Abstract
SM-20 is a novel, evolutionarily conserved “early response” gene originally cloned from a rat aortic smooth muscle cell (SMC) cDNA library. SM-20 encodes a cytoplasmic protein, which is induced by platelet-derived growth factor and angiotensin II in cultured SMC and is upregulated in intimal SMC of atherosclerotic plaques and injured arteries. We have now examined SM-20 expression during differentiation of cultured skeletal myoblasts and during skeletal myogenesis in vivo. Low levels of SM-20 mRNA and protein were expressed in proliferating mouse C2C12 myoblasts. Differentiation by serum withdrawal was associated with a marked induction of SM-20 mRNA and the expression of high levels of SM-20 antigen in myotubes. The induction was partially inhibited by blocking differentiation with bFGF or TGFβ. Similar results were obtained with the nonfusing mouse C25 myoblast line, suggesting that SM-20 upregulation is a consequence of biochemical differentiation and is fusion independent. During mouse embryogenesis, SM-20 was first observed at 8.5E in the dermomyotomal cells of the rostral somites. SM-20 expression progressed in a rostral to caudal pattern, with highest levels seen in the muscle primordia and mature muscles. SM-20 thus represents a novel intracellular protein that is regulated during skeletal muscle differentiation and development.
Keywords: Skeletal muscle, Mouse embryo, Myogenesis, SM-20, Myocyte differentiation, Antibody staining, Somites
SM-20 is a growth factor-responsive gene, originally cloned from a rat aortic smooth muscle cell (SMC) cDNA library (22). The full-length SM-20 mRNA is ≈2.5 kb and encodes a protein that runs with a molecular weight of ≈40 kDa on SDS-polyacrylamide gels. In SMC, the SM-20 protein is found exclusively in the cytoplasm and stains as filaments in a pattern distinct from that of α-actin (23). The SM-20 gene is conserved and has homologues in Drosophila melanogaster (12) and C. elegans (22). In SMC culture, SM-20 is inducible by a variety of agonists, including platelet-derived growth factor (PDGF), angiotensin II, and isoproterenol, and is superinducible in the presence of cycloheximide, suggesting that it is part of the primary response to growth agonists (22). In the normal arterial wall SM-20 is expressed in medial SMC, but not in adventitial fibroblasts or endo-thelial cells. SM-20 is most highly expressed in intimal SMC of atherosclerotic plaques and in the neointimal SMC formed in response to balloon arterial injury (23).
Because SM-20 was cloned from rat SMC, our initial studies focused on its expression in smooth muscle and the vasculature. High levels of SM-20 mRNA and protein were also observed in adult cardiac and skeletal muscle (23). To study further the regulation of SM-20 in sarcomeric muscle, we have now examined its expression during differentiation of cultured myoblasts and during skeletal myogenesis in vivo. We report that SM-20 is upregulated during skeletal muscle differentiation in culture and during skeletal myogenesis in mouse embryos. SM-20 represents a new intracellular protein that may play an important role in sarcomeric muscle differentiation and development.
MATERIALS AND METHODS
Animals
Female FVB mice were housed in the Mount Sinai Medical Center Animal Facility and fed normal animal chow ad lib. Mice were stimulated to ovulate with pregnant mare’s serum gonadotropin (Sigma, St. Louis, MO). After 48 h, the mice were injected with human chorionic gonadotropin (Sigma) and housed with male FVB mice. The following morning, mice were checked for vaginal plugs; positives were considered 0.5 day postcoitum (0.5E). Embryos were excised at stages 5.5 to 16.5E, fixed overnight in 4% paraformaldehyde in phosphate-buffered saline (PBS), pH 7.2, and subsequently embedded in paraffin. Sections (6 μM nominal thickness) were serially cut and used for immunocytochemistry.
