Abstract
High-grade urothelial cancer contains intrinsic molecular subtypes that exhibit differences in underlying tumor biology and can be divided into luminal-like and basallike subtypes. We describe here the first subtype-specific murine models of bladder cancer and show that Upk3a-CreERT2; Trp53L/L; PtenL/L; Rosa26LSL-Luc (UPPL: luminallike) and BBN (basal-like) tumors are more faithful to human bladder cancer than the widely-used MB49 cells. Following engraftment into immunocompetent C57BL/6 mice, BBN tumors were more responsive to PD-1 inhibition than UPPL tumors. Responding tumors within the BBN model showed differences in immune microenvironment composition, including increased ratios of CD8+:CD4+ and memory:regulatory T cells. Finally, we predicted and confirmed immunogenicity of tumor neoantigens in each model. These UPPL and BBN models will be a valuable resource for future studies examining bladder cancer biology and immunotherapy.
INTRODUCTION
In the United States, bladder cancer is the 5th most common malignancy with approximately 79,000 new cases and nearly 17,000 deaths expected in 2017 (1). Bladder cancer is comprised of both low-grade and high-grade tumors. While low-grade tumors are almost uniformly non-invasive (Ta), high-grade tumors can become muscleinvasive and metastatic.
Multiple studies have now identified distinct RNA expression subtypes within both low- and high- grade bladder cancer (2-10). Building upon the work of Hoglund and colleagues (5), we along with others have recently described distinct subtypes of highgrade muscle-invasive urothelial carcinoma, which we have termed luminal-like and basal-like, that have gene expression patterns that appear to be consistent with differentiation states of normal urothelium and reflect gene expression patterns and biology between breast and bladder cancer (2-4, 11).
Cisplatin-based chemotherapy has been the only FDA approved therapy to treat advanced bladder cancer for over two decades until the recent approval of immune checkpoint antibodies targeting the PD-1 / PD-L1 axis. PD-1 axis blockade induces a response in approximately 20-30% of advanced urothelial carcinoma patients, with the premise that activation of immune checkpoint pathways result in active immunosuppression (12-17). Response to PD-1 axis inhibition in urothelial bladder cancer has been associated with a number of intrinsic tumor features such as tumor mutational burden and tumor molecular subtype, as well as tumor microenvironment features such as the presence of PD-L1 expressing tumor-infiltrating immune cells, CD8+ cytotoxic T cells in the tumor, and expression of effector T cell genes by gene expression profiling (13).
Multiple immune competent mouse models of bladder cancer currently exist including the carcinogen-induced models: MB49 (DMBA derived cell line) and BBN [N-butyl-N-(4-hydroxybutyl)nitrosamine] (18, 19) as well as numerous autochthonous, genetically engineered murine (GEM) models (20) some of which progress to muscleinvasive bladder cancer and metastasis (21-24).
We report here the generation of a novel GEM model of high grade, muscle-invasive bladder cancer that faithfully recapitulates the luminal molecular subtype of bladder cancer: Upk3a-CreERT2; Trp53L/L; PtenL/L; Rosa26LSL-Luc (UPPL) mice. This model is characterized by papillary histology and decreased levels of immune infiltration relative to basal tumors derived from BBN-treated animals; a pattern that is similar to human disease (3,5,11). We have generated cell line adoptive transfer models for luminal-like UPPL tumors as well as for basal tumors derived from BBN treated animals. Cell line derived tumors from the UPPL model maintain luminal-like characteristics such as high expression of Pparg and Gata3 gene signatures. Moreover, gene expression profiles from BBN and UPPL models more closely map to human bladder cancer and to normal murine urothelial cells than the commonly used MB49 model, which appears to more closely resemble fibroblasts. As models of bladder cancer biology in immunocompetent mice, these models can be used to interrogate subtype-specific responses to immune checkpoint inhibition and other immunotherapy strategies in vivo.
MATERIALS AND METHODS
Mouse models and establishment of mouse bladder cancer cell lines
All animal studies were reviewed and approved by The University of North Carolina at Chapel Hill Institutional Animal Care and Use Committee (IACUC). For the BBN carcinogen-induced mouse bladder cancer model, C57BL/6 mice (Charles River Laboratories) were continuously exposed to 0.05% N-Butyl-N-(4-hydroxybutyl) nitrosamine (BBN) in drinking water. Trp53 and Pten conditional knockout mice were obtained from Jackson Labs (STOCK: 008462) and Terry Van Dyke (25) respectively and crossed with Upk3a-CreERT2 allele (Jackson Labs STOCK: 015855) and the Rosa26LSL-Luciferase allele (Jackson Labs, STOCK: 005125) (UPPL model) or crossed with Krt5-CreERT2 allele (Gift from Brigid Hogan, Duke University) and Rosa26LSL-tdTomato (Jackson Labs, STOCK: 007914) (KPPT model). In order to induce Cre recombination in the bladder of UPPL or KPPT mice, 5mg of tamoxifen was given orally by gavage in both the UPPL and KPPT model. In the KPPT model, transurethral injection of 4-hydroxy-tamoxifen was also performed. Tumor development was regularly monitored by bladder ultrasonography.
Mice were sacrificed for the humane endpoints as follows. For the autochthonous mouse models, mice were sacrificed for weight loss more than 10% of the initial weight or tumor size diameter of >7mm as evaluated by bladder ultrasound. In our studies all mice were sacrificed because of tumor size. The endpoint for allograft models was tumor volume >500mm3, skin ulcer formation, or weight loss greater than 20% body weight.
Generation of UPPL1541 and BBN963 cell lines.
Once the bladder tumors became >7 mm in diameter, they were harvested for pathological evaluation, in vitro analysis and for establishing cell lines. Tumors were dissociated and digested with collagenase and dispase (Roche). The dissociated tumor cells were resuspended in growth media and plated to a plastic plate as previously described (26). Cell lines were passaged more than 10 times before use.
RNA/DNA extraction, library prep, and RNAseq or Whole Exome Sequencing (WES).
RNA was extracted from the primary tumors and the established cell lines using a RNeasy kit (QIAGEN), and DNA was extracted from primary tumors, established cell lines, and tail clippings using a DNeasy kit (Qiagen). Whole exome and transcriptome library preparation was performed using Agilent SureSelect XT All Exon and Illumina TruSeq Stranded mRNA library preparation kits, respectively. Libraries were sequenced via 2×100 runs on an Illumina HiSeq 2500 at the UNC High Throughput Sequencing Facility (HTSF).
RNAseq analysis
Sequence reads were aligned to the murine genome (mm9), and gene expression was generated as reads per kilobase of exon model per million mapped reads per gene by using MapSplice and upper quartile normalized via RSEM (University of Kentucky Bioinformatics Labs, Lexington, KY) (27).
RNA sequencing data were normalized for variations in read counts, log2 transformed, and median centered before analysis. When combining data sets, we adjusted for batch effects using the surrogate variable analysis R package (version 3.12.0; R Foundation, Vienna, Austria). Subtype calls were made using the BASE47 classification algorithm based on the median-centered expression of Mus musculus homologs of genes found in the classifier (3). Clustering was done using average linkage clustering with a centered correlation similarity metric. Immune gene signature scores were derived as previously described (11).
Gene data were grouped into immune gene signatures, which were murine orthologs of signatures previously identified through unsupervised clustering and gene expression profiling of sorted immune cells (11,28,29). Gene data was matched to predefined immune gene signature clusters via Entrez IDs. Each gene signature was calculated as the average value of all genes included in the signature. Differential expression for each gene signature was analyzed between tumor models and treatment groups via ANOVA (analysis of variance – 1 way anova), adjusted for multiple testing using a false discovery rate of 0.05. To determine the prognostic value of each immune gene signature, linear univariate correlation modeling was used with signature/clinical variable as a continuous variable compared to tumor size. Heat map of the log10 transformed p value of gene signature correlations were displayed with color gradient calculated via:
Gata3 and Pparg gene signatures
The PPARy gene signature was derived by determining the genes that are significantly upregulated (samr package FDR<0.05) in UMUC9 cells treated with rosiglitazone, a PPARy agonist in the GSE47993 dataset (11). The GATA3 gene signature was pulled from the BIOCARTA curated gene signature set in MSidDB.
PvClust
The significance of clustering nodes was determined using the pvclust R package (version 2.0-0, R Foundation, Vienna, Austria) (30). Significance of all nodes was calculated with a correlation distance metric and average linkage clustering.
qPCR normalization for TCR/BCR repertoire profiling
Tumor RNA concentrations were determined using a Qubit RNA BR assay kit, 1:200 in dilution buffer. Using a Qiagen Quantitect Reverse Transcription kit, cDNA was synthesized from 50ng-1μg starting total RNA (typically 1μL per sample). Purified T/B cell RNA was included as a positive control, and DI H2O was included as a negative control. The reaction was carried out according to the manufacturer’s instructions, using a Veriti thermocycler (Applied Biosystems).
