Abstract
The yeast C-type cyclin Ume3p/Srb11p and its cyclin-dependent kinase (Cdk) Ume5p are required for the full repression of genes involved in the stress response or meiosis. This cyclin–Cdk kinase copurifies with the RNA polymerase II holoenzyme complex, suggesting it functions through modification of the transcriptional machinery. This report describes two domains required for Ume3p–RNA Pol II holoenzyme association. One domain contains the highly conserved cyclin box that directs cyclin–Cdk interaction and requires Ume5p for holoenzyme binding. The second domain, termed HAD for holoenzyme associating domain, is located within the amino-terminal region of the cyclin and is sufficient for holoenzyme binding independent of Ume5p or the cyclin box. In addition to its role in RNA Pol II holoenzyme association, the HAD is also required for Ume3p-dependent repression in vivo. Finally, HAD mutations do not affect the ability of the Ume3p–Ume5p kinase to phosphorylate in vitro the carboxy-terminal domain (CTD) of RNA polymerase II, a reported target of cyclin C-Cdk activity. In conclusion, this study demonstrates that the association of the Ume3p to the holoenzyme is complex, involving two independent domains, both of which are required for full Ume3p-dependent repression in vivo. Furthermore, HAD-dependent repression does not appear to involve CTD phosphorylation, suggesting a different role for this domain in directing Ume3p–Ume5p activity.
Keywords: Cyclin C, RNA polymerase II holoenzyme, Protein–protein interaction, Leucine zipper
IN eucaryotes, transcription initiation is controlled by complex interactions between enhancer binding transcription factors, chromatin-modifying enzymes, and the basal transcription machinery [for recent reviews see (2,19,49,52)]. Among other activities, the basal machinery contains RNA polymerase II (RNA Pol II) and additional general factors (e.g., TFIIB, TFIIF, TFIIH) [reviewed in (18)] collectively termed the holoenzyme. The correct localization and assembly of the RNA polymerase II holoenzyme to the promoter is sufficient to promote transcription and has been proposed to be the main role for transcriptional activators (16,25). In addition to RNA Pol II and the general transcription factors, the holoenzyme contains the mediator, an activity required for enhancer-dependent activation both in vivo and in vitro [for reviews, see (3,19)]. The mediator was identified by its ability to associate with the carboxyl-terminal domain (CTD) repeats of RNA Pol II (26,28,39). The CTD is a heptapeptide repeat (26–52 reiterations, depending on the organism) that is essential for activator-dependent RNA Pol II activity [reviewed in (7)]. Different CTD phosphorylation states have been linked to RNA Pol II activity (31). Specifically, the unphosphorylated form of RNA Pol II is predominantly found in the preinitiation complex whereas the elongating RNA Pol II species is multiply phosphorylated (33,41,46,58). These results suggest a role for protein kinases in the transition of RNA Pol II between the inactive and active forms [reviewed in (8)].
Several components of the mediator (e.g., Gal11p, Sin4p, Rgr1p, six Med proteins) (20,37,45) have been identified. Nine additional mediator proteins (Srb for suppressors of RNA polymerase B) were isolated as suppressors of the cold-sensitive growth phenotype associated with RNA Pol II derivatives lacking a portion of the CTD domain (28,38). All the Srb proteins copurify with the RNA Pol II holoenzyme, indicating that they influence RNA Pol II activity in a direct manner. One of the Srb proteins (Srb10p) is identical to Ume5p, a cyclin-dependent kinase or Cdk (60) most closely resembling the Cdk8 subfamily in higher organisms (32,61). Another Srb protein (Ume3p/Srb11p) is a C-type cyclin (38). Several findings support the model that Ume3p and Ume5p form a cyclin–Cdk complex. First, genetic tests revealed that both proteins are involved in the repression of diverse genes including SPO13, SSA1, and SUC2 (4,6,59,66). Moreover, epistasis studies indicated that both Ume3p and Ume5p function in the same regulatory pathway (6). Finally, both two-hybrid (29) and coimmunoprecipitation studies (our unpublished results) demonstrated that Ume3p and Ume5p interact in vivo. Because cyclin C–Cdk8 kinases from both human and Drosophila also copurify with the RNA polymerase II holoenzyme (39,61,62), the function of this class of kinases may be conserved.
Unlike cyclins, which regulate mitotic cell division, Ume3p levels remain constant throughout the cell cycle. Rather, this cyclin is destroyed in response to heat shock or during meiosis (6). This destruction is important to relieve Ume3p-dependent repression as mutants resistant to meiosis-induced degradation fail to fully express SPO13 (6). In addition to its role in repression, the Ume3p–Ume5p kinase has also been implicated in transcriptional activation. Specifically, mutants lacking Ume3p or Ume5p have been reported to exhibit a 5–100-fold reduction in the expression of a galactose-inducible reporter gene (29,38). A role in transcriptional activation is consistent with several reports indicating that cyclin C–Cdk8 kinases from higher systems are able to phosphorylate the CTD repeat in vitro (32,50). Moreover, yeast mutants lacking Ume5p (Srb10p) display an approximate 10-fold reduction in CTD phosphorylation in vivo (38). Although the reduction in CTD phosphorylation may be indirect, these findings raise the possibility that the Ume3p–Ume5p kinase regulates transcription through direct modification of RNA Pol II.
The mediator was first described as an activity required for transcription initiation. However, genetic studies have indicated that several components of the mediator (e.g., Ume3p, Ume5p, Rgr1p, Sin4p) function as transcriptional repressors (4,6,37,59,66). These findings may indicate that the mediator functions in both a positive and negative manner. However, other explanations are also suggested in the literature. Although several components involved in transcriptional activation (e.g., TFIIB, Gal11p) always copurify with the holoenzyme, the presence or absence of other factors appears to depend on the purification protocol utilized. For example, holoenzyme preparations isolated over several chromatography steps lack the Ume3p–Ume5p kinase (45). However, holoenzyme fractions prepared using affinity purification retain this cyclin–Cdk (37). These findings may suggest that more than one “type” of holoenzyme exists in the cell (53) or that the Ume3p–Ume5p kinase association with the holoenzyme is less stable than others. Alternatively, these results may question the physiological relevance of repressor proteins that only associate with the holoenzyme under mild purification protocols.
To explore the functional relationship between the association of the Ume3p–Ume5p kinase with the holoenzyme and its role in transcriptional repression, a combined genetic and biochemical approach was employed. In this study, we report the identification of two domains (cyclin box and holoenzyme associating domain or HAD) that are able to independently direct Ume3p binding to the holoenzyme in coimmunoprecipitation studies. We further demonstrate that the cyclin box domain requires the Cdk for binding whereas the HAD does not. In addition, HAD mutations that destabilize RNA Pol II holoenzyme interaction also reduce Ume3p-dependent repression of the HSP70 heat shock gene SSA1 in vivo. Finally, the HAD mutations do not affect Ume3p–Ume5p-dependent phosphorylation of the CTD in vitro. Taken together, these findings indicate that the interaction of Ume3p to the RNA Pol II holoenzyme is complex, involving at least two domains, both of which are important for Ume3p–Ume5p-dependent repression. Moreover, CTD phosphorylation may not play an important role in Ume3p–Ume5p repression of SSA1 in vivo.
