Summary
Circular RNAs (circRNAs) are a new class of RNAs that can be used as biomarkers in clinical blood samples. However, little is known about circRNAs' diagnostic values for rheumatoid arthritis (RA). In this study, the hsa_circ_0054189, hsa_circ_0008675, hsa_circ_0082689, hsa_circ_0082688, hsa_circ_0010932, hsa_circ_0002473 and hsa_circ_0044235 in peripheral blood were determined by quantitative reverse transcription–polymerase chain reaction (qRT–PCR). For hsa_circ_0044235, only one abnormal expression circRNAs in peripheral blood was selected as a targeted circRNA to explore the diagnostic value for RA. Our work demonstrated that the hsa_circ_0044235 in peripheral blood was decreased significantly in RA patients. The hsa_circ_0044235 in peripheral blood from RA patients did not correlate with C‐reactive protein (CRP), erythrocyte sedimentation rate (ESR), rheumatoid factor (RF), anti‐citrullinated protein antibodies (ACPA) or disease activity score 28 (DAS28). Receiver operating characteristic (ROC) curve analysis suggested that the hsa_circ_0044235 in peripheral blood has significant value in the diagnosis of RA. The risk score based on hsa_circ_0044235 in peripheral blood also distinguished significantly the patients with RA from systemic lupus erythematosus (SLE). This study suggests that the hsa_circ_0044235 in peripheral blood may be a potential biomarker of patients with RA.
Keywords: biomarker, circular RNAs, rheumatoid arthritis
Introduction
Rheumatoid arthritis (RA) is a chronic debilitating systemic autoimmune disease characterized by inflammation and destruction of the joints. Approximately 1% of the population suffers from RA, and many patients develop long‐term joint damage, severe illness and disability 1. Early diagnosis and proper treatment can relieve the pathogenetic condition effectively in RA patients. Current diagnostic methods, including American College of Rheumatology (ACR) classification criteria, show various disadvantages for the early diagnosis of RA. Therefore, new biomarkers aiming to improve the diagnosis and prognosis analysis of RA will be highly valuable.
Although great efforts have been made 2, 3, 4, 5, the detailed mechanisms of RA pathogenesis and development are still unknown. Thus, in‐depth study on the genetic and molecular abnormality of RA is necessary for the screening of new biomarkers. More than 80% of the genome in humans is transcribed into RNA transcripts without protein‐coding potential 6. However, recent studies have revealed that although non‐coding RNAs do not code directly for proteins, they play a regulatory role in the transcription and translation of protein‐coding genes 7. According to the transcript structure, these so‐called non‐coding RNAs (ncRNAs) are divided loosely into two major classes: linear RNA, such as microRNAs (miRNAs), long non‐coding RNAs (lncRNAs) and circular RNAs (circRNAs). Previous studies have shown that miRNAs and lncRNA are involved in the pathogenesis of RA and can be used as biomarkers for the diagnosis of RA 8, 9, 10, 11. Increasing evidence reveals that circRNAs are used widely in various physiological and pathological conditions, such as atherosclerotic vascular diseases, neurological disorders, prion infection and cancer 12, 13, 14, 15. However, little is known about the roles of circRNAs in human RA.
Recently, two studies have revealed that circRNAs involved in the pathogenesis of RA 16, 17 and hsa_circ_104871 in peripheral blood mononuclear cells (PBMCs) could be used as a potential biomarker of RA 16. In one of our previous studies we found some differentially expressed circRNAs in the peripheral blood from SLE patients by circRNA microarray screening, which suggests that circRNAs might play a role in autoimmune diseases. Considering that both SLE and RA are common autoimmune diseases, some differentially expressed circRNAs between SLE and HCs were selected to investigate the possibility of being used as biomarkers for diagnosis of SLE and RA. In this study, hsa_circ_0044235 in peripheral blood was found to have potential for use as a new biomarker for RA diagnosis.
