba-1 expression is enhanced in dKO RPE due to inflammatory processes, Iba-1 analysis was performed in RPE flat mounts. Data in Fig. 5 show a statistically significant 400% increase in the Iba-1 staining intensity in RPE of one-year-old NRF-2/PGC-1α dKO versus WT mice (p < 0.05). Notably, we also observed a clear RPE size increase in our dKO mice compared to their WT counterparts (p < 0.01). During normal aging, an increased RPE cell size in C57BL/6J mouse has been interpreted as a prognosis of cell death [46].
Fig. 5.
Increased immune cell infiltration and the enlargement of RPE cells are present inNRF-2/PGC-1αdKO mice. Representative confocal Iba-1 (green) and Phalloidin (red) immunohistochemical images of retinal flat mounts samples of one-year-old (A) WT and (B) NRF-2/PGC-1α dKO mice. Insets show the low magnification view from RPE layer. The scale bar indicates 20 µm. (C) There was a 400% increase in Iba-1 positive cells in the NRF-2/PGC-1α dKO RPE compared to the WT flat mount samples. (D) The RPE cell size difference in NRF-2/PGC-1α dKO and WT mice. The horizontal black lines represent the average size of the RPE cell. *p < 0.05, Student's t-test (n = 4).
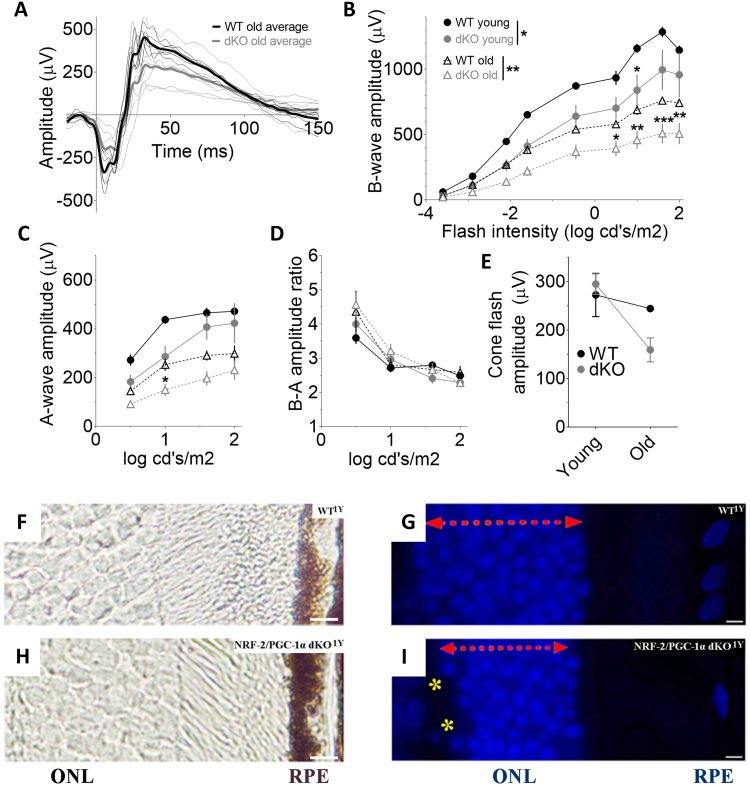
3.7. NRF-2/PGC-1α dKO mice show age-related visual loss and photoreceptor dysmorphology
Given the observed structural changes noted above, we subsequently assessed the in vivo function of rod and cone pathways by performing ERG in the WT and dKO mice. As presented in Fig. 6, the ERGs amplitudes were lower in dKO mice even at a young age, but the difference reached statistical significance only in b-wave amplitudes (a-wave: p = 0.13; b-wave: p < 0.05). The cone response, as determined by the paired-flash paradigm, was comparable between young dKO and WT mice (Fig. 6E). With age, the rod-function significantly decreased in WT mice (a- and b-wave: p < 0.001), while cone-function remained stable (Fig. 6B-E). Group-wise comparison revealed significant differences between old dKO and WT mice in dark-adapted a- and b-wave amplitudes (a-wave: p < 0.05, b-wave: p < 0.01). There was a clear tendency towards age-genotype interaction in cone-flash amplitude (p = 0.06), indicating that also cone-function tends to decrease in dKO mice with age (see Fig. 6E). The ratio between b- and a-wave amplitudes remained unchanged regardless of genotype or age (Fig. 6D, all group-wise comparisons p > 0.28), indicating that a photoreceptor defect was the cause for the ERG amplitude changes. Since impaired photoreceptor function was found in the retina of dKO mice, the next step was to study the photoreceptor layer in more detail. The photoreceptor cells of dKO mice showed decreased thickness of the outer nuclear layer (ONL) and disorganization in the outer portion of the ONL (Fig. 6F-I). The staining of protein aggregation, autophagy and oxidative stress markers and cellular morphology of photoreceptors in one-year-old mice were similar to those observed in RPE cells (Supplementary fig. 5). Electron micrographs, in addition to RPE cells, revealed focal photoreceptor layer degeneration in dKO mice (Supplementary fig. 5).