MATERIALS AND METHODS
Strains and Plasmids
Genotypes for all yeast strains are listed in Table 1. Yeast strain RSY472 is a derivative of EGyl95 (13) and was used for all lexAop-LEU2 activation experiments. The construction of 2μ. high-copy plasmids containing wild-type (pKC342) and mutant (pKC355-KERQKΔ, pKC219-L28A, pKC362-A110V) forms of UME3 has been described elsewhere (6). The single amino acid substitutions L28P (pKC395), W14A (pKC259), and L31A (pKC255) were constructed by site-directed mutagenesis on pKC342 using the oligonucleotides (+72 AGGCAGAAGCCATGGCTATTG +92), (+31 AGG CATCATGCGCAATATACC +51), and (+82 TTATGGCTAGCGGAGTGCCAG +102), respectively. The UME3 single-copy centromere plasmids used for the lacZ assays were constructed by cloning the 3000-bp BamHI fragments containing the ADH1 promoter and UME3 open reading frame (ORF) from pKC342, pKC219, pKC395, and pKC355 (6) into the BamHI site of pRS316 (55) to form pKC243-WT, pKC241-L28A, pKC242-L28P, and pKC244-KERQKΔ, respectively. The plasmids used for the in vivo recruitment assays [lexA, pEG202; positive control, pSH17-4 (13); Ume3p, pKC342, lexA-Ume3p, pEG3] have been described elsewhere (6). The mutant lexA-Ume3p fusion plasmids (Fig. 3) were constructed by fusing the 1500-bp EcoRI-NotI fragments from pKC395, pKC219, pKC355, pKC259, and pKC255 respectively, to the carboxyl end of pEG202 to form lexA-Ume3pL28P, lexA-Ume3pL28A, lexA-Ume3pKERQKΔ, lexA-Ume3pW14A, and lexA-Ume3pL31A, respectively. The HA tagged allele of UME5 (pUM513) was constructed as follows: a single copy of the HA epitope was inserted at the amino-terminus of the UME5 coding region using site-directed mutagenesis (30). Next a BamHI site was engineered 8 bp upstream of the initiator ATG to generate a 1600-bp BamHI fragment containing the HA-tagged UME5 ORF. This fragment was placed under the control of the GPD1 promoter with a CYC1 terminator in the 2μ vector pRS426 (55). This plasmid was able to complement the aberrant mitotic expression phenotype of a spo13-lacZ reporter gene in a ume5Δ strain [RSY440 (6)]. The T7-His6 tagged allele of UME5 (pUM514) was constructed by amplifying the UME5 ORF with 5′ (GCGCGGATCCAAATGTATAATGGC) and 3′ (CGCGGGATCCCTATCTTCTGTTTTT) primers then inserting the PCR product into pGALSET983 (12) under the control of the GAL1 promoter. The T7/His6-Ume5p derivative is able to complement the aberrant mitotic expression phenotype of a spo13-lacZ reporter gene in a ume5Δ strain (data not shown), indicating that the protein is functional. All introduced mutations were verified by DNA sequence analysis (data not shown). The HA-Kin28 plasmid (PGK14-4) used in the CTD kinase assays was a kind gift from Dr. Philip Kaldis, Yale University.
TABLE 1.
YEAST STRAINS
| Strain | Genotype | Source |
|---|---|---|
| RSY10 | MATa ade2 ade6 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | (59) |
| RSY391 | MATa ade2 ade6 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ume3::LEU2 | (6) |
| RSY385 | MATa cyh2r-z ho::lys2 leu2.:hisG lys2 trp1::hisG ura3 ume3Δ::LEU2 | (6) |
| RSY440 | MATa cyh2r-z ho::LYS2 leu2::hisG lys2 trp1::hisG ura3 ume5Δ::LEU2 | (6) |
| RSY472 | MATa his3-11,15 leu2-3,112 lys2-1 trp1-1 ura3-1 2lexAop-LEU2-3 | this study |
FIG. 3.
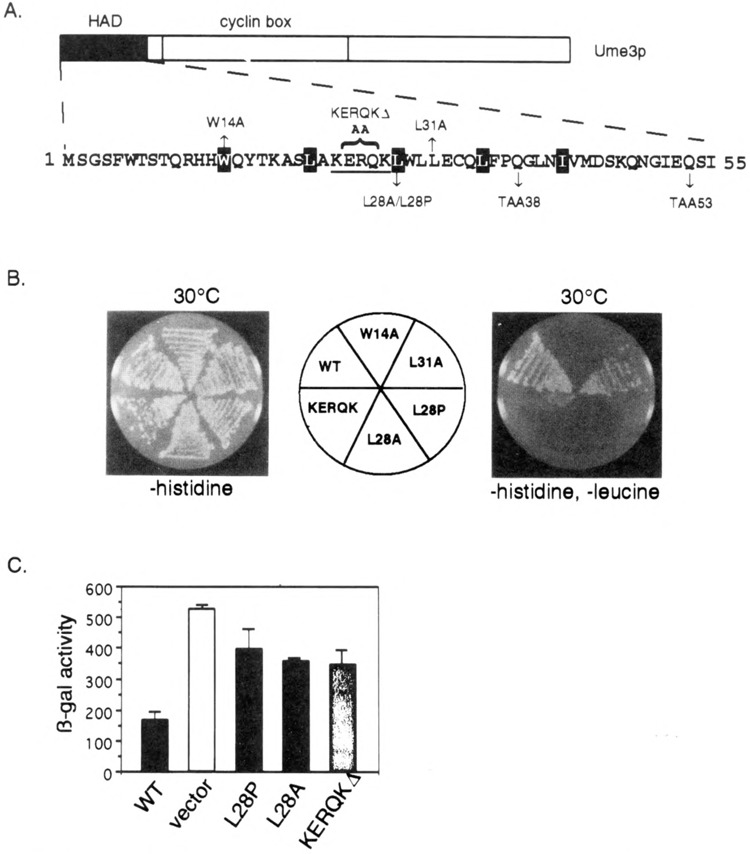
Functional analysis of the HAD mutants. (A) HAD mutant descriptions. The amino-terminal region containing the holoenzyme associating domain (HAD) is presented. Site-directed alterations are shown above and below the primary sequence. The KERQK sequence (bold) was substituted with two alanines to form the KERQKΔ mutation. The stop codons generated by the mutagenesis (TAA38 and TAA53) are shown. The reiterated hydrophobic residues comprising the putative leucine zipper are depicted as white lettering on black. (B) HAD mutations disrupt in vivo recruitment of the holoenzyme. Wild-type (WT) lexA-Ume3p or HAD mutants are tested for their ability to activate the lexAop-LEU2 reporter gene at 30°C as indicated by growth on –histidine, –leucine medium (right). No growth defect is apparent when only plasmid maintenance is selected (–histidine). Positive control is the same as Fig. 1. (C) HAD mutants are defective in Ume3p-dependent repression. The expression of ssa1-lacZ reporter plasmid was assayed in the HAD mutants and wild-type Ume3p constructs (no lexA domain) in a ume3Δ background. Three independent transformants from each group were assayed and β-galactosidase (β-gal) activity is presented in Miller units (42). Error bars indicate standard deviations of the experiments.
Mutagenesis
To generate heat-stable lexA-UME3 mutants, pEG3 was mutagenized in vitro (54) then introduced into RSY472, which contains the LEU2 gene under the control of lexA operators (lexAop-LEU2). Transformants growing at 30°C were replica-plated onto medium selecting for the activation of the LEU2 reporter then incubated at 37°C. Plasmids from colonies able to grow on leucine-deficient medium at 37°C were rescued into E. coli and the phenotype verified. lexA-UME3 clones that produced truncated proteins were subjected to sequence analysis to determine the position of the newly introduced termination codon.
Western Blot Analysis
To visualize lexA-Ume3p fusion proteins (Fig. 2B, Fig. 3C), extracts were prepared and analyzed by Western blot as previously described (6). lexA-Ume3p or lexA was detected with alkaline phosphatase-conjugated secondary antibodies (Sigma) directed against the rabbit polyclonal lexA antibodies (a gift from E. Golemis) and chemiluminescense using CDP-star (Tropix). For the coimmunoprecipitation experiments, extracts were prepared essentially the same way except that the cells were lysed in holoenzyme lysis buffer (45 mM HEPES, pH 7.5, 0.15 M potassium acetate, 30% glycerol, 3 mM EDTA, yeast protease inhibitor cocktail, Sigma). Coimmunoprecipitation studies with lexA-Ume3p (Fig. 2C) or Ume3p utilized 1 mg of protein extract and were incubated with the appropriate antibody overnight at 4°C, collected on protein A-agarose beads (Sigma, 2 h rotating at 4°C), and washed four times in holoenzyme buffer 3 (20 mM HEPES, pH 7.5, 0.25 M potassium acetate, 5 mM EDTA, 0.1% NP-40). His6-tagged Ume5p extracts (5 mg) were treated as described for Ume3p except that nickel beads (Qiagen) were added, incubated overnight at 4°C, then collected by centrifugation. The beads were washed extensively with holoenzyme buffer 3. The immunoprecipitates or nickel beads were resuspended in 2× SDS PAGE loading buffer (40) and separated on either 15% (lexA-Ume3p truncations) or 10% (Ume3p derivatives) SDS polyacrylamide gels. Bands were detected using the primary antibodies indicated followed by either alkaline phosphatase-conjugated secondary antibodies directed against the rabbit polyclonal lexA antibodies and chemiluminescense using CDP-star (Fig. 2C) or 0.4 μCi of 125I-conjugated sheep anti-mouse IgG as the secondary anti-body (ICN) followed by phosphorimaging (FUJI, Inc.).
FIG. 2.
