SUMMARY
Lineage analysis of autoreactive B cells can reveal the origins of autoimmunity. In the autoimmune disease pemphigus vulgaris (PV), desmoglein 3 (DSG3) and DSG1 autoantibodies are predominantly of the IgG4 subclass and less frequently of IgG1 and IgA subclasses, prompting us to investigate whether anti-DSG IgG4 B cells share lineages with IgG1, IgA1, and IgA2. Combining subclass-specific B cell deep sequencing with high-throughput antibody screening, we identified 80 DSG-reactive lineages from 4 PV patients. Most anti-DSG IgG4 B cells lacked clonal relationships to other subclasses and preferentially targeted DSG adhesion domains, whereas anti-DSG IgA frequently evolved from or to other subclasses and recognized a broader range of epitopes. Our findings suggest that anti-DSG IgG4 B cells predominantly evolve independently or diverge early from other subclasses and that IgA is most often not the origin of IgG autoreactivity in PV. These data provide insight into how autoreactivity diversifies across B cell subclasses.
In Brief
Ellebrecht et al. use next-generation sequencing to identify clonal relationships among antigen-specific B cells in the autoimmune disease pemphigus vulgaris. They find that autoreactive IgG4 B cells are largely clonally distinct from autoreactive IgG1 and IgA, thus elucidating the class-switch pathways that diversify and modify an autoimmune response in humans.
Graphical Abstract

INTRODUCTION
The vast sequence diversity of the antibody (Ab) variable region confers protection against a broad range of pathogens while unavoidably creating autoreactive clones that harbor the potential to cause autoimmune disease (Wardemann et al., 2003). Heavy-chain variable regions initially recombine with the immunoglobulin (Ig)M constant region. After activation, naive IgM B cells undergo somatic hypermutation (SHM) and class switch in response to cytokines and co-activating signals (Stavnezer et al., 2008). While the Ab variable region determines antigen reactivity, the constant region (Fc) can fix complement and recruit leukocytes, thus causing or augmenting end-organ damage. Because class switch deletes intervening genomic DNA, it only occurs 5’ to 3’ (IgM > IgG3 > IgG1 > IgA1 > IgG2 > IgG4 > IgE > IgA2). Next-generation sequencing (NGS) has defined class-switch events in normal and allergic individuals (Horns et al., 2016; Looney et al., 2016), but little is known about subclass-specific B cell repertoires in human autoimmunity.
Pemphigus vulgaris (PV) is a potentially fatal disease in which Abs to the adhesion protein desmoglein 3 (DSG3) induce mucosal blistering, and later development of anti-DSG1 Abs leads to skin blistering in its mucocutaneous subtype (Kasperkiewicz et al., 2017). Although the Fc is not required for blistering in PV (Anhalt et al., 1986; Mascaró et al., 1997; Payne et al., 2005), DSG auto-Abs have a characteristic subclass distribution. Patients with active disease have DSG-specific IgG4 over IgG1, while anti-DSG IgG2 and IgG3 are uncommon (Ayatollahi et al., 2004; Futei et al., 2001; Spaeth et al., 2001). The IgG4 autoAb predominance is so pronounced that total serum IgG4 is enriched in PV patients (Funakoshi et al., 2012). Some patients in remission still demonstrate anti-DSG IgG1 (Ayatollahi et al., 2004; Bhol et al., 1994; Spaeth et al., 2001). The majority of patients with active disease also have anti-DSG IgA (Mentink et al., 2007; Spaeth et al., 2001), the dominant isotype in mucosa where PV manifests but whose role in PV is poorly understood.
The occurrence of anti-DSG IgG1, IgG4, and IgA indicates that switch to these subclasses is a stereotyped feature of the autoimmune response in PV. To trace the development of autoreactive B cells across subclasses, we combined NGS of IgG1, IgG4, IgA1, and IgA2 B cell repertoires with Ab phage display (APD) to identify DSG3- and DSG1-reactive lineages. This revealed that anti-DSG IgG4 B cells represent a largely distinct population in PV, with rare connections to IgG1 and IgA2, while IgA1 B cells show a high degree of clonal overlap with IgA2 and can evolve from anti-DSG IgG1. These results provide insight into the clonal relationships of subclass-specific autoreactive B cell repertoires in a model human Ab-mediated disease.
RESULTS
NGS of Subclass-Specific B Cell Repertoires Reproducibly Captures Lineage Diversity
To define subclass-specific anti-DSG B cell repertoires, we combined two methods to obtain large-scale lineage data (NGS) and screen for antigen specificity (APD) (Figure S1A). We performed NGS of IgG1, IgG4, IgA1, and IgA2 variable-heavy (VH) gene transcripts in 4 patients with active PV (Table S1). NGS libraries were produced using indexed constant region primers with a custom signature for every patient subclass. The CH1 sequence internal to the primer binding site and custom library signature, both of which can independently discriminate isotype and/or subclass, were collectively called the isosig (Figure S1B). Ensuring concordance among all 3 elements of the primer index sequence and isosig allowed high-confidence patient-subclass assignment and stringent filtering of misclassified sequences that could be falsely interpreted as cross-subclass lineage relationships. This pipeline yielded 225,970 to 9,678,823 raw paired-end reads per patient subclass, which condensed to 172,101 to 3,828,206 isosig-verified non-redundant sequences (Table S2).
B cell lineages are often defined by grouping sequences with the same VH, joining gene segment, heavy chain (JH), and length of the third complementarity determining region (CDR3) and then setting an 80%–95% threshold for CDR3 nucleotide identity (Hershberg and Luning Prak, 2015; Hoh et al., 2016; Horns et al., 2016; Looney et al., 2016; Tipton et al., 2015), depending on the relative risk of false-positive versus false-negative clonal groupings. We analyzed the distribution of CDR3 nearest-neighbor percent nucleotide differences between patients (who cannot share lineages, although similar CDR3 sequences could occur randomly or by convergent evolution) and within patients (in whom clonal lineages naturally occur). Certain amino-acid motifs are enriched in pathogenic auto-Abs (Yamagami et al., 2010), but convergent CDR3 sequences among PV patients are rare (Chen et al., 2017). Thus, we compared CDR3 sequences among patients to define a per-patient CDR3 nucleotide identity threshold with 99% confidence that two sequences in a presumptive B cell lineage do not belong to different lineages (Figure S1C). CDR3 percent nucleotide similarity thresholds ranged from 80.6% in pemphigus vulgaris patient (PV)8 to 86.1% in PV18. We next analyzed the CDR3 nearest-neighbor percent nucleotide difference within each patient, which revealed two major probability densities (Figure S1D): the left peak representing clonal lineages and the central peak representing unrelated B cell clones. These analyses justify that the CDR3 nucleotide identity thresholds used for clustering reasonably separate clonal lineages from unrelated B cells.
Within each patient subclass, we identified distinct B cell lineages (Table S2). IgG4 had the fewest lineages (3,495–9,966), followed by IgA2 (10,787–21,697), with comparable numbers of IgA1 and IgG1 lineages (22,547–32,045 and 19,728–41,675, respectively). We performed rarefaction on the number of lineages in each patient subclass (Hsieh et al., 2016) to ascertain sampling depth (Figure 1A), which predicts that we captured 71.07%–97.00% of the circulating B cell lineages (Figure 1B). We then estimated the Shannon and Simpson indices to describe lineage abundance and evenness, with the Simpson index being weighted more heavily toward dominant species (Morris et al., 2014). 90.23%–99.97% and 99.62%–100.00% of the diversity measured by the Shannon and Simpson indices was captured (Figure 1B), indicating sufficient sampling of circulating IgG1, IgG4, IgA1, and IgA2 lineages, particularly in regard to dominant clones.
