Abstract
Motivation
In recent years there have been several efforts to generate sensitivity profiles of collections of genomically characterized cell lines to panels of candidate therapeutic compounds. These data provide the basis for the development of in silico models of sensitivity based on cellular, genetic, or expression biomarkers of cancer cells. However, a remaining challenge is an efficient way to identify accurate sets of biomarkers to validate. To address this challenge, we developed methodology using gene-expression profiles of human cancer cell lines to predict the responses of these cell lines to a panel of compounds.
Results
We developed an iterative weighting scheme which, when applied to elastic net, a regularized regression method, significantly improves the overall accuracy of predictions, particularly in the highly sensitive response region. In addition to application of these methods to actual chemical sensitivity data, we investigated the effects of sample size, number of features, model sparsity, signal-to-noise ratio, and feature correlation on predictive performance using a simulation framework, particularly for situations where the number of covariates is much larger than sample size. While our method aims to be useful in therapeutic discovery and understanding of the basic mechanisms of action of drugs and their targets, it is generally applicable in any domain where predictions of extreme responses are of highest importance.
Availability and implementation
The iterative and other weighting algorithms were implemented in R. The code is available at https://github.com/kiwtir/RWEN. The CTRP data are available at ftp://caftpd.nci.nih.gov/pub/OCG-DCC/CTD2/Broad/CTRPv2.1_2016_pub_NatChemBiol_12_109/ and the Sanger data at ftp://ftp.sanger.ac.uk/pub/project/cancerrxgene/releases/release-6.0/.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The emerging development of new cancer therapeutics requires an extended process of experimentation, testing, safety and efficacy evaluation. Human cancer cell lines can help identify associations between molecular subtypes, pathways and drug responses. In recent years, there have been several efforts to generate genomic and gene-expression profiles of collections of cell lines to determine the mechanisms of their responses to panels of candidate therapeutic compounds. These data provide the basis for the development of in silico models of sensitivity based on measured features of cancer cells; see Garnett et al. (2012), Barretina et al. (2012), Basu et al. (2013), Seashore-Ludlow et al. (2015), Rees et al. (2016). Developing classification and prediction methodology from these data profiles will be beneficial in identifying single and combinatorial chemotherapeutic response potential in patients. Advanced computational methodology developed in this chemical-genetics space will be useful not only in therapeutic discovery, but also in understanding the basic mechanisms of action for drugs and putative drug–drug interactions.
A handful of classification and prediction algorithms have been utilized in order to better understand patient responses to drugs, and may facilitate the individualization of patient treatment. For instance, in Costello et al. (2014) the authors, as part of the NCI-DREAM drug-sensitivity prediction challenge, analyzed and compared 44 drug-sensitivity prediction algorithms based on genomic and proteomic profiles of breast cancer cell lines. The complete list of methods applied to the prediction of chemosensitivity thus far is too large to list exhaustively. Some of these methods include random forest described by Breiman (2001) and applied by Riddick et al. (2011), Touw et al. (2013), among others; various regularized regression methods such as ridge and lasso regression described by Hoerl and Kennard (2000) and Tibshirani (1996) respectively and applied by Hoggart et al. (2008), Wu et al. (2009), Ayers and Cordell (2010) and Geeleher et al. (2014); and support-vector machines described by Cortes and Vapnik (1995) and applied by Bao and Sun (2002). A method used more recently in cancer cell-line profiling and genome-wide association studies is elastic net (EN) developed in Zou and Hastie (2005). EN has been applied by Barretina et al. (2012), Garnett et al. (2012), Liang et al. (2013), Neto et al. (2014), Guinney et al. (2014) and Sokolov et al. (2016) among others. The mathematical formulation of elastic net combines the formulations for ridge and lasso regressions. Like ridge and lasso regressions, EN also typically applies to cases where the number of predictors exceeds the number of observations while overcoming some of the limitations of lasso and ridge regression models. In addition, EN encourages the so called grouping effect, where strongly correlated predictors tend to be in or out of the model together (cf. Zou and Hastie, 2005). Example applications of EN include: endophenotypic analysis where 100 SNPs were identified for each chromosome as ‘interesting’ based on having the highest absolute-value regression coefficients (Palejev et al., 2011). Additionally, multiple groups have used regression for chemosensitivity prediction analyzing an integrated set of ‘omics’ data on a limited set of compounds (cf. Shimokuni et al., 2006). Relatedly, the DREAM3 Challenge was launched to use genomic information to build models capable of ranking sensitivity (Wan et al., 2014). Sensitivity data were analyzed using EN to identify a parsimonious model that best predicted response to a single agent, navitoclax (Basu et al., 2013) and to identify biomarkers of small-molecule sensitivity.