Fig. 6.
Dysmorphology of photoreceptors coincides with impaired retinal function inNRF-2-PGC1αdKO mice. (A) Dark-adapted ERG waveforms at 1.60 log cd·s/m2 flash. Thin lines represent individual mouse responses and thick lines group-averaged waveforms. (B) ERG b-wave declined already in three-month-old (3 M) dKO mice (n = 6) as compared to their age-matched WT (n = 6) littermate mice, and the difference became more significant in one-year-old (1Y) mice (dKO: n = 7; WT: n = 6). (C) ERG a-wave tended to decline at both ages but reached statistically significant level only at older age. (D) The ratio between b- and a-wave amplitudes did not change regardless of the genotype or the age. (E) The double-flash paradigm revealed a tendency towards age-related decline in the cone-dominant function in dKO mice. (F) Thin unstained wax section, (G) DAPI staining of outer nuclear layer (ONL) in WT samples compared with (H,I) one-year-old (y) dKO mice, respectively. Photoreceptor atrophy of the ONL (red vertical arrow) is seen in DAPI-labeled dKO retinas cut from sections located 250–350 µm from the optic nerve head. Retinas show disorganization in the outer portion of the ONL (yellow asterics). The scale bar indicates 5 µm. **p < 0.01, *p < 0.05, two-way ANOVA.
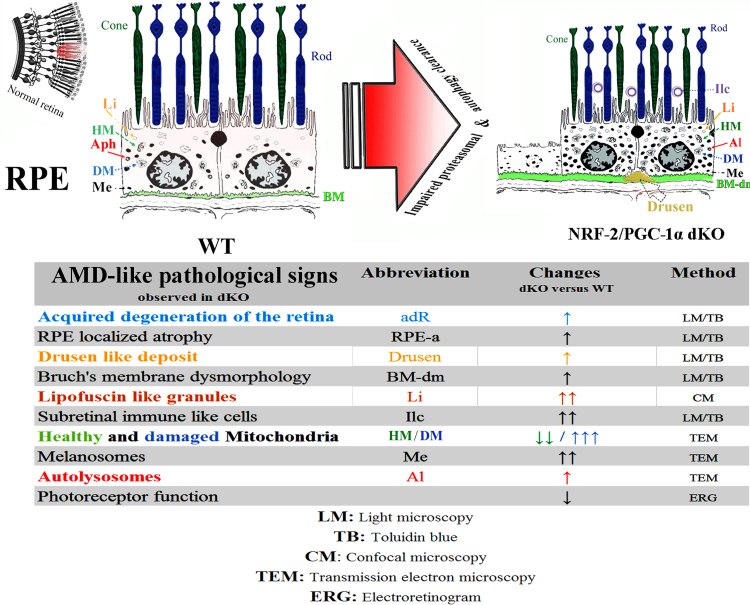
3.8. NRF-2/PGC-1α dKO mouse model shows a dry AMD-like phenotype
Taken together, our dKO mice show pathological signs that are consistent with those observed in patients with dry AMD. The pathological changes observed in these dKO mice include age-related RPE degeneration, intra- and extracellular deposit accumulation, Bruch's membrane dysmorphology, immune cell invasion, damaged mitochondria, impaired autophagy, altered pigmentation and visual loss consistent with a defect in photoreceptors (Fig. 7).
Fig. 7.
