Abstract
We describe the molecular analysis of three families with hypodontia involving primarily molar teeth and report two novel mutational mechanisms. Linkage analysis of two large families revealed that the hypodontia was linked to the PAX9 locus. These two families revealed missense mutations consisting of a glutamic acid substitution for lysine and a proline substitution for leucine within the paired domain of PAX9. A pair of identical twins affected with hypodontia in a third family demonstrated a 288-bp insertion within exon 2 that resulteds in a putative frameshift mutation and a premature stop codon. The insertion was associated with the loss of 7-bp from exon 2. A block of 256-bp of sequence within the insertion was completely identical to downstream sequence from the second intron of the PAX9 gene. These studies extend the spectrum of mutations in PAX9 associated with hypodontia to include heretofore undescribed categories, including missense mutations.
Keywords: hypodontia, cleft lip, PAX9, mutation
INTRODUCTION
Familial hypodontia is a genetically heterogeneous condition characterized by the congenital absence of primary and/or permanent teeth, ranging in severity from mild, involving the absence of one or two teeth, to severe, where the majority of teeth can be missing [Thesleff, 2000]. Autosomal dominant (AD) hypodontia has been localized to at least three chromosomal loci, MSX1 [Vastardis et al., 1996], PAX9 [Stockton et al., 2000], and an unknown locus on chromosome 10 [Liu et al., 2001]. In addition, a fourth locus is implicated [Nieminen et al., 1995; Arte et al., 1996]. A missense mutation in MSX1 has been associated with AD hypodontia involving the second premolars and third molars [Vastardis et al., 1996]. We [Stockton et al., 2000] and others [Frazier-Bowers et al., 2002] have reported frame-shift and nonsense mutations [Nieminen et al., 2001] in PAX9 as well as a submicroscopic deletion involving the PAX9 gene in families with AD hypodontia involving primarily molar teeth [Das et al., 2002]. A third locus associated with hypodontia involving the absence of a few to the entire set of permanent teeth has been mapped to chromosome 10q11.2 [Liu et al., 2001]. A distinct unknown locus underlying hypodontia involving incisors and premolars has been implicated based on exclusion mapping of candidate genes [Nieminen et al., 1995; Arte et al., 1996].
Three classes of mutations within or involving the PAX9 gene have been noted to date. Following a total genome scan of a large family with AD hypodontia, we initially identified a single-base insertion within the paired-domain region of PAX9 that led to a frame-shift mutation at residue 73 of the 341 aa protein [Stockton et al., 2000]. A second frame-shift mutation outside the paired-domain at residue 264 was recently reported [Frazier-Bowers et al., 2002]. Nieminen et al. [2001] reported a nonsense mutation at amino acid (aa) residue 114 within the paired-domain. In addition to these single-base mutations, we recently reported a submicroscopic deletion of >57 kb involving the PAX9 gene in a small nuclear family affected with severe hypodontia of the primary and permanent dentition [Das et al., 2002]. Two genes map within the deletion interval in the latter family: PAX9, the only odontogenic gene within the interval and SLC25A21, encoding mitochondrial oxodicarboxylate carrier. These data, in concert with the previous data, suggested that human PAX9 is a dosage-sensitive gene with haploinsufficiency resulting in hypodontia.
Here we describe two missense mutations and a 288-bp insertion causing a putative frameshift within the second exon of PAX9 encoding the paired-domain of PAX9 in three families with hypodontia.