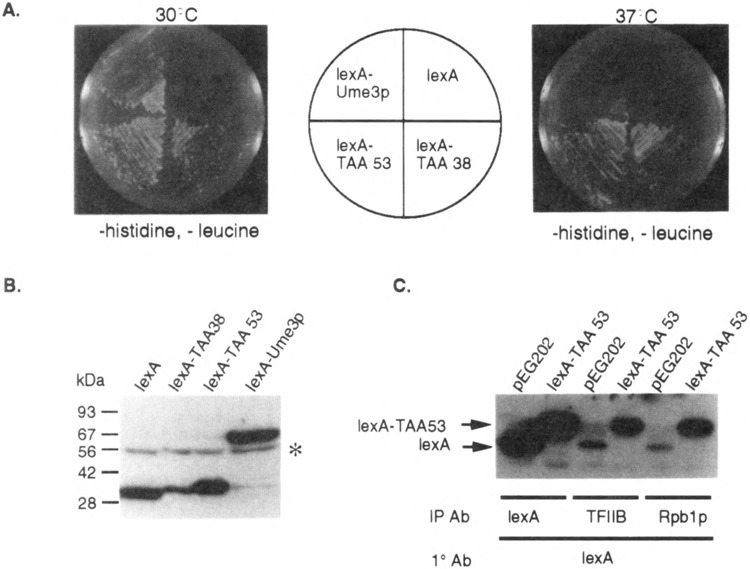
Identification of the Ume3p RNA Pol II holoenzyme associating domain (HAD). (A) Identification of heat-resistant lexA-Ume3p mutants. Transformants harboring mutagenized lexA-UME3 constructs (lexA-TAA53 and lexA-TAA38) were identified by their ability to activate lexAop-LEU2 at 37°C (bottom quadrants) compared to the wild-type lexA-Ume3p control. (B) lexA-TAA38 and lexA-TAA53 are truncated proteins. Western blot analysis of yeast extracts containing lexA and lexA-Ume3p mutants as indicated. Size standards are given at the left. The asterisk indicates nonspecific cross-hybridization. (C) lexA-TAA53 coimmunoprecipitates with the RNA Pol II holoenzyme. Immunoprecipitates of antibodies directed toward either lexA (expression control) or two components of the holoenzyme (TFIIB and Rpb1p) were blotted and probed lexA antibodies. lexA- and lexA-TAA53-specific bands are indicated by the arrows.
CTD Kinase Assays
The extracts for the CTD kinase assays were prepared as follows. RSY10 (WT), RSY391 (ume3Δ), or RSY440 (ume5Δ) cultures harboring the plasmids indicated in the text were lysed in buffer A (0.2 M Tris Base, 0.39 M ammonium sulfate, 10 mM magnesium sulfate, 20% glycerol, 1 mM EDTA, pH 7.9, pH adjusted to 7.3 with acetic acid, yeast protease inhibitor cocktail, Sigma) by vigorous vortexing in the presence of glass beads. Lysates were cleared by centrifugation (1 h at 50,000 × g) and the resulting supernatant concentrated by addition of equal volume of saturated ammonium sulfate. The precipitates were collected by centrifugation, the pellets resuspended in buffer A, then immunoprecipitated with antibodies as described in the text. CTD phosphorylation assays were performed with the immunoprecipitates essentially as described elsewhere (5). The CTD peptide [(YSPTSPS)4] was a kind gift from Dr. Philip Kaldis, Yale University. The remainder of the sample was analyzed by Western blot as described above.
β-Galactosidase Assays
β-Galactosidase activities were determined from at least three independent transformants (RSY391) containing the ssa1-lacZ reporter gene (pZD0425) (56). Cultures (50 ml) were grown to mid-log phase (5 × 106 cells/ml) at 23°C, harvested, and the cell pellet resuspended in 0.25 ml buffer A (20% glycerol, 1 mM DTT, 100 mM Tris-Cl, pH 8.0, 1 mM phenylmethylsulfonyl fluoride). Soluble extracts were prepared by vigorous vortexing with glass beads (4 min) followed by centrifugation (12,000 × g) for 15 min. The extracts were assayed at three enzyme concentrations to ensure that the reactions were in the linear range as described (42). Protein concentrations were determined using the BioRad micro procedure. Units are given in Miller units.
RESULTS
In Vivo Recruitment Assay for Monitoring Heat Shock-Induced Destruction of Ume3p
We developed an in vivo recruitment assay to identify sequences within Ume3p required for heat shock-induced degradation. This assay takes advantage of the observation that components of the RNA polymerase II holoenzyme complex (e.g., Gal11p, Sin4p) can activate transcription when tethered to a simple promoter via a heterologous DNA binding domain (1,24). This activity is most likely due to the ability of holoenzyme components to recruit the entire complex to the promoter with sufficient efficiency to stimulate transcription. Similarly, the lexA-Ume3p fusion protein is able to activate a LEU2 reporter gene under the control of lexA operators (lexAop-LEU2) (21), as evidenced by the ability of a leu2 mutant strain (RSY472) to grow in medium lacking leucine (Fig. 1). This activity requires the presence of the fusion protein as lexA or Ume3p alone are unable to promote growth. However, lexA-Ume3p is unable to activate the LEU2 reporter gene at 37°C due to the Ume3p-dependent destruction of this fusion protein in response to heat shock (6). Therefore, mutations that protect lexA-Ume3p from heat-induced degradation will support growth of RSY472 at 37°C.
FIG. 1.
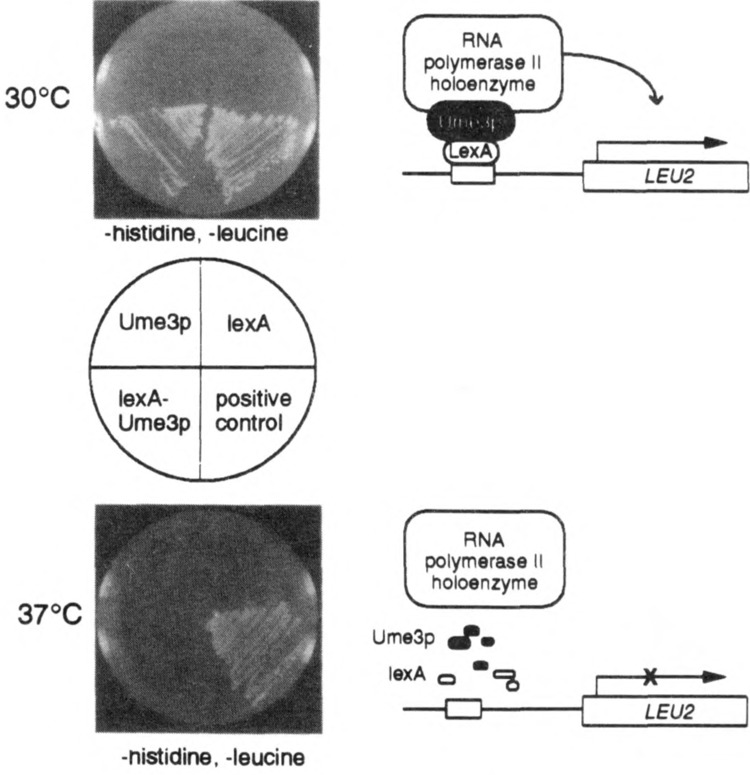
The lexA-Ume3p in vivo recruitment assay. lexA-Ume3p recruitment of the RNA polymerase holoenzyme is indicated by activation of a lexAop-LEU2 reporter gene and growth of RSY472 on medium lacking leucine at 30°C (top panel). Cells harboring either Ume3p or lexA alone are unable to grow. At 37°C (bottom panel), lexA-Ume3p is no longer able to support growth due to the heat-induced destruction of the fusion protein. The positive control, lexA fused to the Ga14p activation domain, demonstrates that this system is not inherently temperature sensitive.
Identification of the Ume3p HAD
A mutagenized lexA-UME3 pool was introduced into RSY472 via transformation, and individual colonies able to grow at 37°C on medium lacking leucine (i.e., able to active lexAop-LEU2, see Fig. 2A) were selected for further study (see Materials and Methods for details). Western blot analyses revealed two classes of stabilized lexA-Ume3p mutants. The first class represented single amino acid substitutions that did not alter the lexA-Ume3p fusion protein size. The analysis of these mutants was described elsewhere (6). The second class contained truncated versions of lexA-Ume3p (Fig. 2B). Eight of the truncated derivatives were subjected to DNA sequence analysis and found to individually contain newly introduced translation termination codons from amino acid 38 to 64. Subsequent screening of additional heat-stable proteins did not uncover any truncated mutant larger than codon 65 nor smaller than codon 38 (data not shown). These findings suggested two possibilities. First, the truncated proteins must lack the signals required for heat-induced degradation. This hypothesis was largely confirmed in a previous study [(6); see Discussion]. Second, because the first 38 residues of Ume3p can function as a transcriptional activator, this region may be sufficient to direct lexA-Ume3p association to the RNA Pol II holoenzyme.