Cell Culture
Fusion-competent mouse C2C12 (1,24) were originally obtained from the American Type Culture Collection (Rockland, MD). A highly myogenic and stable subclone of these cells (C2C12-39) was used for these experiments. The fusion-defective, biochemically competent C25 myoblast cell line was derived from C2M cells (19). This clonal cell line proliferates in high mitogen medium and terminally differentiates in low mitogen medium (10). Differentiation is accompanied by expression of proteins characteristic of embryonic skeletal muscle cells, and initiation of spontaneous contractions. The cells, however, do not form syncitia containing more than two to three nuclei. Proliferation medium consisted of DMEM containing 15% fetal bovine serum and 1% chick embryo extract (Gibco BRL, Gaithersburg, MD). To induce differentiation, cells at 70–80% confluence were switched to DMEM supplemented with 3% horse serum. In some experiments, 5 ng/ml transforming growth factor-β (TGFβ) or 50 ng/ml basic growth factor (bFGF) (R&D Systems, Minneapolis, MN) was added to the differentiation medium. Differentiation was assessed by the morphologic appearance of elongated myoblasts and the induction of troponin I mRNA.
Immunocytochemistry
For immunocytochemistry, sections were deparaffinized and rehydrated. Endogenous peroxidases were blocked by incubation for 30 min at room temperature with 0.3% H2O2 and then washed for 30 min in PBS. Nonspecific immunoreactivity was blocked by incubation for 20 min at room temperature with normal goat serum. Sections were subsequently incubated overnight at 4°C with a 1:50 dilution of an affinity-purified polyclonal antibody raised against the full-length SM-20 protein expressed in E. coli (23). After washing in PBS, the primary antibody was detected using a biotinylated goat anti-rabbit IgG (Zymed, San Francisco, CA) for 15 min at room temperature. The slides were washed in PBS, reacted with horseradish peroxidase-conjugated streptavidin (Zymed), and developed with amino-ethylcarbazol (Zymed). After washing in tap water, all sections were counterstained with hematoxylin, dehydrated, and coverslipped. Negative control sections were prepared by substitution of the primary antibody with PBS or an irrelevant antibody of the same isotype.
Isolation ofMurine SM-20 cDNA
An 11.5E murine cDNA library (Clontech) was screened by hybridization with the full-length rat SM-20 cDNA, using procedures previously described (22). Filters were washed at a final stringency in 1× SSC (0.55 mM sodium citrate, 0.15 M sodium chloride) at 50°C. Three clones were isolated and their inserts subcloned into pBluescript (Stratagene, La Jolla, CA). The largest insert, ≈1.2 kb, was sequenced commercially (Biotechnology Center, Utah State University, Logan, UT). The mouse sequence had 95% identify with nucleotides 328–1515 of the rat sequence (Accession #U06713). The derived amino acid sequence of the longest mouse open reading frame (238 amino acids) had 98% identity with that of the rat. The mouse clone was used for RNA blot analysis.
RNA Blot Analysis
Extraction of total RNA, agarose gel electrophoresis, transfer to nitrocellulose, and hybridization to 32P-labeled DNA were as previously described (22). Pre-hybridization and hybridization were done at 42°C. Final washes were in 0.5× SSC and 0.1% SDS at 60°C for 45 min. The 1.2-kb mouse SM-20 cDNA and the full-length mouse troponin I cDNA (11) were labeled with [32P]dCTP by random oligomer priming to a specific activity of greater than 108 cpm/μg and used at 3 × 106 cpm/ml. Experiments shown in the figures represent blots in which all lanes had equal loading (20 μg/lane) as confirmed by staining of 18s and 28s ribosomal RNA with ethidium bromide.
RESULTS
SM-20 Expression in Cultured Skeletal Myoblasts
To examine SM-20 gene expression during skeletal myocyte differentiation, RNA blot analysis was performed on cultured C2C12 myoblasts. Low levels of SM-20 mRNA were detected in proliferating myoblasts grown in 15% fetal bovine serum (Fig. 1). In contrast, incubation in differentiation medium (3% horse serum) for 48 h induced a marked increase in SM-20 mRNA levels; the induction was coincident with that of troponin I. bFGF or TGFβ blocked C2C12 differentiation, as evidenced by the complete inhibition of troponin I expression (Fig. 1) and by the lack of myocyte fusion. Both agonists also markedly reduced SM-20 expression, although low levels of SM-20 mRNA remained.