MATERIALS AND METHODS
Families With Hypodontia
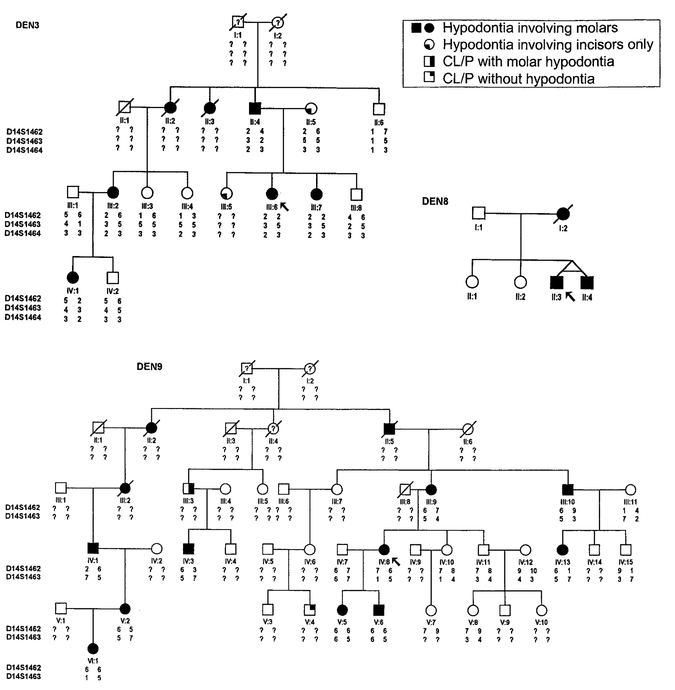
The subjects in this study were from three unrelated Caucasian families with hypodontia (families DEN3, DEN8, and DEN9) (Fig. 1). After informed consent was obtained, dental records were obtained from the referring dentist(s) and blood samples or buccal swabs collected from available members of each family. Dental records were not available for individuals III:3 and IV:3 (DEN9) and their affection status was obtained by taking a history.
Fig. 1.
Pedigrees of hypodontia kindreds DEN3, DEN8, and DEN9 with haplotypes for markers linked to the PAX9 locus. Note that individuals II:5 and III:5 in family DEN3 are missing incisors, which represent a different class of hypodontia. CL/P: cleft lip with cleft palette.
Linkage Analysis
Genomic DNA was extracted from blood [Sambrook et al., 1989] and buccal swabs [Meulenbelt et al., 1995] as described previously. Linkage analysis to determine whether the PAX9 locus was linked to the hypodontia observed in each family was performed using micro-satellite markers, D14S1462 and D14S1463 located ~39 and ~49 kb, respectively upstream of the PAX9 gene [Das et al., 2002]. Polymerase chain reaction (PCR) was performed and the products were analyzed on a 6% sequencing gel. Two-point linkage analysis was carried out between markers D14S1462, D14S1463, and D14S1464 and the disease locus and each other using MLINK and ILINK of the FASTLINK computer package [Cottingham et al., 1993]. An AD mode of inheritance with no phenocopies was used in the analysis. Each marker allele was assumed to have equal frequency. A disease frequency of 0.001 was used in the analysis.
DNA Sequencing
For the detection of mutations, PAX9 exons 1–4 were amplified from affected individuals from each of the three families (III:6 and III:7 in family DEN3, V:5 in family DEN9, and II:3 and II:4 in family DEN8) by PCR with the primer sets and conditions described previously [Stockton et al., 2000]. In addition, new primers that had minimal homology to the related gene PAX1 were developed to allow sequencing of a portion of exon 2. The sequences of all primers used in this study are shown in Table 1.
TABLE 1.