Graphical summary of AMD hallmarks observed in one-year-oldNRF-2/PGC-1αdKO versus wild type mice. All measurements were made from a well-defined retinal region located ~ 250–350 µm from the optic nerve head. During aging, the retinas of dKO mice show more severe age-related degenerative changes compared to wild type aged-matched samples and, importantly, RPE cells from dKO exhibit the pathological signs of AMD. Abbreviations: RPE: retinal pigment epithelium; Li: Lipofuscin-like deposit, HM: healthy Mitochondria, DM: damaged Mitochondria, Aph: autophagosome, Al: Autolysosomes, Me: Melanosomes (pigments); Ilc: Immun-like cells; BM: Bruch's membrane BM-dm: Bruch's membrane dysmorphology.
4. Discussion
Recently, impaired proteolysis in response to chronic and overt oxidative stress has been suggested as a key contributor to age-related retinal degeneration, particularly in AMD [1], [8]. To study proteolysis of age-related retinal degeneration, we sought to develop an animal model that would provide the most robust protein aggregation in RPE cells. Previously, genetic ablation of the NRF-2 and PGC-1α has been shown to induce RPE degeneration [15], [30]. Our findings reveal that knockout of both, NRF-2 and PGC-1α, resulted in the highest increase of markers for protein aggregation (ubiquitin and p62/SQSTM1) and oxidative stress (4-HNE) in RPE cells. Since lack of PGC-1α gene results in mitochondrial ROS production and loss of NRF-2 gene cripples the antioxidant defense system and impairs autophagic clearance system, the RPE cells of dKO mice are expected to suffer more intense oxidative stress due to accumulation of damaged mitochondria producing excessive ROS. Thus, loss of both genes may enhance the deleterious effects of each other by obstructing the two main pathways of mitochondrial homeostasis control, clearance of damaged mitochondria by autophagy and mitochondrial biogenesis. In addition, there is evidence of reduced proteasomal clearance as measured by increased ubiquitin protein conjugates in the dKO mice. Proteins whose turnover is regulated by proteasomes control melanosome differentiation and cellular pigmentation [47], [48]. The reduced proteasome activity could explain the increased number of melanosomes, containing ROS eliminating melanine, in dKO mice.
The presence and the number of autolysosomes suggest the molecular machinery required for autophagic clearance system is maintained in dKO mice. This observation is in line with immunostainings showing high expression of Beclin-1 and LC3. However, the elevated levels of p62 and ubiquitin and the increased size of autolysosomes indicate an insufficient rate in autophagic clearance. This hypothesis is further strengthened by the higher ratio of damaged mitochondria shown in dKO TEM analysis. Although the loss of PGC-1α induces mitochondrial ROS, the NRF-2 deficiency impairs the autophagic degradation system, thereby accelerating the accumulation of damaged mitochondria. This key feature of mitochondrial damage has been implicated in the pathophysiology of AMD [49]. In donors with AMD, reductions in mitochondrial number and area were amplified compared with age-matched controls and in addition, there was a greater loss of cristae and matrix density. Further support for the idea that mitochondrial damage is increased in AMD emerges from studies showing that the increased mitochondrial DNA damage correlates with disease severity [50], [51]. Others have shown that increased oxidative stress in mitochondria induces endoplasmic reticulum stress [52], [53], [54]. Consistent with these findings linking mitochondrial oxidative- and ER-stress, we observed elevated levels of GRP78 and ATF4 and endoplasmic reticulum vacuolization in the dKO mice.
It is generally accepted that autophagy is a cytoprotective mechanism that is increased during aging, but its capacity eventually becomes overwhelmed with advanced age or disease [36], [55], [56], [57], [58]. Recently, autophagy has been the cellular target in many in vitro and in vivo RPE studies. This body of research, where there was a selective elimination of autophagy-related genes, has aided our understanding of the complex interplay between proteins and the consequences associated with disrupted autophagy. Of importance to our work, genetic disruption of autophagy in the RPE can elicit effects that are reminiscent of results in our NRF-2/PGC-1α dKO mice. For example, ATG7 and Beclin-1 knockdown in RPE cultures increased ROS generation, exacerbated oxidative stress-induced reduction of mitochondrial activity, reduced cell viability, and promoted lipofuscin accumulation [55]. Sukseree et al. [59] documented that ATG7 gene deletion upregulated autophagy adaptor p62/SQSTM1 in mice. This observation of a p62/SQSTM1 compensatory response was reported by Zhang et al. [60] after knocking out either ATG5 or ATG7 in their mouse model. Autophagic RB1CC/FIP200 gene deficiency evoked age-dependent RPE degeneration, subretinal migration of microglia cells, accumulation of drusen-like compounds, and expression of choroidal neovascularization foci in their KO mouse model [61]. These results, documenting features in autophagy KO mice that parallel results in our NRF-2/PGC-1α dKO mouse model suggests autophagic flux is disrupted in the dKO mouse.