Though ‘out of the box’ elastic net has been successful at prediction in many cases across different applications (Neto et al., 2014), some limitations still exist. For example, reported mean-squared error mainly reflects prediction of bulk non-responder cell lines in the center of the distribution, whereas the most sensitive cell lines, potentially of greater biological relevance, lie on the tail of the distribution. Thus, when using the default implementation of EN (e.g. ‘out of the box’ glmnet in R) on such datasets, the resulting EN model can selectively underperform on regions of the data that we are most interested in predicting correctly. Other challenges of using EN include more general regularization caveats, such as model over-fitting and under-fitting depending on the dimension and input feature type (binary versus continuous). Noisy genetic sensors that originate from heterogeneous genomic datasets can also pose a challenge to such models.
Here, we apply a weighted-response method to elastic net method for prediction of chemosensitivity profiles to achieve a predictive understanding of sensitivity for two different datasets. In the first dataset from Rees et al. (2016), hereafter referred to as CTRP dataset, we apply our method to a collection of 823 cell lines exposed to 481 compounds. A second dataset from Yang et al. (2012) and Iorio et al. (2016), hereafter referred to as the Sanger dataset, contains data on 985 cell lines that were exposed to 265 different compounds. To accurately report predictive performance results, for both datasets, we divided data for each compound randomly into two groups—a training set and a test set, on which we evaluated the accuracy of our predictions. For both the training and the test sets, we have prior knowledge of the identification of cell lines as sensitive or non-sensitive. We propose several response-weighting schemes that aim to predict the sensitivity score for the sensitive cell lines in the test dataset based on modeling of the training dataset. These static weighting schemes are formulated so as to put increasingly more weights on observations further left in the left tail; this is done in order to better predict more sensitive cell lines. We aim to show that response weighting generally improves the prediction performance for the tail observations as compared to elastic net with equal weighting on all responses, though prediction of observations in the extreme tail leaves some room for improvement.
The identification of observations at the extremeties have gotten a lot of attention in the outlier-detection literature. Rousseeuw and Leroy (2005) discussed the robust regression framework for modeling in presence of outliers. More recent and rather exhaustive surveys of outlier detection were performed by Hodge and Austin (2004) and Chandola et al. (2009) and references therein. Additionally, Meinshausen (2006) has proposed a quantile random-forest based approach that can be used for detecting outliers. Work reported by Cai and Reeve (2013) and by Schaumburg (2010) also use quantile regressions to predict extreme observations. Our proposed iterative weighting method is most similar in philosophy to that of Cheze and Poggi (2006). In Cheze and Poggi (2006), the authors adopted an iterative scheme for identifying outliers. The key idea in that paper was to use boosting (Freund and Schapire, 1997) with regression trees (Breiman et al., 1984). Since boosting works by iteratively identifying and re-weighting badly predicted observations, their algorithm keeps track of the most frequently appearing observations in the bootstrap sample and provides a confidence interval for such an observation to be an outlier. One key way, in which our work differs from Cheze and Poggi (2006), or for that matter any other outlier detection literature, is that we assume that the set of ‘outliers’ as defined by the tail of the distribution is already specified; our goal is to be able to predict the tail observations well—possibly at the cost of the rest of the observations. As in Cheze and Poggi (2006) we keep track of the badly predicted observations and at each iteration apply more weight to those. However, unlike in Cheze and Poggi (2006), we employ elastic net for prediction instead of decision trees.
In the following sections, we first define the relevant error measure and weighting schemes most suited for our purposes (Section 2). Next we apply our proposed methods (Section 3). Specifically in Section 3.1, we perform wide-ranging benchmarking of prediction performance via simulations with synthetic data. By comparing our proposals under a variety of tunable design parameters, we show that iterative response weighting outperforms other methods under a variety of data-generation parameters. In Section 3.2, we apply our methods to real chemosensitivity data and judge its performance. Finally we discuss some key features of our work and describe some directions for future work (Section 4).
