SUMMARY
Complementing long-standing traditions centered on histology, fMRI approaches are rapidly maturing in delineating brain areal organization at the macro-scale. The non-human primate (NHP) provides the opportunity to overcome critical barriers in translational research. Here, we establish the data requirements for achieving reproducible and internally valid parcellations in individuals. We demonstrate that functional boundaries serve as a functional fingerprint of the individual animals and can be achieved under anesthesia or awake conditions (rest, naturalistic viewing), though differences between awake and anesthetized states precluded the detection of individual differences across states. Comparison of awake and anesthetized states suggested a more nuanced picture of changes in connectivity for higher-order association areas, as well as visual and motor cortex. These results establish feasibility and data requirements for the generation of reproducible individual-specific parcellations in NHPs, provide insights into the impact of scan state, and motivate efforts toward harmonizing protocols.
In Brief
Noninvasive fMRI in macaques is an essential tool in translation research. Xu et al. establish the individual functional parcellation of the macaque cortex and demonstrate that brain organization is unique, reproducible, and valid, serving as a fingerprint for an individual macaque.
INTRODUCTION
The non-human primate (NHP) brain model remains one of the most promising vehicles for advancing translational neuroscience (Essen, 2012; Hutchison et al., 2015; Mars et al., 2011; Nelson and Winslow, 2009; Phillips et al., 2014; Rilling, 2014; Vanduffel et al., 2014). This point is particularly true for investigations requiring the definition or manipulation of specific cortical targets and their associated circuitry. Traditionally, such lines of inquiry have relied on histological techniques, which are capable of delineating specific cortical areas based on postmortem cytoarchitectonic properties. Beyond their labor intensive ness, such methodologies are inherently limited in their utility for the longitudinal examinations needed to study neuroplasticity, neuromodulation, and brain development. The availability of histological atlases for various NHP species provides an alternative means of guiding intracranial measurements and interventions. However, such atlases are inherently limited in their ability to account for differences in brain areal organization among animals, which can negatively impact experimentation when not properly considered (Cerliani et al., 2017; Gordon et al., 2017a, 2017b; Van Essen et al., 2016).
Recent fMRI studies in the humans have demonstrated the feasibility of using resting state fMRI (R-fMRI) methodologies to delineate the functional brain organization of the cortex at the macroscale based on intrinsic brain function (Cohen et al., 2008; Craddock et al., 2012; Glasser et al., 2016; Gordon et al., 2016; Wig et al., 2014). Not surprisingly, the postmortem histological parcellation approaches used to differentiate spatially contiguous areas into functional units based on differences in their cytoarchitectural and connectivity properties have served as a model for such efforts. In particular, Cohen et al. (2008) proposed a novel approach to perform areal parcellation in the human brain that measures gradients in intrinsic functional connectivity among neighboring vertices, which can, in turn, be used to identify transitions across distinct functional units. This approach has been extended to the full neocortex, with applications revealing functionally discrete areas at the group-average level that are homogeneous and reproducible across studies (Glasser et al., 2016; Gordon et al., 2016; Schaefer et al., 2017). Several of the borders have been highlighted for their correspondence to task activations and cytoarchitectonically defined areas (Buckner and Yeo, 2014; Gordon et al., 2017a; Wig et al., 2014), though such relations are yet to be fully validated at the individual level. Most recently, this gradient-based boundary mapping approach has been applied at the individual level in humans, successfully mapping full-brain gradient and transition patterns that despite commonalities appear to be individual specific (Gordon et al., 2017a, 2017b; Laumann et al., 2015; Xu et al., 2016) and have moderate to high test-retest reliability using as little as 20 min of data.
Work by several groups has suggested the possibility of parcellating the macaque brain in vivo as well using methods such as fMRI, diffusion tractography, and invasive neural recordings. However, to date, these studies (Mars et al., 2011; Neubert et al., 2015; Schönwiesner et al., 2015; Vanduffel et al., 2014) have primarily focused on more limited sections of cortex, such as visual cortex, including occipito-temporal, parietal, and frontal cortex. The logical extension of this work (fMRI-based parcellation of the neocortex of the macaque in its entirety) would be extremely valuable in evaluating the strengths and limitations of the macaque as a translational model (Hutchison and Everling, 2012, 2014; Hutchison et al., 2012a, 2012b; Miranda-Domínguez et al., 2014; Shen et al., 2012).
In the present work, we pursue this aim using gradient-based fMRI parcellation methods to arrive at a putative map of the functional brain organization of the macaque cortex in its entirety at the individual level. Although promising in human literature, there are several challenges that need to be addressed to achieve this goal, namely (1) the feasibility of the boundary mapping parcellation in an individual macaque, (2) the validity of the parcellation, (3) the reliability of the gradient-based boundary mapping approach across scans in an individual macaque, and (4) the stability of the parcellation and the quantity of individual data required for an accurate estimate of full-brain functional areal organization. To address these challenges, we made use of four adult rhesus macaque datasets from two independent research institutions: Nathan Kline Institute (NKI; n = 2) and Oregon Health and Science University (OHSU; n = 2). Each macaque dataset includes between 294 and 688 min of awake and/or anesthetized fMRI scans. Using these data, we first identified the individual-specific functional boundaries and areal parcellation for each dataset and compared it against the available anatomical borders from previous studies. Then, we assessed the functional homogeneity of the gradient-based areal parcellation and demonstrated the uniqueness of the result obtained for each specific monkey. We also evaluated the stability of functional connectivity (FC) profiles, gradient, and boundary maps to investigate the total quantity of data required for estimating full-brain functional areal organization. In addition, we examined the impact of a monocrystalline iron oxide ferumoxytol (MION) contrast agent on the quality of data needs for parcellation, as prior work has demonstrated its utility in increasing the ability to detect FC patterns for NHPs when scanning at 3.0 T (Grayson et al., 2016). Similarly, we examined the potential influences of anesthesia, which is known to decrease measures of FC somewhat (Hutchison et al., 2014). Together, these analyses establish feasibility and data requirements for the generation of reproducible individual-specific parcellations in the NHP and provide insights into the impact of varying factors (e.g., scan state) on findings.
RESULTS
For each individual macaque, the gradient-based parcellations were calculated for each different condition (awake or anesthesia) and contrast agent status (with or without MION). The awake data were acquired with naturalistic viewing paradigm – movies were playing during the awake scans for the NKI dataset (monkey ID: NKI-R, NKI-W). To investigate the impact of differing awake states (movie and rest), we additionally collected one pure rest session for one animal (NKI-W). In the following section, ‘‘awake’’ states without any specifications refer to naturalistic viewing states.
High Reproducibility of Gradient-Based Parcellations
We first focused on the awake states with MION contrast collected from the NKI site (Figure 1). Within each individual animal, the gradients and edge density detected for the awake state with MION demonstrated high reproducibility across individual scans (see all animals in Figures S2 and S3). Consistent with prior work in humans (Xu et al., 2016; Gordon et al., 2017a), gradient maps showed a higher degree of reproducibility than edge maps when equivalent amounts of data were used. In order to obtain a better assessment of the upper bound for reproducibility, we randomly divided the sessions for each macaque into two subsets (Table S1) and regenerated gradient and edge maps for the data available in each subset. Markedly higher reproducibility was shown for each of the measures, as well as the resultant parcellation maps (see Figure 1 for an edge overlap map). The spatial correlations between two gradient maps for macaque NKI-R and NKI-W while awake were 0.71 and 0.81; for edge density, the correlations were 0.30 and 0.47. The Dice coefficients between two parcellations were 0.68 and 0.71.
Figure 1. Subject-Specific Areal Organization Is Highly Reproducible across Two Independent Subsets of Sessions under Awake and Anesthesia.
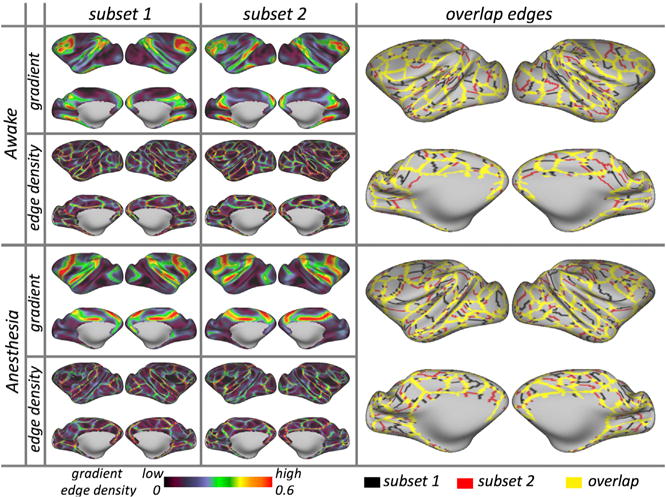
Examples of gradient, edge density, and overlapped edges in NKI-W. See Figure S3 for all animals.