Quantitative PCR was performed in triplicate with 0.5 μM of each forward and reverse primers, 0.1 μM TaqMan probe (TCR: FAM reporter with TAMRA quencher; BCR: VIC reporter with TAMRA quencher), cDNA (2.5 μL), and BioRad SsoAdvanced Universal Probe Supermix (2x) and DI H2O for a final volume of 10 μL per well. Cycling conditions for TCR and BCR were both set for 45 cycles of recommended Taqman conditions for the QuantStudio 6 Flex system.
Purified T/B cell cDNA was used for positive control and calibration curve, and the template free cDNA synthesis reaction was used for negative control. The calibration curve was determined using Ct values from purified T/B cell cDNA, 10 fold serially diluted in nuclease-free water ranging from 1:0 (cDNA: H2O, v/v) to 1:1×1012 For both T- and B- cell calibration curves, Ct values were detectable as dilute as 1:1×105, with a coefficient of determination of > 0.99 for the linear fit of log10(dilution) versus Ct. Each sample’s Ct value was read out as the ratio of T- or B-cell cDNA to total cDNA.
5’ RACE amplification of TCR/BCR sequences
Based on qPCR results, all tumor samples were normalized by T- or B-cell RNA starting template. Using a Clontech SMARTer RACE 5’/3’ kit, cDNA was generated using the manufacturer’s protocol. cDNA was diluted with tricene/EDTA buffer, and 5’ RACE was carried out using the manufacturer’s protocol with 0.5 μM custom barcoded gene-specific reverse primer, using a Veriti thermocycler (Applied Biosystems Veriti 96-well) with the following cycling conditions:
30 cycles:
94°C, 30 sec
68°C, 30 sec
72°C, 3 min
5’ RACE products were pooled, and clean-up/concentration were performed using a Zymogen Genomic DNA Clean & Concentrator. Pools of samples were eluted in 32 μL of nuclease-free water heated to 70°C. DNA concentration was measured using a Qubit DNA HS assay kit. Purity (A260/280nm and A260/230nm ratios) was determined using a ND-1000 spectrophotometer. 1-5 μL of pooled DNA was visualized in a 1.5% agarose gel to confirm presence of proper band sizes (TCR and BCR: 400-500bp).
TCR/BCR repertoire profiling
For TCR/BCR repertoire studies, pooled TCR or BCR amplicons were size selected using a Sage Science Pippin Prep 1.5% agarose cassette (HTC1510). Bands were size selected at 450-650 bp. After size selection, samples were analyzed using either Agilent 2100 Bioanalyzer or Tapestation to ensure purity. Illumina MiSeq library preparation was performed using a KAPA Biosystems DNA Preparation kit. Libraries were run at 6 pM on an Illumina MiSeq using a 600-cycle kit (2×300 paired-end), with 15% PhiX spike-in.
Mouse allograft model and treatment by Anti-PD-1 antibody
BBN963 and UPPL1541 cell lines were injected subcutaneously in C57BL/6J mice at 1×107 and 1×106 cells respectively. Once tumors reached 200 mm3 in tumor volume, treatment either by anti-Pd-1 antibody (clone RMP1-14, Millipore) or isotype control IgG (Sigma) by intraperitoneal injection was started. The treatment was administered once a week at a dose of 10mg/kg. The tumor size was measured by caliper weekly or twice weekly.
Flow Cytometry
For tissue dissociation, tissues were homogenized in cold media using the GentleMACs Dissociator and the samples were passed through a 70 μM cell strainer, followed by homogenization using a 5 mL syringe plunger. The samples were centrifuged for seven minutes at 290 RCP, 4°C, decanting the supernatant. The remaining pellet was resuspended into 1 mL of ACK lysis buffer (150 mM NH4Cl, 10 mM, KHCO3, 0.1 nM Na2EDTA in DPBS, pH 7.3) for 2 minutes at room temperature before quenching with 10 mL of cold media. The samples were centrifuged for seven minutes at 290 RCP, 4°C, resuspended in 10 mL of cold media, and passed through a 40 μM cell strainer. Cell counting was performed by running a diluted aliquot of sample on a MACSQuant flow cytometer, counting lymphocytes as gated by forward scatter area versus side scatter area.
Samples were washed and resuspended in cold DPBS, normalized by count, and transferred onto a 96 well V-bottom plate at 2.5×106 lymphocytes per well. Cells were resuspended in FVS700 viability stain (BD, 1:1000 dilution in 100 μL DPBS) for 40 minutes on ice. Wells not receiving viability staining were resuspended in DPBS. Cells were washed twice in staining buffer (0.02% NaN3, 2% BSA in DPBS), resuspended in 50 μL Fc block (1:50 dilution in staining buffer), and incubated on ice for 15 minutes. Antibody master mix was added to samples at 50 μL per sample with final antibody concentrations as indicated in Supplemental Table 1 (All mAbs from BD Biosciences). Please see Supplemental Table 1 for list of Antibodies.
Cells were incubated on ice in the dark for 45 minutes and washed twice with staining buffer. Cells were fixed in 2% paraformaldehyde overnight. The following morning, a minimum of 100,000 events were collected for each sample on a BD LSRFortessa flow cytometer. FlowJo flow software Version 10 (Treestar) was used for analyses. Fluorescence Minus One (FMO) controls were used to guide gating strategies.
Analysis and statistics
All flow cytometry, TCR/BCR sharing, and Shannon entropy statistics were calculated with Mann-Whitney U test.
For TCR/BCR amplicon sequencing analyses, raw .fastq files were demultiplexed by barcode sequences of the gene-specific primers. Sorted R1 and R2 files were respectively merged. Sequencing quality was confirmed through the FastQC quality control tool. TCR and BCR amplicon data were analyzed via IMGT/HighV-QUEST. Data was converted into standard in-lab format, and downstream analysis was performed with custom scripts as well as the tcR R package.
Neoantigen prediction
C57BL/6 mice were given a single subcutaneous flank injection of BBN963, UPPL1541, or MB49 cells. Tumor growth was monitored until tumors reached 100 mm3, at which point mice were humanely sacrificed with CO2 asphyxiation followed by cervical dislocation. Tumors were dissected for downstream DNA/RNA extraction as described above. Matched normal DNA was extracted from tail-clippings or liver from the mouse in which the cell lines were respectively derived. Library prep and sequencing were performed as described above. Bioinformatics prediction of neoantigens was performed as previously described(11). Predicted neoantigens were filtered on expression in all replicates with >5× read support.
Vaccine/ELISpot assay for neoantigen immunogenicity
For vaccination of mice with predicted neoantigens, peptides were given as a subcutaneous injection of a pool of 8 equimolar peptides (5 nmol total peptides) and 50 μg poly(I:C) in PBS. Peptides were synthesized by New England Peptide, using custom peptide array technology. A second identical injection was repeated 6-7 days after primary injection. Mice were humanely sacrificed with CO2 asphyxiation followed by cervical dislocation 5-6 days after second injection. Spleens were harvested and prepared into a red blood cell lysed, single cell suspension. Splenocytes were plated in triplicate at 500,000 cells per 100 μL media onto an IFN-γ capture antibody coated ELISpot plate (BD Biosciences) for 48 hours, along with 1 nmol of a single peptide against which the respective mouse was vaccinated. IFN-γ expression was compared to splenocytes incubated with vehicle control.
Neoantigen-enriched T cell Coculture
C57BL/6 mice were vaccinated with either a pool of the top 8 predicted BBN963 neoantigens or irrelevant peptide (SIINFEKL) control, with a second identical booster given 7 days after primary vaccine. One week after secondary vaccination, spleens were harvested and prepared into a red blood cell lysed, single cell suspension. T cells were isolated using Miltenyi murine Pan T Cell Isolation Kit II. Using previously described methods (31), T cells were expanded in the presence of bone marrow derived dendritic cells pulsed with a single peptide against which the derivative mouse was vaccinated against. Seven days following ex vivo expansion, 100,000 T cells were cocultured 10:1 with BBN963 cells onto an IFN-γ capture ELISpot plate for 72 hours. Controls included T cell only, BBN963 only, and media only negative controls, as well as antigen-enriched T cells co-cultured with respective peptide-pulsed UPPL1541 cells as positive control. Signal intensity was read out using an ELISpot plate reader. T cell/BBN963 co-culture spot counts were subtracted from their respective T cell only control, and then taken as a percentage of the counts from their respective peptide-pulsed target positive controls.
RESULTS
Inactivation of Pten and Trp53 in Uroplakin3a expressing cells results in muscle-invasive, high-grade urothelial carcinoma.