FIG. 1.
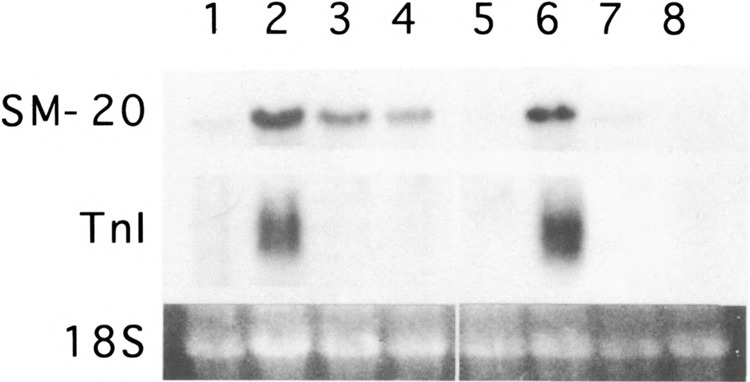
Regulation of SM-20 mRNA in skeletal myocyte culture. RNA blot analysis of mouse C2C12 (lanes 1–4) and C25 (lanes 5–8) myocytes maintained in proliferating conditions (15% fetal bovine serum; lanes 1 and 5) or differentiated for 48 h in 3% horse serum in the absence (lanes 2 and 6) or presence of 50 ng/ml bFGF (lanes 3 and 7) or 5 ng/ml TGFβ (lanes 4 and 8). Blots were hybridized with radiolabeled SM-20 and troponin I. 18S ribosomal RNA is shown as an indicator of equal loading of lanes.
To determine whether myocyte fusion was necessary for SM-20 expression, SM-20 mRNA levels were examined in C25 myoblasts. This cell line, which is derived from C2M cells, undergoes biochemical differentiation, but does not fuse efficiently to form myotubes (10). Similar to that seen in C2C12 cells, low levels of SM-20 mRNA and virtually no troponin I mRNA were observed in proliferating C25 myoblasts (Fig. 1). Incubation in differentiation medium resulted in the induction of SM-20 and troponin I mRNAs. Like in C2C12 cells, bFGF and TGFβ completely blocked the induction of troponin I and markedly inhibited the induction of SM-20. In both cell types, TGFβ appeared more potent than bFGF in blocking SM-20 expression.
To examine the expression of SM-20 protein during skeletal myocyte differentiation, immunohisto-chemical studies were performed using an affinity-purified polyclonal antibody to SM-20. Proliferating C25 myoblasts exhibited low levels of SM-20 antigen, distributed in a granular pattern around the nucleus (Fig. 2A). Differentiation was associated with a substantial increase in SM-20 antigen, distributed in a speckled pattern throughout the cytoplasm (Fig. 2B). Levels of SM-20 antigen in proliferating C2C12 myoblasts (not shown) were similar to those seen in C25. In contrast, high levels of SM-20 were detected after C2C12 myocyte differentiation (Fig. 2C). Most significantly, the induction of SM-20 antigen appeared confined to myotubes and was not increased in isolated cells.
FIG. 2.