Sequence of Primers Used
| Name | Sequence | Location |
|---|---|---|
| hPAX9exl-319 | 5′-CCACTGAGGCGGTGCGGAAAG-3’ | Exon 1 |
| hPAX9exl-518 | 5′-CCTACAACTTGTAGGAACACGAGCAAAG-3′ | |
| hPAX9ex2AF | 5′- AGGCAGCTGTCCCAAGCAGCG- 3′ | Exon2 |
| hPAX9ex2aR | 5′-TGTATCGCGCCAGGATCTTGCTG-3′ | |
| hPAX9ex2bF | 5′-ATCCGACCGTGTGACATCAGCC-3′ | |
| hPAX9ex2bR | 5′-GGAGGGCACATTGTACTTGTCGC-3′ | |
| hPAX9ex2cF | 5′-GCATCTTCGCCTGGGAGATCCG-3′ | |
| hPAX9ex2cR | 5′-GAGCCCCTACCTTGGTCGGTG-3′ | |
| hPAX9ex2alF | 5′-GGGGACAGCCCCAGTAGTTA-3′ | |
| hPAX9ex2a2F | 5′-AGGAGTGTTCGTGAACGGGA-3′ | |
| hPAX9ex2alR | 5′-CACGGTGGGGGTAGTGAC-3′ | |
| hPAX9K91EFa | 5′-CCCAGGCGAAGATGCCGGGGTCTCTCCGCT-3′ | |
| hPAX9K91ER | 5′-CATCAGCCGCCAGCTACGGGTCTCGCA-3′ | |
| hPAX9InsFl | 5′-AAGATCCTGGCGCAGCC-3′ | |
| hPAX9InsR | 5′-CGTCTCGATCTTGGCCTGA-3′ | |
| hPAX9ex3F | 5′-TTTGGGTCCCGTCTCAAGAGTGG-3′ | Exon 3 |
| hPAX9ex3R | 5′-CCTAAATCCCCGCCGCCACG-3′ | |
| hPAX9ex4Fl | 5′- GGAGAGTAGAGTCAGAGCATTGCTG- 3′ | Exon 4 |
| hPAX9ex4F2 | 5′-CCCAAGTGTCGCCTTACATGA-3′ | |
| hPAX9ex4R | 5′-GAGACCTGGGAATTGGGGGA-3′ |
The underlined base represents a change from the wild-type sequence.
RESULTS
Linkage Analysis
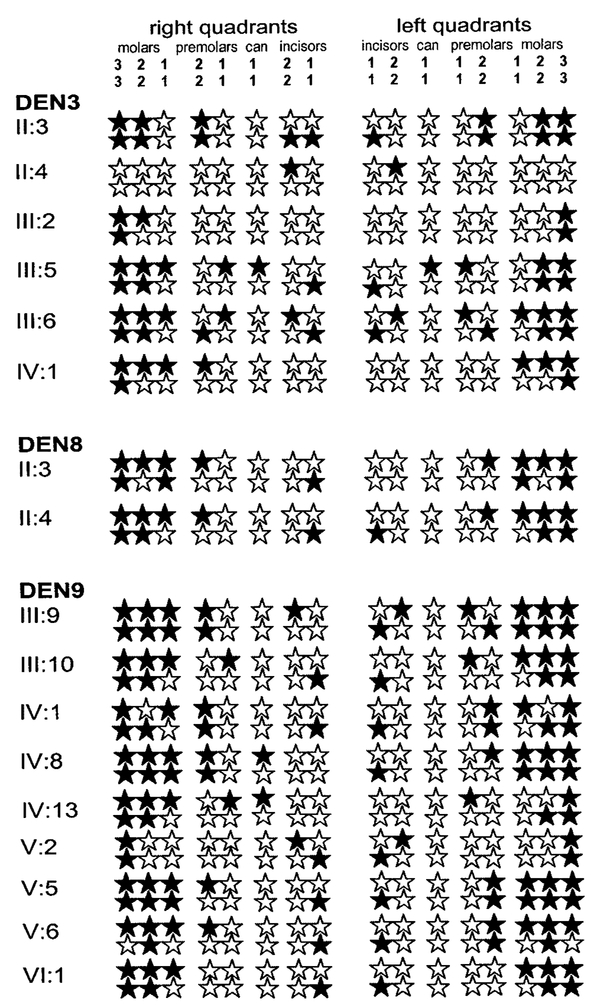
The pedigrees used in this study and haplotypes for chromosome 14 markers linked to PAX9 (where available) are shown in Figure 1. Available data on the identity of congenitally missing permanent teeth in selected affected members are shown in Figure 2. All individuals missing one or more molar teeth were considered affected. As seen in Figure 2, these individuals also lacked incisor(s), premolars, and occasionally canines. Information on the status of the primary dentition was only available on individual VI:1 in DEN9 and was reported as being normal. An AD mode of inheritance was strongly suggested for hypodontia in the larger families DEN3 and DEN9. For pedigree DEN3, there were no recombination events between markers D14S1462, D14S1463, and D14S1464 and the disease locus and the maximum two point lod scores were 2.7, 2.6, and 1.65, respectively, at Θ=0. For pedigree DEN9, a maximum lod score of 3.2 occurred between the disease locus and markers D14S1462 and D14S1463 at Θ=0. Marker D14S1464 was not genotyped in pedigree DEN9. Although there were no recombination events between the disease locus and the marker loci for either of these families, there was an apparent recombination event between marker loci D14S1462 and D14S1463 in pedigree DEN9. This apparent recombination event occurred in meioses transmitted from unaffected spouse III:8 to his offspring.