It is important to note that in the RPE, autophagy proteins participate in both conventional autophagy as well as the non-canonical pathway associated with phagocytosis and degradation of outer segments. This novel autophagy-related function in RPE, coined LC3-associated phagocytosis (LAP), was first described by Kim et al. [62]. They showed that ATG5 is a crucial component in regulating RPE cell phagocytosis and in maintaining normal visual cycle in mice. Thus, elimination of autophagy genes can have a negative consequence on vision. Others have also shown that genetic disruption of genes associated with lysosomal function can also negatively affect both autophagy and phagocytosis since lysosomal degradation of proteins is the final endpoint for both pathways. For example, elimination of the lysosomal pump regulator Cryba1 gene and the subsequent lysosomal dysregulation caused impairment of both autophagy and phagocytosis, coupled with visual loss, in Cryba1 KO mice [63].
The decline in intracellular degradation systems not only increased the amounts of intracellular protein aggregates and oxidative stress, but also induced inflammation [64], [65]. Macrophages are known to be activated by oxidative damage, which plays a role in AMD pathogenesis [66]. In AMD, macrophages accumulate in the subretinal space [67]. Our dKO mouse model shows Iba-1-positive mononuclear phagocyte infiltration into the subretinal space, revealing an immune response that coincides with the increased protein aggregation and RPE size increase, the latter being the result of the expansion of RPE to maintain the integrity of the blood-retina barrier following the death of neighboring RPE [46].
In summary, aging, oxidative stress, protein aggregation and inflammation are significant risk factors for RPE damage and AMD development. Our collective data point to a key role for NRF-2 and PGC-1α in the process of RPE damage that coincides with increased oxidative stress, protein aggregation, accumulation of lipofuscin, mitochondrial damage, and mononuclear phagocyte infiltration into the retina, which also occurs in processes associated with dry AMD [7]. Our findings reveal that both UPS and lysosome/autophagy clearance rates are insufficient in the NRF-2/PGC-1α dKO RPE cells. Functionally, the pathology manifests as an age-related loss of photoreceptor function. We believe that the NRF-2/PGC-1α dKO mouse represents a novel research model for investigating aging processes in RPE cells as well as dry AMD.
Acknowledgements
This work was supported by the Kuopio University Hospital (KK) (Grant Number 5503743), the Finnish Eye Foundation (JJP, KK), the Finnish Funding Agency for Technology and Innovation (AK, KK), the Health Research Council of the Academy of Finland (KK, HS) (Grant Numbers 218050 and 296840), the Päivikki and Sakari Sohlberg Foundation (KK, HS), the Evald and Hilda Nissi Foundation and the Finnish Cultural Foundation-North-Savo (AKoi), the Lindsay Family Foundation and an anonymous donor for AMD research (DAF), National Science Centre, Poland (JB) (Grant number 2017/27/B/NZ3/00872). The researchers are very grateful to Mrs. Anne Seppänen, Mrs. Anne Karppinen, Mrs Virpi Miettinen, Mr. Anthony Rodriguez and Mr. Eric Barron for their skillful technical assistance.
