Abstract
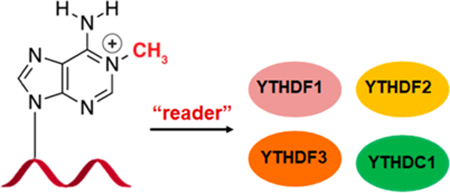
N1-methyladenosine (m1A) is an important post-transcriptionalmodification in RNA; however, the exact biological role of m1A remains to bedetermined. By employing a quantitative proteomics method, we identified multipleputative protein readers of m1A in RNA, including several YTH domain familyproteins. We showed that YTHDF1–3 and YTHDC1, but not YTHDC2, could binddirectly to m1A in RNA. We also found that Trp432 in YTHDF2, a conservedresidue in the hydrophobic pocket of the YTH domain that is necessary for itsbinding to N6-methyladenosine (m6A), is required for its recognition of m1A. An Ananalysis of previously published data revealed transcriptome-wide colocalization of YTH domain-containing proteins and m1A sites in HeLa cells, suggesting that YTH domain-containing proteins can bind to m1A in cells. Together, our results uncovered YTH domain-containing proteins as readers for m1A in RNA and provided new insight into the functions of m1A in RNA biology.
Graphical abstract
RNA is known to contain more than 100 distinct types of post-transcriptional modifications, which can modulate its splicing, localization, stability, and translation.1 It was found that methylation at the N6 position of adenosine (m6A) is involved in the epigenetic control of gene regulation.2−6 Recent transcriptome-wide mapping also revealed the presence of m1A in human mRNA,7−9 though there is some controversy about the accuracy and specificity for some of the m1A mapping methods.10 These studies also suggested the potential role of m1A in modulating mRNA splicing and translation.7−9 TRMT6, TRMT61A, and TRMT10C are three known m1A methyltransferases in human cells (writers),9 and m1A can be demethylated to adenosine by ALKBH1 and ALKBH3 (erasers).8,11 However, it remains unknown which cellular proteins are involved in binding to m1A in RNA (readers).
YTH domain-containing proteins can bind directly to m6A in RNA. YTHDF1 promotes protein synthesis by interacting with the translation machinery;12 YTHDF2 accelerates the degradation of m6A-carrying mRNA and affects the translation of heat shock proteins;13,14 YTHDF3 facilitates the translation and decay of m6A-bearing RNA in synergy with YTHDF1 and YTHDF2;15,16 YTHDC1 regulates mRNA splicing by modulating pre-mRNA splicing factors and mediates nuclear export of m6A-harboring RNA;17,18 YTHDC2 was found to be essential for meiosis in mammalian germline.19−21 Together, the YTH domain-containing proteins modulate various biological processes through binding to m6A-modified RNA.
In this study, we employed a mass spectrometry-based method to identify the m1A-binding proteins in RNA. We revealed that several YTH domain-containing proteins directly interact with m1A in RNA. In addition, functional analysis suggested important roles of YTH domain family proteins in the regulation of m1A-carrying RNA. Our results provided a foundation for further understanding of the biological functions of this important RNA modification.