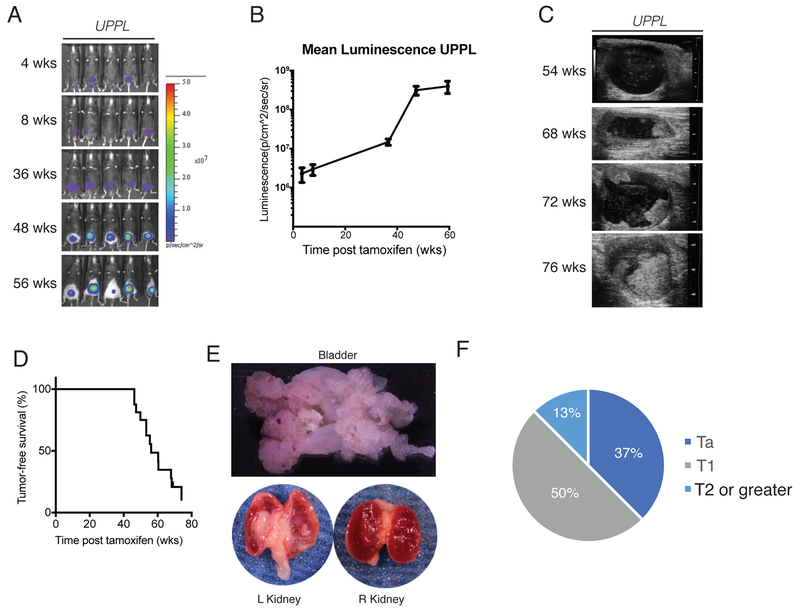
Our previously published studies describing luminal-like and basal-like molecular subtypes of bladder cancer demonstrated that these subtypes reflect the gene expression patterns of the differentiation states of the normal urothelium (3). Basal-like bladder tumors harbor gene expression patterns most similar to basal and intermediate cell layers of the bladder while luminal tumors harbor gene expression patterns most similar to umbrella cells (2,3). To determine whether different cells of origin account for the differential gene expression patterns between basal-like and luminal-like high-grade, muscle-invasive bladder cancer we conditionally inactivated Pten and Trp53 in Keratin5 (K5) or Uroplakin3a (Upk3a) expressing basal/intermediate and umbrella/intermediate cell layers respectively using previously reported K5-CreERT2 (32) and Upk3a-CreERT2 (Jackson Labs) transgenic mice. Dual inactivation of Pten and Trp53 by surgical injection of adenoviral cre into the bladder has been previously shown to induce bladder cancer in mice (24). Using standard animal husbandry, we generated cohorts of Upk3a-CreERT2; Trp53L/L; Pte/L/L; Rosa26LSL-Luc mice (hereafter termed “UPPL”) as well as K5-CreERT2; Trp53L/L; PtenL/L; Rosa26LSL-tdTomato mice (hereafter called “KPPT”) that were backcrossed 10 times to a C57BL/6 background. Both UPPL and KPPT mice were gavaged with tamoxifen every other day for 3 doses starting at 6-8 weeks of age to induce CreERT2 activity. Serial in vivo luminescence (for UPPL mice) and ultrasound of the bladder were used to monitor for tumor development and growth. UPPL mice demonstrated gradually increasing luminescence signal over time in the region of the bladder (Figure 1A and 1B). In addition, by ultrasound, papillary appearing tumors began to be apparent at a median of 58 weeks (Figure 1C and 1D).
Figure 1. Inactivation of Pten and Trp53 in Upk3a expressing cells results in high grade muscle invasive bladder tumors.
A) Bioluminescent images of UPPL mice at indicated timepoints. B) Quantification of luminescence over the region of the bladder. C) Ultrasound images of bladder tumor formation. D) Tumor free survival as detected by ultrasound. E) Gross images of the kidneys and bladder from a tumor bearing UPPL mouse. F) Tumor stage assessed histologically based on human TNM staging.
In contrast, KPPT mice administered tamoxifen by gavage died rapidly of epithelial hyperplasia of the snout, paws, and papillary skin lesions (Supplemental Figure 1A-1C). This likely represents the inactivation of Pten and Trp53 (and pursuant epithelial overgrowth) in K5 expressing basal cells in multiple organs including the epidermis, trachea, and GI tract. In an attempt to activate K5-CreERT2 solely in the K5 expressing basal cells of the bladder, we administered 4-OHT intravesicaly at various concentrations (2000 nM and 200 nM). Mice injected with intravesical 4-OHT at 2000 nM exhibited a similar but attenuated phenotype to KPPT mice that had been gavaged with tamoxifen (Supplemental Figure 1B, 1C) and had a shortened survival. In contrast, KPPT mice injected with intravesical 4-Hydroxytamoxifen (4-OHT) at 200 nM had an extended survival but did not develop bladder tumors despite Cre-mediated recombination as evidenced by increased tdTomato signal over the region of the bladder by IVIS imaging (Supplemental Figure 1D). Moreover, histologic examination of the bladders of mice injected with intravesical 4-OHT at 2000nM or 200nM showed no significant histologic changes of the urothelium (Supplemental Figure 1E).
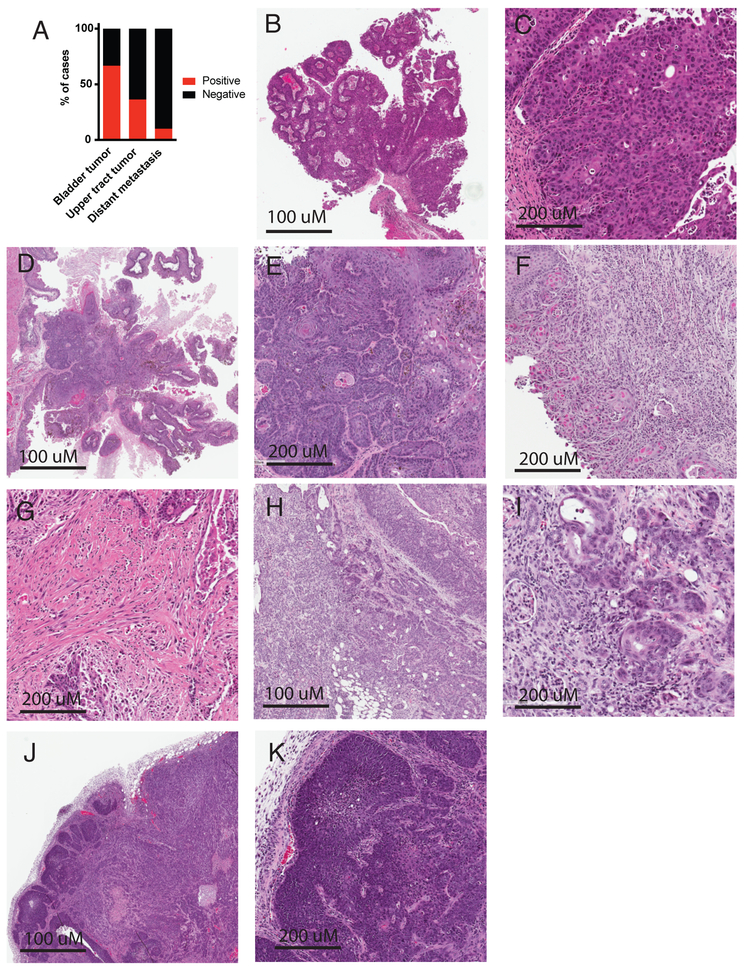
Approximately 95% of UPPL mice developed tumors within 77 weeks (Figure 1D). Grossly and histologically, bladder tumors in UPPL mice appeared to be papillary in nature (Figure 1E and Figure 2B, 2C, 2D), which is a feature documented to be enriched in the luminal-like molecular subtype (2,4). The UPPL tumors were characterized as high-grade by an expert genitourinary pathologist (SW) with some having prominent squamous differentiation (Figure 2D, 2E). In addition, UPPL tumors were noted to have different depths of invasion into the bladder wall including both lamina propria invasion (Figure 1F and 2F) and muscularis propria invasion (Figure 2G). In keeping with the known field defect of urothelial tumors in human disease, tumors were also noted to form in the renal pelvis and ureters of about a third of mice (Figure 1E, Figure 2H, 2I). Finally, rare macroscopic metastases were seen (Figure 2A, 2J, and 2K).
Figure 2. Representative histology of UPPL tumors.
A) Bar graph indicating the percentage of UPPL mice that developed bladder tumors, upper tract tumors, and distant metastases at the time of sacrifice. B) low power view of papillary appearing UPPL tumor. C) High power view of papillary UPPL tumor. D) Low power view of papillary tumor with squamous histology. E) High power view of squamous histology. F) UPPL tumor showing lamina propria invasion. G) UPPL tumor with muscularis propria invasion. H) Upper tract tumor demonstrating invasion into the renal parenchyma. I) High power view of urothelial tumor invading renal parenchyma. J) Cervical lymph node metastases. K) High power view of cervical lymph node metastases.
BBN and UPPL models are basal- and luminal-like models respectively of human bladder cancer.