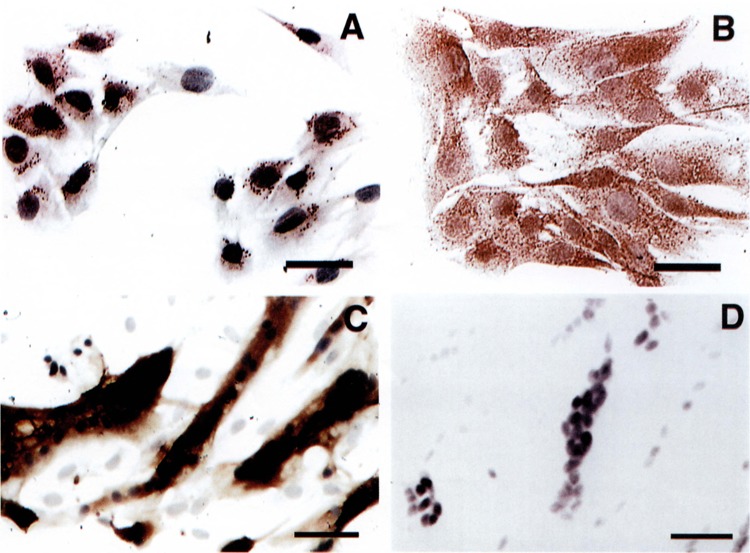
Expression of SM-20 antigen in skeletal myocyte culture. C25 myocytes were stained with antibody to SM-20 under proliferating conditions (A) and after differentiation for 48 h in 3% horse serum (B). C2C12 myocytes were stained after differentiation for 48 h in 3% horse serum (C). A parallel culture of differentiated C2C12 myocytes was stained with preimmune serum (D). Calibration bars: 50 μm for (A) and 100 μm for (C and D).
SM-20 Expression in Mouse Myocyte Development
To examine SM-20 expression during skeletal myogenesis, immunocytochemistry was performed on mouse embryos at developmental stages 7.5E through 16.5E. SM-20 expression was not observed in 7.5E embryos. SM-20 expression was first noted at 8.5E in the dermomyotomal cells of the developing rostral somites (Fig. 3A, B); midbody somites (Fig. 3A) and caudal somites (Fig. 3C) remained negative. At 9.5E, midbody somites became positive for SM-20 (not shown). At 10.5E, virtually all somites, except the most caudal ones, expressed SM-20. A marked difference in staining intensity was observed between the dermomyotomal cells and the myocytes of the myotome, with the myocytes expressing more intensely than the dermomyotomal cells (Fig. 3D, F).
FIG. 3.
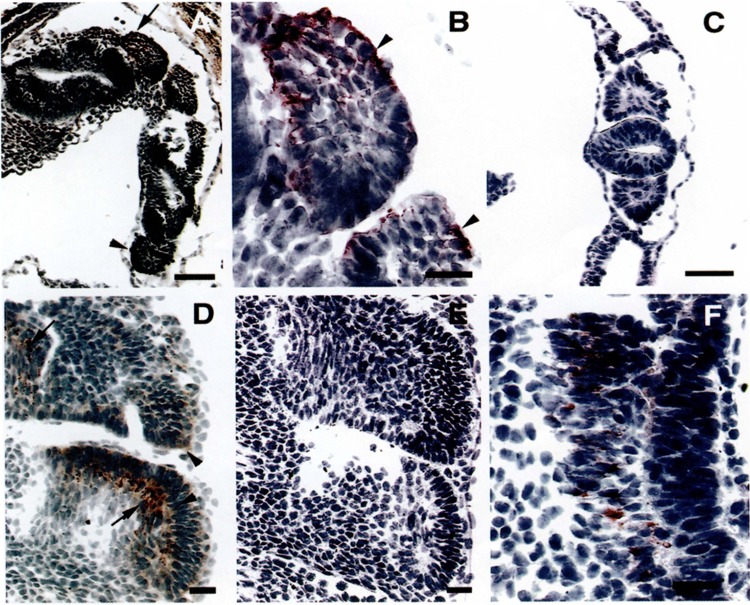
Expression of SM-20 during development of murine skeletal muscle: 8.5 and 10.5E. (A) Frontotangential section, passing through the neural tube, from an 8.5E mouse embryo stained with antibody to SM-20, showing left side rostral (arrow) and midbody somites (arrowhead). Rostral somites are positive for SM-20 antigen. (B) Higher magnification of rostral somites. Columnar epithelial cells (arrowheads) are strongly positive for SM-20 antigen. Because of the section plane some of the columnar cells may be cut transversely. (C) Higher magnification of caudal somites, showing no staining for SM-20. (D) Sagittal section from a 10.5E embryo. SM-20 antigen is detected in dermomyotomal cells (arrowheads) and in elongated myocytes (arrows) located in the myotome. (F) Negative control, consisting of a sagittal section from a 10.5E embryo (serial to section shown in D) stained with secondary antibody alone. (E) High magnification of a 10.5E somite, showing SM-20-positive cells (arrow heads) in the dermomyotomal (asterisk) and myotomal (double asterisk) compartments. An arrow indicates positive dermomyotomal cells. Calibration bars: 100 μm for (A), 20 μm for (B–E), and 10 μm for (F).