To test whether the amino-terminal region of Ume3p is sufficient for RNA Pol II holoenzyme binding, a series of coimmunoprecipitation experiments was performed. Cultures harboring plasmids expressing either lexA (pEG202) or the lexA-UME3 truncation mutant with a stop codon at amino acid 53 (lexA-TAA53) were grown to mid-log phase and protein extracts prepared (see Materials and Methods for details). These extracts were immunoprecipitated with either lexA antibodies or antibodies directed against one of two components of the RNA polymerase holoenzyme (TFIIB or Rpb1p) (18). The immunoprecipitates were collected and the presence or absence of lexA or lexA-TAA53 was determined by Western blot analysis. The control immunoprecipitations with antibodies directed against lexA revealed that lexA and lexA-TAA53 were expressed at similar levels in these strains (Fig. 2C). Antibodies directed against the general transcription factor TFIIB or the largest subunit of RNA Pol II (Rpb1p) were able to precipitate lexA-TAA53. Some nonspecific coimmunoprecipitation of lexA with the holoenzyme components was also detected in the control reactions but to a significantly lesser extent than lexA-TAA53. Identical results were obtained in coimmunoprecipitation studies including the lexA-Ume3p protein truncated at amino acid 38 (lexA-TAA38, data not shown). These results demonstrate that the amino-terminal region is sufficient to direct the association of Ume3p to the holoenzyme. For convenience, this domain will be referred to as the RNA Pol II HAD.
Integrity of the HAD Is Required for In Vivo Recruitment of the RNA Pol II Holoenzyme
The consensus of five protein secondary structure algorithms (DSC, NNPREDICT, PHD, Predator, and SIMPA) predicts that the HAD is α-helical in nature (11,15,27,35,51). In addition, a reiterating heptad repeat of hydrophobic residues is present (white lettering on black, Fig. 3A) similar to those observed in leucine zipper motifs (64). To investigate whether either of these putative structures is important for HAD function, three mutations were introduced into this region. Our strategy was to first disrupt the leucine zipper motif by substituting an alanine for a leucine at position 28 (L28A, see Fig. 3A). The alanine substitution was chosen because this residue significantly destabilizes leucine zipper dimerization without significantly altering α-helical structure (43). This mutation was introduced into the full-length lexA-UME3 fusion gene using oligonucleotide-directed mutagenesis (see Materials and Methods for details) and its ability to bind the RNA Pol II holoenzyme determined by the in vivo recruitment assay. lexA-Ume3pL28A failed to activate the LEU2 reporter gene, indicating that this mutation interfered with lexA-Ume3p-RNA Pol II holoenzyme interaction (Fig. 3B). No effect on overall protein levels was observed with this mutant (data reviewed but not shown), indicating that the failure of lexA-Ume3pL28A to activate LEU2 was not due to changes in protein stability. An identical result was obtained with the W14A mutation (data not shown). These findings suggest that the potential leucine zipper-like motif is important for HAD function. To investigate the requirement of the putative α-helix, a proline was substituted at the L28 position. Prolines disrupt helix formation by preventing the free rotation of the α-carbon bond in the peptide chain (57). As observed with the L28A substitution, lexA-Ume3pL28P also failed to activate LEU2 expression consistent with the requirement of an α-helical structure for HAD function. Finally, the L28 residue was left intact but the periodicity of the helix was shifted by replacing the KERQK residues (bold face type, Fig. 3A) with two alanines (net loss of three amino acids). This mutation (KERQKΔ) places the first half of the putative helix out of register with the second half. In addition, this sequence is highly conserved among C-type cyclins but not observed in other cyclin subfamilies (see Discussion). lexA-Ume3pKERQKΔ was still able to recruit the holoenzyme as indicated by LEU2 activation. However, the severely reduced growth rate of this strain on medium lacking leucine suggests that this mutation significantly reduces the strength of the RNA Pol II holoenzyme interaction. These results indicate that relatively subtle mutations can affect HAD domain function in the in vivo recruitment assay. In addition, these findings suggest that an α-helix/leucine zipper motif is required for HAD function.
HAD Is Required for Full Transcriptional Repression Activity by Ume3p
To determine if the HAD is required for Ume3p repressor function, the ability of the mutants described above to complement a ume3 null mutation was determined. In these studies, the lexA DNA binding domain was removed from Ume3p and the mutant derivatives. Previous studies have found that lexA-Ume3p is unable to substitute for Ume3p (6). To monitor Ume3p repressor activity, the expression of an ssa1-lacZ reporter gene was assayed. Yeast strain RSY385 harboring a chromosomal deletion of UME3 was transformed with the ssa1-lacZ reporter plasmid (pZD0425) (56) and either wild-type UME3 or the HAD mutants on a single-copy centromere plasmid under the control of the ADH1 promoter (see Materials and Methods for construction details). Three independent transformants from each group were picked and grown to mid-log phase (5 × 106 cells/ml) at the repressing temperature (23°C). Extracts were prepared and (β-galactosidase (β-gal) activity determined. Similar to previous findings (6), the vector control resulted in a 3.5-fold derepression of ssa1-lacZ compared to the wild-type UME3 plasmid (Fig. 3C). The analysis of the three HAD mutants (L28A, L28P, and KERQKΔ) revealed a significant increase (two- to threefold) in ssa1-lacZ expression but to a lesser extent that the vector-alone control. Repeating these experiments in a different strain background or with a SPO13 reporter gene produced identical results (data not shown). We conclude that the HAD is required for normal Ume3p-dependent repression in vivo, although these relatively subtle mutations do not appear to completely abolish the activity of this cyclin.
HAD Mutations Confer Ume5p Dependency on Ume3p Association With the Holoenzyme
The results presented above indicate that the HAD is sufficient to direct lexA to the RNA Pol II holoenzyme and is required for Ume3p-dependent transcriptional repression in the context of the full-length cyclin. To test whether the HAD is required for Ume3p-RNA Pol II holoenzyme binding, coimmunoprecipitation experiments were performed as described earlier. For these studies, extracts were prepared from strains expressing either a myc-tagged wild-type Ume3p or HAD mutant derivatives (without the lexA DNA binding domain) under the control of the ADH1 promoter (see Materials and Methods for plasmid construction details). Ume3p-myc expression complements ume3Δ mutations, indicating that the presence of the tag does not significantly alter cyclin activity (6). Similar amounts of protein were observed in the control lanes (myc), indicating that the various mutations do not affect the overall stability of the cyclin (Fig. 4A, top panel). Interestingly, all three HAD mutant proteins were able to coimmunoprecipitate with either Rpb1p or TFIIB, although some relatively small reductions in TFIIB immunoprecipitations were observed. These results indicate that although the HAD is sufficient to direct the association of a lexA truncation mutants to the RNA Pol II holoenzyme, it is not essential for binding in the context of full-length Ume3p.
FIG. 4.