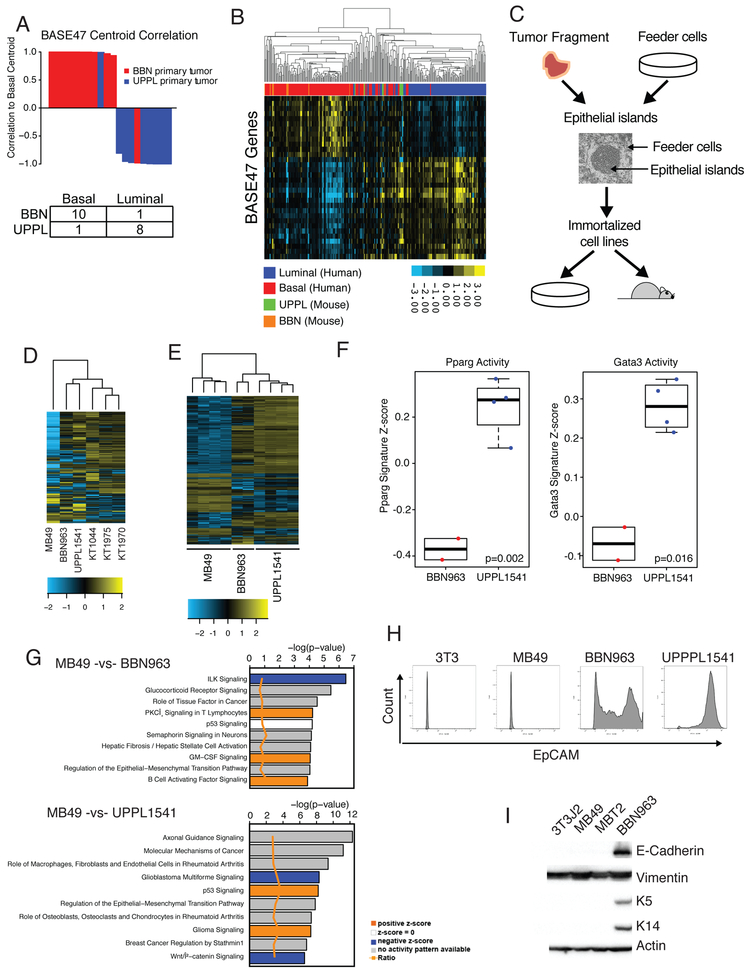
Bladder tumors induced by the carcinogen N-Butyl-N-(4-hydroxybutyl) (BBN) have been previously documented to harbor a number of histologic features (e.g. squamous differentiation, Supplemental Figure 2) and gene expression patterns known to be found in basal-like bladder tumors (7). We therefore established 11 independent BBN-induced bladder tumors by continuously administering 0.05% BBN in drinking water as previously described (19). Given the papillary nature of UPPL tumors we hypothesized that they correspond to a luminal-like molecular subtype. We therefore performed global transcriptome profiling of 9 UPPL and 11 BBN mouse tumors using RNAseq. We first performed molecular subtype classification using our previously published BASE47 (Bladder cancer Analysis of Subtypes by gene Expression) (3) subtype classifier and found that 8 of the 9 UPPL tumors had high correlation to the luminal centroid of gene expression (Figure 3A). To further validate our observation, we co-clustered the UPPL and BBN murine tumors with human tumors from the TCGA (The Cancer Genome Atlas [n=408]) using genes with corresponding homologs across the species and found that the majority of UPPL and BBN tumors co-clustered with human luminal-like tumors and basal-like tumors respectively (Figure 3B).
Figure 3. BBN and UPPL tumors recapitulate the human basal and luminal molecular subtypes of bladder cancer.
A) Waterfall plot of the correlation to the Basal centroid for BBN and UPPL primary tumors, with an accompanying confusion matrix indicating the subtype calls. B) Unsupervised clustering heatmap of BBN and UPPL primary tumor samples with the TCGA BLCA dataset across genes in the BASE47 classifier. C) Representative flowchart of the workflow to transition a primary tumor extracted from a mouse model to a cell line and a cell line derived tumor. D) Unsupervised clustering heatmap of MB49, BBN963, UPPL1541 and normal urothelium (KT) cell lines across the top 10% most differentially expressed genes across the samples. E) Unsupervised clustering heatmap of MB49, BBN963, and UPPL1541 cell line derived tumors across the top10% most differentially expressed genes across the samples. F) Box plots of Pparg and Gata3 gene signature scores from RNAseq data of BBN963 and UPPL1541 cell line derived tumors. G) IPA Analysis plots showing activated pathways in MB49 cell line derived tumors relative to BBN963 and UPPL1541 cell line derived tumors H) Flow Cytometry plot for EpCAM expression in 3T3, MB49, UPPL1541, and BBN963 cell lines I) Western blot of whole cell lysates from 3T3, MB49, MBT2, and BBN963 cell lines blotted for the indicated antibodies.
Currently, very few cell lines exist for modeling bladder cancer in immunocompetent mice; therefore, we set out to generate additional cell lines which could be utilized in future studies. In particular, MB49 cells have long been the workhorse of syngeneic bladder cancer cell lines (18) for studies requiring an immunocompetent host. Given the long latency of tumor formation in the UPPL model, we established tumor cell lines from both UPPL and BBN tumors using the conditional reprogramming of cells (CRC) method described previously (26). Specifically, transplantable cell lines were established from BBN (BBN963) and UPPL (UPPL1541) tumors (Figure 3C) and have been confirmed to grow in C57BL/6 mice. In parallel, using the CRC method, we generated three primary cell lines derived from normal mouse urothelium of tamoxifen treated K5-CreERT2; Rosa26LSL-tdTomato mice, hereafter called KT mice (KT1044, KT1975, and KT1970) as a normal reference for comparison. Interestingly, the vast majority of epithelial cells that grew in vitro from CRC culture of KT mouse bladders expressed tdTomato suggesting they at some point had expressed K5 (Supplemental Figure 3A).
To assess the similarities between our newly generated models and MB49 cells, we performed whole transcriptome profiling on the BBN963 and UPPL1541 cell lines, MB49 cells, 3T3 cells, and three primary mouse urothelial cell lines (KT1044, KT1975, and KT1970). Unsupervised hierarchical clustering of the cell lines on differentially regulated genes across samples (Supplemental Table 2) demonstrated that MB49 cells had transcriptome profiles that differed significantly from the other cell lines (Figure 3D) when tested by multiscale bootstrap resampling (p = 0.0, Supplemental Figure 4A), while BBN963 and UPPL1541 cells had transcriptome profiles that more closely resembled normal urothelial (KT) cells. To ensure that we had not tainted our MB49 cells, we obtained MB49 cells from an independent source (Phil Abbosh, Fox Chase Cancer Center) and performed transcriptome profiling. We found that the transcript levels (across all genes) is highly correlated when comparing “UNC MB49” to “FCCC MB49” (R=0.94 respectively, Supplemental Figure 3B) suggesting our MB49 cells were genuine. Intriguingly, hierarchical clustering of MB49 cells with 3T3 cells, our three primary mouse urothelial cell lines, BBN963 cells, and UPPL1541 cells demonstrated that the MB49 cells co-clustered with 3T3 cells (Supplemental Figure 3) significantly by PVClust (Supplemental Figure 3D). To examine the RNA expression profiles of these cell lines in the context of the tumor microenvironment we generated RNA expression data on cell line derived tumors from MB49, BBN963, and UPPL1541 cells. Clustering of these cell line derived tumors using differentially regulated genes (Supplemental Table 3) again demonstrated that MB49 tumors have significantly divergent transcriptome profiles when tested by multiscale bootstrap resampling (p = 0.0, Supplemental Figure 4B) and demonstrate that this finding is not merely an artifact of cell culture (Figure 3E). Finally, reassuringly, Pparg and Gata3 gene signatures were upregulated in UPPPL1541 tumors relative to BBN963 tumors (Figure 3F), demonstrating that cell line derived UPPL1541 tumors maintain molecular features of a luminal-like molecular subtype.
We next performed Ingenuity Pathway Analysis (IPA) comparing MB49, UPPL1541, and BBN963 cell line derived tumors. General pathways related to cancer were enriched in BBN963 tumors relative to UPPL1541 tumors (Supplemental Figure 5). In contrast, pathways related to fibrosis and epithelial to mesenchymal transition (EMT) appeared to be highly upregulated in MB49 cell line derived tumors compared to either the BBN963 or UPPL1541 tumors (Figure 3G). Based on these observations, we examined the expression of a set of epithelial markers in the mouse bladder cell lines. Assessment of EpCAM by flow cytometry demonstrated that a significant proportion of BBN963 and UPPL1541 cells expressed cell surface EpCAM while MB49 cells had little to no EpCAM expression, similar to the mouse fibroblast line 3T3 (Figure 3H). In keeping with this finding, we also noted that MB49 cells did not express K5 or K14 in immunoblots of whole cell lysates (Figure 3I), implying that MB49 cells have lost characteristic urothelial cytokeratin expression patterns potentially from undergoing epithelial-to-mesenchymal transition (EMT). Furthermore, we noted that MB49 cell line derived tumors had relatively high and low expression of Vimentin and Cdh1 (E-cadherin) respectively consistent with MB49 cells being more mesenchymal than BBN963 and UPPL1541 cell line derived tumors (Figure 3I and Supplemental Figure 6). In aggregate these findings suggest that MB49 cells and tumors more closely resemble fibroblasts than urothelial cells and highlight the potential benefit of our models.