Materials and methods
Patient variables
A total of 77 patients who fulfilled the revised ACR criteria for RA 18 were recruited from the First Affiliated Hospital of Nanchang University from January to September 2017. Among these, eight patients were new‐onset rheumatoid arthritis (< 6 months of disease duration) 19. RA disease activity was measured using the disease activity score 28 (DAS28) 20. All patients received therapy with disease‐modifying anti‐rheumatic drugs (DMARDs). The patient characteristics of RA patients are shown in Table 1. In addition, this study included 50 healthy controls (HC) who were free from autoimmune or inflammatory diseases and who were unrelated to the patients. As an autoimmune disease control, 31 SLE patients fulfilled the revised ACR criteria for SLE 21 were also enrolled from the First Affiliated Hospital of Nanchang University from January to September 2017. All study protocols were approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University (no. 019). All provided signed informed consent.
Table 1.
Clinical characters and laboratory measures of rheumatoid arthritis (RA)
| Age (year), mean (s.d.) | 54·99 ± 12·58 |
|---|---|
| Sex (female/male) | 63/14 |
| DAS28, mean (s.d.) | 5·09 ± 1·43 |
| ESR (mm/h), mean (s.d.) | 57·88 ± 39·22 |
| CRP (mg/l), mean (s.d.) | 34·62 ± 43·30 |
| RF, positive (70 patients) | 43 |
| ACPA (RU/ml), mean (s.d.) (68 patients) | 303·04 ± 428·83 |
| WBC (109/l), mean (s.d.) | 7·29 ± 2·53 |
| RBC (1012/l), mean (s.d.) | 3·91 ± 0·61 |
| HGB (g/l), mean (s.d.) | 110·38 ± 20·71 |
| HCT (l/l), mean (s.d.) | 0·34 ± 0·07 |
| PLT (109/l), mean (s.d.) | 290·02 ± 133·06 |
| Lymphocytes (109/l), mean (s.d.) | 1·64 ± 0·78 |
| Lymphocytes (%), mean (s.d.) | 23·55 ± 9·89 |
| Monocytes (109/l), mean (s.d.) | 0·44 ± 0·20 |
| Monocytes (%), mean (s.d.) | 6·16 ± 2·54 |
| Neutrophils (109/l), mean (s.d.) | 5·13 ± 2·24 |
| Neutrophils (%), mean (s.d.) | 68·91 ± 11·16 |
ACPA = anti‐cyclic citrullinated peptide; DAS28 = disease activity score 28; CRP = C‐reactive protein; ESR = erythrocyte sedimentation rate; HCT = haematocrit; HGB = haemoglobin; Pglyrp1/PGLYRP‐1 = peptidoglycan recognition protein‐1; PLT = platelets; RBC = red blood cell; RF = rheumatoid factor; WBC = white blood cell.
Collection of peripheral blood samples and total RNA extraction
Peripheral blood samples (2 ml) were drawn from the median cubital vein with an ethylenediamine tetraacetic acid (EDTA) anti‐coagulated vacutainer and total RNA was extracted as soon as possible. Total RNA was extracted from 250 l blood using Trizol reagent, according to the manufacturer's instructions. The concentration and quality of the RNA were assessed by absorbance spectrometry measuring absorbance ratios of A260/A280 and A 260/A230 using a NanoDrop ND‐1000 spectrophotomete (Agilent, Santa Clara, CA, USA).
Quantitative reverse transcription–polymerase chain reaction (qRT–PCR) analysis
Reverse transcription (RT) reaction was performed using a PrimeScript™ RT reagent kit (Takara Bio Inc, Japan). The qPCR was then performed on an ABI 7500 Real‐time PCR System (Applied Biosystems, Thermo Fisher Scientific, Inc., Waltham, MA, USA) following the manufacturer's instructions. The primers used in qRT–PCR are shown in Supporting information, Table 1. ‐action was used as an internal control. The 2–∆Ct method was used to analyse the data.
ESR, CRP and autoantibody measurement
Erythrocyte sedimentation rate (ESR) was determined according to the instructions described by the manufacturer. C‐reactive protein (CRP) and rheumatoid factor (RF) were measured by nephelometry. Anti‐citrullinated protein antibodies (ACPA) of immunoglobulin (Ig)G in serum were measured by commercially available enzyme‐linked immunosorbent assay (ELISA) kits (Kexin, Shanghai, China).