2 Materials and methods
Let us consider n observations on chemosensitivity of n cancer cell lines (CCL) treated with a specific compound. For each cell line we also denote by the genetic expression data of each cell line on p specific genetic features. We denote by —the n × p feature matrix for the genetic expression data of the n CCLs on the p features. In the following description, we denote by a prediction for y. For a set A, we denote by the indicator to the set A; so that if the condition in set A is satisfied and 0 otherwise.
2.1 Elastic net prediction
As noted earlier, for the purposes of our model-based prediction in the current paper, we will primarily employ elastic net for squared-error loss. The mathematical formulation of elastic net is expressed as an optimization problem. Let be the parameter of interest. Following, e.g. Hastie and Qian (2014),
| (1) |
In the above formulation in (1), the wi ’s are individual observation weights so that and . We note here that the ‘standard’ weighting scheme for application of elastic net is to have equal weights for all observations:
| (2) |
The parameter α is such that and acts as a bridge between ridge penalty and lasso penalty ; for α = 1, the formulation in (1) is a lasso problem and for α = 0, a ridge problem. The parameter denotes the overall tuning parameter.
Given the parameter estimate , prediction of y will be given by .
2.2 Measure of error
For prediction of continuous data, the ubiquitous measure of error is the root-mean-squared error (RMSE) defined by . As noted earlier, the purpose of this work is to predict the sensitivity score of the sensitive cell-lines that fall on the lower (left) tail of the data distribution. This requirement necessitates a slightly modified definition of error measure that only accounts for the error incurred in the sensitive region of the data. We introduce the following definition:
| (3) |
where , the left curtail point, is assumed to be known in advance. The letter stands for left. Since we are mostly concerned with sensitive cancer cell lines, will be our default choice of performance measure. For the sake of completeness, we also define the following measures of RMSE. For non-responsive cell lines that lie in the right tail defined by some right curtail point , we can define
| (4) |
A similar definition for both sensitive and non-responsive tails also follows directly:
| (5) |
In the following descriptions, we will treat only the measure as we describe in detail our response-weighting strategy for prediction of left-tailed observations.
2.3 Weighting for left-tail predictions
The equal weighting scheme in (2) treats all observation with the same amount of importance. For concentration-response curves, however, smaller values of the area-under-curve (AUC) indicate increased sensitivity to the compounds. As such, we will now focus on predicting the cell lines with smaller AUC values well; i.e. the AUC values falling in the left tail of the data. Contrary to equal-weighting scheme (2), for left-tailed prediction, we put more weights on the left-tail observations which are defined via the curtail point which is assumed to be known. We tested the following weighting schemes: for ,
| (6) |
Above, the notations and denote the sample mean and sample standard deviation respectively, of the observed y values. In all the cases above, we rescale the weights so that they sum to unity.
Remark 1 (choice of ). An important issue is the choice of the left curtail point . Though, the definitions in (6) are applicable for any arbitrary choice of , for the rest of this paper, we adhere to the following data-dependent choice of :
| (7) |
where stands for median absolute deviation defined as
| (8) |
This choice of left cutoff point can be thought of as a robust proxy for the lower limit of a 90% confidence interval for the location parameter of cell-line AUC values. It is so chosen that probabilistically only about 5% of the AUCs will fall below this threshold. More detailed discussion can be found in the Supplement.
Remark 2 (exponentiated weights). It is straightforward to construct exponentiated versions of the weighting schemes in (6). For example, for any positive number , we can define the -order exponentiated weight as, for and rescale to make the weights sum to unity. Note that these weighting schemes can be easily extended for right tail or both tails. For the sake of completeness, we have provided a definition in Supplementary Section S2.
Algorithm 1 Iterative Response Weighted Elastic Net
Data: The following inputs are assumed as given:
The training data where and
An elastic net prediction function that takes as argument the training data and a weight vector and returns the predicted valiue: .
A left curtail point
Left RMSE calculation function: as in (3)
A tolerance level () and a maximum possible number of iterations ).
Initialization: Set t = 0 and Error = . Choose for all and set .
1: while (Error ) do
2: Find
3: Set
Renormalize so that the weights sum to 1.
4: Set and set .
5: Calculate Error =
6: Set
7: end while
2.4 Iterative weighting scheme
Elastic net fits a linear regression line to the mean of the observed response values. As such, it will tend to overestimate the left tail (and underestimate the right tail) observations without any tail-weighting scheme. In our above weighting schemes, we put more emphasis on tail observations by putting more weights on observations in the tail and fitting an elastic net regression. In all the above schemes, more weight is assigned to those observations that are further from the left-tail curtail point.