At 12.5E, elongated cells cluster around focal sites of developing cartilage, forming the muscle primordia of the developing limb. All of the elongated cells were SM-20 positive (Fig. 4A). At higher magnification, a differential staining pattern was noted, with lighter staining cells intermingled with those expressing higher levels of SM-20 (Fig. 4B). By 14.5E, cells in the muscle primordia have differentiated into well-differentiated myotubes. These cells stained for SM-20 with greatest intensity (Fig. 4C, D). Abdominal muscles showed a similar degree of differentiation to that seen in the limb muscles and also stained intensely for SM-20 (Fig. 4C, D). The same pattern was observed at 16.5E (not shown) and in the adult (23).
FIG. 4.
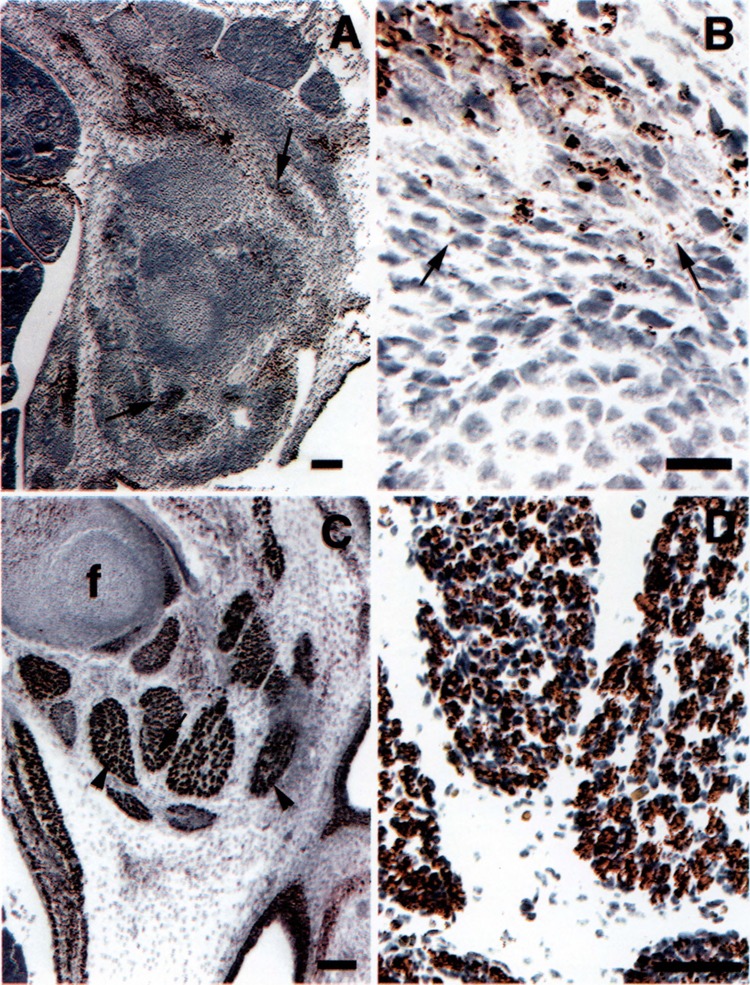
Expression of SM-20 during development of murine skeletal muscle: 12.5 and 14.5E. (A) Sagittal section of a 12.5E embryo, showing primordial muscle masses comprised of SM-20-positive cells (arrows). (B) Higher magnification of the area shown by asterisk in (A). Arrows indicate lighter staining cells. (C) Sagittal section of a 14.5E embryo, showing differentiated limb muscles comprised of strongly stained myotubes (arrowheads). Abdominal muscles (asterisk) show the same degree of differentiation and staining as the limb muscles, f, femur. (D) Higher magnifications of muscle indicated by an arrow in (C) showing prominent staining of myotubes. Calibration bars: 200 μm for (A), 20 μm for (B), 100 μm for (C), and 50 μm for (D).