Figure 1. Rarefaction and Coverage Estimation Reveals Sufficient Sampling of B Cell Lineages in Peripheral Blood.
(A) Rarefaction curves of read number versus number of lineages reveals overall depth of sampling.
(B) Quantification of sampling coverage by asymptotic diversity estimation.The percentage of overall lineage diversity coverage is lower than the percentage of Shannon or Simpson diversity coverage, indicating that sampling of 60 mL peripheral blood captures most of the abundant lineages while losing some rare lineages.
To determine reproducibility, we analyzed biological replicates from two separate 30-mL blood samples drawn at the same time from PV17 and PV18 and technical replicates from resequencing PV16 or re-synthesizing PV17 and PV18 libraries using different cDNA aliquots. Each pair of replicates was compared for overlapping lineages, followed by population size estimation of the fraction of captured lineages (Table S3). Mark-and-recapture analysis using the Chapman estimator (Chapman, 1954) demonstrates that 96.57%–99.97% of B cell lineages with ≥5 reads are captured by technical replicates, and 35.46%–81.99% of lineages are captured by 30-mL biological replicates. Collectively, these data indicate that sequencing subclass-specific B cell repertoires from 60 mL peripheral blood provides substantial representation of the circulating B cell lineages in a given patient and captures the vast majority of the B cell lineage diversity.
High-Throughput Screening of Anti-DSG B Cells in PV Indicates a Predominance of IgG4 Lineages, Followed by IgA1, IgG1, and IgA2
Having established reliable parameters to define and reproducibly capture B cell lineages, we next sought to identify antigenspecific clones. We generated 16 APD libraries, comprising IgG1, IgG4, IgA1, and IgA2 repertoires in each of the 4 patients. Up to 1 billion clones per patient subclass were screened against DSG3 and DSG1 by APD, and up to 200 recombinant monoclonal antibodies (mAbs) per patient (50 per subclass) were purified and analyzed by ELISA to confirm DSG reactivity, as well as indirect immunofluorescence (IIF) on human skin to independently validate the PV phenotype.
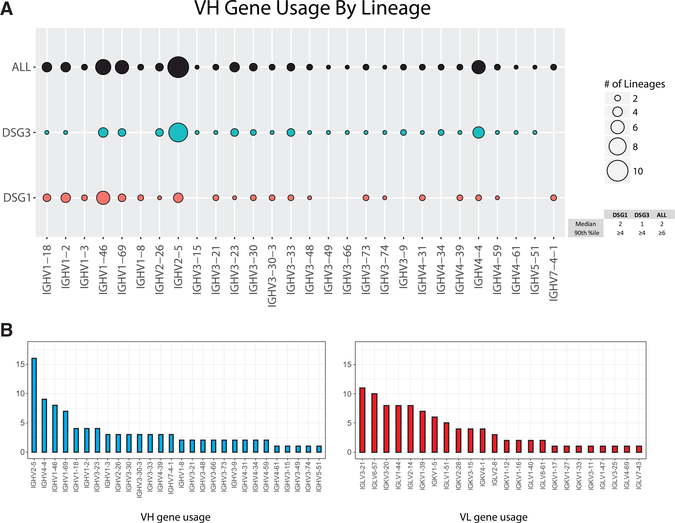
96 distinct anti-DSG B cell clones were identified (Table S4). The plurality of DSG-reactive clones was IgG4 (n = 41), followed by IgA1 (n = 21), IgG1 (n = 18), and IgA2 (n = 16). Comparable distributions across subclasses were observed for DSG3- and DSG1-reactive lineages (Table S5). Anti-DSG clones used 28 VH and 24 variable light-chain (VL) genes; no convergent CDR3 sequences were identified across patients. Because clonality cannot be reliably inferred from L-CDR3 nucleotide identity, subsequent genetic analyses focused on the heavy chain. Anti-DSG1 lineages most often used IGHV1–46, IGHV2–5, and IGHV1–2; anti-DSG3 lineages most often used IGHV2–5, IGHV4–4, and IGHV1–46 (Figure 2A). In subclass-specific lineages (Figure S2), the vast majority (90%) of VH genes were used by 2 or fewer clones. VH genes used at or above the 90th percentile abundance threshold among both subclass-specific and total anti-DSG lineages (found in ≥3 or ≥6 clones, respectively) were IGHV2–5, IGHV1–46, IGHV1–69, and IGHV4–4. More restricted distribution of VH than VL gene usage was observed (Figure 2B), reflecting promiscuous pairing of sequences within the same VH family with different light chains (Tables S4 and S8).
Figure 2. Anti-DSG Repertoires in PV Show Preferential VH Gene Usage.
(A) All anti-DSG, anti-DSG3, or anti-DSG1 lineages were categorized by VH gene usage. Dot size corresponds to number of lineages using each VH gene (legend). The median and 90th percentile for number of lineages using a particular VH gene are indicated.
(B) Graphs indicate the number of DSG-reactive clones using each VH and VL gene.
Although the Majority of IgG4 Lineages Have No Relatives in Other Subclasses, Anti-DSG Lineages Demonstrate Increased Connectivity to Other Subclasses Compared to the Global Repertoire
By matching the 96 APD clones to NGS lineages, we defined 80 distinct anti-DSG lineages, 29 of which span ≥2 subclasses (Figure 3A). Including NGS relatives of anti-DSG clones found in other subclasses, we identified 40 IgG4-, 30 IgA2-, 27 IgA1-, and 26 IgG1-containing anti-DSG lineages. The overlap between subclasses and nearest-neighbor quantitation appears in Figures 3B and 3C. The majority of IgA1- and IgA2-containing anti-DSG lineages demonstrate clonal relatives in other subclasses, particularly between IgA1 and IgA2. IgG1 lineages demonstrate equivalent overlap with IgG4, IgA1, and IgA2. In contrast, only 12 (30%) of 40 IgG4-containing anti-DSG lineages have relatives in other subclasses (Figure 3D). Thus, anti-DSG IgG4 B cells are less related to other subclasses than anti-DSG B cells from the other subclasses (p = 1.2 × 105, two-tailed Fisher’s exact test).
Figure 3. Autoreactive Lineages Bridge Subclasses.
(A) Phage display identified 80 anti-DSG lineages that were mapped to NGSderived lineages within each subclass, highlighted in blue. Asterisk indicates clones found only by APD.
(B) Number of lineages containing every possible pair of subclasses.
(C) Percentage of lineages within a given subclass with relatives in another subclass.
(D) Distribution of clones between single- and multi-subclass lineages. IgG4 was compared to the sum of IgA1, IgA2, and IgG1 by two-tailed Fisher exact test.