Next, we looked at data obtained during the anesthetized state using MION in subjects from each site (NKI and OHSU). Similar to the awake results, we observed a high degree of reproducibility in gradients, edge density, and corresponding parcellations (see representative subjects in Figure 1, bottom and all subjects in Figure S3). The spatial correlations in gradients between subsets were 0.79 (NKI-R), 0.87 (NKI-W), 0.84 (OHSU-1), and 0.90 (OHSU-2); for the edge density, the correlations were 0.30, 0.48, 0.57, and 0.52. The Dice coefficients between parcellations were 0.69, 0.70, 0.70, and 0.72, respectively.
In addition, we examined the reproducibility of gradient, edge density, and parcellations using data obtained without MION (i.e., blood-oxygen-level dependent [BOLD]). Compared with the data using MION, the findings were similar but less reproducible. Specifically, in the two awake NKI datasets (NKI-R, NKI-W), the spatial correlations between two subsets were 0.32 and 0.54 for the gradient maps and 0.20 and 0.34 for the edge density maps; the Dice coefficients were 0.67 and 0.66 for the final parcellations. In the anesthetized data from OHSU, the spatial correlations were 0.69 and 0.78 in gradients and 0.65 and 0.59 in edge density. The Dice coefficients were 0.70 and 0.67 for the final parcellations.
Parcellation Homogeneity
Consistent with prior work in humans (Laumann et al., 2015), we assessed the internal validity of the parcellation maps obtained for each macaque. Specifically, we calculated the homogeneity of FC similarity in the parcels generated from subset 2 (reference subset) using the data from subset 1 (test subset). The mean homogeneity across all parcels in the awake with MION condition was 0.91 for NKI-W and 0.77 for NKI-R, while under anesthesia across the four macaques was in the range of 0.85 to 0.90 (mean = 0.88; SD = 0.019; Figure 2A, red line). Then, we assessed the degree to which this parcellation was more homogeneous than a null distribution of mean homogeneities generated from 1,000 random parcellation maps for the same subject; each of these maps was generated by randomly rotating the parcellation units around the cortical surface for each subject (Gordon et al., 2016). The mean homogeneity of null model parcellations ranged from 0.73 to 0.86 (mean = 0.82; SD = 0.048) for four macaques; in each macaque, the real parcellations were significantly greater than that obtained in null model parcellations (awake: Z score = 5.94, 10.79, p < 0.001; anesthesia: 9.20, 9.56, 9.00, and 7.22, p < 0.001).
Figure 2. Homogeneity of FC in the Parcels.
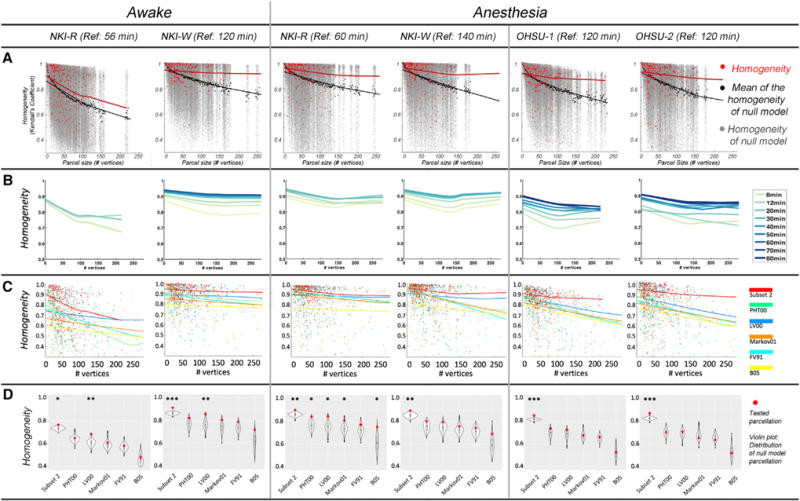
(A) Homogeneity of real parcels (red dots) by parcel size compared to homogeneity of null model parcels (gray dots) under awake and anesthesia states. The lines are the LOWESS (locally weighted scatterplot smoothing) fit represent the effect of parcel size on homogeneity of the subject parcels (red line) and the null model parcels (black line).
(B) The homogeneity of parcellations vary with the amount of test data. The LOWESS fit line plotted the homogeneity at each increment of data used in subset 1 under same condition for each macaque.
(C) The homogeneities tested in subset 1 at each of parcel size for functionally defined parcellation (red line) and five atlases.
(D) The weighted mean homogeneity across parcels of functionally defined parcellations and atlases (red dots) compared with the averaged homogeneity across parcels of each of null model (distribution of 1,000 randomization in violin plot). ***p < 0.001, **p < 0.01, and *p < 0.05 in its 1,000 null model randomizations.
We noted that the parcellation from the awake condition, with MION, was more homogeneous for macaque NKI-W than for macaque NKI-R. This discrepancy arose partially because the total amount of data used for NKI-W was approximately three times that used for NKI-R. Accordingly, we randomly sampled subset 1 and examined the relationship between the homogeneity and amount of scan time. As we expected, the homogeneity increased with the scan time (Figure 2B; the LOWESS [locally weighted scatterplot smoothing] fit line plotted the homogeneity at each increment of data used). Consistent with expectations based on prior work in humans, the homogeneity rapidly increased with 30 min of data (average incensement = 0.09), with relatively modest increases as additional data are added (Figure 2B).
Using the same test subset (subset 1) as above, we also used homogeneity to compare the fit of the individual-specific, functionally derived parcellations in the present work with those of five atlas-based parcellations for the macaque: LV00 (Lewis and Van Essen, 2000), Markov11 (Markov et al., 2011), PHT00 (Paxinos and Franklin, 2000, FV91 (Felleman and Van Essen, 1991) and B05 (Brodmann, 1905). These five atlases were mostly defined upon architectonics in a single animal (LV00 was based on 5 specimens). Van Essen’s group has registered the parcellation to F99 macaque surface, which enabled us to compare the homogeneity with the functionally derived parcellation here (Van Essen et al., 2012). Specifically, we compared the homogeneities of each alternative atlas parcellation against the homogeneities of a null model, which was generated using the same procedure as above (i.e., 1,000 random rotations of the specific parcellations units around the cortical surface). The individual-specific, functionally derived parcellations were more homogeneous than any of the alternative parcellations (Figure 2C). Across the five atlases, only LV00 showed consistently greater homogeneity (than the null model) for both NKI-R and NKI-W, and that was limited to the awake condition with MION (Figure 2D). PHT00, FV91, Markov11, and B05 were not significantly more homogeneous than their null models in the awake condition for either NKI-R or NKI-W. In the anesthetized state, all five atlases failed to show significantly greater homogeneousness than their corresponding null models in at least 3 of the 4 macaques (NKI-W, OHSU-1, and OHSU-2). We also divided parcels into subgroups based on the parcel size and compared the homogeneity among parcellations for each subgroup. The functionally defined parcellations were significantly greater than five atlases across four monkeys (Figure S4).
Additionally, we used homogeneity to confirm that the findings from functionally derived parcellations exhibited individual specificity. Specifically, we compared the homogeneity of an appropriately matched parcellation (i.e., homogeneity calculation and parcellation based on two nonoverlapping data subsets from the same subject) versus inappropriately matched parcellation (i.e., homogeneity calculation and parcellation based on data subsets from different subjects). As part of this examination, we also examined the specificity of findings for an individual with respect to the scan condition and contrast agent status). In 17 of 20 subsets, we found homogeneity to be maximal when the individual, scan condition, and contrast agent status were all appropriately match (red triangles in Figure 3) versus not (gray dots in Figure 3; results from the five atlases are depicted in blue).
Figure 3. Average Homogeneity Was Higher within Individual Specific Functionally Derived Parcellation under the Same Condition (Red Triangle) than Inappropriately Matched Parcellation.