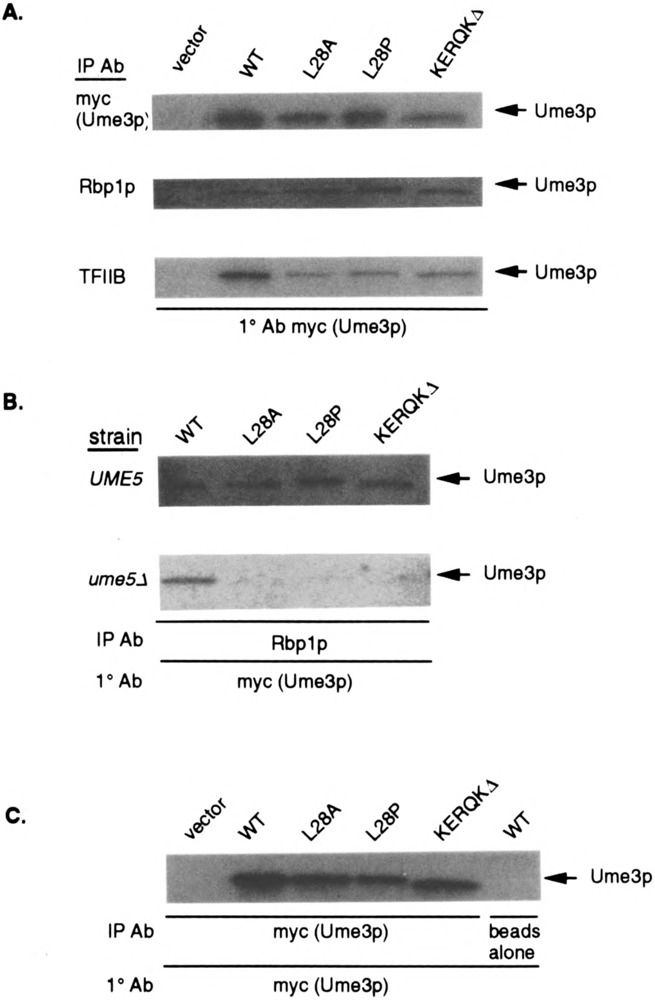
HAD mutant binding to the RNA Pol II holoenzyme requires Ume5p. (A) HAD mutants coimmunoprecipitate with the holoenzyme. Extracts expressing full-length myc-UME3 constructs (lacking the lexA domain) or the different HAD mutants were immunoprecipitated with either myc (to control for protein levels in the extracts), Rpb1p, or TFIIB antibodies, then blotted and probed with the myc monoclonal antibody (mAb). The vector lane controls for nonspecific binding of the myc mAb. (B) HAD mutant binding to the holoenzyme requires Ume5p. Extracts prepared from wild-type (UME5) or ume5 mutant (ume5Δ) strains harboring the HAD mutant expression constructs were immunoprecipitated with Rpb1p, blotted, and probed with the myc mAb to detect Ume3p. The Ume3p and HAD mutant specific bands are indicated by the arrows. (C) HAD mutants do not affect cyclin stability in the ume5Δ mutant strain. Ume3p and the HAD mutant protein levels (arrows) were determined in ume5Δ extracts by Western blot analysis. The vector lane controls for nonspecific epitope recognition by the mAb. The beads alone lane controls for nonspecific association of Ume3p to the protein A sepharose.
The results described above suggest that more than one region may mediate Ume3p–RNA Pol II holoenzyme association. One obvious candidate domain is the highly conserved cyclin box that directs binding to the Cdk partner [reviewed in (48)]. To test this possibility, the ability of Ume3p and the various HAD mutants to associate with the holoenzyme in the absence of Ume5p was examined. UME3 and HAD mutant constructs were transformed into an isogenic wild-type (RSY10) or ume5Δ mutant (RSY440) strains and extracts prepared as described earlier. These extracts were incubated with antibodies directed against Rpb1p and the immunoprecipitates examined for the presence of Ume3p or the HAD mutant derivatives by Western blot analysis. As observed previously, the HAD mutants coimmunoprecipitated with Rpb1p in the wild-type UME5 strain with affinities similar to Ume3p (Fig. 4B, top panel). Repeating these experiments with extracts prepared from the ume5Δ mutant strain revealed that wild-type Ume3p was still able to associate with the holoenzyme (Fig. 4B, bottom panel). However, the HAD mutant proteins did not coimmunoprecipitate with Rpb1p in extracts lacking Ume5p. Longer exposures of this gel revealed a weak signal in the KERQKΔ lane, suggesting that this HAD mutant retains some function in this assay. This finding is consistent with the partial activity observed for this mutant with the in vivo recruitment assay. The overall levels of these HAD mutants were similar in the ume5Δ extracts (Fig. 4C), indicating that the differences in holoenzyme association were not due to changes in protein stability. These experiments allowed several conclusions to be drawn. First, Ume3p is able to associate with the RNA Pol II holoenzyme in the absence of Ume5p. Moreover, this Ume5p-independent binding is mediated through the HAD. Finally, another domain exists that is able to direct Ume3p association to the holoenzyme in a Ume5p-dependent manner. This requirement and the location of the domain strongly suggest that this second activity is provided by the cyclin box.
Ume5p Associates With the RNA Pol II Holoenzyme Independent of Ume3p
The finding that Ume3p can bind the holoenzyme in the absence of Ume5p raises the question of whether Ume5p association is mediated through the cyclin. Previous studies have reported that an inactivated Ume5p is still able to copurify with the holoenzyme (38). Although these experiments indicated that kinase activity is not important, a physical role for the cyclin was not tested. To address this question, a crude protein extract prepared from a mid-log ume3Δ culture (RSY391) overexpressing T7/His6-tagged Ume5p (pUM514) was incubated with nickel beads (see Materials and Methods for details). The beads were collected by centrifugation and analyzed for the presence of the holoenzyme component TFIIS (47,65). Western blot analysis identified the 36-kDa TFIIS (arrow) in the ume3Δ mutant extract (Fig. 5, lane 2) but not in the extract lacking pUM514 (lane 1). The amount of TFIIS associating with Ume5p is significantly less than that observed in straight immunoprecipitation studies (lane 3). This may be due to the presence of different types of holoenzyme complexes in the cell (53) and/or the disruption of Ume5p interactions during the purification process. In either case, these data demonstrate that Ume5p can associate with the RNA Pol II holoenzyme independent of Ume3p.
FIG. 5.

Ume5p associates with the RNA Pol II holoenzyme independent of Ume3p. A nickel bead pull-down experiment was performed with protein extracts prepared from a ume3Δ mutant strain either expressing the T7/His6-tagged Ume5p (lane 2) or not (lane 1). Western blot analysis identified a band (arrow) in lane 2 or in TFIIS immunoprecipitates (lane 3) the size predicted for TFIIS [36 kDa (10)]. Size standards (kDa) are presented on the left. The asterisk indicates the IgG heavy chain contained in the TFIIS immunoprecipitations.
The HAD Domain Is Not Required for CTD Phosphorylation by the Ume3p–Ume5p Kinase
Both direct in vitro experiments and indirect in vivo studies have indicated that one substrate for cyclin C–Cdk8 kinases is the CTD repeat of RNA Pol II (22,32,38,50). We have demonstrated in this study that the HAD is important for Ume3p-dependent repression. A previous study reported that cyclin D–Cdk4 targets the tumor suppressor Rb for phosphorylation through an amino-terminal domain in the cyclin (9). Therefore, we examined the possibility that the HAD is involved in directing CTD phosphorylation by Ume3p–Ume5p. First, the wild-type Ume3p–Ume5p kinase was immunoprecipitated from crude extracts using antibodies directed against the HA-tagged Ume5p (see Materials and Methods for details). This immunoprecipitate was able to phosphorylate the CTD (Fig. 6A, lane 1), demonstrating that, similar to higher eucaryotic C-type cyclin–Cdk kinases, Ume3p–Ume5p can directly phosphorylate the CTD in vitro. Moreover, this activity is specific for both Ume3p and Ume5p as extracts made from either mutant culture failed to exhibit activity (lanes 2 and 3). To examine the role of the HAD domain in CTD phosphorylation, the L28P mutant cyclin (Ume3pL28P) was expressed in a strain deleted for the wild-type copy of UME3 (ume3Δ). Immunoprecipitates from this extract were still able to phosphorylate the CTD with similar efficiency as the wild-type control (Fig. 6B, lane 3). The elevated levels for Ume3pL28P CTD kinase activity observed in this experiment was not reproducible over several different assays. An additional Ume3p control contains a single amino acid substitution in the cyclin box (A110V) that, similar to the L28P mutation, stabilizes this cyclin in response to heat shock, but is fully functional in our repression assays (6). Ume3pA110V is also able to direct phosphorylation of the CTD (Fig. 6B, lane 2). These studies indicate that the HAD is dispensable for CTD phosphorylation in vitro. Moreover, these results suggest that the repression defect observed in HAD mutants is not due to the failure to phosphorylate the CTD.
FIG. 6.