Fig. 2.
Clinical phenotypes of affected individuals in pedigrees DEN3, DEN8, and DEN9. Filled stars represent congenitally missing teeth.
PAX9 Mutations in the Three Families
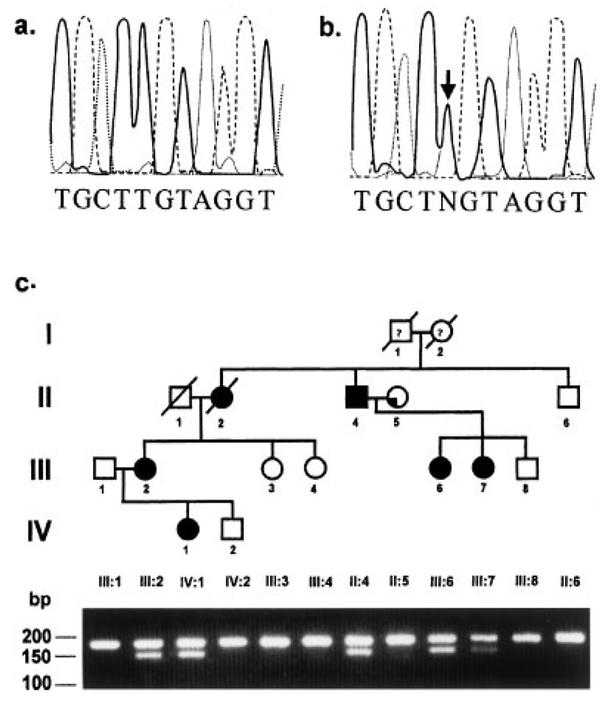
Since the linkage analysis clearly implicated the PAX9 locus, we conducted DHPLC analysis on all PAX9 exons and observed a fragment with altered migration in both patients III:6 and III:7 but not in six other hypodontia patients within a subfragment of exon 2 extending from +1212 through +1460 (+1 is A of the ATG codon) (data not shown). We amplified this region with a new set of PCR primers that had no homology to PAX1 as noted in Figure 3 and observed an A to G transition mutation at position +271 which results in the substitution of glutamic acid for lysine at aa residue 91 (Fig. 3a,b). An allele-specific PCR assay was developed to detect the mutant allele wherein the mutant allele was digested by the enzyme BsrBI while the wild-type allele was undigested (Fig. 3c). The mutation segregated with hypodontia and was not seen in any of the unaffected members of the family or in 108 control chromosomes.
Fig. 3.
DNA sequence chromatograms from a control (a) and an affected individual (b) from pedigree DEN3 showing the point mutation in exon 2 of the PAX9 gene on the non-coding strand. c: Segregation of the mutation in family DEN3. An allele-specific PCR test was developed by substituting a C for a T 4-bp upstream from the mutation in the forward primer, hPAX9K91EF. This mutation in combination with the mutation in DEN3 created a cleavage site for BsrBI. The primers hPAX9K91EF and hPAX9K91ER were used to amplify a 179-bp product containing the mutation. Digestion with BsrBI resulted in a 149-bp fragment and a 30-bp fragment (not visible) in addition to a 179-bp fragment representing the wild-type allele in affected individuals.