DISCUSSION
Skeletal muscle development is characterized by the fusion of myoblasts to terminally differentiated multinucleated myotubes and is accompanied by changes in gene expression required for progression to and maintenance of the differentiated state [reviewed in (8)]. Considerable information has been obtained concerning the regulation of skeletal muscle contractile protein genes during development and differentiation (2,5,13). In addition, great progress has been made in identifying the cascade of regulatory proteins involved in mediating skeletal muscle differentiation (4,14,16,21). We now report that SM-20, a growth factor-responsive gene originally characterized in vascular smooth muscle, is highly regulated during skeletal muscle development and differentiation.
During mouse development, SM-20 expression was first seen at 8.5E in the dermomyotome of somites. At this time, the somites appear as spherical structures comprised of predominantly tall columnar proliferating epithelial cells. It is thought that the dermomyotome is the source of proliferating myogenic cells (3,6,9,25). Eventually, these cells migrate ventrally to become postmitotic myocytes, which form the myotome. The expression of SM-20 thus precedes that reported for MyoD and myogenin and the sarcomeric contractile proteins, which are expressed in postmitotic myocytes (2,8). The presence of SM-20 antigen in the cells of the dermomyotome may be analogous to its expression in proliferating C2Ci2 cells (see below).
Subsequent expression of SM-20 progressed in a rostro-caudal fashion, concomitant with the development of the somites. This pattern of expression is similar to that seen with MyoD and myogenin (20) and contractile proteins (13). Highest levels of SM-20 antigen were found in the postmitotic myocytes of the myotome and of the developing limb bud. The distribution of SM-20 antigen in the developing limb was variable, with cells staining with highest intensity intermingled with those of a lower intensity. The lower intensity staining cells may correspond to proliferating myoblasts that have recently migrated from the corresponding somite. SM-20 levels remained high in adult skeletal muscle. The persistent high expression of SM-20 at the later developmental stages is similar to that seen for the skeletal muscle contractile proteins and thus may be regulated similarly.
In cell culture, low levels of SM-20 mRNA were observed in proliferating C2C12 myoblasts. This differs from most of the sarcomeric contractile protein mRNAs, which are not expressed during proliferation. Conditions that promote differentiation markedly induced SM-20 expression. Inhibition of muscle differentiation by growth factors resulted in lower levels of SM-20. Thus, SM-20 serves as a marker for the state of differentiation of these cells. Although the changes in SM-20 levels following treatment with these growth factors may be indirect and simply a consequence of the inhibition of differentiation, it is also possible that they might act directly on SM-20 transcription. Several consensus sequences for MyoD binding are present in the proximal promoter region of the rat SM-20 gene (K. Menzies and M. B. Taubman, unpublished observations). TGFβ and bFGF maintain the proliferating potential of myoblasts in part by inducing high levels of cyclin D1 (18), which in turn prevents activation of myogenic regulatory factors, such as the myogenic basic helix–loop–helix (myo-bHLH) family of transcription factors, which includes MyoD (7,15,17). Analysis of the SM-20 promoter will ultimately provide further insights into the specific mechanisms for regulation of SM-20 transcription.