To investigate how inter-subclass relationships in the anti-DSG repertoire compare to the global repertoire, we compared the average pairing rate of the anti-DSG and global repertoire (i.e., the normalized proportion of overlapping clonal lineages) for each patient-subclass pair, which showed that the anti-DSG repertoire is significantly more interconnected than the global repertoire (p < 0.0001, two-tailed paired t test; Figure 4A). Separating out the anti-DSG repertoire by each subclass pair indicates that the highest average pairing rate is detected for IgA1-IgA2, both in the global and anti-DSG repertoires (Figure 4B). We subsequently evaluated which inter-subclass relationships demonstrated the highest proportional overlap in the anti-DSG compared to the global B cell repertoire, which would indicate enrichment for specific inter-subclass relationships and, hence, might identify preferential class-switch pathways for anti-DSG B cells. We normalized the average pairing rate of the anti-DSG repertoire to that of the global repertoire to obtain a relative pairing rate (Figure 4C). Surprisingly, anti-DSG IgG4-containing lineages were among the most enriched for inter-subclass connectivity compared to global IgG4 (demonstrated by a steeper slope between relative pairing rates for global and anti-DSG lineages). No significant difference in IgG4 read number between multi-subclass and single-subclass lineages was found by Mann-Whitney U test (Figure 4D), indicating that lineage size and/or sampling depth had minimal effect on finding inter-subclass connectivity. Thus, although the majority of anti-DSG IgG4 lineages identified do not have APD- or NGS-detectable relatives in other subclasses, DSG reactivity is associated with an increase in subclass connectivity of IgG4 B cells compared to the global IgG4 B cell repertoire.
Figure 4. Connectivity of the Anti-DSG and Global Repertoire.
(A) The average pairing rate for each subclass was plotted for both the global and anti-DSG repertoires. Each data point represents a pair of subclasses within a patient, representing 14 (of a maximum 24) pairs of subclasses across the 4 patients. ****p < 0.0001, by paired t test.
(B and C) Shown here: (B) absolute and (C) relative pairing rates (normalized to the global repertoire) for each subclass pair.
(D) Read numbers for single- and multi-subclass IgG4-containing anti-DSG lineages were compared to determine whether pairing rate is dependent upon number of reads mapping to a given lineage. Mann-Whitney test was not significant (n.s.). Circles represent the pairing rate (black = global, red = DSG-reactive) of each patient-subclass population.
Phylogenetic Analysis Indicates that Anti-DSG IgG4 B Cells Are Rarely Derived by Class Switch from IgG1 B Cells
The increased connectivity of anti-DSG B cells with other subclasses compared to the global B cell repertoire prompted us to investigate the clonal development of anti-DSG B cells in greater detail. We constructed maximum parsimony trees of anti-DSG lineages represented in at least two subclasses, which uses SHM to trace the evolution of DSG-reactive clones. As a control for PCR or sequencing errors, we amplified and sequenced a monoclonal template of the same length, which showed that PCR mutations, insertions or deletions (indels), and/or base-call errors occur in the raw NGS reads (Figure S3A) but that almost all reads can be reassigned to the correct sequence by clustering reads within a 97% operational taxonomic unit (OTU) radius and excluding nodes with <5 reads or indels (Figure S3B). Some lineages spanned multiple closely related VH genes, leading to computational assignment of the top-ranked NGS cluster to a different VH gene from the APD-confirmed clone; these lineages were excluded from phylogenetic analysis, since assignment of the APD-confirmed clone to the identical VH gene used by the plurality of the sequences in an NGS lineage allowed better understanding of the evolution of antigen-specific clones. A total of 18 multi-subclass lineages were subjected to finer analysis (Figures 5, S4, S5, S6, S7, S8, S9, and S10; Table S6).
Figure 5. Select Trees from IgG4-Containing Multi-subclass Lineages.
(A) PV16.A1.D1.P4.10.
(B) PV17.G4.D1.P3.15.
(C) PV17.G4.D1.P4.8.
(D) PV17.G1.D1.P4.6.
Read numbers are indicated inside each node. Blue-edged labels indicate nucleotide Hamming distance between nodes. Labeled vertices indicate inferred (white) or NGS-identified (red and blue) intermediates produced as mAbs and tested for DSG reactivity by ELISA.
To determine whether IgG1 serves a precursor for IgG4 B cells, we examined 7 anti-DSG lineages containing both IgG1 and IgG4 (Figures 5, S4, and S5). One lineage suggests an IgG1-to-IgG4 switch, with the IgG1 clone differing from the IgG4 by 1 nt (Figure 5A). Figure 5B shows a lineage with a divergence of IgG1 and IgG4 B cells from a common intermediate with 13 shared mutations, a pattern that is supportive of, but does not provide direct evidence for, an IgG1-to-IgG4 switch. Similarly, Figure 5C indicates a lineage with the co-evolution of IgG1 and IgG4 B cells demonstrating varying degrees of shared mutation but no direct evidence of sequential switch from IgG1 to IgG4. The remaining 4 lineages either have no shared mutations between IgG1 and IgG4 (Figure 5D) or have a few shared mutations before segregated evolution of IgG1 and IgG4 B cells (Figures S4 and S5). Figure S4A shows a lineage in which IgG1 and IgG4 relatives were identified directly by APD, but the IgG1 clone demonstrates a 3-nt CDR1 insertion relative to the IgG4 clone, indicating independent evolution of these anti-DSG IgG1 and IgG4 B cells from a common precursor.
Together with the finding that the majority of anti-DSG IgG4 lineages lack relatives in other subclasses (Figures 3B–3D), these data suggest that class switch from IgG1 to IgG4 represents a minority pathway for the development of anti-DSG IgG4 B cells or that these switch events occur in a temporarily restricted fashion that cannot be captured after the onset of clinically active disease.
Anti-DSG IgG4 B Cells Evolve Independently of IgA1 but Can Directly Give Rise to Anti-DSG IgA2 B Cells
To determine whether IgG4 B cells can arise from IgA1 B cells, which could imply a mucosal origin of autoimmunity in PV, we examined the clonal relationships between anti-DSG IgG4 and IgA B cells. Only 2 lineages contained both IgA1 and IgG4 clonal members with ≥5 reads per subclass. No examples of direct switch from IgA1 to IgG4 were detected by APD or NGS (Figures 5A and 5B). However, evidence of switch from IgG4 to IgA2 was detected in 2 of the 3 IgG4- and IgA2-containing lineages analyzed (Figures S5 and S6). In the other IgG4I-gA2 lineage, IgA2 and IgG4 clones showed shared mutations but independently switched from IgA1 and IgG1, respectively (Figure 5A).
The Majority of Anti-DSG IgA1 B Cells Demonstrate Evidence of Class Switch from IgG1 or to IgA2
In contrast to anti-DSG IgG4 lineages, anti-DSG IgA1 lineages show a high degree of inter-subclass connectivity. In all 10 IgA1- and IgA2-containing lineages analyzed, a switch from IgA1 to IgA2 was observed (Figures 5A, S7A, S7B, S8, S9, and S10). Additionally, anti-DSG IgG1 B cells demonstrate evidence of a switch to IgA1 in 4 of 6 lineages in which they coexist (Figures 5C and S7). In 2 of these lineages (Figures S7A and S7B), a switch from IgG1 to IgA1 as well as IgA2 was observed, suggesting that IgA1 B cells may serve as an intermediate for clonal maturation from IgG1 to IgA2.