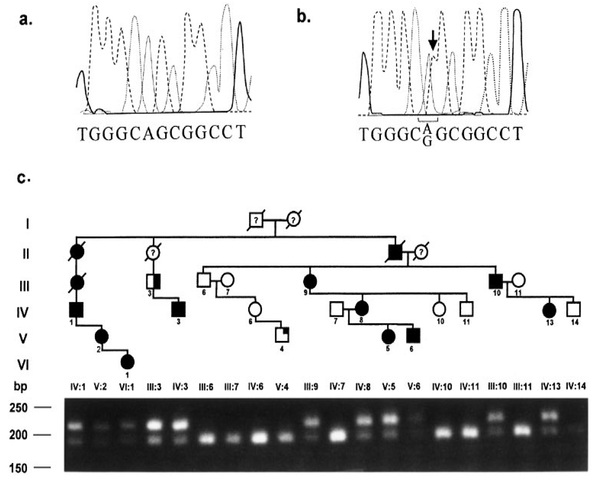
Sequencing of patient V:5 in DEN9 identified a T to C transition mutation at position +62 (Fig. 4a,b). This mutation results in a substitution of leucine by proline at aa position 21 within the PAX9 protein. The mutation abolished an MspA1 site in exon 2 and an allele-specific PCR test was used to assess segregation of the mutation in the family (Fig. 4c), as well as to examine 54 control individuals. The mutation segregated with hypodontia, and was not seen in any of the unaffected members of the family or in 108 control chromosomes. Two members of this family, individuals III:3 and V:4, were also affected with cleft lip and palate. The missense mutation was observed in only III:3 but not in V:4.
Fig. 4.
DNA sequence chromatograms from a control (a) and an affected individual (b) from pedigree DEN9 showing the point mutation in exon 2 of the PAX9 gene on the non-coding strand. c: Segregation of the mutation in DEN9. The mutation in DEN9 destroyed a cleavage site for MspA1. Amplification of a 214-bp PCR product with primers hPAX9ex2a2F and hPAX9ex2a1R was followed by digestion with MspA1. Digestion of the product with MspA1 resulted in a 189-bp product and a 25-bp product (not visible) in unaffected individuals.
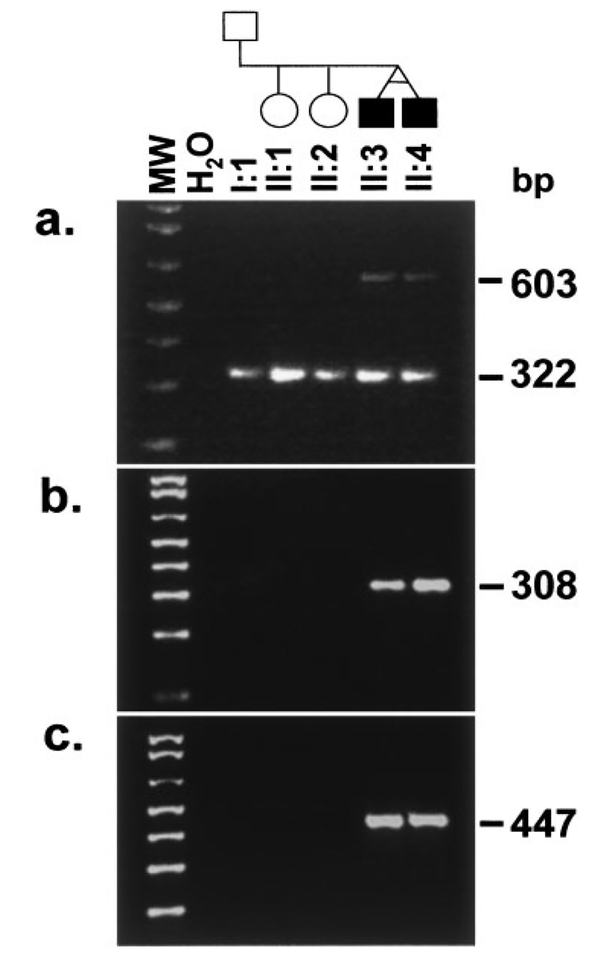
Amplification of the exon 2a subfragment from the twin brothers, II:3 and II:4 in family DEN8 revealed a 322-bp fragment of the expected size as well as a larger ~600-bp fragment (Fig. 5a). Sequencing of the 322-bp fragment revealed exon 2 sequence that was identical to the wild-type sequence. Sequencing of the larger fragment seen only in the affected twins revealed an insertion of 288-bp (Fig. 6a). BLAST analysis of this inserted sequence against the GENBANK databases revealed that the first 13-bp bore no homology to any known human sequence while 19-bp at the 3′-end of the insertion was identical to sequences within several BAC clones mapping to chromosome 2. However, the remaining central 256-bp of sequence bore complete sequence homology to sequence from the second intron of the PAX9 gene located 504-bp downstream from the end of exon 2. Additional homologies limited to 20- to 30-bp segments from within this central 256-bp were found to random genomic sequences. To further confirm the presence of this insertion, primers spanning the 5′- and 3′-junctions of the insertion within exon 2 were designed and used in PCR amplifications with template DNA from all available members of the family. The expected product of 308-bp was only observed in the affected twins and in none of the unaffected family members (Fig. 5b) or in 50 control individuals (data not shown). Additional tests were conducted by PCR using primers spanning the 5′- and 3′-junctions of the insertion in combination with primers within exon 2 downstream and upstream of the insertion, respectively. The expected PCR products of 447 and 374-bp, respectively, were only seen in the affected twins and not in the other family members (Fig. 5c). The inserted sequence results in a shift in reading frame at aa 58, with a stop codon at aa 177, assuming that exons of the mutant PAX9 gene are spliced in a manner similar to that of the wild-type gene.