Statistical analysis
Statistical analysis and graphic presentation were carried out with GraphPad Prism version 5.0 (GraphPad Software, San Diego, CA, USA) and spss version 16.0 (SPSS Inc., Chicago, IL, US). Student's t‐test was used where the normality test passed; otherwise, the non‐parametric Mann–Whitney test was used to analyse the data. Similarly, the Pearson method or the non‐parametric Spearman's method was used for correlation analysis. Receiver operating characteristic (ROC) curves were carried out to assess the diagnostic value of dysregulated circRNAs. A value of P < 0·05 was considered as the significant difference.
Results
Screening of abnormal expression circRNA in peripheral blood from RA patients and HC
The seven selected circRNA (hsa_circ_0054189, hsa_circ_0008675, hsa_circ_0082689, hsa_circ_0082688, hsa_circ_0010932, hsa_circ_0002473 and hsa_circ_0044235) were first tested in peripheral blood from 31 RA patients and 31 HC by qRT–PCR. Compared with HC, the expression level of hsa_circ_0044235 in peripheral blood from RA patients was down‐regulated significantly (P < 0·0001), while the expression of hsa_circ_0054189, hsa_circ_0008675, hsa_circ_0082689, hsa_circ_0082688, hsa_circ_0010932 and hsa_circ_0002473 in peripheral blood did not show any remarkable differences between RA patients and the HC (Fig. 1).
Figure 1.

Quantitative reverse transcription–polymerase chain reaction (qRT–PCR) determined the relative expression levels of circular RNAs (circRNAs) in peripheral blood from rheumatoid arthritis (RA) patients and healthy controls (HC). (a) The average expression of hsa_circ_0010932 did not show any remarkable differences between patients with RA and the HC. (b) The average expression of hsa_circ_0002473 did not show any remarkable differences between patients with RA and the HC. (c) The average expression levels of hsa_circ_0082688 did not show any remarkable differences between patients with RA and the HC. (d) The average expression levels of hsa_circ_0082689 did not show any remarkable differences between patients with RA and the HC. (e) The average expression levels of hsa_circ_0054189 did not show any remarkable differences between patients with RA and the HC. (f) The average expression levels of hsa_circ_0008675 did not show any remarkable differences between patients with RA and the HC. (g) The average expression of hsa_circ_0044235 in patients with RA was significantly more decreased than those of the HC.
Validation the of hsa_circ_0044235 in the second stage in peripheral blood from RA patients and HC
To verify the results of hsa_circ_0044235 in peripheral blood identified in the screening stage, we conducted a validation study in an independent cohort including 46 RA patients and 19 HC. From the data of all the RA patients and HC, the expression level of hsa_circ_0044235 in peripheral blood from 77 RA patients was decreased significantly compared to 50 HC (Fig. 2).
Figure 2.

The expression level of hsa_circ_0044235 in peripheral blood from 77 rheumatoid arthritis (RA) patients was decreased significantly compared to 50 healthy controls (HC).
Potential diagnostic values of hsa_circ_0044235 in peripheral blood in RA
Next, we performed an analysis to evaluate the potential diagnostic value of hsa_circ_0044235 in peripheral blood, and we established a ROC curve. The area under the ROC curve (AUC) was up to 0·779 [95% confidence interval (CI) = 0·700–0·859; P < 0·0001] (Fig. 3). The sensitivity and specificity were 61·04 and 90·0%, respectively. We speculated that the hsa_circ_0044235 in peripheral blood might be used as a biomarker of RA.
Figure 3.

Receiver operating characteristic (ROC) analysis of hsa_circ_0044235 in peripheral blood from rheumatoid arthritis (RA) patients. AUC = area under the curve.
To determine whether the hsa_circ_0044235 in peripheral blood from RA patients were relevant biomarkers for the severity of RA, the correlation test was analysed to evalute the relationship between the clinical features of RA and the level of hsa_circ_0044235. However, the level of hsa_circ_0044235 in peripheral blood from patients with RA did not correlate with DAS28, CRP, ESR, RF or ACPA, which indicates the severity of the disease (data no shown).