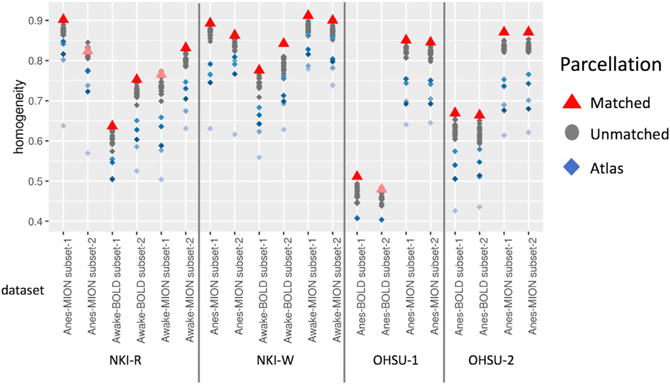
Gray dots indicate the homogeneity calculation and parcellation based on different condition or subjects. Blue dots indicate the average homogeneity based on five atlases.
We noted that the homogeneity scores based on BOLD conditions dataset were lower than MION conditions within each macaque. Thus, we compared the homogeneity across different states (awake versus anesthesia) and contrast types (MION versus BOLD). The MION data exhibited higher homogeneity than BOLD condition regardless of awake (Figure S5, blue line) or anesthetized states (Figure S5, turquoise line); the Z scores between MION and BOLD data (t test) in the awake state were 8.94 (NKI-R) and 11.14 (NKI-W) (both p < 0.001), while those under anesthesia were 22.16 (OHSU-1) and 14.12 (OHSU-2) (both p < 0.001). These differences between awake and anesthesia are examined and discussed in later sections.
Comparison of Parcellation with Known Topographic Areas
To further assess the validity of the parcellation maps using an external reference, the putative boundaries delineated by our functional gradient-based parcellation were overlaid with the areas 3, 4, and 17 borders previously established using postmortem histology (Brodmann, 1905; Van Essen and Dierker, 2007). As depicted in Figure 4, the parcels derived by gradient-based mapping approximately overlapped with areas 3, 4, and 17 borders. This suggests that for these specific areas, gradient-based boundaries appeared to represent differences in function that are captured by histologic and topographic mapping.
Figure 4.
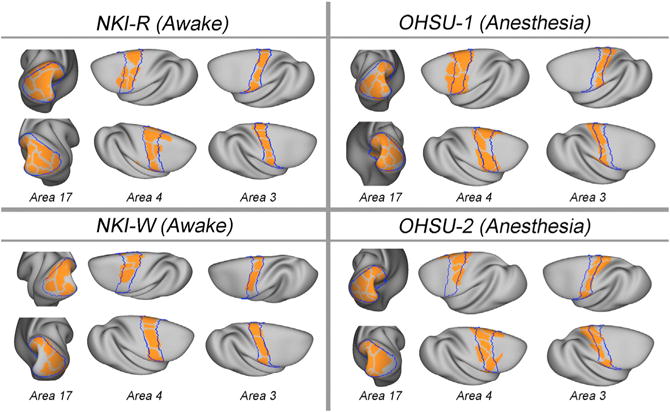
Comparison of Parcellation with Brodmann Areas Subject-specific parcels (orange) overlaid on Brodmann boundaries (blue) of postmortem histologically defined areas (17, 3, and 4).
It is important to note that while there were some clear correspondences between our findings and those of the postmortem map obtained from prior work, some notable differences exist. While such differences likely suggest either greater detectability or the detection of a unique aspect of the functional architecture in the macaque, the possibility of artifactual findings related to the imaging also need to be considered in some cases. For example, in early visual cortices (e.g., the fractionates in area 17), we found additional edges not apparent in the postmortem map; examination of the raw data for these divisions suggests they may reflect an artifact arising from incomplete coverage, though alternatively, they may reflect the presence of further subdivisions. Future work with optimized imaging will be required to help differentiate between these possibilities.
Comparison of Parcellation with fMRI Task Activations (Somatosensory Stimulation)
To provide additional insights into the relative fit of our functionally derived parcellations with those of individual atlases. In this regard, we took advantage of the availability of somatosensory stimulation data obtained for NKI-R and NKI-W using data obtained during BOLD awake scanning. Figure 5 depicts left hemisphere activation during right-hand stimulation with the parcellation overlaid on top of it. Consistent with prior work in humans, visual examination of activations and deactivations associated with stimulation appears to suggest that in many cases, functional activations and deactivations appear to be at least grossly bound by the functional brain organization revealed by parcellation. To further test this point, we compared the fit of activations obtained for each of the animals with their respective parcellations and those of the five atlases. The inhomogeneity was calculated to access the fitness by computing the weighted SD of z-values from the task activation for each parcel (Schaefer et al., 2017). A lower inhomogeneity of a parcellation indicates higher fitness of a given parcellation and activation. In both NKI-R and NKI-W, the parcellation driven by the condition matched functional data (awake BOLD) had better correspondence than atlases.
Figure 5. Task Activations Are Correspondence With Individual- and Condition-Specific Functional Defined Boundaries.
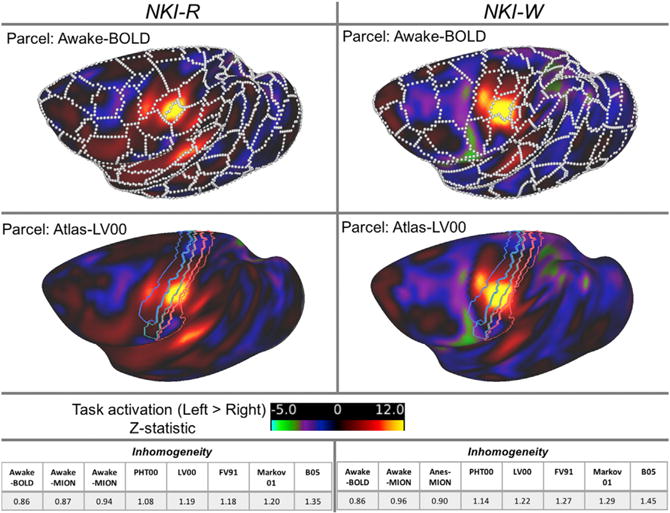
Z-statistic map of left to right contrast of the BOLD condition for NKI-R and NKI-W overlapped with the edges (white borders) defined by naturalistic viewing BOLD sessions of same animal, as well as with sensorimotor areas defined by atlas-PHT00.
Data Requirements for Mapping the Reliable FC, Gradient, and Edges
Here, we first examined the stability of FC, gradient, and edge maps derived from data obtained during the awake state using MION in the NKI dataset. Specifically, for each of the two macaques, we randomly split the data obtained with MION into two independent subsets (Table S1) and used subset 2 to derive reference FC, gradients, and edge density. Next, we randomly sampled subset 1, started at 8-min amount, adding increments (Supplemental Experimental Procedures); the maps generated from each increment were compared to the reference maps in the same macaque. The spatial correlation (Pearson’s r) was used to examine the similarity of FC and gradient, while the Dice coefficient was used to measure the similarity of the parcellations between test and reference dataset.
Consistent with prior human work, the average FC, gradients, and parcellation from awake state (Figure 6, green line) were progressively increased by the amount of data incorporated into the analysis. Focusing on the subject NKI-W, who has more data, the average spatial correlation observed for 8 min from subset 1 with subset 2 (reference) was 0.80 (SD = 0.063) for FC and 0.56 (SD = 0.122) for gradient maps; the mean Dice coefficient was 0.68 (SD = 0.017) for the parcellations. The correlations were rapidly increased to 0.90 (SD = 0.027) for FC and 0.70 (SD =0.071) for gradients with 28 min of data; the mean Dice coefficient increased to 0.70 (SD = 0.016) for parcellations. The averaged correlations increased more slowly up to 88 min. Next, we looked at data obtained during the anesthetized state using MION in subjects from each site, NKI and OHSU. Again, we found that the stability profiles were highly similar to those observed in the awake state (Figure 6, red line); the similarities were uninterruptedly increased from 8 min to 28 min and began to plateau after 40–56 min. The average spatial correlations across four monkey from subset 1 with subset 2 (reference) increased to 0.92 (SD = 0.037) for FC, 0.83 (SD = 0.343) for gradient, and 0.73 (SD = 0.365) for parcellations at 40 min (details in Supplemental Results).
Figure 6. Stability of Subject-Specific Functional Connectivity, Gradient, and Edge Density as a Function of the Amount of Data Used within Different Conditions (Awake versus Anesthesia, with MION versus without MION).