Fig. 5.
An insertion within exon 2 in family DEN8. a: Amplification of a subfragment of exon 2 with primers hPAX9ex2a2F and hPAX9ex2a1R resulted in the expected 322-bp fragment and a novel 603-bp additional fragment only in the affected twin brothers II:3 and II:4 in family DEN8. b: The large fragment was sequenced and a 288-bp insertion detected (see Fig. 6). Primers, hPAX9InsF1 and hPAX9InsR, were designed to span the 5′and 3′ junctions of the insertion. Amplification with these primers resulted in a 308-bp fragment only in the affected brothers. c: Amplification with hPAX9ex2a2F and hPAX9InsR resulted in a 447-bp fragment only in II;3 and II:4.
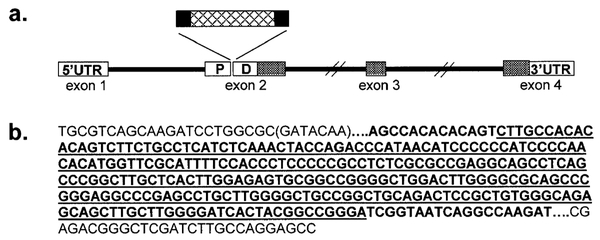
Fig. 6.
a: Schematic diagram showing the insertion within the sequence of the paired domain (PD) of PAX9 in family DEN8. The hatched region within the insertion depicts sequence that has complete homology with sequence from intron 2. b: Sequence of a portion of PAX9 exon 2 showing the inserted sequence. The exonic sequence is in plain type and the inserted sequence is in bold. The exonic sequence deleted during the insertion process is in parentheses. The junctions are identified by a series of dots. The underlined 256-bp sequence bore sequence homology to sequence from the second intron.
DISCUSSION
We have described three novel mutations in PAX9 associated with hypodontia involving predominantly molar teeth. Previously, frame-shift mutations resulting in premature stop codons due to the insertion of a single base have been described by us [Stockton et al., 2000] and others [Frazier-Bowers et al., 2002], as well as a nonsense mutation [Nieminen et al., 2001], all within the paired domain of PAX9. In contrast, two of the mutations we describe here are missense mutations representing a new category of dominant mutations in PAX9 leading to hypodontia. Neither of these missense mutations were observed in 108 control alleles, strongly supporting pathogenicity. The T62C transition mutation codes for an exchange of leucine 21 for proline, while the A271G mutation causes a substitution of positively charged lysine 91 for negatively charged glutamic acid. Both mutated residues are highly conserved within the paired-domain of the human PAX family of genes as well as in the PAX9 gene of various species (Fig. 7), suggesting that they are vital to the function of the protein. Haploinsufficiency of PAX9 appears to be the underlying cause for hypodontia associated with the insertion, nonsense, and deletion mutations described to date. Whether the missense mutations described here result in functional haploinsufficiency due to the deleterious effect of the mutation on the DNA-binding capacity of PAX9 will require further experimental investigation. The possibility that the missense mutations have a dominant-negative effect cannot be ruled out either, although given our previous data that conclusively demonstrates that haploinsufficiency for PAX9 results in hypodontia [Das et al., 2002], we favor the former possibility.