Subsequently, we compared the hsa_circ_0044235 in peripheral blood between new‐ and late‐onset RA patients. Results showed that hsa_circ_0044235 in peripheral blood was decreased in new‐onset compared to late‐onset RA patients, but the difference was not statistically significant (Fig. 4).
Figure 4.

The expression of hsa_circ_0044235 in peripheral blood did not show any remarkable differences between new‐ and late‐onset rheumatoid arthritis (RA) patients.
hsa_circ_0044235 in peripheral blood in patients with RA and SLE
Compared to SLE patients, the level of hsa_circ_0044235 in peripheral blood was clearly decreased in RA patients (P = 0·0003) (Fig. 5).
Figure 5.
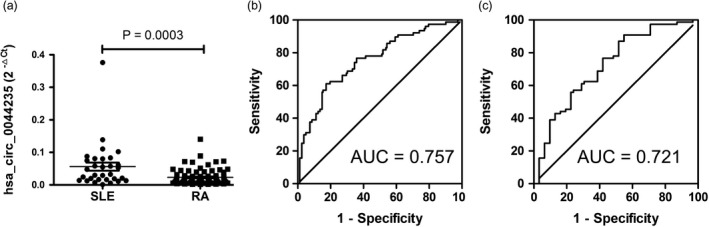
Receiver operating characteristic (ROC) curve analysis of the risk‐score of hsa_circ_0044235 in peripheral blood. A The expression level of hsa_circ_0044235 in peripheral blood in rheumatoid arthritis (RA) patients was decreased significantly compared to those of the systemic lupus erythematosus (SLE) controls. (b) ROC curve analysis hsa_circ_0044235 in peripheral blood for the risk‐score in RA patients versus all controls (HC and SLE). (c) ROC curve analysis of hsa_circ_0044235 in peripheral blood for the risk‐score in RA patients versus SLE patients. AUC = area under the curve;
Subsequently, a risk score based on hsa_circ_0044235 in peripheral blood was analysed in RA patients and all controls (HC+SLE); the AUC for the risk score was 0·757 (95% CI = 0·682–0·832; P < 0·0001) (Fig. 5). The sensitivity and specificity were 61·04 and 82·72%, respectively. The risk score also distinguished RA patients from SLE patients significantly, and the AUC was 0·721 (95% CI = 0·612–0·831; P = 0·0003) (Fig. 5). The sensitivity and specificity were 90·91 and 45·16%, respectively.
MiRNAs targets of hsa_circ_0044235 prediction
To determine the function of hsa_circ_0044235, we predicted the target miRNA by aligning with the miRNA response elements (MREs) of hsa_circ_0044235 using miRanda software. Three putative miRNAs targets, hsa‐miR‐892a, hsa‐miR‐135b‐5p and hsa‐miR‐135a‐5p, were found. As shown in Fig. 6, the expression of hsa‐miR‐892a in peripheral blood was increased significantly in patients of RA than HC. However, the expression of hsa‐miR‐135b‐5p and hsa‐miR‐135a‐5p did not show any remarkable differences between patients with RA and HC.
Figure 6.

Quantitative reverse transcription–polymerase chain reaction (qRT–PCR) determined the relative expression levels of miRNAs in peripheral blood from rheumatoid arthritis (RA) patients and healthy controls (HC). (a) The expression of hsa‐miR‐892a in peripheral blood was increased significantly in RA patients than HC. (b) The expression of hsa‐miR‐135b‐5p did not show any remarkable differences between patients with RA and HC. (c) The expression of hsa‐miR‐135a‐5p did not show any remarkable differences between patients with RA and HC.