Iterative weighting adopts a somewhat different idea: it starts by putting equal weights on all observations and fitting a mean regression line. Then, at each iteration, it assigns more weights to those observations in the tail (defined by ) that deviate more from the original response value. In detail, if yi is a specific observation in the tail and is the prediction for that observation at the iteration, then for the fit, we assign to , the weight,
| (9) |
The observations that are not in the tail are assigned a very small value. In our empirical studies, any small value or even zero works well. This procedure is motivated by the boosting idea (Freund and Schapire, 1997), which is celebrated in the machine-learning literature owing to its ability to reinforce a weak classifier through successive iterations and re-weighting (see also Freund and Schapire, 1996, 1997; Freund, 1995; Friedman, 2001). The detailed algorithm is as shown in Algorithm 1.
In the following section, we will first implement these algorithms on synthetic datasets and then judge their performances.
3 Application and results
3.1 Experiments on synthetic data
We considered a large array of simulation designs whose parameters, we varied in a systematic fashion (see Neto et al., 2014). A detailed description of the simulation designs can be found in the Supplementary Section S3. In brief, we generated data based on the regression framework . The parameters we varied are as follows: i) sample size (n) of both training and test data; ii) number of features (p), i.e. the number of columns of X; the design matrix; iii) saturation (), the higher the value of —the more number of nonzero elements in ; iv) signal-to-noise ratio (η); v) within feature correlation (ρ)—within-group correlation among the columns of X.
3.1.1. Choice of performance measure
One important aspect in the simulation study is the choice of performance measure. We considered the measure. However, owing to the variability in the scale of the response variable, partly due to varying sparsity levels in different designs, we will consider a scaled error measure defined as follows:
| (10) |
We used on the test data to compare the performance. Note that, the order of the performances of different methods for a particular design remains unaffected by this rescaling while enabling comparison of performances across different designs.
3.1.2. Comparison of methods
We considered the following 11 methods: (1) equally weighted; (2–8)-tscore weighted with exponents k = 1/6; 1/4; 1/2; 1; 2; 4; 6; (see Remark 2) for definition of exponentiation; (9) sigmoid-weighted; (10) quartile-curtailed-weighted; (11) iteratively weighted.
3.1.3. Training, validation and prediction
We divided our simulated data for each design parameter combination into two equal parts randomly and used one part to perform 10-fold cross-validation to select optimal λ parameter for elastic net as in (1). Additional details can be found in Supplementary Section S3. We have kept in all scenarios; in our trials we found this value to perform well. On the other hand, cross-validation for both takes up a lot of computational time while providing negligible performance boosts.
Comparison of all methods in a single figure is cumbersome. For the sake of clarity, we summarize the key findings and figures.
3.1.4. Simulation results
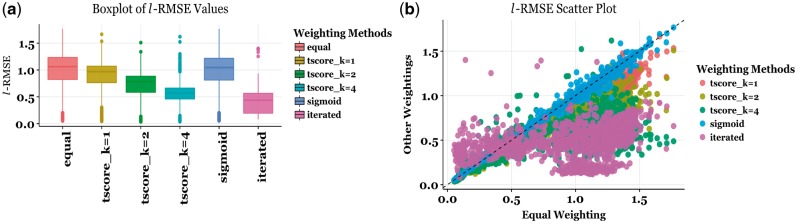
The simulation results demonstrate that response weighting in general, and iterative weighting in particular is very effective in predicting left-tail simulated concentration-response sensitivity scores (area-under-curve; AUC) across all simulation parameters (Fig. 1a). The left-most boxplot represents values for equal weighting as in (2) and the right most box plot represents the values for iterative weighting with Algorithm 1. t-score with exponent k = 4 ( from left) and iterative weighting outperformed the other tested methods and the improvement in prediction performance is substantial.
Fig. 1.
(a) The box plot of values for all weighting methods and all designs. The left most bar plot represents equal weighting while the right most one represents iterative weighting. (b) The scatter plot of values for all weighting methods (vertical axis) as compared to the equal weighting scheme (horizontal axis). Observations below the diagonal signify that another weighting method performs better than equal weighting (Color version of this figure is available at Bioinformatics online.)