IgG4 Lineages Develop DSG Reactivity Early in Development and Preferentially Target Amino-Terminal DSG Epitopes
To determine when DSG reactivity arises during the course of B cell evolution, we produced select NGS-identified and computationally inferred clones from IgG4-containing multi-subclass lineages. DSG ELISA indicated that, in the 7 multi-subclass lineages tested, relatives of anti-DSG clones in other subclasses generally showed similar patterns of DSG reactivity (Figure 6A). In the PV17.G1.D1.P4.8 lineage (Figure 5C), relatives of the anti-DSG1 IgG1 APD clone demonstrated no detectable DSG binding by ELISA, although these same Abs immunoprecipitated DSG1 in epitope-mapping experiments, suggesting that DSG reactivity arose from non- or weakly DSG-reactive clones within this phylogenetic tree. 4 of the 7 most recent common ancestors (MRCAs)—some of which had no SHM and, hence, were unmutated common ancestors (UCAs) (Figure 6A)—and 8 of the 11 additional germline-reverted IgG4 heavy chains (Figure 6B) had DSG reactivity comparable to that of the somatically mutated B cell clone, suggesting that autoreactivity is often acquired early during the development of anti-DSG IgG4 B cells.
Figure 6. DSG ELISA Reactivity of Lineage Intermediates and Germline-Reverted Clones.
(A) Relatives from IgG4-containing multi-subclass lineages were tested for DSG reactivity (10 mg/mL). Figure references for relevant clones are indicated.
(B) DSG reactivity of germline-reverted IgG4 clones (10 mg/mL). GL, germline; MRCA, most recent common ancestor; UCA, unmutated common ancestor. Data are representative of 1–2 independent experiments. Red line indicates cutoff value, mean + 3 SD for negative controls (averaged across 14–15 independent experiments).
We next determined the DSG extracellular cadherin (EC) domain specificity of APD-derived anti-DSG B cell clones (Table 1). Reactivity to EC1, which contains residues important for DSG trans-adhesion, is favored in anti-DSG IgG4 B cells compared to other subclasses (p = 0.0065, Fisher’s exact test). Similar patterns were observed when comparing DSG3- and DSG1-reactive B cell specificities (Table S7), indicating a correlation between the targeting of functional domains of DSGs and the switch to IgG4.
Table 1.
Epitope Mapping of Subclass-Specific Anti-DSG Antibodies
| EC1 | EC2 | EC3 | EC4 | EC5 | |
|---|---|---|---|---|---|
| IgA1 | 7 | 2 | 2 | 1 | 2 |
| IgA2 | 4 | 0 | 3 | 0 | 1 |
| IgG1 | 8 | 0 | 1 | 3 | 1 |
| IgG4 | 27 | 0 | 1 | 3 | 0 |
| Grand total | 46 | 2 | 7 | 7 | 4 |
Data reflect 1–2 independent experiments.
To determine whether targeted DSG epitopes change during B cell diversification, we mapped select NGS and APD clones and computationally inferred intermediates. In most cases, epitope specificity did not change within lineages, despite different heavy- and light-chain pairings (Table S8). The one exception occurred in a lineage in which the IgG1 and IgG4 clones had no shared mutations and targeted different EC epitopes (Figure 5D; Table S8), further supporting their independent evolution from a common precursor. Our data indicate that epitope spreading between EC domains rarely occurs during diversification by SHM or class switch, which is corroborated by prior studies of PV serum auto-Abs (Ohyama et al., 2012).
DISCUSSION
PV presents a valuable opportunity to investigate the lineage relationships of subclass-specific B cell repertoires in a mucosal autoAb-mediated disease. The preferential association of pathogenic auto-Abs with IgG4, the persistence of anti-DSG IgG1 in remission, and the occurrence of anti-DSG IgA in active disease indicate a common class-switch regulatory pathway in PV. We aimed to maximize the depth, rigor, and reproducibility of our approach by using high-volume blood samples; a nonredundant primer index and isosig verification to avoid false subclass assignment; technical and biological replicates; error correction informed by sequencing a monoclonal template; and independent validation of DSG reactivity by ELISA, IIF on human skin, and DSG epitope mapping. 60 mL of blood yields approximately 108 PBMCs, 2 × 106 class-switched B cells, and 10,000 anti-DSG3 B cells (Nishifuji et al., 2000). A total of 96 anti-DSG clones were identified, representing 80 distinct lineages. We used APD to identify anti-DSG lineages due to its superior depth of sampling and its specificity for identifying anti-DSG3 clones in PV but not normal individuals (Cho et al., 2016), but it can be biased by random heavy- and light-chain pairing that may create antibody specificities that did not exist in vivo, relative undersampling of low-affinity clones, and overrepresentation of plasma cells. However, most anti-DSG Abs are produced by plasmablasts, evidenced by the drop in anti-DSG titers to the normal range after anti-CD20 B cell depletion (Joly et al., 2017), so preferential sampling of plasmablasts by APD is highly relevant to PV pathogenesis. Also, mass spectrometry studies have shown that 50% of serum auto-Abs are produced by 1–7 anti-DSG B cell clones (Chen et al., 2017), reinforcing the finding that the immune response in PV is marked by oligoclonal expansions. Simpson diversity indices (Figure 1B) indicate that our approach captures nearly all dominant lineages, suggesting that we are reasonably sampling both global and DSG-reactive lineages. Collectively, the studies performed provide a comprehensive analysis of subclass-specific human autoimmune B cell repertoires.
Our studies indicate that DSG reactivity is enriched in IgG4 B cells, as IgG4 is the most common subclass in anti-DSG lineages but the least common in the global repertoire, which complements past reports showing that DSG reactivity is enriched in serum IgG4 Abs (Funakoshi et al., 2012; Futei et al., 2001). Also consistent with past studies (Chen et al., 2017; Cho et al., 2014), we found that VH1–46 and VH1–69 gene usage is enriched in anti-DSG lineages, as well as VH4–4 and VH2–5 (Figure 2). Preferential VH gene usage has been noted in systemic lupus erythematosus (Tipton et al., 2015), multiple sclerosis (von Büdingen et al., 2012), influenza vaccination (Jackson et al., 2014; Pappas et al., 2014), and rotavirus infection (Tian et al., 2008), suggesting that particular antigens induce stereotyped immune responses. Interestingly, all 4 PV patients demonstrated VH2–5 gene usage in their anti-DSG IgA repertoires, while IgG repertoires more often used VH1–46 and VH1–69 (Figure S2), suggesting that different subclasses react differently to the same antigen. This difference in reactivity is also supported by the finding that IgA clones react across all five EC domains of DSG, while IgG4 preferentially binds EC1 (Table 1). VH2–5 IgA has been reported to be expressed by human ileal plasma cell precursors and skin epidermal cells and can recognize skin pathogens (Jiang et al., 2015; Yuvaraj et al., 2009). Although we did not investigate polyreactivity or cross-reactivity to foreign antigens, these reports raise the possibility that cross-reactivity may stimulate anti-DSG IgA B cell activation, as has previously been suggested for anti-DSG3 VH1–46 B cells reacting to rotavirus VP6 protein (Cho et al., 2016) or anti-DSG1 IgG4 reacting to the sandfly salivary antigen LJM11 (Qian et al., 2016).