BBN963 tumors demonstrate evidence of an antigen-driven T cell response
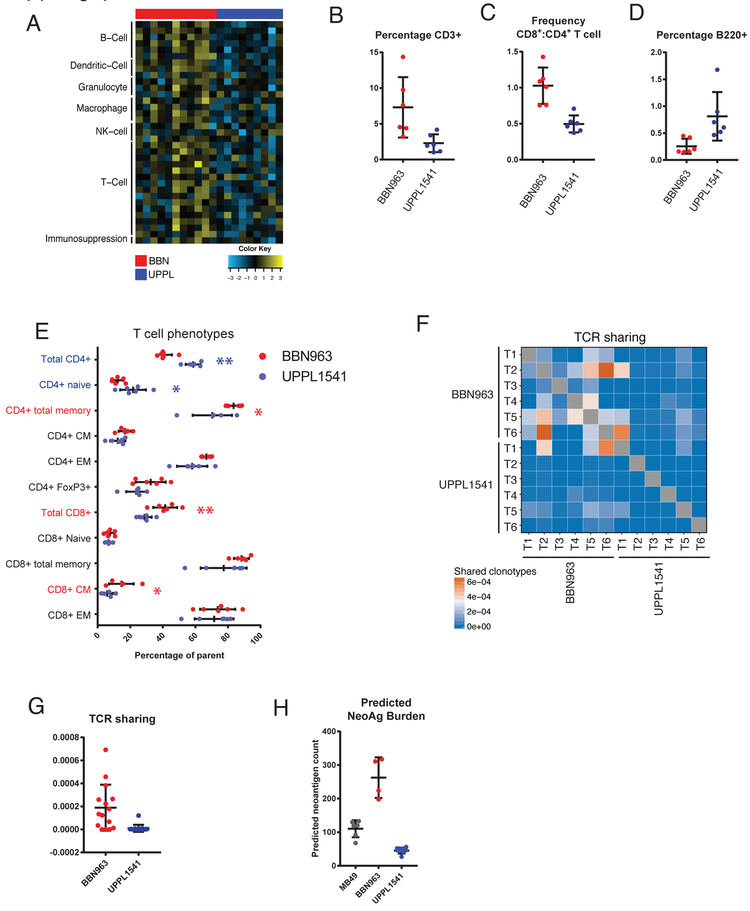
Human basal-like and luminal-like bladder cancers demonstrate different patterns of immune infiltration and are also correlated with differential response to checkpoint inhibitor therapy (11,13), suggesting subtype-specific differences in the tumor-immune microenvironment. Immune gene signature expression derived from previously published studies were compared among 11 BBN and 9 UPPL models (11,28,29,33). Consistent with immune gene signature patterns observed in human tumors, BBN (basal-like) tumors demonstrated greater overall expression of immune gene signatures (Figure 4A and Supplemental Figure 7A) than did UPPL (luminal-like) tumors, including those for T cells, B cells, dendritic cells, other innate immune cells, and immunosuppression (11).
Figure 4: Immune characterization of UPPL1541 and BBN963 subtype specific bladder models.
A) Immune gene signature expression across 9 UPPL and 11 BBN primary tumors. B) Flow cytometric characterization of tumor infiltrating T cells (CD3+) in cell line derived (allograft) BBN963 and UPPL1541 tumors. Each datapoint represents an independent mouse. C) Frequency of CD8+ to CD4+ T cells in cell line derived BBN963 and UPPL1541 tumors. D) Flow cytometric characterization of tumor infiltrating B cells (B220+) in cell line derived BBN963 and UPPL1541 tumors. E) T cell phenotypic subpopulations in cell line derived BBN963 and UPPL1541 tumors, with significantly increased phenotypes highlighted with respective colors (Mann-Whitney U-test, *: p < 0.05, **: p < 0.001). Heatmap F) and respective quantification G) of T cell receptor clonotype sharing within cell line derived BBN963 and UPPL1541 tumors, derived from whole-tumor RNA-based T cell receptor amplicon sequencing. H) Predicted neoantigen burden (class I and II, >500 nM predicted binding affinity) in MB49, BBN963, and UPPL1541 cell line derived tumors.
To further explain the observed immunological differences between BBN and UPPL tumors, we performed flow cytometric analysis in cell line derived BBN963 and UPPL1541 tumors. Comparing the frequency of tumor infiltrating lymphocytes (TILs) by flow cytometry from BBN963 and UPPL1541 tumors, we observed significantly greater frequencies of CD3+ T cells (Figure 4B), as well as increased ratio of CD8+ cytotoxic T cells to CD4+ helper T cells (Figure 4C) in BBN963. Somewhat surprisingly, the proportion of B cells was higher in UPPL tumors; however the overall proportion of B cells in the lymphocytic infiltrate was low (Figure 4D). To further characterize the phenotype of the tumor-infiltrating T cells, expression of CD44 and CD62L were used to identify naïve (CD44−, CD62L+), central memory (CM; CD44+, CD62L+), and effector memory (EM; CD44+, CD62L−) populations. Among CD4+ T cells, UPPL1541 tumors were enriched for naïve T cells, while the frequency of the total memory pool (CD44+) was significantly greater in BBN963 tumors (Figure 4E). Moreover, EM and CM frequencies both trended higher in BBN963. Among CD8+ T cells, the CM frequency was significantly greater in BBN963. Additionally, CD4+ FoxP3+ regulatory T cells (Tregs) trended toward higher frequency in BBN963. Thus, memory subpopulations of both CD8+ and CD4+ T cells had increased frequencies in the BBN tumors suggesting the presence of an antigen driven T cell response in BBN963.
Analyzing BBN963 and UPPL1541 cell line derived tumors by T cell receptor repertoire profiling, BBN963 tumors demonstrated a higher degree of clonotype sharing between animals (Figure 4F, 4G), suggesting that there may be greater convergent repertoire selection in BBN tumor infiltrating T cells in the context of an antigen-driven response. To examine if the increased immune infiltration and TCR repertoire sharing seen in BBN tumors were associated with the number of targetable tumor antigens, we performed neoantigen prediction on BBN963 and UPPL1541 cells and not unexpectedly observed significantly higher neoantigen burden in BBN963 compared to UPPL1541 (Figure 4H and Supplemental Table 4 and 5). This, in combination with the increased TCR repertoire sharing, further supports the hypothesis that the immune infiltration seen in BBN tumors is driven by an antigen-specific immune response.
To investigate the functional significance of immune infiltrating T cell subpopulations, we performed univariable linear regression with frequency of T cell phenotypic subpopulations as a continuous predictor variable and tumor mass as the response variable in untreated mice. In BBN963, the frequency of total and naïve CD8+ T cells was positively associated with tumor mass, and the frequency of CD4+ memory, CD4+ CM, total CD8+ memory, and CD8+ EM T cells were all inversely correlated with tumor mass (Supplemental Figure 7B). In UPPL1541, no features were positively associated with tumor mass while CD8+ total memory and specifically CD8+ CM T cells were both weakly inversely correlated with tumor mass. These associations are suggestive of tumor infiltrating memory T cells being functional and capable of antitumor activity in both BBN and UPPL, with greater functional significance in BBN963.
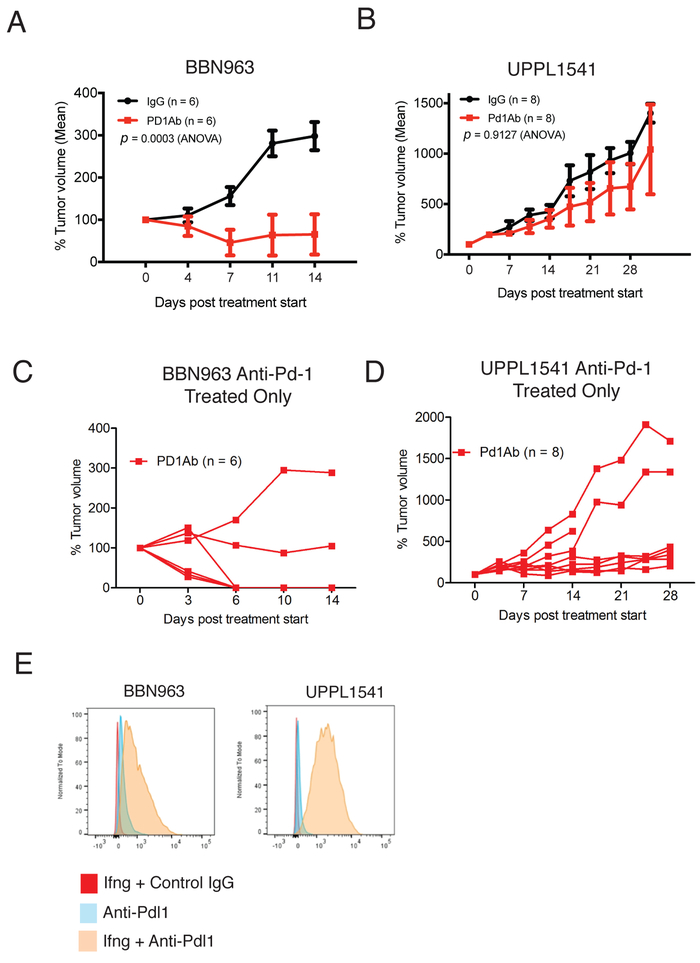
BBN963 tumors stratify by response to PD-1 axis inhibition
The relative overexpression of immune gene signatures and evidence of an antigen driven T cell response in BBN963 cell line derived tumors are suggestive of possible greater responsiveness to immune checkpoint inhibitor therapy in BBN963. Accordingly, we observed dramatic decreases in mean tumor volume in BBN963 following anti-Pd-1 therapy, while UPPL1541 tumors demonstrated only modest control of tumor growth (Figure 5A, 5B). Despite the mean tumor size being substantially controlled in BBN963 following anti-Pd-1 therapy, the growth pattern of individual tumors demonstrated a mixed-response pattern (Figure 5C). There was heterogeneity of anti-Pd-1 response in UPPL1541 tumors as well (Figure 5D). The difference in responsiveness between BBN963 and UPPL1541 tumors did not appear to be secondary to differential expression of Pd-L1 as both BBN963 and UPPL1541 cell lines upregulated Pd-L1 expression when exposed to IFN-γ (Figure 5E).