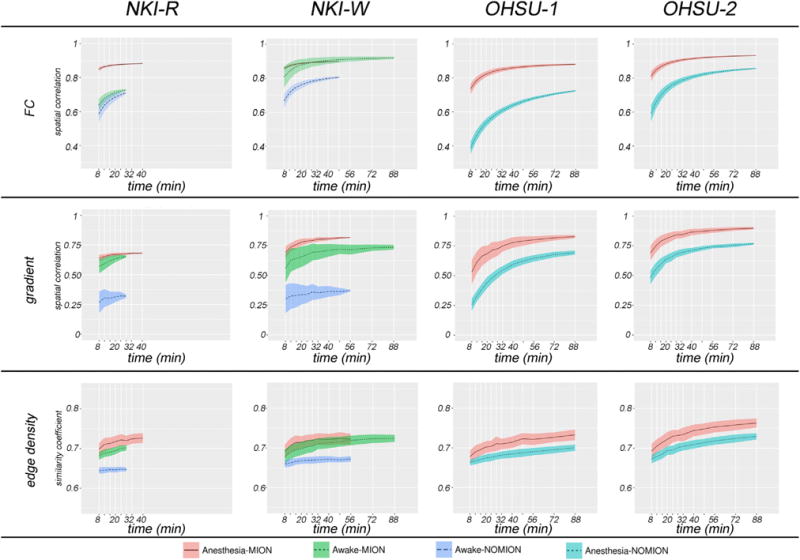
Each line represents the mean (black line) and SD (colored patch) of similarity within each condition for each monkey; the similarity between maps generated from each incremental amount of data (from subset 1) and those from the other half of data (reference: subset 2).
In order to establish the impact of MION, for each of the four macaques, we also examined the stability of FC, gradient, edge density generated using the data obtained without MION (Figure 6; awake, blue line; anesthesia, turquoise line). When the data obtained for each monkey without the use of MION was divided into two subsets, the correspondence remains notably lower than obtained with MION, regardless of whether anesthesia was used (OHSU-1, OHSU-2) or not (NKI-W, NK-R).
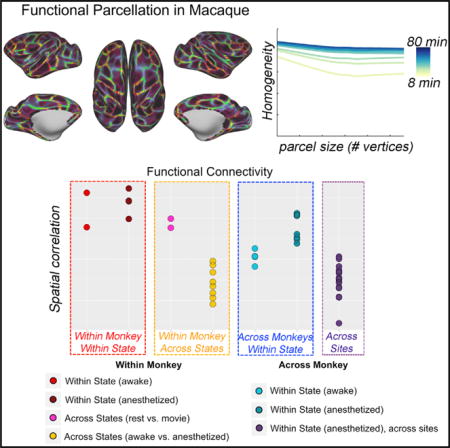
Identification of Unique Individual Areal Organization (Fingerprinting)
Next, we worked to confirm that despite apparent commonalities, the parcellations obtained were unique to each individual. To accomplish this, we assessed the similarity of the FC, gradients, and edge density generated across the four macaques and subsets (i.e., subset 1 and subset 2 for each macaque); Figure S6 depicts the spatial correlations between 21 FC, gradient, and edge density maps (2 macaque × 2 subsets × 3 conditions from NKI and 2 macaques × 2 subsets × 2 conditions from OHSU). To facilitate the visualization, we highlighted the spatial correlations of FC from MION data in Figure 7A. The correlations between subsets within a given subject in the same state (awake or anesthetized, red and dark red dots, respectively) are notably greater than those between different subjects (blue and turquoise dots). When looking within the same state, an individual can be successfully identified using half of dataset for each macaque. Of note, the correlations in gradient and edge density across the monkey while under anesthesia (cadet blue dots) is higher than while awake (turquoise dots), possibly suggesting that functional boundaries are more prone to variation while awake. Across states, measures obtained from data in the same macaque under two different awake states (i.e., naturalistic viewing, rest) exhibited a relatively high degree of similarity (fuchsia dots). However, the correlations in findings between awake and anesthetized (yellow dots) states in the same macaque were much less than those obtained in the same state. In addition, we also observed site effects, with correlations between monkeys across two sites (purple dots) being lower than within the same site while under the same circumstances (e.g., anesthesia with MION). Details of MION effect can be seen in Figure S6.
Figure 7. FC across Subsets, Conditions, and Monkeys.
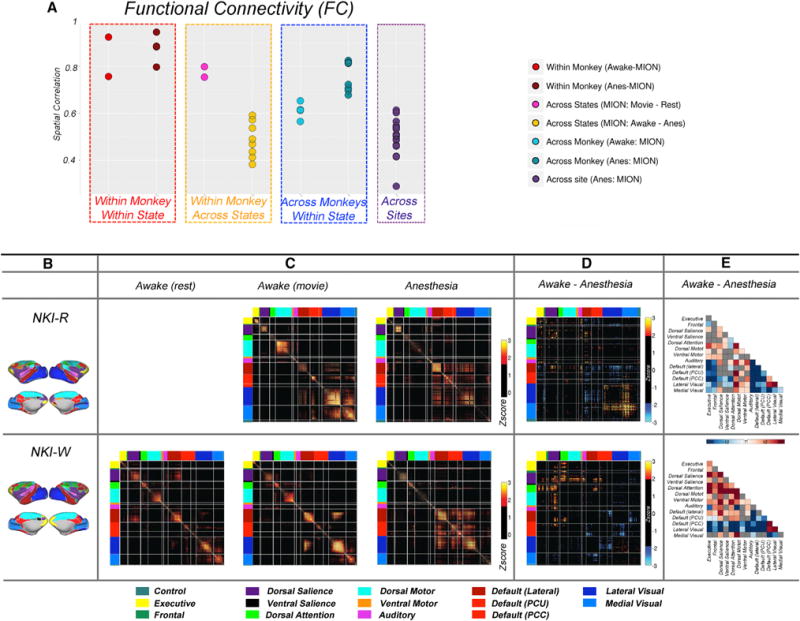
(A) Dots plots of the spatial correlations in FC (see Figure S6 for gradient and edge density) from two subsets (subset 1 and subset 2), three conditions (awake movie, awake rest, anesthesia), and four monkeys from two sites.
(B) The network level of parcellations.
(C) Functional correlation matrices for the awake states (movie and rest) and anesthesia. The correlations were transformed to Z scores (subtract the mean and divide by SD) and threshold at Z > 1.645 (p < 0.05, one tailed).
(D) The differences of correlation matrices (r[movie] r[anesthesia]) that were transformed to Z scores and threshold at Z > 1.960 (p < 0.05, two tailed).
(E) The significant differences of FC between awake (movie) and anesthetized states at the network level. The significance was defined by randomization of paired t test (p < 0.05, Bonferroni corrected).
Effect of State During Awake Imaging
Here, we examined the impact of differences among awake states by comparing FC, gradient, and edge maps produced during the viewing of movies with those obtained during rest. Human imaging studies have drawn attention to the overall stability of FC patterns across differing awake conditions, despite more fine-grained modifications in connectivity associated with manipulations of state (e.g., rest, movies, and task performance). Consistent with the findings from humans, we found a high degree of similarity between FC patterns (r = 0.79) associated with rest and those observed with movie viewing. At the network level, a similar connection profile was found within and between networks, except lateral visual cortex, which showed greater within network connectivity in movie viewing than rest. We also found a relatively high similarity for each, the gradient maps and the parcellations, generated using the two awake states (movie and rest). The spatial correlation of gradient was 0.59 and Dice coefficient of parcellations was 0.68. As the similarity of FC, gradient and edge density between two awake states (movie and rest) within each monkey (see Figures 7A and S6, fuchsia dots) is higher than across monkeys in the awake movie state, this suggests that naturalistic viewing, which is more favorably tolerated by NHPs, can be used as a flexible paradigm for awake imaging.
Effect of Anesthesia
The FC, gradient, and edge density generated from subsets of data obtained under same state showed a high degree of correspondence. However, the correspondence of areal organization was substantially lower when compared across the awake and anesthetized states in the same subjects (NKI-R, NKI-W). Specifically, the spatial correlations in FC between awake and anesthesia was 0.52 (SD = 0.08) (Figure 7A, yellow dots). The gradient and edge density showed even less similarity between awake and anesthetized, with the spatial correlations across states with half subsets of data in NKI dataset being 0.25 (SD = 0.14) and 0.12 (SD = 0.02), respectively (Figure S6, yellow dots).
Visual examination of the FC matrices for the awake states (rest and movies) and anesthesia revealed notable regional connectivity differences that appeared to be specific to anesthesia (Figures 7B–7E). We found evidence of anesthesia-related decreases within and between most networks, with specific findings differing somewhat between the two monkeys. Decreases in within-network FC for the dorsal salience, dorsal motor, lateral visual, and medial visual networks were consistent across these two subjects. Complementing these within-network findings were anesthesia-related increases in between network FC for the visual networks; these increases were most consistent for FC with default network and fronto-parietal components. Of note, while comparison of awake and anesthetized states was not possible for the datasets collected at OHSU, the sparsity of the data collected there was more similar to that obtained under anesthesia at NKI, suggesting some level of consistency.