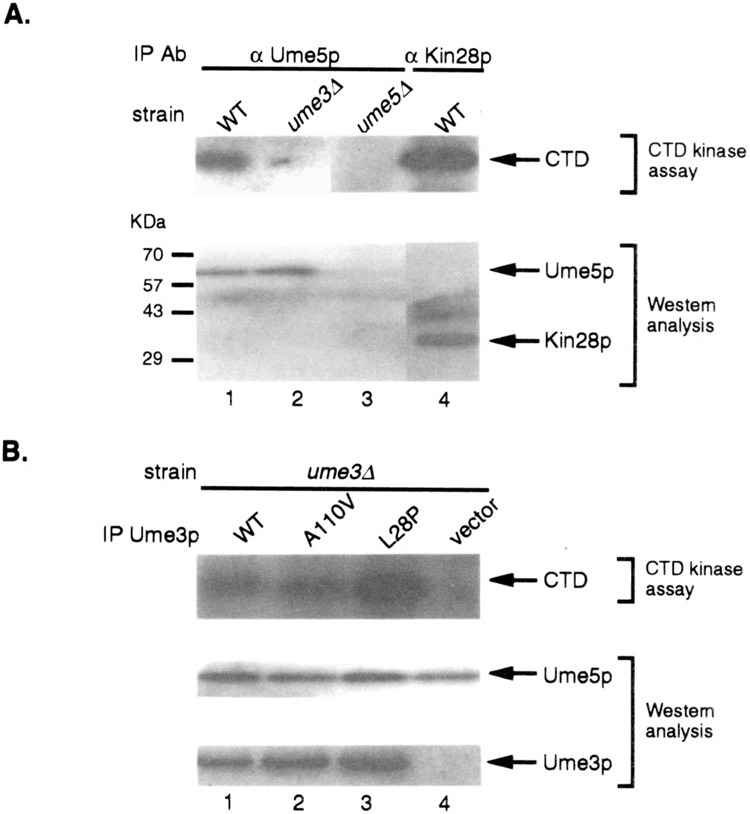
The HAD domain is not required for CTD phosphorylation by the Ume3p–Ume5p kinase. (A) Ume5p phosphorylates the CTD in a Ume3p-dependent manner. Protein extracts prepared from strains with the indicated genotypes harboring either HA tagged Ume5p or Kin28p were immunoprecipitated using the HA mAb. Immunoprecipitates were incubated with CTD peptide and radiolabeled ATP. The phosphorylated CTD was visualized using SDS-PAGE and autoradiography (top panel). Aliquots of the immunoprecipitates were probed by Western blot analysis to verify the recovery of Ume5p or Kin28p (arrows, lower panel). (B) The HAD is dispensable for CTD phosphorylation. A ume3Δ strain harboring HA-tagged Ume5p and the indicated UME3 expression constructs were assayed for CTD kinase activity as described above. The levels of Ume5p and the Ume3p derivatives in the immunoprecipitates were determined by Western blot analysis.
DISCUSSION
Identification of the HAD
Ume3p–Ume5p is a cyclin–Cdk required for the full repression of genes involved in the stress response (SSA1) or meiosis (e.g., SP013). Previous studies found that Ume3p–Ume5p copurifies with the RNA polymerase II holoenzyme (38). This report investigates the functional relationship between the association of Ume3p–Ume5p to the RNA Pol II holoenzyme and its role in transcriptional repression. Two domains, HAD and the cyclin box, were identified that are able to independently target Ume3p to the holoenzyme. The cyclin box is a highly conserved domain that mediates binding of the cyclin to the Cdk [reviewed in (48)]. The requirement of Ume5p for cyclin box-directed association to the holoenzyme suggests that this domain functions indirectly through the Cdk. The second domain, HAD, is located in the amino-terminal 38 amino acids. The analysis of single amino acid substitutions introduced into this region suggests that the HAD requires a leucine zipperlike motif and an α-helical structure for holoenzyme binding. These results indicate the association of the Ume3p–Ume5p kinase to the holoenzyme is complex, involving two domains on the cyclin, and at least one on the kinase. Previous studies have shown that mutations in the cyclin box destroy Ume3p repression activity (6). Similarly, HAD mutants fail to fully repress a reporter gene regulated by Ume3p. Therefore, although either the HAD or cyclin box can direct Ume3p association to the holoenzyme, neither interaction is sufficient on its own to mediate full transcriptional repression. Finally, the HAD is not required for the in vitro phosphorylation of the CTD, a substrate of C-type cyclin–Cdk kinases. These findings suggest a different role for the HAD in mediating SSA1 and SP013 repression.
Role of the RxxL Motif in Ume3p Degradation
We previously identified three regions that are required for heat-induced destruction of Ume3p (6).
One region, termed RxxL, resembles the destruction box involved in the degradation of B-type cyclins (17,44). This motif is located at residue 25 in Ume3p and is contained in the lexA-TAA38 and lexA-TAA53 truncation mutant proteins (Fig. 3A). The presence of RxxL in these proteins, combined with their stability in response to heat shock, is paradoxical. Two possibilities appear most likely to explain these findings. First, we have previously demonstrated that the multiple cis-acting destruction signals function in overlapping, but not identical, capacities. Specifically, mutating the RxxL and PEST region elements individually results in a three- to fivefold stabilization in response to heat shock (6). However, combining these mutations results in a dramatic (> 15-fold) increase in half-life, suggesting that these two regions are functioning synergistically. Therefore, in isolation, the RxxL region may function at a reduced capacity or not at all. Alternatively, the RxxL element may be inactive due to the nature of the truncation mutants. All destruction boxes identified to date are found in the amino portion of the protein. The truncation mutation places the RxxL element within 35 residues from the end of the protein. Perhaps this location forbids necessary secondary structure requirements for recognition by the destruction machinery. This possibility is supported by the failure to isolate truncated alleles longer than residue 64. Longer derivatives of lexA-Ume3p may have been destablized in response to heat shock by providing sufficient sequence carboxyl to the RxxL element to allow its recognition by the destruction machinery.
CTD Phosphorylation and Transcriptional Repression
This study identified two domains that mediate Ume3p association with the holoenzyme. Based on its location and Ume5p dependence, one region is likely to be the cyclin box. The second domain (HAD) is found in the amino portion of the protein. The HAD mediates Ume5p-independent association to the holoenzyme and is required for full Ume3p-dependent repression activity. The finding that HAD mutants are defective in repression even though they can still associate with the RNA Pol II holoenzyme (through the cyclin box) suggests that the HAD performs another function in addition to localization. One possible role is suggested by studies involving cyclin D-dependent phosphorylation of the tumor suppressor Rb. Phosphorylation of Rb by cyclin D/Cdk2 or Cdk4 complexes is required to inactivate Rb and allow cell cycle progression [reviewed in (23)]. The region of cyclin D1, D2, or D3 required for binding Rb is also located in the amino-terminal region of the cyclin (9). If a similar organization has been maintained with C-type cyclins, then the HAD may bind Ume5p substrates and thus help direct Ume5p-dependent phosphorylation. A recent study (22) reported that Ume3p–Ume5p phosphorylation of the CTD in preinitiation complexes can inhibit transcription in vitro. However, because CTD phosphorylation is not affected in HAD mutants, this domain may be performing another role in mediating SSA1 or SP013 repression in vivo. These findings suggest two possibilities. First, several studies examining the impact of CTD phosphorylation in vivo and in vitro have revealed that some promoters are strongly affected whereas others exhibit little or no effect [reviewed in (8)]. Therefore, in the case of SSA1 and SP013, the ability of HAD mutant kinases to phosphorylate the CTD may be inconsequential to the repression of these loci. Rather, the protein that directly binds the HAD to mediate its holoenzyme association may represent an important substrate for the Ume3p–Ume5p kinase to repress SP013 and SSA1 transcription. The second possibility predicts that the HAD targets the Ume3p–Ume5p kinase to a specific site within the holoenzyme, which in turn stimulates CTD phosphorylation. Because the in vitro assays utilize free CTD peptide as substrate, this specialized function of the HAD is not necessary and therefore does not affect kinase activity. Studies are currently under way to identify the binding target for the HAD and determine if it is indeed a substrate for Ume5p or whether is serves as a specific tether for Ume3p–Ume5p within the RNA Pol II holoenzyme.
Conservation of the HAD
Sequence inspection revealed a significant conservation of amino acids in human, rat, and Drosophila cyclin C (Fig. 7A). Most notably, the K/RERQK/H motif (Fig. 7A, asterisks) is retained in the amino-terminal region of all C-type cyclins identified to date that associate with the RNA Pol II holoenzyme (32,34,36,39). This motif is not found in the closely related human cyclin H (14) or the yeast Cc11p (63), suggesting that K/RERQK/H is specific to the cyclin C subfamily. This report demonstrates that deleting this motif affects both RNA Pol II holoenzyme association and repression activity, indicating that, at least in yeast, this sequence is important for function. The results from this study also indicated the requirement of a leucine zipper-like motif and an α-helical structure for HAD function. Secondary structure algorithms (11,15,27,35,51) also predict the presence of an α-helix in the same location in the human cyclin C (HsCycC, Fig. 7B). Furthermore, the HsCycC sequence does contain three hydrophobic heptad repeats (white lettering on black) shifted only one amino acid from the motif observed in Ume3p. In addition to primary and secondary structure similarities, a lexA-HsCycC fusion protein can activate the lexAop-LEU2 reporter gene in the in vivo recruitment assay (unpublished results), indicating the presence of a functional HAD in HsCycC. Taken together, these observations are consistent with the retention of HAD function in other C-type cyclins.