Fig. 7.
Comparison of the sequence of a portion of the paired domain region from all human PAX family of proteins and PAX9 proteins from various species with the PAX9 mutant sequence from family DEN9 (a) and family DEN3 (b).
The twin boys in family DEN8 revealed an unusual mutation in the second exon of the PAX9 gene (Figs. 5 and 6). The insertion resulted in the loss of 7-bp of exonic sequence at the site of the insertion. The first 13-bp of the insertion consisted of sequence with no homology to any known human sequence, while 19-bp at the 3′ end of the insertion had homology to the sequence of several BAC clones mapping to chromosome 2. Dot matrix analysis of the inserted sequence and 200 kb of sequence encompassing the PAX9 gene did not reveal any direct or inverted repeats that might have played a role in the insertion. There was no molecular evidence for a retro-transposition event leading to the insertion. However, the loss of 7-bp from the exon suggests that the retrotransposon sequence could have undergone deletion as well, resulting in the absence of a “signature” that might have allowed elucidation of the mechanism of the insertional event.
We observed an apparent recombination event between marker D14S1462 and D14S1463 which are ~10 kb apart. In this regard, it is interesting to note that in the hypodontia family with a submicroscopic deletion of PAX9 that we previously reported [Das et al., 2002], one of the deletion breakpoints mapped to this same interval, suggesting that this could be a “hot-spot” for such events.
Two members of family DEN9 displayed cleft lip and palate but only one of them carried the Leu21Pro missense mutation. Nonsyndromic cleft lip with or without cleft palate (CL/P, MIM 119530) and isolated cleft palate (CPO, MIM 119540) are common developmental anomalies. Orofacial clefts can occur at a frequency of 1 in 500 to 1 in 2,500 live-born infants and the frequency is dependent on a number of factors including geographical location, ethnicity, and socio-economic status. Complex inheritance involving multiple genes as well as gene–environment interactions, are considered to underlie the cause of these conditions. van den Boogaard et al. [2000] have reported the association of a nonsense mutation (S105X) in MSX1 with orofacial clefting and tooth agenesis. Twelve members of a single family who showed various combinations of cleft lip, cleft palate, and tooth agenesis carried the nonsense mutation, and the authors concluded that MSX1 was a candidate gene for orofacial clefting, and that the observed phenotype was similar to that seen in the Msx1 knock-out mice. In contrast, an Arg239Pro missense mutation in MSX1 was associated with tooth agenesis involving the second premolars and third molars [van den Boogaard et al., 2000], and a S202X nonsense mutation was associated with Witkop syndrome [Jumlongras et al., 2001] but no orofacial clefting was reported. The orofacial clefting observed in patient III:3 in DEN9 who carried the Leu21Pro mutation in PAX9 was associated with hypodontia involving molar teeth while that seen in patient V:4 in the same family was not. Given the small sample size, it is difficult to derive any conclusions about the possible role of the Leu21Pro mutation in PAX9 in the orofacial clefting observed in III:3, but it is possible that the presence of this mutation may have decreased the threshold for orofacial clefting in this individual. It is also interesting to note that like MsxI null mice, Pax9 knock-out mice also display orofacial clefting. Detailed investigation of the possible role of PAX9 alleles in human orofacial clefting will require knowledge of the major susceptibility loci for this condition.