RESULTS AND DISCUSSION
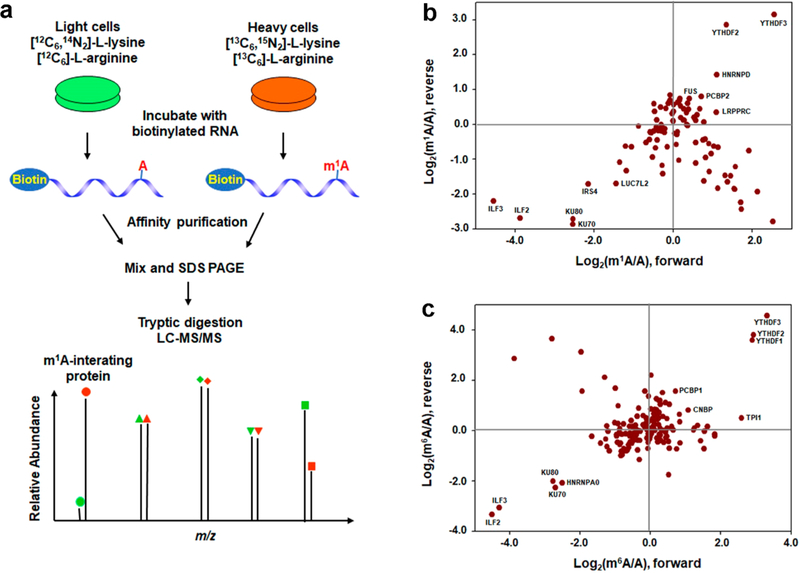
To understand better the biological functions of m1A in RNA, we employed the stable isotope labeling by amino acid in cell culture (SILAC)22-based quantitative proteomics method to screen systematically for cellular proteins that bind to m1A-bearing RNA (Figure 1a). To this end, we used a 5′-biotin-labeled m1A-carrying RNA sequence derived from human SOX18 gene, which was previously shown to harbor an m1A site near the start codon in both HEK293T and HeLa cells,7,8 as the probe bait and the corresponding unmethylated sequence as the control bait. We cultured the cells separately in heavy and light culture media and extracted proteins from these cells. An equal amount of proteins from the heavy- and light-labeled cells were incubated with biotin-conjugated m1A-bearing RNA and the corresponding unmethylated RNA, respectively, which was designated as the forward SILAC experiments. The opposite incubations in reverse SILAC experiments were conducted to remove experimental bias. After the incubation, the RNA-conjugated beads were extensively washed to remove nonspecific proteins, and the bound proteins were isolated from the beads, digested with trypsin, and analyzed using LC−MS/MS (Figure 1a).
Figure 1.
Identification of m1A-binding proteins. (a) Schematic overview of the SILAC-based quantitative proteomics method for discovering m1A-binding proteins. Shown is the workflow for a forward SILAC labeling experiment. (b,c) Scatterplot of the proteins identified from pull-down assays using m1A- (b) or m6A- (c) carrying RNA over the corresponding unmodified RNA and the whole-cell protein lysate of HEK293T cells. The proteins present in top right quadrant are m1A- (b) or m6A (c)-binding proteins, whereas those in the bottom left quadrant are proteins which bind more weakly to the m1A- or m6A-bearing RNA than the control RNA probe. The proteins that fall in the bottom right quadrant represent those proteins displaying large light/heavy ratios in both forward and reverse SILAC labeling experiments, which are likely due to incomplete SILAC labeling (for those proteins with slow turnover rate). The data were based on results from two forward and one reverse SILAC experiments.
The LC−MS/MS results revealed multiple proteins exhibiting preferential binding toward the m1A-bearing RNA over the corresponding rA-containing RNA (Figure 1b, Figure S1). In HEK293T cells, these proteins include YTH domain family members YTHDF3 and YTHDF2 and heterogeneous nuclear ribonucleoprotein hnRNPD, with the SILAC protein ratios (m1A/rA) being 6.9 ± 1.7, 4.1 ± 2.7, and 2.3 ± 0.3, respectively (Figure 1b, Table S2). In HeLa cells, the proteins which bind more strongly to m1A-bearing RNA probe over rA-bearing RNA probe encompass TAR DNA-binding protein (TARDBP), DEAD-box helicase DDX56, YTHDF proteins, and others (Figure S1, Table S2). Among them, YTHDF1, YTHDF2, and YTHDF3 displayed SILAC ratios (m1A/rA) of 2.3 ± 0.7, 1.9 ± 0.3, and 2.1 ± 0.4, respectively (Table S2). It is worth noting that our LC−MS/MS results also identified some proteins that bind more strongly to rA-containing RNA probe over m1A-bearing RNA probe, such as interleukin enhancer binding factors ILF2 and ILF3, which form a complex and negatively regulates microRNA processing,23 KU70 and KU80, which forms a heterodimer and functions in DNA double strand break repair via the nonhomologous end-joining pathway,24 etc. (Figure 1b and Table S2).