DISCUSSION
Functional Boundaries Are Highly Reproducible in Individual Macaques with Sufficient Data
Our findings regarding reproducibility and data requirements are not surprising given recent work carried out in humans (Gordon et al., 2017a; Laumann et al., 2015; Xu et al., 2016). Similar to the findings from humans, more data are generally better when addressing the reproducible FC (Laumann et al., 2015), though with the largest gains being observed as the data used increases up to ~32 min, a finding that echoes the findings from humans, where the most substantial gains were seen up to ~27–30 min. It should be noted that the data needs increase with each additional step of processing (i.e., from FC to gradient and edge calculation). Improvements in reliability for gradients and parcellation slowly but continuously increased as data increased to 40 min, while the largest reproducibility values (spatial correlation and Dice coefficient) were still relatively lower than what can be achieved for FC. These findings suggest that extended data are required to achieve a similar level of reproducibility of parcellations. Additionally, our findings extend recent works emphasizing the importance of contrast agents when scanning NHPs at 3.0 T by demonstrating that the use of MION is a prerequisite to achieving maximal reproducibility for FC, gradients, and functional boundaries maps (Gautama et al., 2003; Grayson et al., 2016; Leite et al., 2002); this is true regardless of whether anesthesia was used or not.
Functional Parcellations Show Internal Validity
Directly relevant to the present study, prior studies in humans suggest that the boundary map-based parcellation had highly homogeneous FC patterns at the group and individual level (Gordon et al., 2017a; Laumann et al., 2015; Xu et al., 2016). In the current study, we tested the homogeneity of boundary map-derived parcellations using subsets of the data (subset 1), such that the parcels were generated completely independently from the other subset of the data (subset 2) for each status within each macaque. On average, the parcel homogeneity was above 0.9 with 120 min of awake data. This high degree of homogeneity indicates that most parcels represented areas of similar FC pattern. It is notable that the homogeneity varied across parcels. As discussed in a previous study (Gordon et al., 2016), the homogeneity of parcels is associated with parcel size, where small parcels are more likely to have higher homogeneity than larger parcels. A few parcels had low homogeneity, and some of them (e.g., in the frontal pole and anterior inferior temporal lobe) may be caused by the low signal-to-noise ratio (SNR) or distortion signal in those areas. Accordingly, it is necessary to use a null model to truly evaluate the homogeneity. By creating the null model from 1,000 random rotations of the identical parcellations around the cortical surface, we found that the homogeneities of parcels were significantly higher than the null model in both awake and anesthetized states. Of note, the homogeneity of the cortical parcellations derived from functional data also exhibited a higher degree of homogeneity than those from five established cortical atlases, further increasing confidence in our findings.
A key challenge for any effort focused on cortical parcellation, whether in the human or NHP, is determination of the ‘‘correct’’ number of parcels. Regardless of which modality or parcellation method is used, a number of differing criteria can be used to make such determinations. In the current study, the watershed flood algorithm yielded between 112 and 141 parcels per hemisphere (mean = 129, SD = 8), for each macaque. An advantage of this methodology, is the lack of need for a priori specification of the number of parcels to be produced. However, the results can be impacted by properties of the data, such as spatial resolution and smoothness; in this regard, a priori specification of parameters (e.g., number of parcellations) can have advantages. Our estimate for number of parcels converge with current estimates generated from histology-based studies (i.e., cytoarchitecture, myeloarchitecture, and chemoarchitecture), which have increased from 78–88 (Felleman and Van Essen, 1991; Lewis and Van Essen, 2000; Markov et al., 2011; Van Essen et al., 2012) to 130–160 in each hemisphere (Paxinos and Franklin, 2000; Van Essen et al., 2012). Human work suggested that the number of parcels might increase when we examine gradient across multiple modalities (e.g., area 55b was more distinctive in task activation) and a selective seed connectivity (Glasser et al., 2016). Future work will benefit from integrative examinations across assessment modalities (e.g., fMRI, diffusion, and histology) in the same animals.
Nonequivalence of Functional Parcels and Traditional Cortical Areas
It is important to note that functional parcels or ‘‘areas’’ identified in the present work are intended to represent basic units in the macroscale functional brain organization; defined using FC, they exhibit abrupt changes in connectivity profiles from one parcel to the next (Cohen et al., 2008). Unlike traditionally defined neocortical areas, which are grounded in architectonics, they are not necessarily expected to be stable across all conditions, as FC patterns are known to vary as a function of state. Previous studies have demonstrated that FC patterns are sculpted more by structural connectivity under deep anesthesia than the awake state (Barttfeld and Uhrig, 2015; Wu et al., 2016). In anesthesia states, when isoflurane levels were varied, the signal fluctuations and connectivity also exhibited distinct profiles at each level within the same animal (Smith et al., 2017; Gao et al., 2017; Peltier et al., 2005; Hutchison et al., 2014). While it is expected that the parcels revealed will in many cases exhibit a gross similarity to traditionally defined neocortical areas, they are not intended to be 1:1. As demonstrated by recent work, the functional areas revealed by gradient-based approaches do appear to exhibit good correspondence with task activations localizers.
A common theme in the parcellation literature is the attempt to establish validity by comparing the parcels obtained with those in atlases derived from more traditional approaches. In this regard, we compared the full brain parcellations derived from our analyses with five atlases, four of which were defined using his tological methodologies and one with multiple modality (e.g., architecture, connections, and myeloarchitecture). First, using parcel homogeneity as a metric of fit, we found that the functionally derived cortical parcellations exhibited superior homogeneity to any of the five atlases. While comparability would be the minimum standard we were aiming for, the superiority was suggestive of three possibilities (or any combination of these): (1) the atlas is not being optimally registered to each individual, (2) our functional parcellations successfully accounted for individual-specific variations that cannot be accounted for when applying an atlas, or (3) functionally derived parcels are capturing aspects of functional brain organization that are not directly related to traditional cortical areas (Reveley et al., 2017; Palomero-Gallagher et al., 2015).
Beyond parcel homogeneity, we compared the specific parcels generated from our work with those in the five atlases. While prior work in the human literature has emphasized the ability to recapitulate traditionally defined cortical areas (Gordon et al., 2016; Laumann et al., 2015), the present work raises caveats. While the gross similarity between our parcellations and those of the five atlases was clear, our parcellations did not recapitulates the areas in high detail, regardless of whether one we focused on awake or anesthetized data. This is not necessarily surprising given the many in the literature arguing that structure and function are not 1:1 and showing examples of functional activations failing to follow cortical areas in a precise manner. Intriguingly, for each of the two NKI monkeys, the parcel obtained in the finger area appeared to bound activations observed during somatosensory stimulation, with notably better fit than any of the traditional cortical atlases. As such, we assert the possibility that functionally derived atlases may be more relevant for functional imaging analyses than traditional and afford the opportunity to generate individual-specific parcellations in vivo. Further validation using a broader range of mapping approaches is needed to more definitively make this point.
Rest and Naturalistic Viewing during Awake Imaging Show Similar Functional Properties
Our findings regarding the relative stability of FC patterns across awake and sensitivities to level of arousal (i.e., sleep or anesthesia) help to synthesize previous findings from the human and NHP literatures. Over the past decade, numerous studies have demonstrated the ability to extract highly similar intrinsic FC networks, regardless of whether imaging is carried out during an active task state or rest (Cole et al., 2014; Fair et al., 2007; Vanderwal et al., 2017). Recent work has increasingly highlighted the potential value of using non-rest states, particularly naturalistic viewing, for assessing FC, as head motion appears to be lower and tolerability higher. While systematic differences in connectivity patterns are undoubtedly present across states, as revealed by within-subject comparisons, differences in connectivity patterns across individuals can be similar, at least for static FC patterns (Finn et al., 2015; O’Connor et al., 2017; Vanderwal et al., 2017; Wang et al., 2017). Given the various behavioral demands of awake imaging for NHP, we have found that naturalistic viewing conditions can be particularly valuable, at least for full-brain parcellations. For those interested in more fine-grained parcellations or temporal dynamics, it is important to further investigate the temporal stability versus state dependence of more subtle distinctions within cortical areas (e.g., polar angle and eccentricity mapping in early visual cortex).