Acknowledgments
Conflict of interest
The authors declare no conflicts of interest. The funding sponsors had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript; nor in the decision to publish the results.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.09.011
Appendix A. Supplementary material
Supplementary material
References
- 1.Datta S., Cano M., Ebrahimi K., Wang L., Handa J.T. The impact of oxidative stress and inflammation on RPE degeneration in non-neovascular AMD. Prog. Retin. Eye Res. 2017;60:201–218. doi: 10.1016/j.preteyeres.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kauppinen A., Paterno J.J., Blasiak J., Salminen A., Kaarniranta K. Inflammation and its role in age-related macular degeneration. Cell Mol. Life Sci. 2016;73:1765–1786. doi: 10.1007/s00018-016-2147-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boya P., Esteban-Martínez L., Serrano-Puebla A., Gómez-Sintes R., Villarejo-Zori B. Autophagy in the eye: development, degeneration, and aging. Prog. Retin. Eye Res. 2016;55:206–245. doi: 10.1016/j.preteyeres.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Smith S.B. Mechanisms of ER stress in retinal disease. In: Ryan S.J., editor. Retina. 5th ed. Elsevier; New York, NY, USA: 2013. pp. 529–536. [Google Scholar]
- 5.Feher J., Kovacs I., Artico M., Cavallotti C., Papale A., Balacco Gabrieli C. Mitochondrial alterations of retinal pigment epithelium in age-related macular degeneration. Neurobiol. Aging. 2006;27:983–993. doi: 10.1016/j.neurobiolaging.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 6.Terluk M.R., Kapphahn R.J., Soukup L.M., Gong H., Gallardo C., Montezuma S.R. Investigating mitochondria as a target for treating age-related macular degeneration. J. Neurosci. 2015;35:7304–7311. doi: 10.1523/JNEUROSCI.0190-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaarniranta K., Sinha D., Blasiak J., Kauppinen A., Veréb Z., Salminen A. Autophagy and heterophagy dysregulation leads to retinal pigment epithelium dysfunction and development of age-related macular degeneration. Autophagy. 2013;9:973–984. doi: 10.4161/auto.24546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrington D.A., Sinha D., Kaarniranta K. Defects in retinal pigment epithelial cell proteolysis and the pathology associated with age-related macular degeneration. Prog. Retin. Eye Res. 2016;51:69–89. doi: 10.1016/j.preteyeres.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinha D., Valapala M., Shang P., Hose S., Grebe R., Lutty G.A. Lysosomes: regulators of autophagy in the retinal pigmented epithelium. Exp. Eye Res. 2016;144:46–53. doi: 10.1016/j.exer.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Höhn A., Jung T., Grimm S., Catalgol B., Weber D., Grune T. Lipofuscin inhibits the proteasome by binding to surface motifs. Free Radic. Biol. Med. 2011;50:585–591. doi: 10.1016/j.freeradbiomed.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Anderson D.H., Talaga K.C., Rivest A.J., Barron E., Hageman G.S., Johnson L.V. Characterization of beta amyloid assemblies in drusen: the deposits associated with aging and age-related macular degeneration. Exp. Eye Res. 2004;78:243–256. doi: 10.1016/j.exer.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 12.Hyttinen J.M.T., Błasiak J., Niittykoski M., Kinnunen K., Kauppinen A., Salminen A. DNA damage response and autophagy in the degeneration of retinal pigment epithelial cells-Implications for age-related macular degeneration (AMD) Ageing Res. Rev. 2017;36:64–77. doi: 10.1016/j.arr.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Blasiak J., Petrovski G., Veréb Z., Facskó A., Kaarniranta K. Oxidative stress, hypoxia, and autophagy in the neovascular processes of age-related macular degeneration. Biomed. Res. Int. 2014 doi: 10.1155/2014/768026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takayama K., Kaneko H., Kataoka K., Kimoto R., Hwang S.J., Ye F. Nuclear factor (erythroid-derived)-related factor 2-associated retinal pigment epithelial cell protection under blue light-induced oxidative stress. Oxid. Med. Cell Longev. 