Discussion
CircRNAs are a special class of endogenous RNAs. Recent studies have revealed that abnormal circRNAs are associated with several human diseases, such as atherosclerotic vascular disease, cancer and autoimmune disease 22, 23, 24, 25, 26. Many dysregulated expression circRNAs are demonstrated to associate with RA 16, 17. Zheng et al. found that the levels of circRNAs (104194, 104593, 103334, 101407 and 102594) in PBMCs were expressed differentially in RA patients versus HC 17. Ouyang et al. showed that the expression of hsa_circRNAs_092516, hsa_circRNAs_003524, hsa_circRNAs_103047, hsa_circRNAs_104871 and hsa_circRNAs_101873 was up‐regulated significantly in PBMCs from RA patients 16. However, little was known about the expression of circRNAs in peripheral blood in RA. In the present study, we we utilized qRT–PCR to detect the expression of circRNAs (hsa_circ_0054189, hsa_circ_0008675, hsa_circ_0082689, hsa_circ_0082688, hsa_circ_0010932, hsa_circ_0002473 and hsa_circ_0044235) in peripheral blood in RA patients and HC, and only hsa_circ_0044235 in peripheral blood was decreased significantly in RA patients. The results indicated that hsa_circ_0044235 in peripheral blood associate with RA.
CircRNAs are a type of closed circular RNAs that do not have 5– or 3– ends; they are free of exonuclease‐mediated degradation and more stable than most linear RNAs 27. Also, circRNA often show tissue/developmental stage‐specific expression 28. These characteristics give circRNAs the potential of being ideal biomarkers for various diseases. For example, Zhao et al. 29 found that hsa_circ_0054633 in peripheral blood shows promise as a candidate biomarker for prediabetes and type 2 diabetes mellitus; Zhao et al. 30 identified that hsa_circ_0124644 in peripheral blood could be used as a new biomarker of coronary artery disease; and Ouyang et al. 16 determined circRNA_104871 in PBMCs as a potential biomarker of RA. However, the diagnostic value of peripheral blood circRNAs in RA has never been explored. Our results showed the AUC of hsa_circ_0044235 in peripheral blood was up to 0·779. The risk score based on hsa_circ_0044235 in peripheral blood also demonstrated that it can distinguish RA patients effectively from SLE patients. This indicated that hsa_circ_0044235 in peripheral blood has potential as a diagnostic biomarker of RA.
In accordance with other reports showing that the expression of circRNAs was not correlated with biomarkers for disease severity 16, our study found that the hsa_circ_0044235 in peripheral blood of RA patients was not correlated with DAS28, CRP, ESR, RF or ACPA, which mirrors the severity of RA 31, 32. These results indicated that the hsa_circ_0044235 in peripheral blood may not be used as markers for systemic inflammation or disease activity in RA.
Bioinformation analysis showed that hsa_circ_0044235 locates at chr17:45247282‐45259003 and is spliced from cell division cycle 27 (cdc27) gene. It was reported that CDC27 might play a role in controlling the timing of mitosis by interacting with mitotic checkpoint proteins, including mitotic arrest deficient (Mad2), p55CDC and butyl bromide (BUBR) 34. However, to our knowledge, no study on the role of CDC27 in RA has been published until now. Our preliminary study showed that the expression of cdc27 in peripheral blood was decreased significantly in RA patients when compared to HC (data not shown). However, the detailed role of CDC27 in RA needs further investigation.
It was reported that circRNAs could function as miRNA ‘sponges’, and thus naturally sequester or competitively suppress the activity of target miRNA 35. To investigate the function of hsa_circ_0044235, the potential miRNA targets of hsa_circ_0044235 were predicted by using Arraystar's miRNA target prediction software. Three putative miRNAs targets of hsa_circ_0044235, hsa‐miR‐892a, hsa‐miR‐135b‐5p and hsa‐miR‐135a‐5p were found, and their levels in peripheral blood of RA patients and HCs were detected and compared. The data showed that the levels of hsa‐miR‐892a in peripheral blood were increased significantly in RA patients, suggesting that hsa_circ_0044235 might play a role in RA by interacting with hsa‐miR‐892a. However, further research is needed to confirm this hypothesis.
There were many limitations in this research. First, because of the relatively small sample size of new‐onset RA patients, the study was restricted by the number of subjects from only one hospital, which may limit the universality of our results. Secondly, because of lack of information on RF and ACPA in HC, we were unable to assess the values of hsa_circ_0044235 in peripheral blood plus RF/ACPA in RA diagnosis. Thirdly, we did not investigate the exact role of hsa_circ_0044235 in RA pathogenesis.