Individual Brain Organization Is Unique While the Awake and Anesthetized States Show Distinct Profiles
As suggested by prior work in humans (Miranda-Domínguez et al., 2014; Xu et al., 2016), we found that individual brain organization can be potentially used for fingerprinting in the monkey. Within the same condition (awake-awake, anesthesia-anesthesia), the within-individual spatial correlations of gradient and edge density maps are explicitly higher than between-individual correlations. Consistent with prior work in humans (Miranda-Domínguez et al., 2014; Finn et al., 2015; O’Connor et al., 2017; Vanderwal et al., 2017), the functional brain organization is similar across different awake conditions (i.e., movie and rest), though some decrements in similarities for FC, gradients, and edge maps were noted. Importantly, the ability to identify individuals using data collected with anesthesia was notably decreased when attempting to match the same subject across the awake and anesthetized states. This is not surprising, as the similarities for full-brain FC, gradient maps, and edge maps were dramatically reduced when looking across the awake and anesthetized states (e.g., Ȉ40% in whole-brain FC similarity). Of note, the current findings were limited in a small sample, and the ability to fingerprint individual brain organization in macaque needs to be confirmed with large datasets in future work.
While the presence of differences in FC properties between awake and anesthetized states was not surprising based on prior work (Hutchison et al., 2013, 2014; Vincent et al., 2007), some of the specific findings suggest a more nuanced picture than previously appreciated. Consistent with prior studies varying levels of anesthesia, we found evidence of anesthesia-related compromises in within- and between-network connectivity for higher-order associative cortices; the specific networks affected differed somewhat across the two monkeys, with the most consistent findings being observed in the salience and dorsal attention networks. However, unlike prior studies, which largely relied on variations of depth of anesthesia, our comparison of awake states and anesthesia also suggested that the visual networks and their connectivity with default and frontoparietal networks actually showed anesthesia-related increases in connectivity. Overall, these findings echo recent studies of the impact of anesthetics on brain differences in humans, which suggested a loss of complexity in the functional architecture of the brain (Chang et al., 2016; Hutchison et al., 2014; Peltier et al., 2005; Smith et al., 2017; Wu et al., 2016). Further work will be required to rule out other possibilities, such as differences in respiration associated with differing states (awake or anesthesia).
In sum, the present work suggests that the awake state and the anesthetized state, as well as their differences, are highly stable. Future work would benefit from more detailed examinations of anesthesia effects that include awake imaging, as well as possibly natural sleep. Additionally, an increased focus on potential confounds will be important as the field moves forward; this will require additional monitoring.
Effects of Sites
Finally, it is worth noting a unique aspect of the present work: the examination of monkey datasets across two institutions. A significant advantage of this strategy was the ability to confirm the reproducibility of findings across two independently collected datasets. Not surprisingly, while we were successful in demonstrating the generality of our findings, we did find evidence of site-related differences. Specifically, we found that the correspondence of gradient and edge maps among subjects was higher within a site than between. Possible factors that might explain site-related variation include differences in (1) anesthesia protocols (e.g., the knockout agent and delay duration after the time of anesthetics administration), (2) head coils (the OHSU dataset was acquired by a knee coil, while NKI data-set was with a surface coil; prior work has demonstrated differences in signal properties, including distortion, of the data obtained from monkeys using these coils; Grayson et al., 2016), and (3) rearing histories. Future work would benefit from more coordinated efforts focused on multisite imaging in the NHP as a means of maximizing the reproducibility of findings between laboratories.
Limitations
It is important to note that despite the various successes of the present work, a number of limitations and potential areas for future optimization exist. As previously discussed, a key limitation of any parcellation methodology is the lack of a gold standard benchmark to help establish validity. The present work made use of comparisons to previously established histologic parcellations to provide insights into validity, as well as the examination of parcel properties (i.e., homogeneity). Future work would benefit from examination of multiple methods of parcellation in the same subjects (e.g., fMRI and diffusion imaging, fMRI and myelin maps, and fMRI and histology). Another consideration for future work is examination of the potential impact of pre-processing strategy decisions on the resultant parcels and their stability. Optimization of preprocessing may be particularly useful if focused on bringing down data needs for achieving the same parcellation results. In addition, although the fingerprinting analyses suggest that the functional brain organization is unique and appears to be stable over time, the number of samples in the present study was small, and large datasets would need to extend and confirm the findings. Finally, the present work relied on the FC similarity measure used in prior studies for gradient-based parcellation. Prior work demonstrated this measure to have favorable properties relative to several other possible measures. However, an exhaustive examination of measures to be used for gradient-based parcellation has yet to be carried out; this would be of benefit both to determine whether some measures may be able to lead to more fine-grained parcellations and to find a less computationally costly measure, if possible. On a related note, a range of alternatives exists for the final parcellation step in the gradient-based strategy; future work determining optimality is merited.
Conclusions and Future Directions
In summary, the present work demonstrates the ability to achieve highly reproducible, individual-specific cortical parcellations in the NHP. Our findings also highlight the many factors that can contribute to stability, in particular the need for sufficient data and contrast agents. Our findings also emphasized the need for consideration of state (i.e., awake versus anesthetized) when interpreting findings and their reproducibility across studies, as well as during the study design process. Transformation of our NHP findings into a common space demonstrated a future direction that can be further optimized to facilitate comparative and translational studies.
EXPERIMENTAL PROCEDURES
Dataset 1: NKI Macaque
The NKI dataset consisted of two rhesus monkeys (Macaca mulatta, one male, age 6 years, 6.4 kg, marked as NKI-R; one female, age 7 years, 4.5 kg, marked as NKI-W), which was collected from the Nathan Kline Institute. Seven fMRI sessions were acquired for NKI-R (294 min in total) and 11 sessions (668 min) for NKI-W. Each session included 4–7 scans (8–10 min per scan). We collected both anesthetized and awake data. MION solution was injected at iron doses of 10 mg/kg intravenously (i.v.) prior to the magnetic resonance imaging (MRI) scanning for all MION sessions. Additionally, we collected somatosensory task (2 sessions for NKI-R and 6 sessions for NKI-W) while the monkeys were awake without MION contrast agent. See Table S1 for details. All methods and procedures were approved by the NKI Institutional Animal Care and Use Committee (IACUC) protocol.
Dataset 2: OHSU Macaque
A second dataset consisted of two male rhesus macaques (M. mulatta, one male, age 5 years, 8.6 kg, marked as OHSU-1; one male, age 5 years, 7.6 kg, marked as OHSU-2), which were collected at the OHSU. Eight sessions were acquired for each monkey under anesthesia. For each session, a 30-min BOLD scan without MION and a 30-min scan with MION were acquired on the same day. The BOLD scan started 45 min after the monkey was anesthetized, followed by the MION scan. The MION solution was injected at iron doses of 8 mg/kg IV. See Table S1 for details. Animal procedures were in accordance with the National Institutes of Health guidelines on the ethical use of animals and approved by the Oregon National Primate Research Center (ONPRC) Institutional Animal Care and Use Committee.
Gradient-Based Parcellation
The structural and functional data were preprocessed in FreeSurfer, FSL, ANTs, and workbench (Figure S1). The native surfaces were reconstructed and down-sampled to 10k_fs_LR surface (Donahue et al., 2016). The volumetric fMRI data were aligned to the anatomic space, projected to the native surface, and down-resampled to the 10 kilo (k) surface. For each individual macaque, the gradient-based parcellations were calculated on the native 10k surface for each different condition (awake or anesthesia) and contrast agent status (with or without MION). The gradient evaluates the spatial transitions in the similarity of neighboring FC profiles across the native cortical surface. Consistent with prior work, a gradient map was generated for the FC map of vertex; for each of these maps, a corresponding edge map was generating using the ‘‘watershed’’ algorithm (Gordon et al., 2016). The final functional boundaries (i.e., edge densities) were then calculated by averaging across the set of edge maps so that the value at each vertex represented the possibility that the vertex was identified as an edge. The watershed algorithm was then used to generate parcels from the edge density map. To characterize a large-scale system of parcellations, we conducted a matching procedure to assign a network identity to parcels using previously established network (Ghahremani et al., 2017). The details of this method were described in previous studies (Laumann et al., 2015; Xu et al., 2016), and a description is presented in Supplement Experimental Procedures.