Figure 5. Anti-Pd-1 treatment of BBN963 and UPPL1541 cell line derived tumors.
A) Mean tumor volume of C57BL/6 mice bearing BBN963 tumors treated with either control IgG or anti-Pd-1 antibody. Anti-Pd-1 treatment was begun when tumors reached 200mm3 B) Mean tumor volume of C57BL/6 mice bearing UPPL1541 tumors treated with either control IgG or anti-Pd-1 antibody. Anti-Pd-1 treatment was begun when tumors reached 200mm3 C) Tumor volume of individual mice from A). D) Tumor volume of individual mice from B). E) BBN963 and UPPL1541 cells were treated with IFN-γ and flow cytometry was used to detect cell surface Pd-L1 expression.
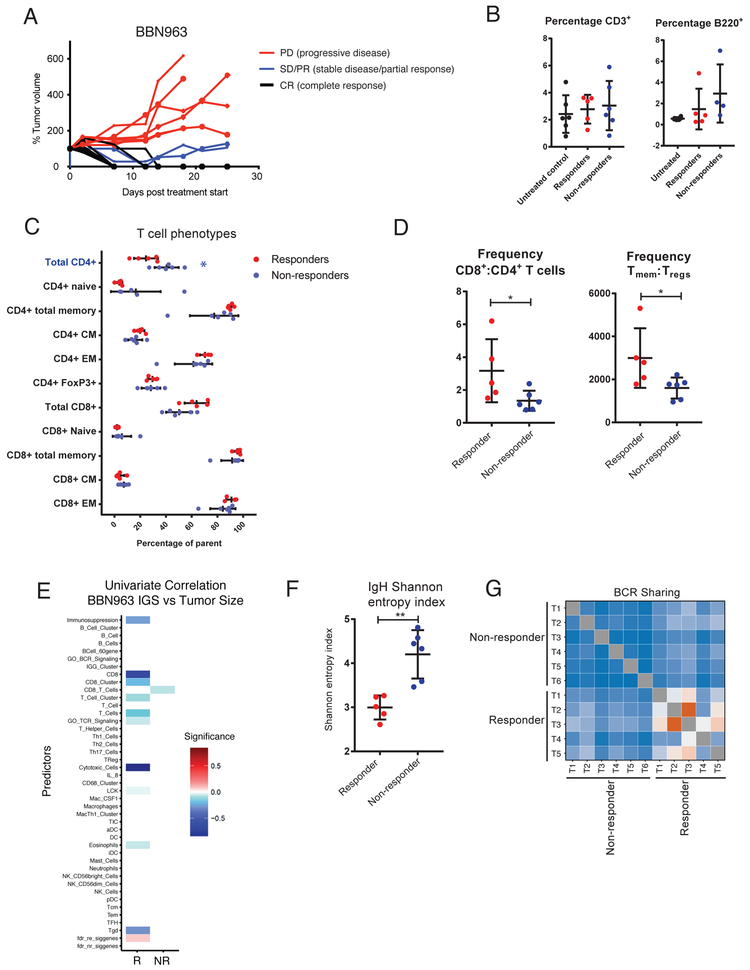
In order to elucidate the immune correlates of these two phenotypes in response to anti-PD-1 therapy, we repeated anti-Pd-1 antibody treatments and analyzed the TIL populations among anti-Pd-1 responder and non-responder BBN963 tumors once response class could be determined on day +14 following tumor inoculation. Surprisingly, no significant changes were observed in the overall T and B cell infiltration frequencies by flow cytometry in responders versus non-responders (Figure 6B). Additionally, phenotyping of tumor infiltrating T cells demonstrated only significantly greater frequencies of total CD4+ among non-responders, with subtle, non-significant variations among other T cell phenotypic subpopulations (Figure 6C). Despite these minimal differences, comparison of subpopulation ratios demonstrated an overall significant increase in the frequency of total memory-to-regulatory T cells as well as significantly higher ratios of CD8+ to CD4+ T cells in responders (Figure 6D). To further examine the role of TILs among responder and non-responder tumors, we calculated immune gene signatures derived from total tumor RNA-seq and independently correlated these signatures to tumor mass. While non-responders only demonstrated modest inverse correlation between a single CD8+ T cell immune signature and tumor mass, responder tumor mass was inversely correlated with multiple immune cell signatures, most significantly with cytotoxic T cell and CD8+ T cell signatures (Figure 6E). This data in aggregate demonstrates the potential importance of the balance of effector to suppressor T cell subpopulations, rather than just absolute numbers, in mediating an antitumor response in responding BBN963 tumors.
Figure 6. Description and immune characterization of BBN963 mixed-response phenotype.
A) Tumor growth curves of anti-Pd-1 treated, cell line derived BBN963 tumors, showing responders (black) and non-responders (red) to therapy. B) Flow characterization of tumor infiltrating T cells (CD3+) and B cells (B220+) in untreated (black), responder (red), and non-responder (blue) BBN963 tumors. C) T cell phenotypic subpopulations in cell line derived BBN963 and UPPL1541 tumors, with significantly increased phenotypes highlighted with respective colors (Mann-Whitney U-test, *: p < 0.05). D) Frequency of memory to regulatory T cells (CD3+CD44+: CD3+CD4+FoxP3+) and CD8+ to CD4+ T cells in responder versus non-responder BBN963 tumors. E) Univariable correlation of tumor size to immune gene signature expression in responder (left) and non-responder (right) BBN963 tumors. Shannon entropy index F) and receptor clonotype sharing G) of tumor infiltrating B cell receptor heavy chain expression in responder and non-responder BBN963 tumors.
To address the role of clonality of the tumor infiltrating lymphocytes in controlling tumor growth in anti-PD-1 responsive BBN963 tumors, we performed T and B cell receptor repertoire profiling of responder and non-responder whole tumor RNA. We observed a modest, non-significant increase in T cell receptor clonotype sharing (Supplemental Figure 8A, 8B) and no differences in Shannon entropy (Supplemental Figure 8C) between responder and non-responder tumors. In contrast, B cell receptor profiling demonstrated significantly lower Shannon entropy indices (Figure 6F) and significantly greater clonotype sharing among responders (Figure 6G) suggesting a potentially important role for B cell clonal shift in mediating response in the BBN963 model.
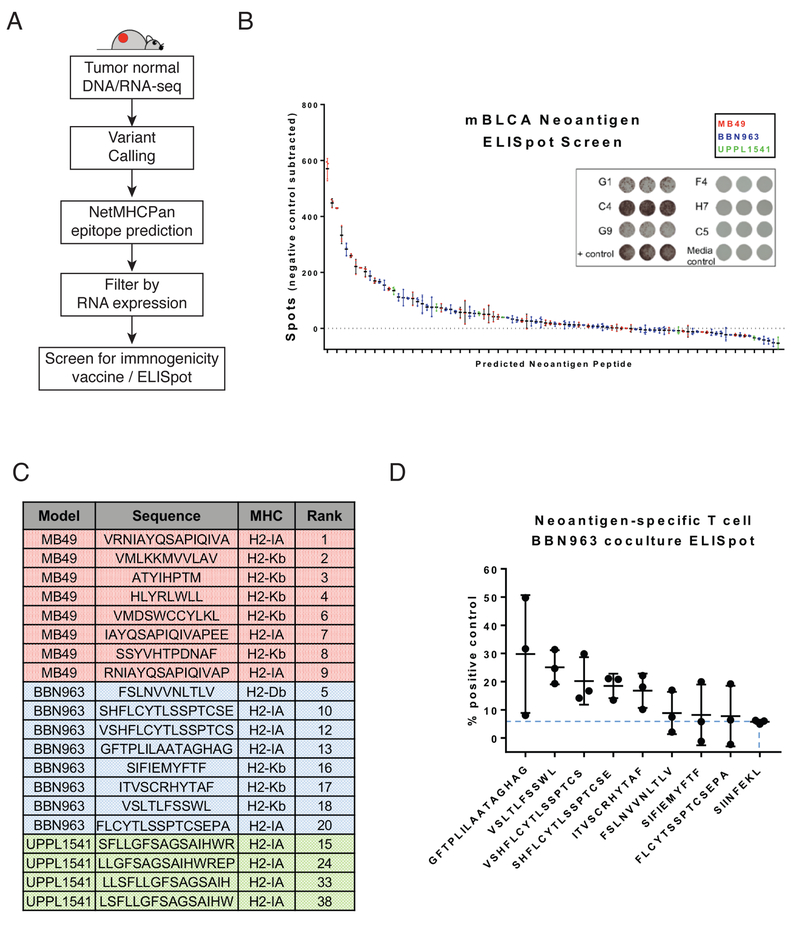
BBN963 and UPPL1541 models express targetable, immunogenic neoantigens.