In addition to its induction in C2C12 cells, SM-20 mRNA and protein were also induced in low serum in fusion-deficient C25 myoblasts. This suggests that, like many of the sarcomeric contractile protein genes (5), upregulation of SM-20 occurs as a concomitant of biochemical differentiation and is not fusion dependent.
We have not yet established whether SM-20 is a member of a larger multigene family. The SM-20 peptide shares sequence homology with the translations of several uncharacterized expressed sequence tags in the Genbank; however, their corresponding mRNAs would not be expected to hybridize with SM-20 cDNA under the stringency of the RNA blots shown in this study. In addition, RNAase protection analyses have suggested that the same SM-20 mRNA is found in smooth, skeletal, and cardiac muscle (not shown), and SM-20 cDNA clones isolated from a rat cardiac library were identical to that cloned in rat smooth muscle. Although these data lend support to the notion that the SM-20 cDNA is identifying a single isoform on RNA blots, they do not rule out the possibility that there is a distinct SM-20 isoform that is expressed at low levels during proliferation and is distinct from the growth factor-inhibited form.
SM-20 was initially cloned from a rat aortic SMC cDNA library. Subsequent studies revealed that in the arterial wall, SM-20 expression was limited to the SMC and therefore served as a novel marker for normal medial SMC as well as SMC found in the neointima of injured vessels. The present study demonstrates that SM-20 is also a novel marker for skeletal muscle differentiation. The function of SM-20 remains to be determined. Preliminary studies involving expression of full-length antisense mRNA in rat aortic SMC suggest that SM-20 may play an important role in maintaining normal cell size (i.e., SM-20 inhibition resulted in a ≈33% increase in cell size and a ≈50% decrease in saturation density) (E. Cavusoglu and M. B. Taubman, unpublished observations). The studies presented above raise the possibility that SM-20 may play a role in establishing or maintaining the normal differentiated state of skeletal muscle.
ACKNOWLEDGMENTS
This research was supported by National Institutes of Health Grant R01 HL43302 (M.B.T.) and R01 CA72775 (D.S.K.). K.M. was supported in part by a National Institutes of Health MARC Predoctoral Fellowship F31 GM15732. M.E.L. is supported by a National Institutes of Health Award K08 HL03741.
REFERENCES
- 1. Blau H. M.; Pavlath G. K.; Hardeman E. C.; Chiu C. P.; Silberstein L.; Webster S. G.; Miller S. C.; Webster C. Plasticity of the differentiated state. Science 230:758–766; 1985. [DOI] [PubMed] [Google Scholar]
- 2. Buckingham M. Making muscle in mammals. Trends Genet. 8:144–148; 1992. [DOI] [PubMed] [Google Scholar]
- 3. Buckingham M. Molecular biology of muscle development. Cell 78:15–21; 1994. [DOI] [PubMed] [Google Scholar]
- 4. Buckingham M. Skeletal muscle development and the role of the myogenic regulatory factors. Biochem. Soc. Trans. 24:506–509; 1996. [DOI] [PubMed] [Google Scholar]
- 5. Buckingham M. E. Actin and myosin multigene families: Their expression during the formation of skeletal muscle. Essays Biochem. 20:77–109; 1985. [PubMed] [Google Scholar]
- 6. Cossu G.; Tajbakhsh S.; Buckingham M. How is myogenesis initiated in the embryo? Trends Genet. 12:218–223; 1996. [DOI] [PubMed] [Google Scholar]
- 7. Edmondson D. G.; Olson E. N. Helix-loop-helix proteins as regulators of muscle-specific transcription. J. Biol. Chem. 268:755–758; 1993. [PubMed] [Google Scholar]