ACKNOWLEDGMENTS
We are most grateful to the members of the families for their willingness to participate in the study. We thank Mr. Michael Bull, Dr. Grant Willcox, and Dr. Everett Rushing for referring the families and for sharing clinical information on patients. We thank Joy Gumin for technical assistance, and Theresa Bower-man, Dalia Angel, and Irving Ortiz for assistance with graphics. This work was supported by NIH grants DE13632 and DE14102 (PIP), Texas Higher Education Applied Technology Program grant 004949–0143-1999 (PIP), a grant from the American Association of Orthodontists Foundation (DTB), and the Baylor MRRC.
Grant sponsor: NIH; Grant numbers: DE13632, DE14102; Grant sponsor: Texas Higher Education Applied Technology Program; Grant number: 004949–0143-1999; Grant sponsor: The American Association of Orthodontists Foundation.
REFERENCES
- Arte S, Nieminen P, Pirinen S, Thesleff I, Peltonen L. 1996. Gene defect in hypodontia: Exclusion of EGF, EGFR, and FGF-3 as candidate genes. J Dent Res 76:1346–1352. [DOI] [PubMed] [Google Scholar]
- Cottingham RW, Idury RM, Schaffer AA. 1993. Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263. [PMC free article] [PubMed] [Google Scholar]
- Das P, Stockton DW, Bauer C, Shaffer LG, D’Souza RN, Wright JT, Patel PI. 2002. Haploinsufficiency of PAX9 is associated with autosomal dominant hypodontia. Hum Genet 110:371–376. [DOI] [PubMed] [Google Scholar]
- Frazier-Bowers SA, Guo DC, Cavender A, Xue L, Evans B, King T, Milewicz D, D’Souza RN. 2002. A novel mutation in human PAX9 causes molar oligodontia. J Dent Res 81:129–133. [PubMed] [Google Scholar]
- Jumlongras D, Bei M, Stimson JM, Wang WF, DePalma SR, Seidman CE, Felbor U, Maas R, Seidman JG, Olsen BR. 2001. A nonsense mutation in MSX1 causes Witkop syndrome. Am J Hum Genet 69:67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Wang H, Zhao S, Zhao W, Bai S, Zhao Y, Xu S, Wu C, Huang W, Chen Z, Feng G, He L. 2001. The novel gene locus for agenesis of permanent teeth (He-Zhao deficiency) maps to chromosome 10q11.2. J Dent Res 80:1716–1720. [DOI] [PubMed] [Google Scholar]
- Meulenbelt I, Droog S, Trommelen GJ, Boomsma DI, Slagboom PE. 1995. High-yield noninvasive human genomic DNA isolation method for genetic studies in geographically dispersed families and populations. Am J Hum Genet 57:1252–1254. [PMC free article] [PubMed] [Google Scholar]
- Nieminen P, Arte S, Pirinen S, Peltonen L, Thesleff I. 1995. Gene defect in hypodontia: Exclusion of MSX1 and MSX2 as candidate genes. Hum Genet 96:305–308. [DOI] [PubMed] [Google Scholar]
- Nieminen P, Arte S, Tanner D, Paulin L, Alaluusua S, Thesleff I, Pirinen S. 2001. Identification of a nonsense mutation in the PAX9 gene in molar oligodontia. Eur J Hum Genet 9:743–746. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Frisch EF, Maniatis T. 1989. Molecular cloning: A laboratory manual Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Stockton DW, Das P, Goldenberg M, D’Souza RN, Patel PI. 2000. Mutation of PAX9 is associated with oligodontia. Nat Genet 24:18–19. [DOI] [PubMed] [Google Scholar]
- Thesleff I 2000. Genetic basis of tooth development and dental defects. Acta Odontol Scand 58:191–194. [DOI] [PubMed] [Google Scholar]
- van den Boogaard MJ, Dorland M, Beemer FA, van Amstel HK. 2000. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat Genet 24:342–343. [DOI] [PubMed] [Google Scholar]
- Vastardis H, Karimbux N, Guthua SW, Seidman IG, Seidman CE. 1996. A human MSX 1 homeodomain missense mutation causes selective tooth agenesis. Nat Genet 13:417–421. [DOI] [PubMed] [Google Scholar]