Although anti-DSG IgG4 B cells demonstrate a higher degree of clonal relatedness to other subclasses than non-DSG-reactive IgG4 (Figure 4), the majority of anti-DSG IgG4 B cells have no relatives detectable by APD or NGS in other subclasses. Nearest-neighbor (Figure 3C) and phylogenetic analyses (Figures 5, S4, and S5) support that anti-DSG IgG1, although only rarely associated with IgG4 through direct class switch, demonstrates the greatest overlap with IgG4. However, in the majority of anti-DSG IgG1- and IgG4-containing lineages, we observed independent evolution of IgG1 and IgG4 from a common precursor or divergent and mostly segregated evolution of IgG1 and IgG4.
The paucity of IgG1-to-IgG4 class-switch events could be due to factors such as undersampling; temporal or spatial restriction of switch events; or switch from IgM, IgG2, or IgG3. Although incomplete sampling is unavoidable in human studies, our observation of class-switch events between anti-DSG IgG1, IgA1, and IgA2 B cells suggests that the predominance of IgG4 without clonal relatives is not solely due to an inability to detect inter-subclass relationships. Prior studies in normal human B cell repertoires found that direct class-switch events to IgG4 were most commonly from IgG1 (45% of events), followed by IgG2 (21%), IgM (17%), IgG3 (16%), and rarely from IgA1 (1%) (Horns et al., 2016). We did not examine IgG2 and IgG3 B cell repertoires in our study, but the potential for direct IgM-to-IgG4 switch as the dominant pathway for anti-DSG IgG4 B cell development in PV is intriguing, since that pathway is characteristic of short-lived plasmablasts, which produce the majority of anti-DSG IgG in PV. This model is also consistent with the early development of DSG reactivity and lack of epitope spreading observed in anti-DSG IgG4 lineages (Figure 6; Table S8). A previous report showed that germline-reverted anti-DSG Abs can retain DSG binding—in particular, those using VH1–46—implying that autoreactivity arose in naive B cells (Cho et al., 2014). In this study, we note early development of DSG reactivity in the majority of IgG4 multi-subclass lineages analyzed (Figure 6A)— including germline reactivity in VH1–46, 2–5, 2–26, 3–9, 3–23, 3–30, and 3–49 IgG4 lineages (Figure 6B)—suggesting that anti-DSG IgG4 lineages may more broadly develop autoreactivity early in their evolution.
The class switch pathways leading to IgG4 are of fundamental interest not only for the pathogenesis of PV but also for the mechanism of other IgG4-mediated diseases (Huijbers et al., 2015) and states of chronic immune stimulation. PV is characterized by a Th2 response (Veldman et al., 2003) and a cytokine milieu (interleukin (IL)-4, IL-10, and IL-13) that promotes class switch to IgG4 (Cho et al., 2015; Hertl et al., 2006; Jeannin et al., 1998; Takahashi et al., 2007). Beekeepers, patients with chronic helminth infections, and patients undergoing allergen desensitization therapy also develop IgG4 responses (Aalberse and Schuurman, 2002; Hussain et al., 1992; Jutel et al., 2005). Novice beekeepers initially develop IgG1 to bee venom antigens, while experienced beekeepers tolerized to stings have IgG4 to the same antigens (Aalberse et al., 1983). Similarly, in endemic pemphigus foliaceus, unaffected individuals can display anti-DSG1 IgG1, while patients with active disease have anti-DSG1 IgG1 and IgG4 (Warren et al., 2003). The finding that IgG4 B cells in PV preferentially target DSG EC1 epitopes (Table 1), where functional residues important for intercellular adhesion reside (Boggon et al., 2002; Harrison et al., 2016), raises the question of whether the ability to cause tissue blistering and damage preferentially induces the class switch to IgG4. In patients undergoing allergen desensitization therapy and in tolerized beekeepers, antigen-specific IgG4 has been reported to be exclusively produced by IL-10-secreting regulatory B cells (van de Veen et al., 2013). However, in PV, the role of regulatory B cells is not well defined. Expansions of IL-10-secreting, CD24hiCD38hi B cells in PV have been associated with complete remission after B cell-depletion therapy (Colliou et al., 2013) but have alsobeenshown todemonstrate an impairedability to suppress Th1 responses and interferongamma production (Zhu et al., 2015). Future studies are necessary to better define the cell of origin for anti-DSG IgG4 and its potential regulatory function.
We have also characterized the lineage relationships of antiDSG IgA in PV, a disease that affects the mucosa where IgA immune responses are predominant. Anti-DSG IgA1 cells demonstrated a class switch from IgG1 (Figures 5C and S7) and frequently switch to IgA2 (Figures 5A, S7A, S7B, and S8, S9, and S10), the latter being consistent with the spatially restricted development of IgA1 and IgA2 in mucosal-associated lymphatic tissues (Cerutti, 2008). High-throughput analysis of class-switch landscapes in normal human B cells indicates that IgG1-IgA1 relationships are common, representing 41% of all class-switch events to IgA1, and that IgA1 is the most frequent origin of direct class switches to IgA2 (Horns et al., 2016), corroborating our findings. Furthermore, we identified a class switch of DSG-reactive lineages from IgG4 to IgA2 (Figures S5 and S6), a relationship that represents only 0.04% of class-switch events to IgA2 in normal human B cell repertoires (Horns et al., 2016) but that may be more common in PV due to the cytokine milieu associated with active disease. Class switch to IgA1 from IgM or IgG1 depends on transforming growth factor (TGF)-beta and CD40 activation in cultured human B cells (Zan et al., 1998) and can occur in a T cell-independent manner on mucosal surfaces by TLR4 engagement and dendritic cell secretion of BAFF and APRIL (Cerutti, 2008). IgA2 production occurs in the lamina propria via secretion of IL-10 and APRIL from colonic epithelium in response to normal intestinal flora in both an IgM-to-IgA2 and an IgA1-to-IgA2 fashion (He et al., 2007). Given our data showing that anti-DSG IgA B cells either develop independently of or switch directly from anti-DSG IgG B cells, and literature showing that TGF-beta, BAFF, and APRIL are not elevated in PV (Asashima et al., 2006; D’Auria et al., 1997; Giordano and Sinha, 2012), anti-DSG IgA1 and IgA2 are likely an epiphenomenon in PV, formed in mucosal lymphoid tissues or epithelia by IgA class-switch mediators. This seems especially likely for anti-DSG IgA2, as its 3° position in the IgH locus precludes it from serving as a reservoir for IgG development.
In sum, our study has elucidated the complex interrelationships between four functionally diverse Ab subclasses during an ongoing autoimmune response. In PV, the majority of anti-DSG IgG4 cells show no evidence of arising from other subclasses, while anti-DSG IgA1, IgG1, and IgA2 show more extensive clonal relationships. Thus, despite the co-existence of IgG1 and IgG4 autoAbs in PV sera, class switch from IgG1 is not a dominant pathway for IgG4 production. Our data also indicate that the PV IgA repertoire is less skewed toward pathogenic DSG epitopes and often arises from IgG, rather than being an origin for IgG autoreactivity. These data provide key insight into how autoreactivity develops and diversifies across Ab subclasses. Ultimately, our work in PV may help elucidate the developmental pathway of IgG4 in other conditions, potentially leading to better therapeutic strategies that target these populations in IgG4-mediated disease or promote IgG4 B cell development in allergen tolerization protocols.