FIG. 7.

HAD domain conservation with other cyclins. (A) Primary sequence comparisons. Members of the C-type and H-type cyclin subfamilies are indicated on the left. Identical residues or conserved substitutions are boxed. The asterisks indicate the conserved KERQK motif. (B) Secondary structure comparison. The primary sequence of each cyclin is presented as indicated. The KERQK motif in Ume3p and the human cyclin C (HsCycC) is in bold face type. The hydrophobic heptad repeats are indicated by the reverse lettering. The secondary structure predictions for each region are indicated below the primary sequence. H = α-helix, spaces indicate no helix, β-sheet, or coiled-coil predicted. Only the consensus of five algorithms (NNPREDICT, PHD, SIMPA, PREDATOR, DSC) is presented. The LxCxE motif that directs HsCycD1 binding to Rb is in bold face type (see text for details).
The amino-terminal region of the human cyclin D1 has also been extensively analyzed with respect to its role in targeting Rb for phosphorylation. This domain has been delimited to include a conserved motif (LxCxE) found in all D-type cyclins (Fig. 7B, bold face type). Altering either the leucine or cysteine residue eliminates cyclin D1–Cdk4 phosphorylation of Rb (9). A similar, but not identical, motif is observed at amino acid 31 in Ume3p (LxCxL). A mutation altering the first leucine (L31A) did not affect Ume3p association with the holoenzyme in the in vivo recruitment assay (unpublished results), suggesting that this sequence is not important for Ume3p HAD function. Secondary structure predictions do indicate the presence of an α-helix in the amino-terminal region of cyclin D1 (Fig. 7B), although a hydrophobic heptad repeat is not observed. If the Ume3p HAD does contribute to identifying substrates for its Cdk, these results would imply that a functional organization has been conserved between different cyclin subfamilies. Moreover, these findings would suggest two criteria for a functional HAD-like domain. The first is an α-helix that may resemble a leucine zipper, a motif known to direct protein–protein interactions in several biological contexts. The second parameter is a unique primary sequence motif that would provide the specificity to insure that the correct substrates were selected. If these rules are conserved, such information may provide a valuable tool to help identify the elusive targets of cyclin–Cdk kinase activity.
ACKNOWLEDGMENTS
We thank J. Burch, R. Perry, and J. Jaehning for critical reading of this manuscript. We thank J. Jaehning, University of Colorado Health Science Center, for Rpb1p, TBP, TFIIS, and TFIIB antibodies and for helpful discussions during the course of this work. We acknowledge M. Mallory for the construction of pUM513. We thank E. Golemis (Fox Chase Cancer Center) for antibodies directed against lexA and P. Kaldis (Yale University) for CTD peptide. K.F.C. was supported by NIH Grant CA-09035-23. This work was supported by NSF grant MCB-9513479 to R.S. and an appropriation from the Commonwealth of Pennsylvania.
REFERENCES
- 1. Barberis A.; Pearlberg J.; Simkovich N.; Farrell S.; Reinagel P.; Bamdad C.; Sigal G.; Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell 81:359–368; 1995. [DOI] [PubMed] [Google Scholar]
- 2. Bjorklund S.; Kim Y.-G. Mediator of transcriptional regulation. Trends Biol. Sci. 21:335–337; 1996. [DOI] [PubMed] [Google Scholar]
- 3. Carlson M. Genetics of transcriptional regulation in yeast: Connection of the RNA polymerase II CTD. Annu. Rev. Cell Dev. 13:1–23; 1997. [DOI] [PubMed] [Google Scholar]
- 4. Carlson M.; Osmond B. C.; Neigeborn L.; Botstein D. A suppressor of snf1 mutations causes constitutive high-level invertase synthesis in yeast. Genetics 107: 19–26; 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cismowski M. J.; Laff G. M.; Solomon M. J.; Reed S. I. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol. Cell. Biol. 15:2983–2992; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cooper K. F.; Mallory M. J.; Smith J. S.; Strich R. Stress and developmental regulation of the yeast C-type cyclin UME3 (SRB11/SSN8) . EMBO J. 16:4665–4675; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Corden J. L. Tails of RNA polymerase II. Trends Biol. Sci. 15:383–387; 1990. [DOI] [PubMed] [Google Scholar]
- 8. Dahmus M. The role of multisite phosphorylation in the regulation of RNA polymerase II activity. Prog. Nucleic Acid Res. Mol. Biol. 48:143–179; 1994. [DOI] [PubMed] [Google Scholar]
- 9. Dowdy S. F.; Hinds P. W.; Louie K.; Reed S. I.; Arnold A.; Weinberg R. A. Physical interaction of the retinoblastoma protein with human D cyclins. Cell 73: 499–511; 1993. [DOI] [PubMed] [Google Scholar]
- 10. Dykstra C. C.; Hamatake R. K.; Sugino A. DNA strand transfer protein β from yeast mitotic cells differs from strand transfer protein a from meiotic cells. J. Biol. Chem. 265:10968–10973; 1990. [PubMed] [Google Scholar]
- 11. Edwards M. S.; Sternberg J. E.; Thornton J. M. Structural and sequence patterns in the loops of beta alpha beta units. Protein Eng. 3:173–181; 1987. [DOI] [PubMed] [Google Scholar]
- 12. Enomoto S.; Chen G.; Berman J. Vectors for expressing T7 epitope- and His6 affinity-tagged fusion proteins in S. cerevisiae . Biotechnology 24:782–786; 1998. [DOI] [PubMed] [Google Scholar]
- 13. Estojak J.; Brent R.; Golemis E. A. Correlation of two-hybrid affinity data with in vitro measurements. Mol. Cell. Biol. 15:5820–5829; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fisher R. P.; Morgan D. O. A novel cyclin associates with M015/CDK7 to form the CDK-activating kinase. Cell 78:713–724; 1994. [DOI] [PubMed] [Google Scholar]
- 15. Frishman D.; Argos P. Seventy-five percent accuracy in protein secondary structure prediction. Proteins 3: 329–335; 1997. [DOI] [PubMed] [Google Scholar]
- 16. Gaudreau L.; Schmid A.; Glaschke D.; Ptashne M.; Horz W. RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PH05 promoter. Cell 89:55–62; 1997. [DOI] [PubMed] [Google Scholar]
- 17. Glotzer M.; Murray A. W.; Kirschner M. W. Cyclin is degraded by the ubiquitin pathway. Nature 349:132–138; 1991. [DOI] [PubMed] [Google Scholar]
- 18. Goodrich J. A.; Tjian R. TBP-TAF complexes: Selectivity factors for eukaryotic transcription. Curr. Opin. Cell Biol. 6:403–409; 1994. [DOI] [PubMed] [Google Scholar]
- 19. Greenblat J. RNA polymerase II holoenzyme and trancriptional regulation. Curr. Opin. Cell Biol. 9:310–319; 1997. [DOI] [PubMed] [Google Scholar]
- 20. Gustafsson C.; Myers L.; Li Y.; Redd M.; Lui M.; Erdjument-Bromage H.; Tempst P.; Kornberg R. Identification of Rox3 as a component of mediator and RNA polymerase II holoenzyme. J. Biol Chem. 272:48–50; 1997. [DOI] [PubMed] [Google Scholar]
- 21. Gyuris J.; Golemis E. A.; Chertkov H.; Brent R. Cdil, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791–803; 1993. [DOI] [PubMed] [Google Scholar]
- 22. Hengartner C. J.; Myer V. E.; Liao S.-M.; Wilson C. J.; Koh S. S.; Young R. A. Temporal regulation of RNA Polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell. 2:43–53; 1998. [DOI] [PubMed] [Google Scholar]
- 23. Hollingsworth R. E. J.; Chen P.-L.; Lee W.-H. Integration of cell cycle control with transcriptional regulation by the retinoblastoma protein. Curr. Opin. Cell Biol. 5:194–200; 1993. [DOI] [PubMed] [Google Scholar]
- 24. Jiang Y.; Stillman D. Involvement of the SIN4 global transcriptional regulator in the chromatin structure of Saccharomyces cerevisiae . Mol. Cell. Biol. 12:4503–4514; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Keaveney M.; Struhl K. Activator-mediated recruitment of the RNA polymerase II machinery is the predominant mechanism for transcriptional activation in yeast. Mol. Cell. 1:917–924; 1998. [DOI] [PubMed] [Google Scholar]