We also performed the SILAC experiments using the corresponding m6A and rA probes in HEK293T cells. As expected, the SILAC-based interaction screening resulted in the identification of YTHDF3, YTHDF2, and YTHDF1 as proteins displaying preferential binding toward the m6A-containing RNA over the rA-containing RNA substrate, with the SILAC protein ratios (m6A/rA) being 14.6 ± 7.9, 9.7 ± 3.6, and 9.0 ±2.7, respectively (Figure 1c, Table S3). This result is consistent with the previous finding,13 validating that our SILAC-based method is capable of identifying specific RNA-binding proteins. Aside from YTH domain family proteins, our SILAC-based interaction screening also identified several other m6A-binding proteins, such as PCBP1 and CNBP, which exhibited the SILAC ratios (m6A/rA) of 2.1 ± 0.8 and 1.9 ± 0.3, respectively (Figure 1c, Table S3). Moreover, our results showed that some proteins exhibit stronger binding affinities to the rA-containing RNA probe over the corresponding m6A-bearing RNA probe, such as heterogeneous nuclear ribonucleoprotein hnRNPA0, and the aforementioned ILF2, ILF3, KU70, and KU80, etc.
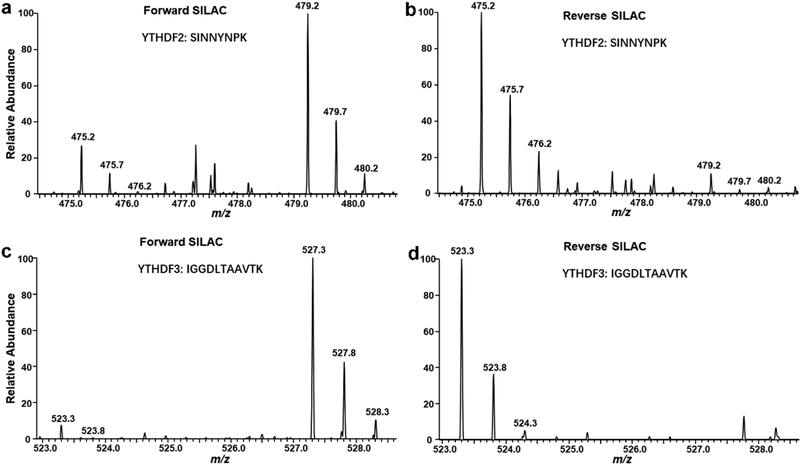
Representative LC−MS results for a tryptic peptide derived from YTHDF2, SINNYNPK are shown in Figure 2, which clearly revealed the stronger binding of YTHDF2 to the m1A-containing RNA than the corresponding unmethylated probe in both forward and reverse SILAC experiments (MS/MS shown in Figure S2). The selective binding of YTHDF3 toward m1A-containing RNA also found its support from a representative tryptic peptide derived from YTHDF3, i.e., IGGDLTAAVTK (Figure 2, MS/MS shown in Figure S3).
Figure 2.
Representative ESI-MS for the [M + 2H]2+ ions of a tryptic peptide of YTHDF2, SINNYNPK (a, b) and a tryptic peptide of YTHDF3, IGGDLTAAVTK (c, d) revealing the preferential binding of YTHDF2 and YTHDF3 toward the m1A-containing RNA probe in both forward (a, c) and reverse (b, d) SILAC experiments.