To establish reproducibility of the parcellation, we randomly split our dataset into two independent subsets (see Table S1) and examined the correspondence between the two subsets. Specifically, for each of the macaques, we used subset 2 to derive a reference and randomly sampled subset 1 to create subsamples of differing time lengths between 8 and 88 min (1,000 random samplings at each length). FC, gradient, and edge density maps were calculated for each sample generated from subset 1. For each time length, spatial similarity between these maps and those obtained from the reference sample was evaluated by Pearson’s r and averaged across the 1,000 random samplings of subset 1. The validity of each parcellation was evaluated by the homogeneity using the Kendall coefficient (Xu et al., 2016). Consistent with prior work in humans, we also measured the fitness of functionally derived parcellations using data obtained during a somatosensory task; this involves calculation of the weighted standard deviation of z-values from the task activation within each parcel (Schaefer et al., 2017). Detailed are provided in Supplement Experimental Procedures. All data are available at the PRIMatE Data Exchange (PRIME-DE; http://fcon_1000.projects.nitrc.org/indi/indiPRIME.html; Milham et al., 2017).
Supplementary Material
Highlights.
Individual functional parcellation is unique, reproducible, and valid in macaques
The awake and anesthetized states show distinct brain organization
Findings suggest ‘‘batch’’ effects across imaging sites
Acknowledgments
This work was supported by gifts from Joseph P. Healey, Phyllis Green, and Randolph Cowen to the Child Mind Institute and grants from the NIH (BRAIN Initiative R01-MH111439 to C.E.S. and M.P.M.; P50MH109429 to C.E.S.; R01-MH107508 to E.L.S. and D.A.F.; and P60-AA010760, R01-MH115357, R01-MH096773, and P50-MH100029 to D.A.F.). This work was also supported in part by NIH grant P01AG026423 and the Yerkes National Primate Research Center base grant (Office of Research Infrastructure Programs; OD P51OD11132).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Results, Supplemental Experimental Procedures, and six figures and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.03.049.
AUTHOR CONTRIBUTIONS
T.X., C.E.S., D.A.F., and M.P.M. designed the study. T.X. analyzed the data with support from J.S.B.R., E.F., and D.S. A.F., G.L., D.R., A.O., J.B., E.E., O.M.-D., A.P., R.C.C., and S.C. performed the experiments and provided support. T.X. and M.P.M. wrote the manuscript with contributions from E.L.S., C.E.S., and D.A.F. All authors commented on the manuscript.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Barttfeld P, Uhrig L. Correction for Barttfeld et al., Signature of consciousness in the dynamics of resting-state brain activity. Proc Natl Acad Sci USA. 2015;112:E5219–E5220. doi: 10.1073/pnas.1515029112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodmann K. Vergleichende Lokalisationslehre der Grosshirnrinde in ihren Prinzipien dargestellt auf Grund des Zellenbaues. Barth; 1905. [Google Scholar]
- Buckner RL, Yeo BTT. Borders, map clusters, and supra-areal organization in visual cortex. Neuroimage. 2014;93:292–297. doi: 10.1016/j.neuroimage.2013.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerliani L, D’Arceuil H, Thiebaut de Schotten M. Connectivity-based parcellation of the macaque frontal cortex, and its relation with the cytoarchitectonic distribution described in current atlases. Brain Struct Funct. 2017;222:1331–1349. doi: 10.1007/s00429-016-1280-3. [DOI] [PubMed] [Google Scholar]
- Chang C, Leopold DA, Schölvinck ML, Mandelkow H, Picchioni D, Liu X, Ye FQ, Turchi JN, Duyn JH. Tracking brain arousal fluctuations with fMRI. Proc Natl Acad Sci USA. 2016;113:4518–4523. doi: 10.1073/pnas.1520613113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AL, Fair DA, Dosenbach NUF, Miezin FM, Dierker D, Van Essen DC, Schlaggar BL, Petersen SE. Defining functional areas in individual human brains using resting functional connectivity MRI. Neuroimage. 2008;41:45–57. doi: 10.1016/j.neuroimage.2008.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MW, Bassett DS, Power JD, Braver TS, Petersen SE. Intrinsic and task-evoked network architectures of the human brain. Neuron. 2014;83:238–251. doi: 10.1016/j.neuron.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock RC, James GA, Holtzheimer PE, 3rd, Hu XP, Mayberg HS. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum Brain Mapp. 2012;33:1914–1928. doi: 10.1002/hbm.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue CJ, Sotiropoulos SN, Jbabdi S, Hernandez-Fernandez M, Behrens TE, Dyrby TB, Coalson T, Kennedy H, Knoblauch K, Van Essen DC, Glasser MF. Using diffusion tractography to predict cortical connection strength and distance: a quantitative comparison with tracers in the monkey. J Neurosci. 2016;36:6758–6770. doi: 10.1523/JNEUROSCI.0493-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essen DV. Surface-based analyses of human, macaque, and chimpanzee cortical organization. J Vis. 2012;12:1377. [Google Scholar]
- Fair DA, Schlaggar BL, Cohen AL, Miezin FM, Dosenbach NUF, Wenger KK, Fox MD, Snyder AZ, Raichle ME, Petersen SE. A method for using blocked and event-related fMRI data to study ‘‘resting state’’ functional connectivity. Neuroimage. 2007;35:396–405. doi: 10.1016/j.neuroimage.2006.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex. 1991;1:1–47. doi: 10.1093/cercor/1.1.1-a. [DOI] [PubMed] [Google Scholar]
- Finn ES, Shen X, Scheinost D, Rosenberg MD, Huang J, Chun MM, Papademetris X, Constable RT. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat Neurosci. 2015;18:1664–1671. doi: 10.1038/nn.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YR, Ma Y, Zhang Q, Winder AT, Liang Z, Antinori L, Drew PJ, Zhang N. Time to wake up: Studying neurovascular coupling and brain-wide circuit function in the un-anesthetized animal. Neuroimage. 2017;153:382–398. doi: 10.1016/j.neuroimage.2016.11.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautama T, Mandic DP, Van Hulle MM. Signal nonlinearity in fMRI: a comparison between BOLD and MION. IEEE Trans Med Imaging. 2003;22:636–644. doi: 10.1109/TMI.2003.812248. [DOI] [PubMed] [Google Scholar]
- Ghahremani M, Matthew Hutchison R, Menon RS, Everling S. Frontoparietal functional connectivity in the common marmoset. Cereb Cortex. 2017;27:3890–3905. doi: 10.1093/cercor/bhw198. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M, et al. A multi-modal parcellation of human cerebral cortex. Nature. 2016;536:171–178. doi: 10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Huckins JF, Kelley WM, Petersen SE. Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb Cortex. 2016;26:288–303. doi: 10.1093/cercor/bhu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Petersen SE. Individual variability of the system-level organization of the human brain. Cereb Cortex. 2017a;27:386–399. doi: 10.1093/cercor/bhv239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Gilmore AW, Nelson SM, Dosenbach NUF, Petersen SE. Individual-specific features of brain systems identified with resting state functional correlations. Neuroimage. 2017b;146:918–939. doi: 10.1016/j.neuroimage.2016.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson DS, Bliss-Moreau E, Machado CJ, Bennett J, Shen K, Grant KA, Fair DA, Amaral DG. The rhesus monkey connectome predicts disrupted functional networks resulting from pharmacogenetic inactivation of the amygdala. Neuron. 2016;91:453–466. doi: 10.1016/j.neuron.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Everling S. Monkey in the middle: why nonhuman primates are needed to bridge the gap in resting-state investigations. Front Neuroanat. 2012;6:29. doi: 10.3389/fnana.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Everling S. Broad intrinsic functional connectivity boundaries of the macaque prefrontal cortex. Neuroimage. 2014;88:202–211. doi: 10.1016/j.neuroimage.2013.11.024. [DOI] [PubMed] [Google Scholar]