EXPERIMENTAL PROCEDURES
Study Approval
Written informed consent was obtained from participants according to an institutional-review-board-approved protocol. Study participant demographics were as follows: PV8 (28-year-old woman), PV16 (61-year-old man), PV17 (63-year-old woman), and PV18 (72-year-old woman).
Sample Processing
50–60 mL heparinized blood was collected by venipuncture. Diagnosis of PV was established by clinical presentation, histology, and DSG ELISA. For PV17 and PV18, two 30-mL blood samples were collected on the same day and processed independently as biological replicates. PBMCs were isolated using Ficoll gradient centrifugation and snap-frozen in methanol prior to storage (80°C).
Library Preparation
APD libraries were produced and screened as described previously (Cho et al., 2016; Payne et al., 2005), with modified primers listed in Table S9. IgG1, IgG4, IgA1, and IgA2 amplicons were first amplified with hinge-specific primers and then APD primers with a unique 3° library signature per patient subclass. For NGS, second-round PCR primers were flanked by a transposase sequence, the 3’ library signature, and an Illumina primer index. More details are found in the Supplemental Experimental Procedures.
Data Processing
Libraries were sequenced using Illumina MiSeq 2 3 300 bp (Institute for Genome Science, University of Maryland; NGS Core, University of Pennsylvania). Reads were quality trimmed and merged using PEAR (v0.9.6) (Zhang et al., 2014) and collapsed into non-redundant (NR) sequences and filtered by isosig match. IMGT/HighV-QUEST was used for variable-diversity-joining (VDJ) assignment of productive sequences.
Clonality was defined by VH, JH, and CDR3 length, and a per-patient CDR3 distance threshold with 99% confidence of excluding unrelated sequences (Figures S1C and S1D). Sequences in each patient-subclass pair were clustered using a customized Perl script and an unweighted pair group method centroid (UPGMC) linkage function from the Algorithm::Cluster module. To reduce VDJ misassignment due to PCR error or SHM, we grouped sequence clusters within a CDR3 distance threshold, ignoring VH or JH, into superclusters ranked by read count, and downstream analyses were performed using the top-ranked VH cluster.
Estimation of Clonal Richness and Diversity and Total Population Size
Sampling for each patient-subclass pair was estimated using rarefaction analysis and asymptotic diversity estimation using the iNEXT R package (Hsieh et al., 2016), with modifications as described previously (Chiu and Chao, 2016). Reproducibility among replicates was conducted using the Chapman estimator (Chapman, 1954) on lineages with ≥5 reads in a replicate. More details are found in the Supplemental Experimental Procedures.
Subclass Connectivity Quantitation
Average pairing rates between subclasses were computed as the harmonic mean of the frequency of shared lineages to normalize for differing subclass sizes. If n lineages are shared between subclasses of read size A and B, the pairing rate is:
For comparing the anti-DSG and global repertoire, the pairing rate was calculated from samples bootstrapped down to the size of the anti-DSG repertoire for a given subclass pair.
Lineage Analysis
Phylogenetic trees were constructed from multi-subclass lineages containing APD-derived heavy-chain sequences mapping to NGS superclusters with identical VH, excluding lineages with indels or subclasses with <5 reads. Primer-trimmed sequences within each subclass were subject to full-length centroid clustering at a 97% OTU radius using the cluster_otus command in USEARCH (Edgar, 2010). To ease visualization, nodes with <5 reads or indels were excluded, except where indicated. Maximum parsimony trees were made using Change-O (Gupta et al., 2015), which uses dnapars.exe from PHYLIP (Felsenstein, 1989). Similar analysis was conducted on a CD8CD137 monoclonal template (Figure S3). To construct germline-reverted or NGS lineage intermediates, codon-optimized heavy chains were synthesized (Integrated DNA Technologies) and subcloned with light chains used in the same lineage. More details are in the Supplemental Experimental Procedures.
Epitope Mapping
Epitope mapping was performed as described previously (Cho et al., 2014; Ohyama et al., 2012), using chimeric DSG3-DSG2 or DSG1-DSG2 domainswapped molecules in High Five or HEK293 supernatants. DSG3- or DSG1reactive clones were evaluated by immunoprecipitation of DSG3-DSG2 or DSG1-DSG2 molecules followed by immunoblotting using an anti-E tag horseradish peroxidase (anti-E-HRP) Ab.
Statistics
Single-subclass versus multi-subclass lineages in IgG4 and non-IgG4 were compared using a two-tailed Fisher exact test in Figure 3D. The average pairing rates were compared using a two-tailed paired t test in Figure 4A. IgG4 reads in single-versus multi-subclass lineages were compared via the Mann-Whitney U test in Figure 4D.
Supplementary Material
Highlights.
High-throughput methods identify antigen- and subclass-specific B cell repertoires
Most autoreactive IgG4 B cells lack clonal relationships to other subclasses
IgG4 lineages acquire autoreactivity early in development
IgA1 evolves from IgG1 and to IgA2 but is not an origin of IgG4 autoreactivity
ACKNOWLEDGMENTS
This project was supported in part by Deutsche Forschungsgemeinschaft grant EL711/1–1 to C.T.E.; National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) grants T32-AR007465 and F30-AR065870 to E.M.M. and AR057001 and AR064220 to A.S.P.; and the Penn Skin Biology and Diseases Resource-based Center (P30-AR069589). The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIAMS or the NIH.
Footnotes
DECLARATION OF INTERESTS
A.S.P. and C.T.E. are inventors on patents related to cellular immunotherapy of pemphigus vulgaris and have equity interest in Cabaletta Bio, focused on targeted cellular immunotherapy of pemphigus vulgaris. A.S.P. serves on the Scientific Advisory Board of Cabaletta Bio.
DATA AND SOFTWARE AVAILABILITY
The accession number for the data reported in this paper is NCBI: PRJNA437136 in the BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/).
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, ten figures, and nine tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.07.093.