With the recent interest in neoantigens as biomarkers of immunotherapy response and therapeutic targets for personalized immunotherapy, we sought to identify and validate neoantigen targets in our subtype-specific bladder models. Using previously described methods (11), neoantigens were predicted in BBN963, MB49, and UPPL1541 cell line derived tumors (Figure 7A), selecting for predicted class I and II binders by NetMHCpan and NetMHCIIpan (affinity < 500 nM). Using 5× RNA-seq coverage and expression in all replicates as cutoffs, we observed BBN963 to have the greatest number of class I (48) and class II (18) predicted neoantigens, followed by MB49 (I: 29; II: 8), and lastly by UPPL1541 (I: 2; II: 5). To validate the immunogenic potential of these predicted neoantigens, we synthesized 96 of 110 predicted neoantigens and performed vaccination/ELISpot analyses to identify the ability of each peptide to induce an IFN-γ response in T cells stimulated by neoantigen peptide-pulsed dendritic cells. Mice were vaccinated with a pool of eight random, equimolar peptides, and splenocytes derived from vaccinated mice were subsequently pulsed with one of the eight peptides on an IFN-γ capture ELISpot plate (Figure 7B, 7C). Based on number of spots induced by each peptide, we observed MB49 to contain the most highly immunogenic class I and II peptides, holding eight of the top ten neoantigens by IFN-γ response. This is followed by BBN963, and lastly by UPPL1541 which contained only one peptide within the top 15. Class I and II neoantigens were equally represented among the top binders, with four and six of ten top neoantigens predicted as class II and class I, respectively. Lastly, to test the potential for these peptides to generate neoantigen-enriched T cell populations, we performed two rounds of vaccination in wild-type C57BL/6 mice using the top eight BBN963 neoantigens, followed by ex vivo stimulation using one of the respective peptides. Co-culture of these neoantigenenriched T cells with BBN963 tumor cells demonstrated an IFN-γ response over that of irrelevant peptide control in five of eight peptides, emphasizing the potential of these peptides for use in neoantigen-based BBN963 treatment models (Figure 7D).
Figure 7. Neoantigen prediction and validation in BBN963, UPPL1541, and MB49.
A) Schema for neoantigen prediction workflow, using tumor DNA, tumor RNA, and matched normal DNA to call mutations via UNSeqr, epitope prediction to identify predicted class I and II binders, and vaccine/ELISpot validation. B) Summary of vaccine/ELISpot results in MB49, BBN963, and UPPL1541, with background subtracted counts ranked by number of spots and representative figures of highly immunogenic wells (G1, C4, G9), weakly immunogenic wells (F4, H7, C5), and controls. C) Summary of top eight predicted neoantigens in MB49 (red), BBN963 (blue) and top four predicted neoantigens in UPPL1541 (green), including sequence, predicted MHC class, and rank among all screened peptides within all three models. D) IFN-γ ELISpot results of BBN963 neoantigen-enriched T cells co-cultured with BBN963 tumors, as a percentage peptide-pulsed target positive control. Blue dashed line marks IFN-γ intensity of irrelevant peptide (SIINFEKL) enriched T cells co-cultured with BBN963.
DISCUSSION
We describe here the first molecular subtype-specific murine model of luminallike bladder cancer. The BBN model was derived from carcinogen-treated mice that develop spontaneous bladder tumors and exhibit a basal phenotype as previously described by others (7). We extend these findings by demonstrating that the BBN model also has an immune infiltration pattern that is consistent with that of human basal tumors (11). The UPPL model was derived from mice with directed knockout of Trp53 and Pten in the urothelial umbrella cells under the control of the Uroplakin-3a promoter. UPPL tumors have papillary histology, decreased immune infiltration, and decreased response to Pd-1 inhibition relative to BBN tumors. Characterized by gene expression profiling, these models reflect human bladder cancer and normal urothelium more closely than does the commonly used MB49 model, which appears to more closely resemble fibroblasts. These results imply the BBN and UPPL models will prove to be valuable resources in studying bladder cancer in immunocompetent animals, and they will be a unique resource with which to study molecular subtype specific biology and treatment effects.
While dual inactivation of Trp53 and Pten in Upk3a expressing cells led to robust bladder tumor formation, inactivation of these same genes in K5 expressing cells did not result in any apparent neoplasia or preneoplastic changes. While we had set out with the goal of answering whether inactivation of Trp53 and Pten in K5 expressing basal and intermediate urothelial cells was permissive for tumorigenesis, we are hesitant to conclude too much from our negative findings given the significant technical differences between how Trp53 and Pten were inactivated in Upk3a expressing cells (systemic tamoxifen by gavage) and K5 expressing cells (local 4-OHT administration into the bladder). Nonetheless, at face value, our results suggest that dual inactivation of Trp53 and Pten are sufficient to initiate bladder tumors in Upk3a expressing but not K5 expressing urothelium.
The immune microenvironments of the mouse models also reflect patterns seen in human disease, with increased overall immune infiltration seen in the BBN basal-like model (11). In parallel with the active immune response, BBN tumors showed increased expression of genes associated with immunosuppression, which is presumably an adaptive response to suppress and/or evade anti-tumor immunity. In the BBN (basal-like) model we noted increased memory polarization, which is complimented by significantly higher expression of effector memory T cell immune gene signatures in human basal versus luminal tumors among TCGA BLCA samples (Supplemental Figure 9). These patterns mirror those observed in our prior study of human bladder cancer (11) and imply that tumor immunobiology and mechanisms of resistance to immunotherapy may differ by tumor molecular subtype. Importantly, there were a number of aspects noted in the described mouse models that have not been as yet examined in human tumors. In the BBN (basal-like) model we noted increased T cell clonotype sharing, suggesting the presence of an active antigen-driven response in these tumors. In contrast, UPPL (luminal-like) tumors showed decreased overall immune infiltration along with an increased frequency of naive T cells, consistent with immune exclusion and lack of antigen experience, respectively.
Our study also highlights several limitations of the widely used MB49 murine bladder cancer model. We demonstrate that MB49 tumors show lack of characteristic urothelial cytokeratins, lack of EpCAM expression, and a profound skewing towards having undergone epithelial-to-mesechymal cell transition. Compared to BBN and UPPL tumors, MB49 had a wide transcriptomic distance from both normal murine urothelial cells and more closely resemble immortalized fibroblasts. These results suggest that while MB49 may be adequate (or in some cases preferred) for studying some aspects of bladder cancer biology (e.g. the post-EMT state), the models reported in our study should gain wide use in the translational bladder cancer research community, both for their subtype-specificity and increased fidelity to human bladder cancer gene expression profiles.
While recent work highlights the mutational faithfulness of the BBN model to human bladder cancer (34) one limitation of the UPPL1541 and BBN963 models is the lack of driver mutations that are also known drivers in human basal-like and luminal-like bladder tumors. Major bladder cancer driver mutations such as MLL, FGFR3, and ARID1A are not mutated in either model, and suspected driver mutations in our BBN963 line such as TCF4, FGFR2, and ITK are not seen at high frequencies in human disease (Supplemental Figure 10). This limitation is not unique to our models, as it is also a feature of MB49. As RNA transcription is downstream of genetic events such as mutations and gene fusions and upstream of protein translation, we feel the transcriptomic fidelity of our models to human bladder cancer is evidence that these models can be used to faithfully study bladder cancer biology in general and subtype-specific biology in particular. As both BBN and UPPL tumors exist as transplantable cell lines syngeneic with the C57BL/6 background and are able to grow both subcutaneously and orthotopically in the bladder, genetic manipulation strategies such as CRISPR/Cas9 may be used to manipulate these tumors should researchers desire to study effects of specific mutations in the context of subtype-specific tumors. A second limitation of the UPPL model, especially for tumor immunology studies, is accounting for the potential effect of PTEN inactivation on the immune microenvironment. PTEN has been found to promote nuclear import of the transcription factor Interferon Regulatory Factor 3 (IRF3), thereby positively regulating type I interferon induction (35). PTEN loss has also been associated with impaired T cell tumor ingress and T cell mediated cytotoxicity in a preclinical model of melanoma where human tumor cell lines engineered with PTEN silenced and to express the murine MHC class I molecule H-2Db were injected into immunocompromised mice followed by transfer of murine antigenspecific T cells and antigen-presenting cells 7 days later (36). Thus it is possible that the UPPL model may be skewed towards an immune evaded or suppressed phenotype due to PTEN loss.
In the ImVigor 210 study, Rosenberg et. al. showed that a subset of luminal tumors were more likely to respond to PD-1 axis inhibition using an anti-PD-L1 antibody (13). We report here that response to monotherapy using a monoclonal antibody against Pd-1 was effective in the BBN but not the UPPL model. This represents a potential discrepancy between our murine models and their homonymous human subtypes, however that conclusion is tempered by two considerations: 1) Our group’s luminal vs basal predictor is different from that used by the ImVigor investigators, and unfortunately despite publication the ImVigor RNA sequencing data have yet to be made public, and 2) The subset of luminal tumors more likely to respond was also more heavily immune infiltrated and skewed towards effector T cell expression, which may have marked them as basal by our classifier and/or represent a subset of luminal tumors not modeled by UPPL.