Scatter plots provide a facile means of visualizing values for all designs (Fig. 1b). On scatter-plot vertical axis we have for weighting methods via t-score with exponents 1, 2 and 4, sigmoid weighting and iterative weighting (indexed by different colors), and horizontal axis we have values with equal weighting. An overwhelming majority of the points lie below the diagonal, indicating that most weighting methods considered have an advantage over no weighting. One possible exception is the sigmoid weighting (blue) which has almost similar performance as no weighting; sigmoid points lie mostly on the diagonal. The iterative weighting scheme overwhelmingly lies below the diagonal (except a few at the bottom left corner) indicating large improvements over equal weighting as well as other weighting schemes.
We tabulated the overall performance of all methods (Supplementary Table S1). For a better understanding of the performance of the methods, we considered error box plots when several design parameters are varied. For clarity, we only compared equal weighting with three best-performing methods, namely t-score with exponents k = 2, 4, and iterative weighting. The results show that iterative weighting works consistently across variations of all design parameters considered.
3.2 Application in chemosensitivity data
We next applied our method to several compounds using two gene-expression datasets. We wanted to assess whether weighting functions (see Section 3.1.2) would help achieve a higher predictive accuracy in predicting the tail for our compound-sensitivity data.
3.2.1. CTRP dataset
Small-molecule sensitivity. An Informer Set of 481 small molecules was tested for sensitivity in 823 publicly available human CCLs (see Rees et al., 2016; data available from the NCI CTD2 Data Portal at: ftp://caftpd.nci.nih.gov/pub/OCG-DCC/CTD2/Broad/CTRPv2.1_2016_pub_NatChemBiol_12_109/). The effects of small molecules were measured over a 16-point concentration range (2-fold dilution) in duplicate. As a surrogate for viability, cellular ATP levels were assessed 72 h after compound transfer using CellTiterGlo (Promega), and the area under curve (AUC) for each compound—cancer cell line pair was calculated by numerically integrating under the 16-point concentration-response curve.
Features. Our feature dataset contains basal gene-expression data for the same 823 cell lines (Barretina et al., 2012); it contains a total of 18 543 features. For each compound, our goal is to predict the value of area-under-curve (AUC) concentration-responses for each the CCLs using the gene-expression data. In particular, lower-tail values of the CCL AUCs suggest greater sensitivity to the compound while values at the upper tail indicate non-responsiveness. Of greater interest is the prediction of the lower-tail values. Basu et al. (2013) and others previously applied elastic net (EN) penalized regression for prediction of the small-molecule sensitivity; here we study the better estimation of tails of the CCL-AUCs.
3.2.2. Sanger dataset
Dataset Description. This dataset is from Yang et al. (2012). The gene expression dataset contains data on 17 737 gene ensembles for 1018 cell lines. This is matched to a separate dataset on area under the curve for dose-response curves for 265 drugs (identified via DRUG_ID) and a total of 1074 cell lines (identified via COSMIC_ID). We combined the AUC and gene-expression datasets in order to obtain a final dataset where we had, for each drug, AUC values corresponding to a (varying) number of cell lines and matching gene-expression values.
3.2.3. Data preparation
For each of these datasets, we further selected those drugs for which there were at least 30 observations in the left tail (as defined by in (7)) of the overall dataset; this led to a further reduction to 185 compounds for both CTRP and Sanger datasets. Thus, in effect, for both CTRP and Sanger, we applied our proposed methods to 185 different compound datasets. For each compound dataset, we trained on 80% of the data, and used cross-validation to select optimal tuning parameters. We report prediction performance on the remaining 20% of the data. Here, we present the results we obtained by applying our proposals to the test split. We refer to Supplementary Section S5 for further details on data pre-processing for each dataset.
3.2.4. Comparison of methods
For application of our proposed methods to real-world data, we have compared the following methods: (i) equally weighted; (ii) t-score with k = 4, (iii) iteratively weighted (iv) random forests (RF) and (v) gradient boosting machines (GBM). The implementation details for these methods can be found on Supplementary Section S5 including summarizing the ideas behind these methods with some key supplemental references.
Implementation. For both datasets, we used glmnet package as implemented in R (see Friedman et al., 2010) for the implementation of elastic net. For implementation of random forest we used the randomForest package in R (see Liaw and Wiener, 2002). For gradient boosting, we used the gbm package (see Ridgeway, 2010).