Conclusions
For the first time, to our knowledge, our study detected circRNA expression in peripheral blood from RA patients and from HC, and indicated that the hsa_circ_0044235 in peripheral blood from RA was decreased. In addition, our results provides novel empirical evidence that the hsa_circ_0044235 in peripheral blood may specifically identify patients with RA and could provide better diagnostic accuracy.
Disclosure
The authors declare no conflicts of interest.
Supporting information
Table 1. Specific circRNA primers used for quantitative qRTPCR
Acknowledgements
We would like to acknowledge the help from Dr Rui Wu at the Department of Rheumatology, The First Affiliated Hospital of Nanchang University, Nanchang, Jiangxi, China. This work was supported by the National Natural Science Foundation of China (81360459), Jiangxi Provincial Natural Science Foundation of China (20151BAB215031, 20171BAB205113), the Science and Technology Project of Health and Family Planning Commission of Jiangxi Province of China (20165094) and the Foundation for Distinguished Young Scientists of Jiangxi Province of China (20171BCB23087)
Contributor Information
Z. Huang, Email: 491353062@qq.com
J. Li, Email: lisir361@163.com
References
- 1. Goekoop‐Ruiterman YP, Huizinga TW. Rheumatoid arthritis: can we achieve true drug‐free remission in patients with RA? Nat Rev Rheumatol 2010; 6:68–70. [DOI] [PubMed] [Google Scholar]
- 2. Luo Q, Deng Z, Xu C et al Elevated expression of immunoreceptor tyrosine‐based inhibitory motif (TIGIT) on T lymphocytes is correlated with disease activity in rheumatoid arthritis. Med Sci Monitor 2017; 10:1232–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luo Q, Zeng L, Mei H et al PD‐L1‐expressing neutrophils as a novel indicator to assess disease activity of rheumatoid arthritis. Int J Clin Exp Med 2017; 10:7716–24. [Google Scholar]
- 4. Huang SH, Liu GW, Li JH et al Expression of TREM‐2 and its inhibitory effects on TNF‐α induced inflammation in fibroblast‐like synoviocytes via inhibiting p38 pathway activation. Clin Exp Rheumatol 2018;36:185–94. [PubMed] [Google Scholar]
- 5. Sujitha S, Rasool M. MicroRNAs and bioactive compounds on TLR/MAPK signaling in rheumatoid arthritis. Clin Chim Acta 2017; 24:106–15. [DOI] [PubMed] [Google Scholar]
- 6. The ENCODE Project Consortium . An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cech TR, Steitz JA. The noncoding RNA revolution‐trashing old rules to forge new ones. Cell 2014; 157:77–94. [DOI] [PubMed] [Google Scholar]
- 8. Abdul‐Maksoud RS, Sediq AM, Kattaia A et al Serum miR‐210 and miR‐155 expression levels as novel biomarkers for rheumatoid arthritis diagnosis. Br J Biomed Sci 2017;7:1–5. [DOI] [PubMed] [Google Scholar]
- 9. Wang W, Zhang Y, Zhu B et al Plasma microRNA expression profiles in Chinese patients with rheumatoid arthritis. Oncotarget 2015; 6:42557–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lu MC, Yu HC, Yu CL et al Increased expression of long noncoding RNAs LOC100652951 and LOC100506036 in T cells from patients with rheumatoid arthritis facilitates the inflammatory responses. Immunol Res 2016; 64:576–83. [DOI] [PubMed] [Google Scholar]
- 11. Zhang HJ, Wei QF, Wang SJ et al LncRNA HOTAIR alleviates rheumatoid arthritis by targeting miR‐138 and inactivating NF‐κB pathway. Int Immunopharmacol 2017; 50:283–90. [DOI] [PubMed] [Google Scholar]
- 12. Burd CE, Jeck WR, Liu Y et al Expression of linear and novel circular forms of an INK4/ARF‐associated non‐coding RNA correlates with atherosclerosis risk. PLOS Genet 2010;6: e1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR‐7 in cancer. Cancer Res 2013; 73:5609–12. [DOI] [PubMed] [Google Scholar]
- 14. Li F, Zhang L, Deng J et al Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β‐catenin pathway. Oncotarget 2015; 6:6001–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li LJ, Zhao W, Tao SS et al Competitive endogenous RNA network: potential implication for systemic lupus erythematosus. Exp Opin Ther Targets 2017; 21:639–48. [DOI] [PubMed] [Google Scholar]