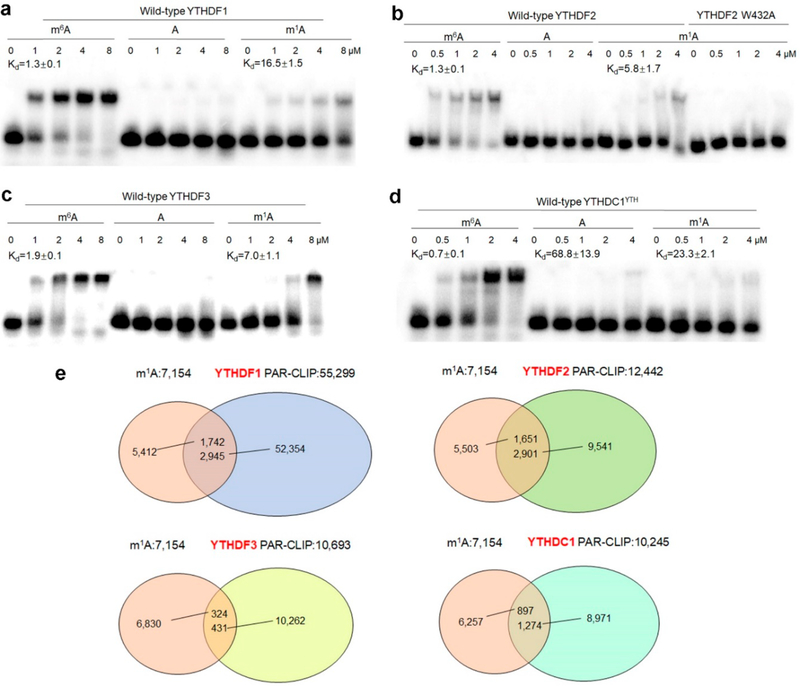
SILAC-based quantitative proteomic approach may also result in the discovery of those proteins that interact indirectly with m1A-bearing bait through protein−protein interaction(s). Thus, we examined whether YTH domain-containing proteins can interact directly with m1A in RNA. Our results from electrophoretic mobility shift assay (EMSA) showed that YTHDF1–3 and the YTH domain of YTHDC1 (YTHDC1YTH), but not the YTH domain of YTHDC2 (YTHD C2YTH), bind more strongly to m1A-carrying RNA substrate than its unmethylated counterpart (Figure 3a–d, Figure S4). We also compared the binding difference of YTH domain-containing proteins toward m1A and m6A-bearing RNA baits. The EMSA results showed that YTHDF1–3 and YTHDC1YTHbind more strongly to m6A-carrying RNA than m1A-carrying RNA, with the Kd values being 1.3 ± 0.1/16.5 ± 1.5, 1.3 ± 0.1/5.8 ± 1.7, 1.9 ± 0.1/7.0 ± 1.1, and 0.7 ± 0.1/23.3 ± 2.1 μM (m6A/m1A), respectively (Figure 3a–d). Together, our results support that YTHDF1–3 and YTHDC1YTH can bind directly to m1A-containing RNA substrate, albeit at affinities that are lower than those toward the m6A-containing RNA probe, which are in keeping with the findings made from SILAC-based interaction screening.
Figure 3.
YTH domain-containing proteins bind directly to m1A-carrying RNA. (a−d) Electrophoretic mobility shift assay (EMSA) for measuring the binding affinity of YTHDF1 (a), YTHDF2 and W432A mutant proteins (b), YTHDF3 (c), and YTH domain of YTHDC1 (d) with m6A, A, and m1A-carrying RNA probes. The Kd values (in μM) are listed in (a)−(d), and error bar represents the SEM (n = 3). (e) Overlap between the m1A sites (200 bp window centered on the m1A sites) and YTHDF1–3, and YTHDC1 PAR-CLIP binding sites in HeLa cells.
The X-ray crystal structure of YTHDF2 revealed three aromatic amino acid residues in the hydrophobic pocket of the YTH domain of YTHDF2 that are crucial for its binding toward m6A.13,25 To explore whether this hydrophobic pocket also assumes an important role in binding toward m1A, we conducted EMSA assay with a mutant form of YTHDF2 protein where the conserved Trp432 was mutated to an alanine (W432A). The result indeed showed that the mutation led to a pronounced attenuation in binding affinity toward the m1A-containing probe (Figure 3b), suggesting that m1A may occupy the same hydrophobic pocket of YTHDF2 that is required for m6A recognition.
We further interrogated the publicly available transcriptome-wide m1A mapping data7 and photoactivatable ribonucleoside cross-linking and immunoprecipitation (PAR-CLIP) data12,16,18,26 of YTH domain-containing proteins in HeLa cells to assess whether YTH domain-containing proteins bind to m1A-bearing RNA in cells. In this aspect, we analyzed the overlap between the m1A sites (200 bp window centered on the m1A sites) and YTHDF1–3, and YTHDC1 PAR-CLIP binding sites in HeLa cells. The result showed that 24.4%, 23.1%, 4.5%, and 12.5% of the m1A sites overlapped with the YTHDF1, YTHDF2, YTHDF3, and YTHDC1 PAR-CLIP peaks, respectively (Figure 3e, Table S4). Therefore, these results suggest that YTH domain-containing proteins can recognize m1A in transcripts in cells. Analysis of the ribosome profiling data and RNA lifetime data12,16,26 in HeLa cells revealed that, upon YTHDF2 knockdown, the YTHDF2-bound m1A-containing mRNA targets experienced a significant increase in lifetime compared with other targets (p = 1.0 × 10−28, Mann− Whitney U-test, Figure S5, Table S5), whereas the YTHDF1-binding m1A-containing targets exhibited a significant decrease in translation efficiency compared with other targets upon genetic depletion of YTHDF1 (p = 8.9 × 10−12, Mann− Whitney U-test, Figure S6, Table S6), suggesting that YTHDF1 may promote translation whereas YTHDF2 mainly accelerates the degradation of m1A-carrying transcripts.