3.2.5. Compound sensitivity prediction results
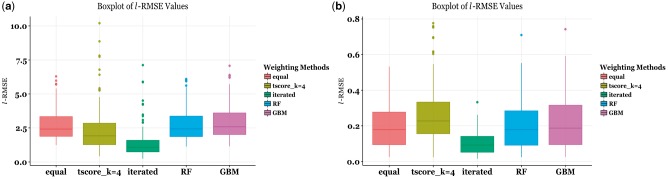
For both datasets, we carried out response-weighted model-building for 185 compounds across five best-performing weighting methods (Fig. 2a and b). We observed that while weighting helps, specific weighting methods such as the t-score (k = 4) and iterative weighting help the performance significantly in the extremely sensitive tail regions of the CTRP data across all compounds. Specifically, the medians of all values over all compounds for equal weighting, RF and GBM are 2.41, 2.43 and 2.59 respectively while the same for t-score with k = 4 is 1.92 and for iterative weighting it is1.07, the lowest. On the other hand, for Sanger data, on average, across all compound datasets, RF and GBM beat t-score with k = 4 while iterative weighting still out performs all other methods. Specifically, the median values for equal weighting, RF and GBM are 0.18, 0.18 and 0.19 respectively while the median for t-score with k = 4 weighting is 0.23. For iterative weighting, this value is 0.09, again out-performing the other methods.
Fig. 2.
Boxplots for ℓ – RMSE values for all compounds for different weighting methods as applied to real chemosensitivity data. (a) the distribution of values for CTRP dataset; (b) the distribution of values for Sanger dataset. In both panels, the methods compared, from left to right, are equal weighting, t-score weighting with k = 4, iterative weighting, random forest (RF), and gradient-boosting machines (GBM) (Color version of this figure is available at Bioinformatics online.)
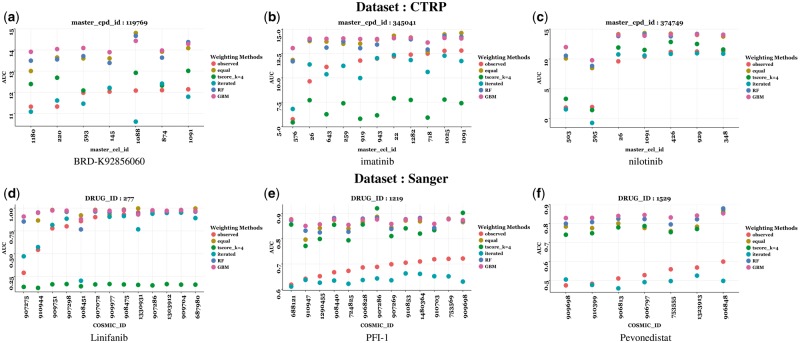
Figure 3(a–c) display prediction results for specific drugs for the CTRP dataset and Figure 3(d–f) show examples of drugs for the Sanger dataset. In Supplementary Table S2 in Supplementary Section S6, we provide the left RMSE values for these weighting methods corresponding to these same six compounds. As seen in the simulation studies, the iterative method outperforms most of the other weighting methods as well as random forest and gradient boosting methods applied here.
Fig. 3.
Predicted and actual AUCs for cell lines in the validation set. The red points are the actual AUC values which are plotted here in increasing order from left to right. The turquoise points represent the predicted CCL AUC values from iterated weighting. Also plotted here are predicted values from equal weighting, t-score with k = 1, 4, random forest and gradient boosting machines. Prediction results for 6 compounds are given here (Color version of this figure is available at Bioinformatics online.)
The rationale for the superior performance of iterative weighting can be understood as follows: the first stage equal-weighted elastic net fits a mean linear-regression line resulting in worse predictions for the outlying observations. In the following steps, the weighting is proportional to the error in prediction in the earlier fitting, which is higher for those observations predicted poorly, thus giving the outliers more importance in subsequent fitting. This improvement in prediction of the outliers is obtained at the cost of poorer predictions for cell lines that are not outliers. Our iterative weighting algorithm is a variant of the well-known boosting algorithm in machine learning that is used to strengthen the performance of otherwise weak classifiers.
We re-iterate that our proposed weighting methods (especially the iterative weighting method) is geared towards predicting left-tail sensitive values. As such, these methods will fail to predict non-sensitive cell lines. Nonethless, in Supplementary Section S7 we compare the overall RMSE values for for all cell lines—not just the sensitive ones. We also show the prediction of all cell lines for compound BRD-K92856060 in the CTRP dataset in order to bring home this point. The left tail predictions for compound BRD-K92856060 are shown in Figure 3(a).