2016 doi: 10.1155/2016/8694641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Z., Chen Y., Wang J., Sternberg P., Freeman M.L., Grossniklaus H.E. Age-related retinopathy in NRF-2-deficient mice. PLoS One. 2011:6. doi: 10.1371/journal.pone.0019456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kansanen E., Kuosmanen S.M., Leinonen H., Levonen A.L. The Keap1-NRF-2 pathway: mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1:45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirotsu Y., Katsuoka F., Funayama R., Nagashima T., Nishida Y., Nakayama K. NRF-2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res. 2012;40:10228–10239. doi: 10.1093/nar/gks827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau A., Wang X.J., Zhao F., Villeneuve N.F., Wu T., Jiang T. A noncanonical mechanism of NRF-2 activation by autophagy deficiency: direct interaction between keap1 and p62. Mol. Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y. The selective autophagy substrate p62 activates the stress responsive transcription factor NRF-2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 20.Jain A., Lamark T., Sjøttem E., Larsen K.B., Awuh J.A., Øvervatn A. p62/SQSTM1 is a target gene for transcription factor NRF-2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pajares M., Jiménez-Moreno N., García-Yagüe Á., Escoll M., de Ceballos M., Van Leuven F. Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy. 2016;12:1902–1916. doi: 10.1080/15548627.2016.1208889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei Y., Gong J., Yoshida T., Eberhart C., Xu Z., Kombairaju P. Nrf2 has a protective role against neuronal and capillary degeneration in retinal ischemia-reperfusion injury. Free Radic. Biol. Med. 2011;52:216–224. doi: 10.1016/j.freeradbiomed.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baldelli S., Aquillano K., Ciriolo M.R. PGC-1α buffers ROS-mediated removal of mitochondria during myogenesis. Cell Death Dis. 2014:5. doi: 10.1038/cddis.2014.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aquilano K., Baldelli S., Pagliei B., Cannata S., Rotilio G., Ciriolo M. P53 orchestrates the PGC-1α-mediated antioxidant response upon mild redox and metabolic imbalance. Antioxid. Redox Signal. 2013:18. doi: 10.1089/ars.2012.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marmolino D., Manto M., Acquaviva F., Vergara P., Ravella A., Monticelli A. PGC-1alpha down-regulation affects the antioxidant response in Friedrich's ataxia. PLoS One. 2010:5. doi: 10.1371/journal.pone.0010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salminen A., Kaarniranta K., Kauppinen A. Crosstalk between oxidative stress and SIRT1: impact on the aging process. Int. J. Mol. Sci. 2013;14:3834–3859. doi: 10.3390/ijms14023834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodgers J.T., Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc. Natl. Acad. Sci. USA. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jäger S., Handschin C., St-Pierre J., Spiegelman B.M. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc. Natl. Acad. Sci. USA. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iacovelli J., Rowe G.C., Khadka A., Diaz-Aguilar D., Spencer C., Arany Z. PGC-1α induces human RPE oxidative metabolism and antioxidant capacity. Investig. Ophthalmol. Vis. Sci. 2016;57:1038–1051. doi: 10.1167/iovs.15-17758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Egger A., Samardzija M., Sothilingam V., Tanimoto N., Lange C., Salatino S. PGC-1α determines light damage susceptibility of the murine retina. PLoS One. 2012:7. doi: 10.1371/journal.pone.0031272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korovila I., Hugo M., Castro J.P., Weber D., Höhn A., Grune T. Proteostasis, oxidative stress and aging. Redox Biol. 2017;13:550–567. doi: 10.1016/j.redox.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koga H., Kaushik S., Cuervo A.M. Protein homeostasis and aging: the importance of exquisite quality control. Ageing Res. Rev. 2011;10:205–215. doi: 10.1016/j.arr.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demishtein A., Fraiberg M., Berko D., Tirosh B., Elazar Z., Navon A. SQSTM1/p62-mediated autophagy compensates for loss of proteasome polyubiquitin recruiting capacity. Autophagy. 2017;9:1–12. doi: 10.1080/15548627.2017.1356549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johansson I., Monsen V.T., Pettersen K., Mildenberger J., Misund K., Kaarniranta K. The marine n-3 PUFA DHA evokes cytoprotection against oxidative stress and protein misfolding by inducing autophagy and NFE2L2 in human retinal pigment epithelial cells. Autophagy. 