REFERENCES
- Aalberse RC, and Schuurman J (2002). IgG4 breaking the rules. Immunology 105, 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aalberse RC, van der Gaag R, and van Leeuwen J (1983). Serologic aspects of IgG4 antibodies. I. Prolonged immunization results in an IgG4-restricted response. J. Immunol 130, 722–726. [PubMed] [Google Scholar]
- Anhalt GJ, Till GO, Diaz LA, Labib RS, Patel HP, and Eaglstein NF (1986). Defining the role of complement in experimental pemphigus vulgaris in mice. J. Immunol 137, 2835–2840. [PubMed] [Google Scholar]
- Asashima N, Fujimoto M, Watanabe R, Nakashima H, Yazawa N, Okochi H, and Tamaki K (2006). Serum levels of BAFF are increased in bullous pemphigoid but not in pemphigus vulgaris. Br. J. Dermatol 155, 330–336. [DOI] [PubMed] [Google Scholar]
- Ayatollahi M, Joubeh S, Mortazavi H, Jefferis R, and Ghaderi A (2004). IgG4 as the predominant autoantibody in sera from patients with active state of pemphigus vulgaris. J. Eur. Acad. Dermatol. Venereol. 18, 241–242. [DOI] [PubMed] [Google Scholar]
- Bhol K, Mohimen A, and Ahmed AR (1994). Correlation of subclasses of IgG with disease activity in pemphigus vulgaris. Dermatology (Basel) 189 (Suppl 1), 85–89. [DOI] [PubMed] [Google Scholar]
- Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, and Shapiro L (2002). C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science 296, 1308–1313. [DOI] [PubMed] [Google Scholar]
- Cerutti A (2008). The regulation of IgA class switching. Nat. Rev. Immunol 8, 421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman DG (1954). The Estimation of Biological Populations. Ann. Math. Statist 25, 1–15. [Google Scholar]
- Chen J, Zheng Q, Hammers CM, Ellebrecht CT, Mukherjee EM, Tang HY, Lin C, Yuan H, Pan M, Langenhan J, et al. (2017). Proteomic Analysis of Pemphigus Autoantibodies Indicates a Larger, More Diverse, and More Dynamic Repertoire than Determined by B Cell Genetics. Cell Rep. 18, 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CH, and Chao A (2016). Estimating and comparing microbial diversity in the presence of sequencing errors. PeerJ 4, e1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MJ, Lo AS, Mao X, Nagler AR, Ellebrecht CT, Mukherjee EM, Hammers CM, Choi EJ, Sharma PM, Uduman M, et al. (2014). Shared VH1–46 gene usage by pemphigus vulgaris autoantibodies indicates common humoral immune responses among patients. Nat. Commun 5, 4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MJ, Ellebrecht CT, and Payne AS (2015). The dual nature of interleukin-10 in pemphigus vulgaris. Cytokine 73, 335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MJ, Ellebrecht CT, Hammers CM, Mukherjee EM, Sapparapu G, Boudreaux CE, McDonald SM, Crowe JE Jr., and Payne AS (2016). Determinants of VH1–46 cross-reactivity to pemphigus vulgaris autoantigen desmoglein 3 and rotavirus antigen VP6. J. Immunol 197, 1065–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colliou N, Picard D, Caillot F, Calbo S, Le Corre S, Lim A, Lemercier B, Le Mauff B, Maho-Vaillant M, Jacquot S, et al. (2013). Long-term remissions of severe pemphigus after rituximab therapy are associated with prolonged failure of desmoglein B cell response. Sci. Transl. Med 5, 175ra30. [DOI] [PubMed] [Google Scholar]
- D’Auria L, Bonifati C, Mussi A, D’Agosto G, De Simone C, Giacalone B, Ferraro C, and Ameglio F (1997). Cytokines in the sera of patients with pemphigus vulgaris: interleukin-6 and tumour necrosis factor-alpha levels are significantly increased as compared to healthy subjects and correlate with disease activity. Eur. Cytokine Netw 8, 383–387. [PubMed] [Google Scholar]
- Edgar RC (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1989). PHYLIP - Phylogeny Inference Package (Version 3.2). Cladistics 5, 164–166. [Google Scholar]
- Funakoshi T, Lunardon L, Ellebrecht CT, Nagler AR, O’Leary CE, and Payne AS (2012). Enrichment of total serum IgG4 in patients with pemphigus. Br. J. Dermatol 167, 1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futei Y, Amagai M, Ishii K, Kuroda-Kinoshita K, Ohya K, and Nishikawa T (2001). Predominant IgG4 subclass in autoantibodies of pemphigus vulgaris and foliaceus. J. Dermatol. Sci 26, 55–61. [DOI] [PubMed] [Google Scholar]
- Giordano CN, and Sinha AA (2012). Cytokine networks in pemphigus vulgaris: An integrated viewpoint. Autoimmunity 45, 427–439. [DOI] [PubMed] [Google Scholar]
- Gupta NT, Vander Heiden JA, Uduman M, Gadala-Maria D, Yaari G, and Kleinstein SH (2015). Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31, 3356–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison OJ, Brasch J, Lasso G, Katsamba PS, Ahlsen G, Honig B, and Shapiro L (2016). Structural basis of adhesive binding by desmocollins and desmogleins. Proc. Natl. Acad. Sci. USA 113, 7160–7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Xu W, Santini PA, Polydorides AD, Chiu A, Estrella J, Shan M, Chadburn A, Villanacci V, Plebani A, et al. (2007). Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelialcell secretion of the cytokine APRIL. Immunity 26, 812–826. [DOI] [PubMed] [Google Scholar]
- Hershberg U, and Luning Prak ET (2015). The analysis of clonal expansions in normal and autoimmune B cell repertoires. Philos. Trans. R. Soc. Lond. B Biol. Sci 370, 20140239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertl M, Eming R, and Veldman C (2006). T cell control in autoimmune bullous skin disorders. J. Clin. Invest 116, 1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoh RA, Joshi SA, Liu Y, Wang C, Roskin KM, Lee JY, Pham T, Looney TJ, Jackson KJL, Dixit VP, et al. (2016). Single B-cell deconvolution of peanut-specific antibody responses in allergic patients. J. Allergy Clin. Immunol 137, 157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horns F, Vollmers C, Croote D, Mackey SF, Swan GE, Dekker CL, Davis MM, and Quake SR (2016). Lineage tracing of human B cells reveals the in vivo landscape of human antibody class switching. eLife 5, e16578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TC, Ma KH, and Chao A (2016). iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol. Evol 7, 1451–1456. [Google Scholar]
- Huijbers MG, Querol LA, Niks EH, Plomp JJ, van der Maarel SM, Graus F, Dalmau J, Illa I, and Verschuuren JJ (2015). The expanding field of IgG4-mediated neurological autoimmune disorders. Eur. J. Neurol 22, 1151–1161. [DOI] [PubMed] [Google Scholar]
- Hussain R, Poindexter RW, and Ottesen EA (1992). Control of allergic reactivity in human filariasis. Predominant localization of blocking antibody to the IgG4 subclass. J. Immunol 148, 2731–2737. [PubMed] [Google Scholar]
- Jackson KJ, Liu Y, Roskin KM, Glanville J, Hoh RA, Seo K, Marshall EL, Gurley TC, Moody MA, Haynes BF, et al. (2014). Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell Host Microbe 16, 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeannin P, Lecoanet S, Delneste Y, Gauchat JF, and Bonnefoy JY (1998). IgE versus IgG4 production can be differentially regulated by IL-10. J. Immunol. 160, 3555–3561. [PubMed] [Google Scholar]