Overall, our results indicate that weighting helps achieve a higher predictive accuracy on the tail regions of a response distribution for both CTRP and Sanger datasets.
4 Discussion
Our model improvement for elastic net, termed response-weighted elastic net (RWEN) can be applied across various datasets where outlier prediction is critical for performance evaluation. Our method can be used in conjunction with other machine-learning algorithms such as support-vector machines for feature selection and used across a broad range of applications. An alternate way to use this method is with other recent elastic net methods such as Bayesian elastic net models (Li and Lin, (2010)) or the adaptive elastic net (Zou and Zhang, 2009). A more computationally expensive two-dimensional cross-validation may also be used to search for an optimal pair of . In our analysis, this step yielded only marginal benefit while increasing the computation time considerably. Owing to this performance shortcoming, we have not included results of such an analysis. The presence of non-linear effects, or non-independent observations, may lower the performance of regularized linear models. In such cases, performance may be enhanced by using lasso type models or other procedures able to efficiently handle large numbers of interactions, such as random forests or boosted regression trees. We note that while identifying sensitive cell lines based on gene-expression features is an interesting problem in and of itself, it is much easier to handle; standard classification techniques such as logistic regression, decision trees or support-vector machines etc., can be employed for such purposes. On the other hand, predicting the actual values in the tail of a distribution is a much less-well-studied problem. A simple classification-based technique also does not distinguish between the degrees of sensitivity, putting all sensitive cell lines on equal footing.
Our current implementation of the algorithm can be used when we have an understanding of whether the cell line is sensitive or unresponsive since we use a cutoff to distinguish the cell lines prior to applying our proposed methods. Thus the prediction for the unresponsive values (observations to the right of the left-tail cut off) are not considered in measuring the performance of the proposed models. Through our method, we can achieve a predictive understanding of the putative biomarkers of sensitivity and this can help distinguish the most critical features of response from the characteristics of the cell line that are neutral. Note that we can also incorporate the process of identification of sensitive cell lines in the analysis pipeline in a two-step procedure as follows: in the first step we dichotomize the training observations depending on whether they fall to the left or right of the left-tail threshold. A simple classification algorithm can be employed to identify the tail. This identification step is then followed by our proposed response-weighted estimation step. We defer the study of this combined procedure to future work.
5 Conclusion
Response weighted elastic net (RWEN) works well in predicting the chemosensitivity profiles of sensitive compounds in a linear model framework. In particular, ourv iterative weighting scheme is able to capture more extreme values accurately compared to other methods. This procedure has general applicability in any scenario where accurate prediction of extreme observations is sought.
Supplementary Material
Acknowledgements
We acknowledge Trevor Hastie for his helpful suggestions on this work. We also acknowledge the late Joshua Gilbert for his contributions.
Funding
This work was supported by the National Institutes of Health (NIH) via the National Cancer Institute (NCI) Cancer Target Discovery and Development Network [grant number U01CA217848]. S.L.S. is an Investigator at the Howard Hughes Medical Institute.
Conflict of Interest: none declared.
References
- Ayers K.L., Cordell H.J. (2010) Snp selection in genome-wide and candidate gene studies via penalized logistic regression. Genet. Epidemiol., 34, 879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Sun Z. (2002) Identifying genes related to drug anticancer mechanisms using support vector machine. FEBS Lett., 521, 109–114. [DOI] [PubMed] [Google Scholar]
- Barretina J. et al. (2012) The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature, 48, S5–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A. et al. (2013) An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell, 154, 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. (2001) Random forests. Mach. Learn., 45, 5–32. [Google Scholar]
- Breiman L. et al. (1984) Classification and Regression Trees. CRC Press. [Google Scholar]
- Cai Y., Reeve D.E. (2013) Extreme value prediction via a quantile function model. Coastal Eng., 77, 91–98. [Google Scholar]
- Chandola V. et al. (2009) Anomaly detection. A survey. ACM Comput. Surv. (CSUR), 41, 1. [Google Scholar]
- Cheze N., Poggi J.-M. (2006) Iterated boosting for outlier detection In: Data Science and Classification. Springer, pp. 213–220. [Google Scholar]
- Cortes C., Vapnik V. (1995) Support-vector networks. Mach. Learn., 20, 273–297. [Google Scholar]
- Costello J.C. et al. (2014) A community effort to assess and improve drug sensitivity prediction algorithms. Nat. Biotechnol., 32, 1202–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund Y. (1995) Boosting a weak learning algorithm by majority. Inf. Comput., 121, 256–285. [Google Scholar]
- Freund Y., Schapire R.E. (1996) Experiments with a New Boosting Algorithm. Machine Learning: Proceedings of the Thirteenth International Conference, Vol. 96, Morgan Kauffman, San Francisco, pp. 148–156.