- 16. Ouyang Q, Wu J, Jiang Z et al Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from rheumatoid arthritis patients. Cell Physiol Biochem 2017; 42:651–9. [DOI] [PubMed] [Google Scholar]
- 17. Zheng F, Yu X, Huang J, Dai Y. Circular RNA expression profiles of peripheral blood mononuclear cells in rheumatoid arthritis patients, based on microarray chip technology. Mol Med Rep 2017; 16:8029–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Arnett FC, Edworthy SM, Bloch DA et al The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum 1988; 31:315–24. [DOI] [PubMed] [Google Scholar]
- 19. Wang J, Shan Y, Jiang Z et al High frequencies of activated B cells and T follicular helper cells are correlated with disease activity in patients with new‐onset rheumatoid arthritis. Clin Exp Rheumatol. 2013; 174:212–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Prevoo ML, van't Hof MA, Kuper HH, van Leeuwen MA, van de Putte LB, van Riel PL. Modified disease activity scores that include twenty‐eight‐joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum 1995; 38:44–8. [DOI] [PubMed] [Google Scholar]
- 21. Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1997; 40:1725. [DOI] [PubMed] [Google Scholar]
- 22. Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF‐associated non‐coding RNA correlates with atherosclerosis risk. PLOS Genet 2010; 6:e1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR‐7 in cancer. Cancer Res 2013; 73:5609–12. [DOI] [PubMed] [Google Scholar]
- 24. Li F, Zhang L, Li W et al Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β‐catenin pathway. Oncotarget 2015; 6:6001–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zheng J, Li Z, Wang T. Microarray expression profile of circular RNAs in plasma from primary biliary cholangitis patients. Cell Physiol Biochem 2017; 44:1271–81. [DOI] [PubMed] [Google Scholar]
- 26. Cardamone G, Paraboschi EM, Rimoldi V, Duga S, Soldà G, Asselta R. The characterization of GSDMB splicing and backsplicing profiles identifies novel isoforms and a circular RNA that are dysregulated in multiple sclerosis. Int J Mol Sci 2017; 18:pii:E576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jeck WR, Sorrentino JA, Wang K et al Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19:141–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Memczak S, Jens M, Elefsinioti A et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495:333–8. [DOI] [PubMed] [Google Scholar]
- 29. Zhao Z, Li X, Jian D, Hao P, Rao L, Li M. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre‐diabetes and type 2 diabetes mellitus. Acta Diabetol 2017; 54:237–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao Z, Li X, Gao C et al Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep 2017; 7:39918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kedar MP, Acharya RV, Prakashini K. Performance of the 2010 American College of Rheumatology/European League against Rheumatism (ACR/EULAR) criteria for classification of rheumatoid arthritis in an Indian population: an observational study in a single centre. Ind J Med Res 2016; 144:288–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Falkenburg WJJ, Kempers AC, Dekkers G et al Rheumatoid factors do not preferentially bind to ACPA‐IgG or IgG with altered galactosylation. Rheumatology (Oxf) 2017; 56:2025–30. [DOI] [PubMed] [Google Scholar]
- 33. Schett G. The role of ACPAs in at‐risk individuals: Early targeting of the bone and joints. Best Pract Res Clin Rheumatol 2017; 31:53–8. [DOI] [PubMed] [Google Scholar]
- 34. Guo H, Chen W, Ming J et al Association between polymorphisms in cdc27 and breast cancer in a Chinese population. Tumour Biol 2015; 36:5299–304. [DOI] [PubMed] [Google Scholar]
- 35. Hansen TB, Jensen TI, Clausen BH et al Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384–88. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1. Specific circRNA primers used for quantitative qRTPCR