- 8. Hauschka S. D. The embryonic origin of muscle. In: Engel A.; Franzini-Armstrong C., eds. Myology. New York: McGraw Hill; 1994:3–96. [Google Scholar]
- 9. Keynes R. J.; Stern C. D. Mechanisms of vertebrate segmentation. Development 103:413–29; 1988. [DOI] [PubMed] [Google Scholar]
- 10. Kohtz J. D. The role of c-myc in mouse myoblast differentiation. New York City: University of New York; 1991. [Google Scholar]
- 11. Koppe R. I.; Hallauer P. L.; Karpati G.; Hastings K. E. cDNA clone and expression analysis of rodent fast and slow skeletal muscle troponin I mRNAs. J. Biol. Chem. 264:14327–14333; 1989. [PubMed] [Google Scholar]
- 12. Lieb M.; Moschella M.; Pick L.; Taubman M. A Drosophila homologue of the novel mammalian gene SM-20. In: 39th Annual Drosophila Research Conference. Bethesda, MD/Washington, DC: The Genetics Society of America; 1998:388B. [Google Scholar]
- 13. Lyons G. E.; Ontell M.; Cox R.; Sassoon D.; Buckingham M. The expression of myosin genes in developing skeletal muscle in the mouse embryo. J. Cell Biol. 111:1465–1476; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Molkentin J. D.; Olson E. N. Defining the regulatory networks for muscle development. Curr. Opin. Genet. Dev. 6:445–453; 1996. [DOI] [PubMed] [Google Scholar]
- 15. Olson E. N.; Klein W. H. bHLH factors in muscle development: Dead lines and commitments, what to leave in and what to leave out. Genes Dev. 8:1–8; 1994. [DOI] [PubMed] [Google Scholar]
- 16. Olson E. N.; Perry M.; Schulz R. A. Regulation of muscle differentiation by the MEF2 family of MADS box transcription factors. Dev. Biol. 172:2–14; 1995. [DOI] [PubMed] [Google Scholar]
- 17. Rao S. S.; Chu C.; Kohtz D. S. Ectopic expression of cyclin D1 prevents activation of gene transcription by myogenic basic helix-loop-helix regulators. Mol. Cell. Biol. 14:5259–5267; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rao S. S.; Kohtz D. S. Positive and negative regulation of D-type cyclin expression in skeletal myoblasts by basic fibroblast growth factor and transforming growth factor beta. A role for cyclin D1 in control of myoblast differentiation. J. Biol. Chem. 270:4093–4100; 1995. [DOI] [PubMed] [Google Scholar]
- 19. Richler C.; Yaffe D. The in vitro cultivation and differentiation capacities of myogenic cell lines. Dev. Biol. 23:1–22; 1970. [DOI] [PubMed] [Google Scholar]
- 20. Sassoon D.; Lyons G.; Wright W. E.; Lin V.; Lassar A.; Weintraub H.; Buckingham M. Expression of two myogenic regulatory factors myogenin and MyoDl during mouse embryogenesis. Nature 341:303–307; 1989. [DOI] [PubMed] [Google Scholar]
- 21. Sassoon D. A. Myogenic regulatory factors: Dissecting their role and regulation during vertebrate embryo-genesis. Dev. Biol. 156:11–23; 1993. [DOI] [PubMed] [Google Scholar]
- 22. Wax S. D.; Rosenfield C. L.; Taubman M. B. Identification of a novel growth factor-responsive gene in vascular smooth muscle cells. J. Biol. Chem. 269:13041–13047; 1994. [PubMed] [Google Scholar]
- 23. Wax S. D.; Tsao L.; Lieb M. E.; Fallon J. T.; Taubman M. B. SM-20 is a novel 40-kd protein whose expression in the arterial wall is restricted to smooth muscle. Lab. Invest. 74:797–808; 1996. [PubMed] [Google Scholar]
- 24. Yaffe D.; Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 270:725–727; 1977. [DOI] [PubMed] [Google Scholar]
- 25. Yun K.; Wold B. Skeletal muscle determination and differentiation: Story of a core regulatory network and its context. Curr. Opin. Cell Biol. 8:877–889; 1996. [DOI] [PubMed] [Google Scholar]