- Freund Y., Schapire R.E. (1997) A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci, 55, 119–139. [Google Scholar]
- Friedman J. et al. (2010) Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw., 33, 1.. [PMC free article] [PubMed] [Google Scholar]
- Friedman J.H. (2001) Greedy function approximation: a gradient boosting machine. Ann. Stat., 29, 1189–1232. [Google Scholar]
- Garnett M.J. et al. (2012) Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature, 483, 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeleher P. et al. (2014) Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol., 15, R47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinney J. et al. (2014) Modeling ras phenotype in colorectal cancer uncovers novel molecular traits of ras dependency and improves prediction of response to targeted agents in patients. Clin. Cancer Res., 20, 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T., Qian J. (2014) Glmnet Vignette, http://web.stanford.edu/∼hastie/glmnet/glmnet_alpha.html (17 April 2018, date last accessed).
- Hodge V., Austin J. (2004) A survey of outlier detection methodologies. Artif. Intell. Rev., 22, 85–126. [Google Scholar]
- Hoerl A.E., Kennard R.W. (2000) Ridge regression: biased estimation for nonorthogonal problems. Technometrics, 42, 80–86. [Google Scholar]
- Hoggart C.J. et al. (2008) Simultaneous analysis of all snps in genome-wide and re-sequencing association studies. PLoS Genet., 4, e1000130.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio F. et al. (2016) A landscape of pharmacogenomic interactions in cancer. Cell, 166, 740–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Lin N. (2010) The bayesian elastic net. Bayesian Anal., 5, 151–170. [Google Scholar]
- Liang Y. et al. (2013) Sparse logistic regression with a l 1/2 penalty for gene selection in cancer classification. BMC Bioinformatics, 14, 198.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A., Wiener M. (2002) Classification and regression by randomforest. R. News, 2, 18–22. [Google Scholar]
- Meinshausen N. (2006) Quantile regression forests. J. Mach. Learn. Res., 7, 983–999. [Google Scholar]
- Neto E.C. et al. (2014) Simulation studies as designed experiments: the comparison of penalized regression models in the large p, small setting. PloS One, 9, e107957.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palejev D. et al. (2011) An application of the elastic net for an endophenotype analysis. Behav. Genet., 41, 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees M.G. et al. (2016) Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol., 12, 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddick G. et al. (2011) Predicting in vitro drug sensitivity using random forests. Bioinformatics, 27, 220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgeway G. (2010) Package ‘gbm’. Available on-line at http://cran.rproject.org/web/packages/gbm/index.html.
- Rousseeuw P.J., Leroy A.M. (2005) Robust Regression and Outlier Detection. Vol. 589. John Wiley & sons.
- Schaumburg J. (2010) Predicting extreme VaR: nonparametric quantile regression with refinements from extreme value theory, SFB 649 Discussion Paper, 2010–009.
- Seashore-Ludlow B. et al. (2015) Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov., 5, 1210–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokuni T. et al. (2006) Chemosensitivity prediction in esophageal squamous cell carcinoma: novel marker genes and efficacy-prediction formulae using their expression data. Int. J. Oncol., 28, 1153–1162. [PubMed] [Google Scholar]
- Sokolov A. et al. (2016) Pathway-based genomics prediction using generalized elastic net. PLoS Comput. Biol., 12, e1004790.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. (1996) Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B (Stat. Methodol.), 58, 267–288. [Google Scholar]
- Touw W.G. et al. (2013) Data mining in the life sciences with random forest: a walk in the park or lost in the jungle? Brief. Bioinform., 14, 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q. et al. (2014) Distinctive subcellular inhibition of cytokine-induced src by salubrinal and fluid flow. PloS One, 9, e105699.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.T. et al. (2009) Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics, 25, 714–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. et al. (2012) Genomics of drug sensitivity in cancer (gdsc): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res., 41, D955–D961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H., Hastie T. (2005) Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B, 67, 301–320. [Google Scholar]
- Zou H., Zhang H.H. (2009) On the adaptive elastic-net with a diverging number of parameters. Ann. Stat., 37, 1733.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.