2015;11:1636–1651. doi: 10.1080/15548627.2015.1061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 36.Viiri J., Amadio M., Marchesi N., Hyttinen J.M., Kivinen N., Sironen R. Autophagy activation clears ELAVL1/HuR-mediated accumulation of SQSTM1/p62 during proteasomal inhibition in human retinal pigment epithelial cells. PLoS One. 2013:8. doi: 10.1371/journal.pone.0069563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu W.J., Ye L., Huang W.F., Guo L.J., Xu Z.G., Wu H.L. p62 links the autophagy pathway and the ubiqutin-proteasome system upon ubiquitinated protein degradation. Cell Mol. Biol. Lett. 2016:2. doi: 10.1186/s11658-016-0031-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin J., Wu H., Tarr P.T., Zhang C.Y., Wu Z., Boss O. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 39.Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 40.Schnell S.A., Staines W.A., Wessendorf M.W. Reduction of lipofuscin-like autofluorescence in fluorescently labeled tissue. J. Histochem. Cytochem. 1999;47:719–730. doi: 10.1177/002215549904700601. [DOI] [PubMed] [Google Scholar]
- 41.Leinonen H., Rossi M., Salo A.M., Tiainen P., Hyvärinen J., Pitkänen M. Lack of P4H-TM in mice results in age-related retinal and renal alterations. Hum. Mol. Genet. 2016;25:3810–3823. doi: 10.1093/hmg/ddw228. [DOI] [PubMed] [Google Scholar]
- 42.Weymouth A.E., Vingrys A.J. Rodent electroretinography: methods for extraction and interpretation of rod and cone responses. Prog. Retin. Eye Res. 2008;27:1–44. doi: 10.1016/j.preteyeres.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 43.Mehalow A.K., Kameya S., Smith R.S., Hawes N.L., Denegre J.M., Young J.A. See comment in PubMed commons belowCRB1 is essential for external limiting membrane integrity and photoreceptor morphogenesis in the mammalian retina. Hum. Mol. Genet. 2013;12:2179–2189. doi: 10.1093/hmg/ddg232. [DOI] [PubMed] [Google Scholar]
- 44.Mattapallil M., Wawrousek E., Chan C.-C., Zhao H., Roychoudhury J., Ferguson T. The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes. Investig. Ophthalmol. Vis. Sci. 2012;53:2921–2927. doi: 10.1167/iovs.12-9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lückoff A., Scholz R., Sennlaub F., Xu H., Langmann T. Comprehensive analysis of mouse retinal mononuclear phagocytes. Nat. Protoc. 2017;12:1136–1150. doi: 10.1038/nprot.2017.032. [DOI] [PubMed] [Google Scholar]
- 46.Chen M., Rajapakse D., Fraczek M., Luo C., Forrester J.V., Xu H. Retinal pigment epithelial cell multinucleation in the aging eye - a mechanism to repair damage and maintain homoeostasis. Aging Cell. 2016;15:436–445. doi: 10.1111/acel.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Juuti-Uusitalo K., Koskela A., Kivinen N., Viiri J., Hyttinen J.M.T., Reinisalo M. Autophagy regulates proteasome inhibitor-induced pigmentation in human embryonic stem cell-derived retinal pigment epithelial cells. Int. J. Mol. Sci. 2017:18. doi: 10.3390/ijms18051089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeong H.S., Choi H.R., Yun H.Y., Baek K.J., Kwon N.S., Park K.C. Ceramide PC102 inhibits melanin synthesis via proteasomal degradation of microphthalmia-associated transcription factor and tyrosinase. Mol. Cell. Biochem. 2013;375:81–87. doi: 10.1007/s11010-012-1530-5. [DOI] [PubMed] [Google Scholar]
- 49.Feher J., Kovacs I., Artico M., Cavallotti C., Papale A., Balacco Gabrieli C. Mitochondrial alterations of retinal pigment epithelium in age-related macular degeneration. Neurobiol. Aging. 2006;27:983–993. doi: 10.1016/j.neurobiolaging.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 50.Ferrington D.A., Kapphahn R.J., Leary M.M., Atilano S.R., Terluk M.R., Karunadharma P. Increased retinal mtDNA damage in the CFH variant associated with age-related macular degeneration. Exp. Eye Res. 2016;145:269–277. doi: 10.1016/j.exer.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terluk M.R., Kapphahn R.J., Soukup L.M., Gong H., Gallardo C., Montezuma S.R. Investigating mitochondria as a target for treating age-related macular degeneration. J. Neurosci. 2015;35:7304–7311. doi: 10.1523/JNEUROSCI.0190-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dou G., Kannan R., Hinton D.R. Endoplasmic reticulum response to oxidative stress in RPE. In: Stratton R.D., Hauswirth W.W., Gardner T.W., editors. Studies on Retinal and Choroidal Disorders. Springer Science+Business Media, LLC; New York, NY, USA: 2012. pp. 241–258. [Google Scholar]