- Jiang D, Ge J, Liao Q, Ma J, Liu Y, Huang J, Wang C, Xu W, Zheng J, Shao W, et al. (2015). IgG and IgA with potential microbial-binding activity are expressed by normal human skin epidermal cells. Int. J. Mol. Sci 16, 2574–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly P, Maho-Vaillant M, Prost-Squarcioni C, Hebert V, Houivet E, Calbo S, Caillot F, Golinski ML, Labeille B, Picard-Dahan C, et al. ; French study group on autoimmune bullous skin diseases (2017). First-line rituximab combined with short-term prednisone versus prednisone alone for the treatment of pemphigus (Ritux 3): a prospective, multicentre, parallelgroup, open-label randomised trial. Lancet 389, 2031–2040. [DOI] [PubMed] [Google Scholar]
- Jutel M, Jaeger L, Suck R, Meyer H, Fiebig H, and Cromwell O (2005). Allergen-specific immunotherapy with recombinant grass pollen allergens. J. Allergy Clin. Immunol. 116, 608–613. [DOI] [PubMed] [Google Scholar]
- Kasperkiewicz M, Ellebrecht CT, Takahashi H, Yamagami J, Zillikens D, Payne AS, and Amagai M (2017). Pemphigus. Nat. Rev. Dis. Primers 3, 17026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looney TJ, Lee JY, Roskin KM, Hoh RA, King J, Glanville J, Liu Y, Pham TD, Dekker CL, Davis MM, et al. (2016). Human B-cell isotype switching origins of IgE. J. Allergy Clin. Immunol. 137, 579–586.e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascaró JM Jr., Espan˜ a, A., Liu Z, Ding X, Swartz SJ, Fairley JA, and Diaz LA (1997). Mechanisms of acantholysis in pemphigus vulgaris: role of IgG valence. Clin. Immunol. Immunopathol. 85, 90–96. [DOI] [PubMed] [Google Scholar]
- Mentink LF, de Jong MC, Kloosterhuis GJ, Zuiderveen J, Jonkman MF, and Pas HH (2007). Coexistence of IgA antibodies to desmogleins 1 and 3 in pemphigus vulgaris, pemphigus foliaceus and paraneoplastic pemphigus. Br. J. Dermatol 156, 635–641. [DOI] [PubMed] [Google Scholar]
- Morris EK, Caruso T, Buscot F, Fischer M, Hancock C, Maier TS, Meiners T, Mu€ller C, Obermaier E, Prati D, et al. (2014). Choosing and using diversity indices: insights for ecological applications from the German Biodiversity Exploratories. Ecol. Evol 4, 3514–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishifuji K, Amagai M, Kuwana M, Iwasaki T, and Nishikawa T (2000). Detection of antigen-specific B cells in patients with pemphigus vulgaris by enzyme-linked immunospot assay: requirement of T cell collaboration for autoantibody production. J. Invest. Dermatol 114, 88–94. [DOI] [PubMed] [Google Scholar]
- Ohyama B, Nishifuji K, Chan PT, Kawaguchi A, Yamashita T, Ishii N, Hamada T, Dainichi T, Koga H, Tsuruta D, et al. (2012). Epitope spreading is rarely found in pemphigus vulgaris by large-scale longitudinal study using desmoglein 2-based swapped molecules. J. Invest. Dermatol 132, 1158–1168. [DOI] [PubMed] [Google Scholar]
- Pappas L, Foglierini M, Piccoli L, Kallewaard NL, Turrini F, Silacci C, Fernandez-Rodriguez B, Agatic G, Giacchetto-Sasselli I, Pellicciotta G, et al. (2014). Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature 516, 418–422. [DOI] [PubMed] [Google Scholar]
- Payne AS, Ishii K, Kacir S, Lin C, Li H, Hanakawa Y, Tsunoda K, Amagai M, Stanley JR, and Siegel DL (2005). Genetic and functional characterization of human pemphigus vulgaris monoclonal autoantibodies isolated by phage display. J. Clin. Invest 115, 888–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Jeong JS, Ye J, Dang B, Abdeladhim M, Aoki V, Hans-Filhio G, Rivitti EA, Valenzuela JG, and Diaz LA (2016). Overlapping IgG4 Responses to Self- and Environmental Antigens in Endemic Pemphigus Foliaceus. J. Immunol 196, 2041–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaeth S, Riechers R, Borradori L, Zillikens D, Budinger L, and Hertl M (2001). IgG, IgA and IgE autoantibodies against the ectodomain of desmoglein 3 in active pemphigus vulgaris. Br. J. Dermatol 144, 1183–1188. [DOI] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JEJ, and Schrader CE (2008). Mechanism and regulation of class switch recombination. Annu. Rev. Immunol 26, 261–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Amagai M, Tanikawa A, Suzuki S, Ikeda Y, Nishikawa T, Kawakami Y, and Kuwana M (2007). T helper type 2-biased natural killer cell phenotype in patients with pemphigus vulgaris. J. Invest. Dermatol 127, 324–330. [DOI] [PubMed] [Google Scholar]
- Tian C, Luskin GK, Dischert KM, Higginbotham JN, Shepherd BE, and Crowe JE Jr. (2008). Immunodominance of the VH1–46 antibody gene segment in the primary repertoire of human rotavirus-specific B cells is reduced in the memory compartment through somatic mutation of nondominant clones. J. Immunol 180, 3279–3288. [DOI] [PubMed] [Google Scholar]
- Tipton CM, Fucile CF, Darce J, Chida A, Ichikawa T, Gregoretti I, Schieferl S, Hom J, Jenks S, Feldman RJ, et al. (2015). Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nat. Immunol 16, 755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Veen W, Stanic B, Yaman G, Wawrzyniak M, Sollner S, Akdis DG, Ru€ckert B, Akdis CA, and Akdis M (2013). IgG4 production is confined to human IL-10-producing regulatory B cells that suppress antigen-specific immune responses. J. Allergy Clin. Immunol 131, 1204–1212. [DOI] [PubMed] [Google Scholar]
- Veldman C, Stauber A, Wassmuth R, Uter W, Schuler G, and Hertl M (2003). Dichotomy of autoreactive Th1 and Th2 cell responses to desmoglein 3 in patients with pemphigus vulgaris (PV) and healthy carriers of PV-associated HLA class II alleles. J. Immunol 170, 635–642. [DOI] [PubMed] [Google Scholar]
- von Büdingen HC, Kuo TC, Sirota M, van Belle CJ, Apeltsin L, Glanville J, Cree BA, Gourraud PA, Schwartzburg A, Huerta G, et al. (2012). B cell exchange across the blood-brain barrier in multiple sclerosis. J. Clin. Invest 122, 4533–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, and Nussenzweig MC (2003). Predominant autoantibody production by early human B cell precursors. Science 301, 1374–1377. [DOI] [PubMed] [Google Scholar]
- Warren SJ, Arteaga LA, Rivitti EA, Aoki V, Hans-Filho G, Qaqish BF, Lin MS, Giudice GJ, and Diaz LA (2003). The role of subclass switching in the pathogenesis of endemic pemphigus foliaceus. J. Invest. Dermatol 120, 104–108. [DOI] [PubMed] [Google Scholar]
- Yamagami J, Payne AS, Kacir S, Ishii K, Siegel DL, and Stanley JR (2010). Homologous regions of autoantibody heavy chain complementaritydetermining region 3 (H-CDR3) in patients with pemphigus cause pathogenicity. J. Clin. Invest 120, 4111–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuvaraj S, Dijkstra G, Burgerhof JG, Dammers PM, Stoel M, Visser A, Kroese FG, and Bos NA (2009). Evidence for local expansion of IgA plasma cell precursors in human ileum. J. Immunol. 183, 4871–4878. [DOI] [PubMed] [Google Scholar]
- Zan H, Cerutti A, Dramitinos P, Schaffer A, and Casali P (1998). CD40 engagement triggers switching to IgA1 and IgA2 in human B cells through induction of endogenous TGF-beta: evidence for TGF-beta but not IL-10dependent direct S mu/S alpha and sequential S mu/S gamma, S gamma/S alpha DNA recombination. J. Immunol 161, 5217–5225. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kobert K, Flouri T, and Stamatakis A (2014). PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu HQ, Xu RC, Chen YY, Yuan HJ, Cao H, Zhao XQ, Zheng J, Wang Y, and Pan M (2015). Impaired Function of CD19 CD24 CD38 Regulatory B Cells in Pemphigus Patients. Br. J. Dermatol 172, 101–110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.