- 26. Kim Y.-J.; Bjorklund S.; Li Y.; Sayre M. H.; Kornberg R. D. A mulitprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77:599–608; 1994. [DOI] [PubMed] [Google Scholar]
- 27. Kneller D. G.; Cohen F. E.; Langridge R. Improvements in protein secondary structure prediction by an enhanced neural network. J. Mol. Biol. 214:171–182; 1990. [DOI] [PubMed] [Google Scholar]
- 28. Koleske A. J.; Young R. A. An RNA polymerase II holoenzyme responsive to activators. Nature 368:466–469; 1994. [DOI] [PubMed] [Google Scholar]
- 29. Kuchin S.; Yeghiayan P.; Carlson M. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc. Natl. Acad. Sci. USA 92:4006–4010; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82:488–492; 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laybourn P. J.; Dahmus M. E. Phosphorylation of RNA polymerase IIA occurs subsequent to interaction with the promoter and before the initiation of transcription. J. Biol. Chem. 5:13165–13173; 1990. [PubMed] [Google Scholar]
- 32. Leclerc V.; Tassan J.-P.; O’Farrell P. H.; Nigg E. A.; Leopold P. Drosophila Cdk8, a kinase partner of cyclin C that interacts with the large subunit of RNA polymerase II. Mol. Biol. Cell. 7:505–513; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee J. M.; Greenleaf A. L. Modulation of RNA polymerase II elongation efficiency by C-terminal heptapeptide repeat domain kinase I. J. Biol. Chem. 272: 10990–10993; 1997. [DOI] [PubMed] [Google Scholar]
- 34. Leopold P.; O’Farrell P. H. An evolutionarily conserved cyclin homolog from Drosophila rescues yeast deficient in G1 cyclins. Cell 66:1207–1216; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Levin J. M. Exploring the limits of nearest neighbour secondary structure prediction. Protein Eng. 7:771–776; 1997. [DOI] [PubMed] [Google Scholar]
- 36. Lew D. J.; Dulic V.; Reed S. I. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell 66:1197–1206; 1991. [DOI] [PubMed] [Google Scholar]
- 37. Li Y.; Bjorklund S.; Jiang Y.; Kim Y.; Lane W.; Stillman D.; Kornberg R. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 92:10864–10868; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liao S.-M.; Zhang J.; Jeffery D. A.; Koleske A. J.; Thompson C. M.; Chao D. M.; Viljoen M.; van Vuuren H. J. J.; Young R. A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature 374:193–196; 1995. [DOI] [PubMed] [Google Scholar]
- 39. Maldonado E.; Shiekhattar R.; Sheldon M.; Cho H.; Drapkin R.; Rickert P.; Lees E.; Anderson C.; Linn S.; Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature 381:86–89; 1996. [DOI] [PubMed] [Google Scholar]
- 40. Maniatis T.; Fritsch E. F.; Sambrook J. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 41. Marshall N. F.; Peng J.; Xie Z.; Price D. H. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 271: 27176–27183; 1996. [DOI] [PubMed] [Google Scholar]
- 42. Miller J. H. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 43. Moitra J.; Szilak L.; Krylov D.; Vinson C. Leucine is the most stabilizing aliphatic amino acid in the d position of a dimeric leucine zipper coiled coil. J. Biochem. 14:12567–12573; 1997. [DOI] [PubMed] [Google Scholar]
- 44. Murray A. W.; Kirschner M. W. Cyclin synthesis drives the early embrionic cell cycle. Nature 339:275–286; 1989. [DOI] [PubMed] [Google Scholar]
- 45. Myers L.; Gustafsson C.; Bushnell D.; Lui M.; Erdjument-Bromage H.; Tempst P.; Kornberg R. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 12:45–54; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. O’Brien T.; Hardin S.; Greenleaf A.; Lis J. T. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature 370:75–77; 1994. [DOI] [PubMed] [Google Scholar]
- 47. Pan G.; Greenblatt A. T. J. Interaction of elongation factors TFIIS and elongin A with a human RNA polymerase II holoenzyme capable of promoter-specific initiation and responsive to transcriptional activators. J. Biol. Chem. 272:24565–24571;1997. [DOI] [PubMed] [Google Scholar]
- 48. Pines J. Cyclins and cyclin-dependent kinases: Take your partners. Trends Biol. Sci. 18:195-197; 1993. [DOI] [PubMed] [Google Scholar]
- 49. Reece R. J.; Platt A. Signaling activation and repression of RNA polymerase II transcription in yeast. Bioessays 19:1001–1010; 1997. [DOI] [PubMed] [Google Scholar]
- 50. Rickert P.; Seghezzi W.; Shanahan F.; Cho H.; Lees E. Cyclin C/CDK8 is a novel CTD kinase associated with RNA polymerase II. Oncogene 12:2631–2640; 1996. [PubMed] [Google Scholar]
- 51. Rost B.; Sander C. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 232:584–599; 1993. [DOI] [PubMed] [Google Scholar]
- 52. Sauer F.; Tjian R. Mechanisms of transcriptional activation: Differences and similarities between yeast, Drosophila, and man. Curr. Opin. Genet. Dev. 7:176–181; 1997. [DOI] [PubMed] [Google Scholar]
- 53. Shi X.; Chang M.; Wolf A. J.; Chang C. H.; Frazer-Abel A. A.; Wade P. A.; Burton Z. F.; Jaehning J. A. Cdc73p and Paflp are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol. Cell. Biol. 17:1160–1169; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sikorski R. S.; Boeke J. D. In vitro mutagenesis and plasmid shuffling: From cloned gene to mutant yeast. In: Guthrie C.; Fink G. R., eds. Methods in enzymology. New York: Academic Press, Inc.; 1991: 302–318. [DOI] [PubMed] [Google Scholar]
- 55. Sikorski R. S.; Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics 122:19–27; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Slater M. R.; Craig E. A. Transcriptional regulation of an hsp70 heat shock gene in the yeast Saccharomyces cerevisiae . Mol. Cell. Biol. 7:1906–1916; 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Solomons T. W. G. Organic chemistry. New York: John Wiley & Sons, Inc.; 1978. [Google Scholar]
- 58. Sterner D. E.; Lee J. M.; Hardin S. E.; Greenleaf A. L. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-cyclin-dependent kinase complex. Mol. Cell. Biol. 15:5716–5724; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Strich R.; Slater M. R.; Esposito R. E. Identification of negative regulatory genes that govern the expression of early meiotic genes in yeast. Proc. Natl. Acad. Sci. USA 86:10018–10022; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Surosky R. T.; Strich R.; Esposito R. E. The yeast UME5 gene regulates the stability of meiotic mRNAs in response to glucose. Mol. Cell. Biol. 14:3446–3458; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tassan J.-P.; Jaqueoud M.; Leopold P.; Schultz S. J.; Nigg E. A. Identification of human cyclin-dependent kinase 8, a putative protein kinase partner for cyclin C. Proc. Natl. Acad. Sci. USA 92:8871–8875; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tassan J.-P.; Schultz S. J.; Bartek J.; Nigg E. A. Cell cycle analysis of the activity, subcellular localization, and subunit composition of human CAK (CDK-activating kinase). J. Cell Biol. 127:467–478; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Valay J. G.; Simon M.; Faye G. The Kin28 protein kinase is associated with a cyclin in Saccharomyces cerevisiae . J. Mol. Biol. 234:307–310; 1993. [DOI] [PubMed] [Google Scholar]
- 64. Vinson C. R.; Sigler P. B.; McKnight S. L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science 246:911–916; 1989. [DOI] [PubMed] [Google Scholar]
- 65. Wade P. A.; Werel W.; Fentzke R. C.; Thompson N. E.; Leykam J. F.; Burgess R. R.; jaehning J A.; Burton Z. F. A novel collection of accessory factors associated with yeast RNA polymerase II. Prot. Exp. Pruif. 8:85–90; 1996. [DOI] [PubMed] [Google Scholar]
- 66. Wahi M.; Johnson A. D. Identification of genes required for alpha 2 repression in Saccharomyces cerevisiae . Genetics 140:79–90; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]