CONCLUSIONS
In summary, we identified by using an unbiased quantitative proteomic method, for the first time, YTH domain-containing proteins as the readers of m1A in RNA, which expands the repertoire of RNA modifications that can be regulated by this protein family. We demonstrated that YTH domain-containing proteins directly interact with m1A-bearing RNA. Furthermore, we identified multiple other putative m1A-binding proteins by using SILAC-based quantitative proteomic screening. Further characterizations of these proteins in m1A recognition and RNA metabolism will improve our understanding about the biological functions of m1A in RNA.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Prof. Chuan He for providing pGEX-4T1-YTHDF2 and pGEX-4T1-YTHDF3 plasmids. This work was supported by the National Institutes of Health (Grant R01 ES025121). X.D. was supported in part by an NRSA institutional training grant (Grant T32 ES018827).
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.anal-chem.8b01703.
Detailed experimental procedures, list of PCR primers, LC−MS and MS/MS data, effects of knock-down of YTHDF1 and YTHDF2 on the lifetime and translation efficiency of m1A-containing RNA, and quantitative proteomic data (PDF)
List of proteins quantified from the SILAC-based interaction screening for m1A-carrying RNA and the corresponding adenosine-carrying RNA using lysates from HEK293T cells and HeLa cells (XLSX)
List of proteins quantified from the SILAC-based interaction screening for m6A-carrying RNA and the corresponding adenosine-carrying RNA using lysates from HEK293T cells (XLSX)
m1A sites which overlap with YTHDF1–3 and YTHDC1 PAR-CLIP binding sites in HeLa cells (XLSX)
Comparison of RNA lifetime in cells with and without siRNA-mediated knockdown of YTHDF1 or YTHDF2 (XLSX)
Translation efficiency of mRNA in cells with and without siRNA-mediated knockdown of YTHDF1, YTHDF2, and YTHDF3 (XLSX)
REFERENCES
- (1). Cantara WA; Crain PF; Rozenski J; McCloskey JA; Harris KA; Zhang X; Vendeix FA; Fabris D; Agris PF Nucleic Acids Res. 2011, 39, D195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2). Jia G; Fu Y; Zhao X; Dai Q; Zheng G; Yang Y; Yi C; Lindahl T; Pan T; Yang YG; He C Nat. Chem. Biol 2011, 7, 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3). Zheng G; Dahl JA; Niu Y; Fedorcsak P; Huang CM; Li CJ; Vagbo CB; Shi Y; Wang WL; Song SH; Lu Z; Bosmans RP; Dai Q; Hao YJ; Yang X; Zhao WM; Tong WM; Wang XJ; Bogdan F; Furu K; et al. Mol. Cell 2013, 49, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4). Jia G; Fu Y; He C Trends Genet. 2013, 29, 108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5). Schwartz S; Agarwala SD; Mumbach MR; Jovanovic M; Mertins P; Shishkin A; Tabach Y; Mikkelsen TS; Satija R; Ruvkun G; Carr SA; Lander ES; Fink GR; Regev A Cell 2013, 155, 1409–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6). Liu N; Dai Q; Zheng G; He C; Parisien M; Pan T Nature 2015, 518, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7). Dominissini D; Nachtergaele S; Moshitch-Moshkovitz S; Peer E; Kol N; Ben-Haim MS; Dai Q; Di Segni A; Salmon-Divon M; Clark WC; Zheng G; Pan T; Solomon O; Eyal E; Hershkovitz V; Han D; Dore LC; Amariglio N; Rechavi G; He C Nature 2016, 530, 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8). Li X; Xiong X; Wang K; Wang L; Shu X; Ma S; Yi C Nat. Chem. Biol 2016, 12, 311–316. [DOI] [PubMed] [Google Scholar]