- 53.Jing G., Wang J.J., Zhang S.X. ER stress and apoptosis: a new mechanism for retinal cell death. Exp. Diabetes Res. 2012 doi: 10.1155/2012/589589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He S., Yaung J., Kim Y.H., Barron E., Ryan S.J., Hinton D.R. Endoplasmic reticulum stress induced by oxidative stress in retinal pigment epithelial cells. Graefes Arch. Clin. Exp. Ophthalmol. 2008;246:677–683. doi: 10.1007/s00417-008-0770-2. [DOI] [PubMed] [Google Scholar]
- 55.Mitter S.K., Song C., Qi X., Mao H., Rao H., Akin D. Dysregulated autophagy in the RPE is associated with increased susceptibility to oxidative stress and AMD. Autophagy. 2014;10:1989–2005. doi: 10.4161/auto.36184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodríguez-Muela N., Koga H., García-Ledo L., de la Villa P., de la Rosa E.J., Cuervo A.M. Balance between autophagic pathways preserves retinal homeostasis. Aging Cell. 2013;13:478–488. doi: 10.1111/acel.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaarniranta K., Tokarz P., Koskela A., Paterno J., Blasiak J. Autophagy regulates death of retinal pigment epithelium cells in age-related macular degeneration. Cell Biol. Toxicol. 2017;33:113–128. doi: 10.1007/s10565-016-9371-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rogińska D., Kawa M.P., Pius-Sadowska E., Lejkowska R., Łuczkowska K., Wiszniewska B. Depletion of the third complement component ameliorates age-dependent oxidative stress and positively modulates autophagic activity in aged retinas in a mouse model. Oxid. Med. Cell. Longev. 2017 doi: 10.1155/2017/5306790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sukseree S., Chen Y.T., Laggner M., Gruber F., Petit V., Nagelreiter I.M. Tyrosinase-cre-mediated deletion of the autophagy gene Atg7 leads to accumulation of the RPE65 variant M450 in the retinal pigment epithelium of C57BL/6 mice. PLoS One. 2016:11. doi: 10.1371/journal.pone.0161640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y., Cross S.D., Stanton J.B., Marmorstein A.D., Le Y.Z., Marmorstein L.Y. Early AMD-like defects in the RPE and retinal degeneration in aged mice with RPE-specific deletion of Atg5 or Atg7. Mol. Vis. 2017;23:228–241. [PMC free article] [PubMed] [Google Scholar]
- 61.Yao J., Jia L., Khan N., Lin C., Mitter S.K., Boulton M.E. Deletion of autophagy inducer RB1CC1 results in degeneration of the retinal pigment epithelium. Autophagy. 2015;11:939–953. doi: 10.1080/15548627.2015.1041699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim J.Y., Zhao H., Martinez J., Doggett T.A., Kolesnikov A.V., Tang P.H. Noncanonical autophagy promotes the visual cycle. Cell. 2013;154:365–376. doi: 10.1016/j.cell.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Valapala M., Wilson C., Hose S., Bhutto I.A., Grebe R., Dong A. Lysosomal-mediated waste clearance in retinal pigment epithelial cells is regulated by CRYBA1/βA3/A1-crystallin via V-ATPase-MTORC1 signaling. Autophagy. 2014;10:480–496. doi: 10.4161/auto.27292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu J., Copland D.A., Theodoropoulou S., Chiu H.A., Barba M.D., Mak K.W. Impairing autophagy in retinal pigment epithelium leads to inflammasome activation and enhanced macrophage-mediated angiogenesis. Sci. Rep. 2016:6. doi: 10.1038/srep20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Piippo N., Korkmaz A., Hytti M., Kinnunen K., Salminen A., Atalay M. Decline in cellular clearance systems induces inflammasome signaling in human ARPE-19 cells. Biochim. Biophys. Acta. 2014;1843:3038–3046. doi: 10.1016/j.bbamcr.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 66.Cruz-Guilloty F., Saeed A.M., Duffort S., Cano M., Ebrahimi K.B., Ballmick A. T cells and macrophages responding to oxidative damage cooperate in pathogenesis of a mouse model of age-related macular degeneration. PLoS One. 2014:9. doi: 10.1371/journal.pone.0088201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ambati J., Atkinson J.P., Gelfand B.D. Immunology of age-related macular degeneration. Nat. Rev. Immunol. 2013;13:438–451. doi: 10.1038/nri3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material