The BBN model exhibited a mixed response to anti-Pd-1 therapy, which allowed us to evaluate differences in the tumor immune microenvironment between responders and non-responders. Responsive tumors showed higher degrees of CD8+-to-CD4+ T cell infiltration and memory-to-regulatory polarization of the tumor infiltrating T cells. The former finding is consistent with prior results in human bladder cancer (13), whereas the latter is to our knowledge the first report of memory T cell polarization associating with response to immune checkpoint inhibition in bladder cancer. This is consistent with the hypothesis that tumor clearance is augmented by generation of T cell memory. While multiple T cell immune gene signatures were strongly correlated with tumor mass in responders, we observed an overall lack of TCR clonotype sharing increase in tumor infiltrating T cells in responders compared to non-responders. Additionally, significant changes to the B cell clonotype sharing and diversity were observed, suggesting B cells may play an important role in the response to checkpoint inhibitor therapy. Presumably, this pattern of T and B cell receptor expression could be explained by several hypotheses: 1) That effector T cell clones capable of promoting anti-tumor immunity are only present in responders but their frequencies are too low to result in discriminating differences in global T cell diversity changes, or 2) That effector T cell clones capable of promoting anti-tumor immunity are present in both responders and non-responders, but the presence of specific B cell clones are necessary to mediate their function. Ongoing studies are evaluating pre-treatment tumor features and on-treatment features measurable from the peripheral blood that associate with eventual response to therapy, as well as to test novel combinations of immunotherapy agents. Thus this model provides a novel mixed-response platform to study efficacy and mechanisms of immunotherapy in bladder cancer.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the members of the Kim, Vincent, and Serody labs for useful discussions as well as Lineberger Bioinformatics Core, the UNC Mouse Phase 1 Unit (MP1U) for technical support. We thank Phil Abbosh for sharing MB49 cells. This work was supported by the Bladder Cancer Advocacy Network (BCAN) Innovation Award (WYK), University Cancer Research Fund (UCRF) [WYK and BGV], NIH K08 Award K08DE026537 (KMB), and UNC Oncology Clinical Translational Research Training Program, 5K12CA120780 (BGV).
Footnotes
Conflicts of interest: William Kim and Jeffrey S. Damrauer are inventors on the BASE47 gene classifier.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 3rd ed. 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25:152–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Damrauer JS, Hoadley KA, Chism DD, Fan C, Tiganelli CJ, Wobker SE, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proceedings of the National Academy of Sciences. 2014;111:3110–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sjödahl G, Lauss M, Lövgren K, Chebil G, Gudjonsson S, Veerla S, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res. 2012;18:3377–86. [DOI] [PubMed] [Google Scholar]
- 6.Volkmer J-P, Sahoo D, Chin RK, Ho PL, Tang C, Kurtova AV, et al. Three differentiation states risk-stratify bladder cancer into distinct subtypes. Proc Natl Acad Sci USA. 2012;109:2078–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rebouissou S, Bernard-Pierrot I, de Reyniès A, Lepage M-L, Krucker C, Chapeaublanc E, et al. EGFR as a potential therapeutic target for a subset of muscle-invasive bladder cancers presenting a basal-like phenotype. Science Translational Medicine. 2014;6:244ra91–1. [DOI] [PubMed] [Google Scholar]
- 8.Dyrskjøt L, Thykjaer T, Kruhøffer M, Jensen JL, Marcussen N, Hamilton-Dutoit S, et al. Identifying distinct classes of bladder carcinoma using microarrays. Nat Genet. 2003;33:90–6. [DOI] [PubMed] [Google Scholar]
- 9.Hedegaard J, Lamy P, Nordentoft I, Algaba F, Høyer S, BP Ulhøi, et al. Comprehensive Transcriptional Analysis of Early-Stage Urothelial Carcinoma. Cancer Cell. 2016;30:27–42. [DOI] [PubMed] [Google Scholar]
- 10.Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell. 2017;171:540–556.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kardos J, Chai S, Mose LE, Selitsky SR, Krishnan B, Saito R, et al. Claudin-low bladder tumors are immune infiltrated and actively immune suppressed. JCI Insight. 2016;1:e85902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powles T, Eder JP, Fine GD, Braiteh FS, Loriot Y, Cruz C, et al. MPDL3280A (anti-PD-LI) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515:558–62. [DOI] [PubMed] [Google Scholar]
- 13.Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma P, Callahan MK, Bono P, Kim J, Spiliopoulou P, Calvo E, et al. Nivolumab monotherapy in recurrent metastatic urothelial carcinoma (CheckMate 032): a multicentre, open-label, two-stage, multi-arm, phase 1/2 trial. Lancet Oncol. 2016;17:1590–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massard C, Gordon MS, Sharma S, Rafii S, Wainberg ZA, Luke J, et al. Safety and Efficacy of Durvalumab (MEDI4736), an Anti-Programmed Cell Death Ligand- 1 Immune Checkpoint Inhibitor, in Patients With Advanced Urothelial Bladder Cancer. Journal of Clinical Oncology. 2016;34:3119–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellmunt J, de Wit R, Vaughn DJ, Fradet Y, Lee J-L, Fong L, et al. Pembrolizumab as Second-Line Therapy for Advanced Urothelial Carcinoma. N Engl J Med. 2017;376:1015–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apolo AB, Infante JR, Balmanoukian A, Patel MR, Wang D, Kelly K, et al. Avelumab, an Anti-Programmed Death-Ligand 1 Antibody, In Patients With Refractory Metastatic Urothelial Carcinoma: Results From a Multicenter, Phase Ib Study. Journal of Clinical Oncology. 2017;:JCO2016716795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Summerhayes IC, Franks LM. Effects of donor age on neoplastic transformation of adult mouse bladder epithelium in vitro. J Natl Cancer Inst. 1979;62:1017–23. [PubMed] [Google Scholar]
- 19.Vasconcelos-Nóbrega C, Colaço A, Lopes C, Oliveira PA Review: BBN as an urothelial carcinogen. In Vivo. 2012;26:727–39. [PubMed] [Google Scholar]
- 20.Kobayashi T, Owczarek TB, McKiernan JM, Abate-Shen C. Modelling bladder cancer in mice: opportunities and challenges. Nat Rev Cancer. 2014;15:42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang ZT, Pak J, Shapiro E, Sun TT, Wu XR. Urothelium-specific expression of an oncogene in transgenic mice induced the formation of carcinoma in situ and invasive transitional cell carcinoma. Cancer Research. 1999;59:3512–7. [PubMed] [Google Scholar]
- 22.Grippo PJ, Sandgren EP. Highly invasive transitional cell carcinoma of the bladder in a simian virus 40 T-antigen transgenic mouse model. The American Journal of Pathology. 2000;157:805–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rampias T, Vgenopoulou P, Avgeris M, Polyzos A, Stravodimos K, Valavanis C, et al. A new tumor suppressor role for the Notch pathway in bladder cancer. Nat Med. 2014. [DOI] [PubMed] [Google Scholar]
- 24.Puzio-Kuter AM, Castillo-Martin M, Kinkade CW, Wang X, Shen TH, Matos T, et al. Inactivation of p53 and Pten promotes invasive bladder cancer. Genes & Development. 2009;23:675–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trotman LC, Niki M, Dotan ZA, Koutcher JA, Di Cristofano A, Xiao A, et al. Pten Dose Dictates Cancer Progression in the Prostate. Plos Biol. 2003;1:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. The American Journal of Pathology. 2012;180:599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Research. 20l0;38:e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–95. [DOI] [PubMed] [Google Scholar]
- 29.Iglesia MD, Vincent BG, Parker JS, Hoadley KA, Carey LA, Perou CM, et al. Prognostic B-cell signatures using mRNA-seq in patients with subtype-specific breast and ovarian cancer. Clin Cancer Res. American Association for Cancer Research; 2014;20:3818–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. - PubMed - NCBI. Bioinformatics. 2006;22:1540–2. [DOI] [PubMed] [Google Scholar]
- 31.Wölfl M, Greenberg PD. Antigen-specific activation and cytokine-facilitated expansion of naive, human CD8+ T cells. Nature Protocols. 2014;9:950–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA. 2009;106:12771–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iglesia MD, Parker JS, Hoadley KA, Serody JS, Perou CM, Vincent BG. Genomic Analysis of Immune Cell Infiltrates Across 11 Tumor Types. - PubMed - NCBI. J Natl Cancer Inst. 2016;108:djw144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fantini D, Glaser AP, Rimar KJ, Wang Y, Schipma M, Varghese N, et al. A Carcinogen-induced mouse model recapitulates the molecular alterations of human muscle invasive bladder cancer. Oncogene. 2018;66:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li S, Zhu M, Pan R, Fang T, Cao Y-Y, Chen S, et al. The tumor suppressor PTEN has a critical role in antiviral innate immunity. Nat Immunol. 2016;17:241–9. [DOI] [PubMed] [Google Scholar]
- 36.Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, et al. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discovery. 2016;6:202–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.