- Hutchison RM, Gallivan JP, Culham JC, Gati JS, Menon RS, Everling S. Functional connectivity of the frontal eye fields in humans and macaque monkeys investigated with resting-state fMRI. J Neurophysiol. 2012a;107:2463–2474. doi: 10.1152/jn.00891.2011. [DOI] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Gati JS, Leung LS, Menon RS, Everling S. Resting-state connectivity identifies distinct functional networks in macaque cingulate cortex. Cereb Cortex. 2012b;22:1294–1308. doi: 10.1093/cercor/bhr181. [DOI] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Gati JS, Everling S, Menon RS. Resting-state networks show dynamic functional connectivity in awake humans and anesthetized macaques. Hum Brain Mapp. 2013;34:2154–2177. doi: 10.1002/hbm.22058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Hutchison M, Manning KY, Menon RS, Everling S. Isoflurane induces dose-dependent alterations in the cortical connectivity profiles and dynamic properties of the brain’s functional architecture. Hum Brain Mapp. 2014;35:5754–5775. doi: 10.1002/hbm.22583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Culham JC, Flanagan JR, Everling S, Gallivan JP. Functional subdivisions of medial parieto-occipital cortex in humans and nonhuman primates using resting-state fMRI. Neuroimage. 2015;116:10–29. doi: 10.1016/j.neuroimage.2015.04.068. [DOI] [PubMed] [Google Scholar]
- Laumann TO, Gordon EM, Adeyemo B, Snyder AZ, Joo SJ, Chen MY, Gilmore AW, McDermott KB, Nelson SM, Dosenbach NUF, et al. Functional system and areal organization of a highly sampled individual human brain. Neuron. 2015;87:657–670. doi: 10.1016/j.neuron.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite FP, Tsao D, Vanduffel W, Fize D, Sasaki Y, Wald LL, Dale AM, Kwong KK, Orban GA, Rosen BR, et al. Repeated fMRI using iron oxide contrast agent in awake, behaving macaques at 3 Tesla. Neuroimage. 2002;16:283–294. doi: 10.1006/nimg.2002.1110. [DOI] [PubMed] [Google Scholar]
- Lewis JW, Van Essen DC. Mapping of architectonic subdivisions in the macaque monkey, with emphasis on parieto-occipital cortex. J Comp Neurol. 2000;428:79–111. doi: 10.1002/1096-9861(20001204)428:1<79::aid-cne7>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Markov NT, Misery P, Falchier A, Lamy C, Vezoli J, Quilodran R, Gariel MA, Giroud P, Ercsey-Ravasz M, Pilaz LJ, et al. Weight consistency specifies regularities of macaque cortical networks. Cereb Cortex. 2011;21:1254–1272. doi: 10.1093/cercor/bhq201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars RB, Jbabdi S, Sallet J, O’Reilly JX, Croxson PL, Olivier E, Noonan MP, Bergmann C, Mitchell AS, Baxter MG, et al. Diffusion-weighted imaging tractography-based parcellation of the human parietal cortex and comparison with human and macaque resting-state functional connectivity. J Neurosci. 2011;31:4087–4100. doi: 10.1523/JNEUROSCI.5102-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milham MP, Ai L, Koo B, Xu T, Balezeau F, Baxter MG, Croxson PL, Damatac CG, Harel N, Freiwald W, et al. An open resource for nonhuman primate imaging. bioRxiv. 2017 doi: 10.1101/227462. [DOI]
- Miranda-Domínguez O, Mills BD, Grayson D, Woodall A, Grant KA, Kroenke CD, Fair DA. Bridging the gap between the human and macaque connectome: a quantitative comparison of global interspecies structure-function relationships and network topology. J Neurosci. 2014;34:5552–5563. doi: 10.1523/JNEUROSCI.4229-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson EE, Winslow JT. Non-human primates: model animals for developmental psychopathology. Neuropsychopharmacology. 2009;34:90–105. doi: 10.1038/npp.2008.150. [DOI] [PubMed] [Google Scholar]
- Neubert FX, Mars RB, Sallet J, Rushworth MFS. Connectivity reveals relationship of brain areas for reward-guided learning and decision making in human and monkey frontal cortex. Proc Natl Acad Sci USA. 2015;112:E2695–E2704. doi: 10.1073/pnas.1410767112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor D, Potler NV, Kovacs M, Xu T, Ai L, Pellman J, Vanderwal T, Parra LC, Cohen S, Ghosh S, et al. The Healthy Brain Network Serial Scanning Initiative: a resource for evaluating inter-individual differences and their reliabilities across scan conditions and sessions. Gigascience. 2017;6:1–14. doi: 10.1093/gigascience/giw011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero-Gallagher N, Eickhoff SB, Hoffstaedter F, Schleicher A, Mohlberg H, Vogt BA, Amunts K, Zilles K. Functional organization of human subgenual cortical areas: Relationship between architectonical segregation and connectional heterogeneity. Neuroimage. 2015;115:177–190. doi: 10.1016/j.neuroimage.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. Gulf Professional Publishing; 2000. [Google Scholar]
- Peltier SJ, Kerssens C, Hamann SB, Sebel PS, Byas-Smith M, Hu X. Functional connectivity changes with concentration of sevoflurane anesthesia. Neuroreport. 2005;16:285–288. doi: 10.1097/00001756-200502280-00017. [DOI] [PubMed] [Google Scholar]
- Phillips KA, Bales KL, Capitanio JP, Conley A, Czoty PW, ’t Hart BA, Hopkins WD, Hu SL, Miller LA, Nader MA, et al. Why primate models matter. Am J Primatol. 2014;76:801–827. doi: 10.1002/ajp.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveley C, Gruslys A, Ye FQ, Glen D, Samaha J, Russ BE, Saad Z, Seth AK, Leopold DA, Saleem KS. Three-dimensional digital template atlas of the macaque brain. Cereb Cortex. 2017;27:4463–4477. doi: 10.1093/cercor/bhw248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rilling JK. Comparative primate neuroimaging: insights into human brain evolution. Trends Cogn Sci. 2014;18:46–55. doi: 10.1016/j.tics.2013.09.013. [DOI] [PubMed] [Google Scholar]
- Schaefer A, Kong R, Gordon EM, Laumann TO, Zuo XN, Holmes AJ, Eickhoff SB, Yeo BTT. Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb Cortex. 2017;18:1–20. doi: 10.1093/cercor/bhx179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönwiesner M, Dechent P, Voit D, Petkov CI, Krumbholz K. Parcellation of human and monkey core auditory cortex with fMRI pattern classification and objective detection of tonotopic gradient reversals. Cereb Cortex. 2015;25:3278–3289. doi: 10.1093/cercor/bhu124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Bezgin G, Hutchison RM, Gati JS, Menon RS, Everling S, McIntosh AR. Information processing architecture of functionally defined clusters in the macaque cortex. J Neurosci. 2012;32:17465–17476. doi: 10.1523/JNEUROSCI.2709-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JB, Liang Z, Watson GDR, Alloway KD, Zhang N. Interhemispheric resting-state functional connectivity of the claustrum in the awake and anesthetized states. Brain Struct Funct. 2017;222:2041–2058. doi: 10.1007/s00429-016-1323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Dierker DL. Surface-based and probabilistic atlases of primate cerebral cortex. Neuron. 2007;56:209–225. doi: 10.1016/j.neuron.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Glasser MF, Dierker DL, Harwell J. Cortical parcellations of the macaque monkey analyzed on surface-based atlases. Cereb Cortex. 2012;22:2227–2240. doi: 10.1093/cercor/bhr290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Donahue C, Dierker DL, Glasser MF. Parcellations and connectivity patterns in human and macaque cerebral cortex. In: Kennedy H, Van Essen DC, Christen Y, editors. Micro-, Meso- and Macro-Connectomics of the Brain. Springer; 2016. pp. 89–106. [PubMed] [Google Scholar]
- Vanderwal T, Eilbott J, Finn ES, Craddock RC, Turnbull A, Castellanos FX. Individual differences in functional connectivity during naturalistic viewing conditions. Neuroimage. 2017;157:521–530. doi: 10.1016/j.neuroimage.2017.06.027. [DOI] [PubMed] [Google Scholar]
- Vanduffel W, Zhu Q, Orban GA. Monkey cortex through fMRI glasses. Neuron. 2014;83:533–550. doi: 10.1016/j.neuron.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, Van Essen DC, Zempel JM, Snyder LH, Corbetta M, Raichle ME. Intrinsic functional architecture in the anaesthetized monkey brain. Nature. 2007;447:83–86. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- Wang J, Ren Y, Hu X, Nguyen VT, Guo L, Han J, Guo CC. Test-retest reliability of functional connectivity networks during naturalistic fMRI paradigms. Hum Brain Mapp. 2017;38:2226–2241. doi: 10.1002/hbm.23517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wig GS, Laumann TO, Petersen SE. An approach for parcellating human cortical areas using resting-state correlations. Neuroimage. 2014;93:276–291. doi: 10.1016/j.neuroimage.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TL, Mishra A, Wang F, Yang PF, Gore JC, Chen LM. Effects of isoflurane anesthesia on resting-state fMRI signals and functional connectivity within primary somatosensory cortex of monkeys. Brain Behav. 2016;6:e00591. doi: 10.1002/brb3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Opitz A, Craddock RC, Wright MJ, Zuo XN, Milham MP. Assessing variations in areal organization for the intrinsic brain: from fingerprints to reliability. Cereb Cortex. 2016;26:4192–4211. doi: 10.1093/cercor/bhw241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