- (9). Safra M; Sas-Chen A; Nir R; Winkler R; Nachshon A; Bar-Yaacov D; Erlacher M; Rossmanith W; Stern-Ginossar N; Schwartz S Nature 2017, 551, 251–255. [DOI] [PubMed] [Google Scholar]
- (10). Grozhik AV; Jaffrey SR Nat. Chem. Biol 2018, 14, 215–225. [DOI] [PubMed] [Google Scholar]
- (11). Liu F; Clark W; Luo G; Wang X; Fu Y; Wei J; Hao Z; Dai Q; Zheng G; Ma H; Han D; Evans M; Klungland A; Pan T; He C Cell 2016, 167, 1897. [DOI] [PubMed] [Google Scholar]
- (12). Wang X; Zhao BS; Roundtree IA; Lu Z; Han D; Ma H; Weng X; Chen K; Shi H; He C Cell 2015, 161, 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13). Zhu T; Roundtree IA; Wang P; Wang X; Wang L; Sun C; Tian Y; Li J; He C; Xu Y Cell Res. 2014, 24, 1493–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14). Zhou J; Wan J; Gao X; Zhang X; Jaffrey SR; Qian SB Nature 2015, 526, 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15). Li A; Chen YS; Ping XL; Yang X; Xiao W; Yang Y; Sun HY; Zhu Q; Baidya P; Wang X; Bhattarai DP; Zhao YL; Sun BF; Yang YG Cell Res. 2017, 27, 444–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16). Shi H; Wang X; Lu Z; Zhao BS; Ma H; Hsu PJ; Liu C; He C Cell Res. 2017, 27, 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17). Xiao W; Adhikari S; Dahal U; Chen YS; Hao YJ; Sun BF; Sun HY; Li A; Ping XL; Lai WY; Wang X; Ma HL; Huang CM; Yang Y; Huang N; Jiang GB; Wang HL; Zhou Q; Wang XJ; Zhao YL; et al. Mol. Cell 2016, 61, 507–519. [DOI] [PubMed] [Google Scholar]
- (18). Roundtree IA; Luo GZ; Zhang Z; Wang X; Zhou T; Cui Y; Sha J; Huang X; Guerrero L; Xie P; He E; Shen B; He C eLife 2017, 6, No. e31311, DOI: 10.7554/eLife.31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19). Hsu PJ; Zhu Y; Ma H; Guo Y; Shi X; Liu Y; Qi M; Lu Z; Shi H; Wang J; Cheng Y; Luo G; Dai Q; Liu M; Guo X; Sha J; Shen B; He C Cell Res. 2017, 27, 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20). Wojtas MN; Pandey RR; Mendel M; Homolka D; Sachidanandam R; Pillai RS Mol. Cell 2017, 68, 374–387. [DOI] [PubMed] [Google Scholar]
- (21). Bailey AS; Batista PJ; Gold RS; Chen YG; de Rooij DG; Chang HY; Fuller MT eLife 2017, 6, No. e26116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22). Ong SE; Blagoev B; Kratchmarova I; Kristensen DB; Steen H; Pandey A; Mann M Mol. Cell. Proteomics 2002, 1, 376– 386. [DOI] [PubMed] [Google Scholar]
- (23). Sakamoto S; Aoki K; Higuchi T; Todaka H; Morisawa K; Tamaki N; Hatano E; Fukushima A; Taniguchi T; Agata Y Mol. Cell. Biol 2009, 29, 3754–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24). Walker JR; Corpina RA; Goldberg J Nature 2001, 412, 607–614. [DOI] [PubMed] [Google Scholar]
- (25). Li F; Zhao D; Wu J; Shi Y Cell Res. 2014, 24, 1490–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26). Wang X; Lu Z; Gomez A; Hon GC; Yue Y; Han D; Fu Y; Parisien M; Dai Q; Jia G; Ren B; Pan T; He C Nature 2014, 505, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.